Abstract
Twenty-two human extraintestinal isolates (11 from blood) and three isolates recovered from patients with diarrhea were genetically characterized as Aeromonas aquariorum, a novel species known only from ornamental fish. The isolates proved to bear a considerable number of virulence genes, and all were resistant to amoxicillin (amoxicilline), cephalothin (cefalotin), and cefoxitin. Biochemical differentiation from the most relevant clinical species is provided.
Members of the genus Aeromonas are responsible for producing intestinal and extraintestinal infections worldwide (1, 10). The most common clinical presentation of Aeromonas is diarrhea followed by localized soft-tissue infections and bacteremia. Bacteremia mainly occurs in patients with underlying diseases, i.e., hepatobiliary disorders, cancer, and diabetes. In patients with hepatobiliary disorders, especially in those with cirrhosis, fatal necrotizing fasciitis can be produced despite surgical intervention and antibiotic therapy (1, 10).
The pathogenicity of Aeromonas has been associated with numerous virulence factors, including the aerolysin/hemolysin group of genes, the cytotonic enterotoxins Ast and Alt (4, 9, 19), the cytotoxin encoded by the act gene (9), and a type III secretion system (TTSS) (8, 20, 26). The TTSS is a virulence mechanism that delivers toxins (AexT among others) directly into the host cell and induces apoptosis (8, 27).
Currently, the genus contains 22 species (A. Alperi, A. J. Martínez-Murcia, W. C. Ko, A. Monera, M. J. Saavedra and M. J. Figueras, submitted for publication). Although A. hydrophila is the most referenced species in the clinical literature, recent studies using molecular identification methods have demonstrated that A. veronii and A. caviae are more prevalent (3, 10, 18, 24).
In a set of extraintestinal isolates received in our laboratory from Taiwan for genetic identification, we have identified 22 as belonging to A. aquariorum, a recently described species from aquarium water and the skin of ornamental fish (17). In addition, three clinical strains isolated from patients with diarrhea in Spain were also identified as belonging to this species. Since this is a poorly known species, we have considered it of interest to study the molecular characterization, antibiotic susceptibility, and virulence potential of these strains in order to provide complementary data to differentiate A. aquariorum from other, commonly encountered clinical species.
Strains and molecular identification.
A total of 150 biochemically identified extraintestinal clinical isolates (including 86 A. hydrophila, 28 A. sobria, 20 A. caviae, and 16 Aeromonas spp.) were received in our laboratory from Wen-Chien Ko (Cheng Kung University Hospital, Tainan, Taiwan) to be genetically identified with a previously described 16S rRNA gene-restriction fragment length polymorphism (RFLP) method (6, 11). In brief, the method consists of an initial digestion of 1,503 bp of the amplified 16S rRNA genes with enzymes AluI and MboI, which produce species-specific patterns for 10 Aeromonas species, including the most relevant clinical species (A. caviae, A. veronii, and A. hydrophila). Only 138 (92%) isolates were identified as Aeromonas species: 73 (52.9%) as A. caviae, 17 (12.3%) as A. hydrophila, 16 (11.6%) as A. veronii, 5 (3.6%) as A. media, and 3 (2.1%) as A. trota. The remaining 24 (17.5%) isolates showed an RFLP pattern similar to that described for A. caviae (Fig. 1). To confirm the identities of all these strains, the rpoD and gyrB genes were sequenced using previously described primers and conditions (23). Two of these isolates were proposed as two new species, A. taiwanensis and A. sanarellii (A. Alperi et al., submitted for publication), and the remaining 22 (15.9%) were shown to belong to A. aquariorum (Fig. 2). Three Spanish strains (MDC562, MDC573, and MDC671) isolated in the Hospital Comarcal Vega Baja (Orihuela, Spain) were also recognized, using the same genes, to belong to A. aquariorum (Fig. 2). In agreement with the results of previous studies (6, 10, 22) only a few strains (17 A. hydrophila and 15 A. caviae) were originally correctly identified using biochemical methods.
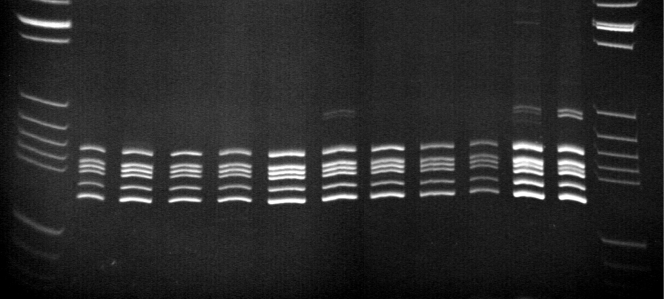
FIG. 1.
Polyacrylamide gel showing almost the same 16S rRNA gene-RFLP pattern for some of the investigated A. aquariorum strains, the type strain of this species, A. hydrophila subsp. dhakensis, and A. caviae. Lanes: 1st and 13th, pBR322 DNA/BsuRI (HaeIII) marker 5; 2nd, A. hydrophila subsp. dhakensis LMG19559; 3rd, A. hydrophila subsp. dhakensis LMG 19562T; 4th, A. aquariorum DSM18362T; 5th, A. caviae CECT838T; 6th to 10th, A. aquariorum clinical strains; 6th to 8th, strains isolated from feces (MDC562, MDC563, and MDC671); 9th, strain from a wound infection after a flame burn (A2-004); 10th, strain isolated from ascites in a case of acute appendicitis with peritonitis (A2-96); 11th, A. taiwanensis; and 12th, A. sanarellii. In the 7th, 11th, and 12th lanes, notice the atypical pattern showing some extra bands.
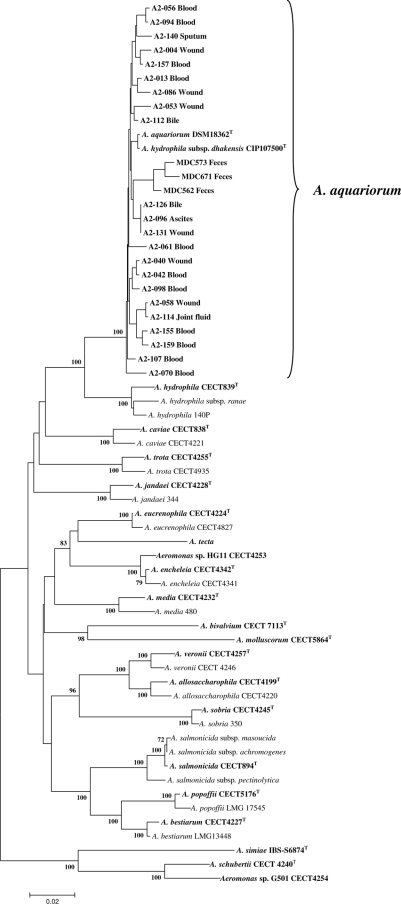
FIG. 2.
Unrooted neighbor-joining phylogenetic tree based on the combined stretch of gyrB (450 bp)-rpoD (790 bp) gene sequences showing the relationship within the genus Aeromonas of 25 clinical A. aquariorum strains. The sources of isolates are given. Type strains are in bold font. Numbers at nodes indicate bootstrap values (1,000 replicates). The scale bar represents 1 nucleotide substitution per 50 nucleotides.
Antimicrobial susceptibility.
The susceptibility of the 25 strains identified as A. aquariorum was evaluated against a panel of 27 antimicrobial agents by using drugs and conditions described previously (17). All the isolates were resistant to amoxicillin (amoxicilline), cephalothin (cephalotin), and cefoxitin, but they varied in their resistance to other antimicrobial agents (Table 1). These responses were similar to those found in the environmental strains of A. aquariorum (17), with the exception of the 68% resistance to amoxicillin-clavulanic acid that contrasted with the 32% encountered in the environmental strains. It has been demonstrated that empirical treatment for Aeromonas bacteremia is normally inappropriate in ca. 20% of the cases and has an important impact on mortality (10).
TABLE 1.
Antimicrobial resistance found in 25 clinical Aeromonas aquariorum strains
| Antimicrobial agent | % of strains with resistance |
|---|---|
| Amoxicillin | 100 |
| Cephalothin | 100 |
| Cefoxitin | 100 |
| Erythromycin | 96 |
| Amoxicillin-clavulanic acid | 68 |
| Ticarcillin | 48 |
| Tetracycline | 28 |
| Ticarcillin-clavulanic acid | 20 |
| Nalidixic acid | 16 |
| Streptomycin | 12 |
| Piperacillin | 12 |
| Piperacillin-tazobactam | 12 |
| Ceftriaxone | 12 |
| Cefoperazone | 12 |
| Ceftazidime | 8 |
| Cefotaxime | 8 |
| Kanamycin | 8 |
| Trimethoprim-sulfamethoxazole | 4 |
| Chloramphenicol | 4 |
| Imipenem | 4 |
| Aztreonam | 0 |
| Amikacin | 0 |
| Ciprofloxacin | 0 |
| Cefepime | 0 |
| Fosfomycin | 0 |
| Gentamicin | 0 |
| Tobramycin | 0 |
Detection of virulence genes.
The strains were also screened for the presence of virulence genes, including the aerolysin/hemolysin genes, ast, alt, act, the TTSS genes (ascF-ascG), and aexT, using primers and conditions described earlier (2, 7, 8, 12). The genes that encode the pore-forming toxin aerolysin/hemolysin were present in all of the strains (Table 2). The two cytotoxic enterotoxin genes, the heat-stable ast and the heat-labile alt, were present in 84% and 28% of the strains, respectively, ast being much more prevalent than was found in previous studies (2). The three strains isolated from feces of patients with diarrhea and a considerable number of the strains from wounds (4/6, 66.6%) and blood (7/11, 63.3%) possessed the ascF-ascG genes. The aexT gene, which encodes one of the best-characterized toxins secreted by this system (27), was present in all stool strains and in ca. 30% of the blood and wound strains (Table 2). Interestingly, eight of the strains simultaneously possessed the act and the TTSS genes, which probably reflects the synergistic role of these genes in quorum sensing (20).
TABLE 2.
Distribution of virulence genes among 25 clinical Aeromonas aquariorum strains
| Origin (no. of strains) | No. (%) of strains with gene |
|||||
|---|---|---|---|---|---|---|
| aerolysin/hemolysina | actb | altc | astc | aexTd | ascF-ascGd | |
| Blood (11) | 11 (100) | 4 (36.6) | 3 (27.2) | 8 (72.7) | 4 (36.3) | 7 (63.6) |
| Wound (6) | 6 (100) | 4 (66.6) | 1 (16.6) | 5 (83.3) | 2 (33.3) | 4 (66.6) |
| Feces (3) | 3 (100) | 0 | 3 (100) | 3 (100) | 3 (100) | 3 (100) |
| Bilis (2) | 2 (100) | 1 (50.0) | 0 | 2 (100) | 0 | 0 |
| Joint fluid (1) | 1 (100) | 1 (100) | 0 | 1 (100) | 0 | 1 (100) |
| Sputum (1) | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 |
| Ascitis (1) | 1 (100) | 1 (100) | 0 | 1 (100) | 0 | 1 (100) |
| Total (25) | 25 (100) | 11 (44) | 7 (28) | 21 (84) | 9 (36) | 16 (64) |
Differentiation of A. aquariorum from other species.
The 16S rRNA gene-RFLP pattern obtained for the Taiwan and Spanish isolates was compared with those of reference strains of A. aquariorum, i.e., the type strain (DSM18362T); two strains previously identified as A. hydrophila subsp. dhakensis (LMG19559 and LMG15962T [15], now considered a synonym of A. aquariorum [16]); and the type strain of A. caviae (CECT838T). All the strains produced either the A. caviae RFLP pattern or a slightly similar pattern with extra bands (Fig. 1). This made us suspect that those strains from Taiwan identified as A. caviae by the 16S rRNA gene-RFLP method or other clinical strains of this species identified at our laboratory and received from Spanish hospitals could in fact belong to A. aquariorum. To evaluate this, we screened the 73 A. caviae strains from Taiwan, as well as 65 strains received from Spanish hospitals, that showed the A. caviae RFLP pattern for the production of acid from d-cellobiose (positive for A. caviae, negative for A. aquariorum [17]). None of the 73 Taiwan strains and only 6 of the Spanish strains, recovered from the feces of patients with diarrhea, were suspected to be A. aquariorum due to their negative production of acid from d-cellobiose. However, the rpoD gene sequences of these strains (data not shown) confirmed only two strains (33%) as belonging to A. aquariorum, while the remaining four belonged to A. caviae. These results showed that some A. caviae strains did not produce acid from d-cellobiose (1, 18) and, furthermore, that A. aquariorum could be hidden under A. caviae strains. The positive production of lysine decarboxylase, gas from d-glucose, and H2S from cysteine of A. aquariorum are also traits that differentiate it from A. caviae, which is negative for these tests. Separation from A. hydrophila can be obtained by the fact that the new species does not utilize l-lactate (17). Strains with such characteristics should be suspected to belong to A. aquariorum.
The sequences of 16S rRNA genes were considered a reliable and straightforward tool for characterizing Aeromonas spp. (10) and have increasingly been used for identifying clinical strains (3, 13, 18, 24). However, we now know that this gene is unreliable for identification below the genus level (5 and A. Alperi et al., submitted). This is due to the high similarity between the sequences of the 16S rRNA genes of the different species and to the existence of nucleotide polymorphisms among the rrn operons of this gene (i.e., microheterogeneities) reported to occur in 8% of strains (5). Furthermore, these microheterogeneities are more prevalent in clinical strains of A. caviae and A. veronii than in environmental strains (5). In fact, the 16S rRNA gene sequence of the type strain of A. aquariorum only differs in three nucleotides from that of A. caviae (17), and microheterogeneities have been described in all three of these positions for strains of the latter species (5). In the present study, these limitations have been overcome by the use of housekeeping genes, such as rpoD and gyrB.
Epidemiologically related strains.
Three A. aquariorum isolates (A2-126, A2-131, and A2-96) showed identical gyrB and rpoD sequences (Fig. 2), indicating that they may be derived from the same clone. Genotyping with enterobacterial repetitive intergenic consensus-PCR using the primers and conditions described previously (21) confirmed that they belonged to the same genotype (data not shown). These strains also bear the same virulence genes and showed resistance to amoxicillin, amoxicillin-clavulanic acid, cephalothin, cefoxitin, erythromycin, and tetracycline. In a retrospective search of the origin of those strains, we noticed that two of them (A2-126 and A2-131) were isolated from a 69-year-old patient with acute cholangitis, one from a bile sample obtained from endoscopic nasobiliary drainage and the other 14 days later from a surgical wound of the abdominal wall after a cholecystectomy. This represents a new case of surgical site infection (SSI). Aeromonas SSIs have been poorly studied but are not rare, as we demonstrated in a recent review (25). The third strain (A2-96) was isolated from an ascites culture 7 months earlier in a 70-year-old patient who presented acute appendicitis complicated with peritonitis. The fact that both patients show the same genotype suggests a probable common source of infection, but the epidemiological relationship of the patients could not be established. Cholangitis is the most common Aeromonas infection of the hepatobiliary systems, with an incidence of isolation from bile samples ranging from 1.3 to 2.9% (10). Cases of secondary Aeromonas peritonitis due to appendix perforation have also been described (10, 14).
The present study is the first description of the implication of the new species, A. aquariorum, in clinical extraintestinal infections. Previously, as was mentioned above, this species had been reported from patients with diarrhea in Bangladesh under the name A. hydrophila subsp. dhakensis (15).
The medical relevance of these findings lies in the fact that the clinical strains of A. aquariorum possess many virulence genes that can potentially play an important role in the development of the infection. Clinicians should be aware of this new important Aeromonas species, which can be associated with diarrhea; bacteremia; wound infections, including SSI; and several other extraintestinal infections, as can be seen from the diversity of clinical samples in which it was recovered. It is likely that A. aquariorum could be distributed globally, probably unrecognized under A. hydrophila or A. caviae. However, more studies will be necessary to confirm this.
Acknowledgments
The MDC's work has been supported by grant no. IMIDTA/2007/68 from IMPIVA, Generalitat Valenciana, Spain. M. J. Saavedra was the recipient of a grant (no. SFRH/BSAB/774/2008) from the Fundação para a Ciênciae Tecnologia. Part of this work was also supported by funds from the European Commission for the HEALTHY WATER Project (FOOD-CT-2006-036306).
The authors are solely responsible for the content of this publication and it does not represent the opinion of the European Commission. The European Commission is not responsible for any use that might be made of data appearing herein.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Abbott, S. L., W. K. Cheung, and J. M. Janda. 2003. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 41:2348-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera-Arreola, M. G., C. Hernández-Rodríguez, G. Zúñiga, M. J. Figueras, R. A. Garduño, and G. Castro-Escarpulli. 2007. Virulence potential and genetic diversity of Aeromonas caviae, Aeromonas veronii, and Aeromonas hydrophila clinical isolates from Mexico and Spain: a comparative study. Can. J. Microbiol. 53:877-887. [DOI] [PubMed] [Google Scholar]
- 3.Al-Benwan, K., S. Abbott, G. Huys, and M. J. Albert. 2007. Cystitis caused by Aeromonas caviae. J. Clin. Microbiol. 45:2348-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert, M. J., M. Ansaruzzaman, K. A. Talukder, A. K. Chopra, I. Kuhn, M. Rahman, A. S. Faruque, M. S. Islam, R. B. Sack, and R. Mollby. 2000. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 38:3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alperí, A., M. J. Figueras, I. Inza, and A. J. Martinez-Murcia. 2008. Analysis of 16S rRNA gene mutations in a subset of Aeromonas strains and their impact in species delineation. Int. Microbiol. 11:185-194. [PubMed] [Google Scholar]
- 6.Borrell, N., S. G. Acinas, M. J. Figueras, and A. J. Martínez-Murcia. 1997. Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S rRNA genes. J. Clin. Microbiol. 35:1671-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chacón, M. R., M. J. Figueras, G. Castro-Escarpulli, L. Soler, and J. Guarro. 2003. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie van Leeuwenhoek 84:269-278. [DOI] [PubMed] [Google Scholar]
- 8.Chacón, M. R., L. Soler, E. A. Groisman, J. Guarro, and M. J. Figueras. 2004. Type III secretion system genes in clinical Aeromonas isolates. J. Clin. Microbiol. 42:1285-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopra, A. K., X. Xu, D. Ribardo, M. Gonzalez, K. Kuhl, J. W. Peterson, and C. W. Houston. 2000. The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect. Immun. 68:2808-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueras, M. J. 2005. Clinical relevance of Aeromonas. Rev. Med. Microbiol. 16:145-153. [Google Scholar]
- 11.Figueras, M. J., L. Soler, M. R. Chacon, J. Guarro, and A. J. Martínez-Murcia. 2000. Extended method for discrimination of Aeromonas spp. by 16S rDNA RFLP analysis. Int. J. Syst. Evol. Microbiol. 50:2069-2073. [DOI] [PubMed] [Google Scholar]
- 12.Figueras, M. J., A. Suarez-Franquet, M. R. Chacón, L. Soler, M. Navarro, C. Alejandre, B. Grasa, A. J. Martínez-Murcia, and J. Guarro. 2005. First record of the rare species Aeromonas culicicola from a drinking water supply. Appl. Environ. Microbiol. 71:538-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua, H. T., C. Bollet, S. Tercian, M. Drancourt, and D. Raoult. 2004. Aeromonas popoffii urinary tract infection. J. Clin. Microbiol. 42:5427-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, L. J., H. P. Chen, T. L. Chen, L. K. Siu, C. P. Fung, F. Y. Lee, and C. Y. Liu. 2006. Secondary Aeromonas peritonitis is associated with polymicrobial ascites culture and absence of liver cirrhosis compared to primary Aeromonas peritonitis. APMIS 114:772-778. [DOI] [PubMed] [Google Scholar]
- 15.Huys, G., P. Kämpfer, M. J. Albert, I. Kühn, R. Denys, and J. Swings. 2002. Aeromonas hydrophila subsp. dhakensis subsp. nov., isolated from children with diarrhoea in Bangladesh, and extended description of Aeromonas hydrophila subsp. hydrophila (Chester 1901) Stanier 1943 (approved lists 1980). Int. J. Syst. Evol. Microbiol. 52:705-712. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Murcia, A., A. Monera, A. Alperi, M. J. Figueras, and M. J. Saavedra. 2009. Phylogenetic evidence indicated that Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 is a synonym of Aeromonas aquariorum sp. nov. Martínez-Murcia et al. Curr. Microbiol. 58:76-80. [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Murcia, A. J., M. J. Saavedra, V. R. Mota, T. Maier, E. Stackebrandt, and S. Cousin. 2008. Aeromonas aquariorum sp. nov., isolated from aquaria of ornamental fish. Int. J. Syst. Evol. Microbiol. 58:1169-1175. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, M. T., D. A. Enoch, K. A. Harris, and J. A. Karas. 2006. Aeromonas veronii biovar sobria bacteraemia with septic arthritis confirmed by 16S rDNA PCR in an immunocompetent adult. J. Med. Microbiol. 55:241-243. [DOI] [PubMed] [Google Scholar]
- 19.Sha, J., E. V. Kozlova, and A. K. Chopra. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sha, J., L. Pillai, A. A. Fadl, C. L. Galindo, T. E. Erova, and A. K. Chopra. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 73:6446-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soler, L., M. J. Figueras, M. R. Chacón, J. Guarro, and A. J. Martínez-Murcia. 2003. Comparison of three molecular methods for typing Aeromonas popoffii isolates. Antonie van Leeuwenhoek 83:341-349. [DOI] [PubMed] [Google Scholar]
- 22.Soler, L., F. Marco, J. Vila, M. R. Chacón, J. Guarro, and M. J. Figueras. 2003. Evaluation of two miniaturized systems, MicroScan W/A and BBL Crystal E/NF, for identification of clinical isolates of Aeromonas spp. J. Clin. Microbiol. 41:5732-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soler, L., M. A. Yañez, M. R. Chacón, M. G. Aguilera-Arreola, V. Catalan, M. J. Figueras, and A. J. Martínez-Murcia. 2004. Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 54:1511-1519. [DOI] [PubMed] [Google Scholar]
- 24.Steinfeld, S., C. Rossi, N. Bourgeois, I. Mansoor, J. P. Thys, and T. Appelboom. 1998. Septic arthritis due to Aeromonas veronii biotype sobria. Clin. Infect. Dis. 27:402-403. [DOI] [PubMed] [Google Scholar]
- 25.Tena, D., C. Aspíroz, M. J. Figueras, A. González-Praetorius, M. J. Aldea, A. Alperí, and J. Bisquert. 2009. Surgical site infection due to Aeromonas species: report of nine cases and literature review. Scan. J. Infect. Dis. 41:164-170. [DOI] [PubMed] [Google Scholar]
- 26.Vilches, S., C. Urgell, S. Merino, M. Chacón, L. Soler, G. Castro-Escarpulli, M. J. Figueras, and J. M. Tomás. 2004. Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl. Environ. Microbiol. 70:6914-6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilches, S., M. Wilhelms, H. B. Yu, K. Y. Leung, J. M. Tomás, and S. Merino. 2008. Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system. Microb. Pathog. 44:1-12. [DOI] [PubMed] [Google Scholar]