Abstract
Cryptococcus neoformans and Cryptococcus gattii are closely related pathogenic fungi. Cryptococcus neoformans is ecologically widespread and affects primarily immunocompromised patients, while C. gattii is traditionally found in tropical climates and has been reported to cause disease in immunocompetent patients. l-Canavanine glycine bromothymol blue (CGB) agar can be used to differentiate C. neoformans and C. gattii, but there are few reports of its performance in routine clinical practice. Growth of C. gattii on CGB agar produces a blue color, indicating the assimilation of glycine, while C. neoformans fails to cause a color change. Using reference and clinical strains, we evaluated the ability of CGB agar and D2 large ribosomal subunit DNA sequencing (D2 LSU) to differentiate C. neoformans and C. gattii. One hundred two yeast isolates were screened for urease activity, melanin production, and glycine assimilation on CGB agar as well as by D2 sequencing. Seventeen of 17 (100%) C. gattii isolates were CGB positive, and 54 of 54 C. neoformans isolates were CGB negative. Several yeast isolates other than the C. gattii isolates were CGB agar positive, indicating that CGB agar cannot be used alone for identification of C. gattii. D2 correctly identified and differentiated all C. gattii and C. neoformans isolates. This study demonstrates that the use of CGB agar, in conjunction with urea hydrolysis and Niger seed agar, or D2 LSU sequencing can be reliably used in the clinical laboratory to distinguish C. gattii from C. neoformans. We describe how CGB agar and D2 sequencing have been incorporated into the yeast identification algorithm in our laboratory.
Cryptococcus gattii (formerly Cryptococcus neoformans var. gattii) differs from the closely related C. neoformans in several ways, including a contrasting host profile and a reduced susceptibility to certain antifungal agents (19, 20). Cryptococcus gattii has traditionally been thought to be geographically restricted to tropical and subtropical climates, although recent reports indicate that it can be found worldwide, including in regions with distinctly nontropical climates, like the Pacific Northwest (2, 4, 8, 12, 14, 17, 19). Clinically, the majority of cryptococcosis cases that occur in AIDS patients and other immunocompromised hosts are caused by C. neoformans var. neoformans or C. neoformans var. grubii. In contrast, C. gattii has been reported to cause meningoencephalitis and pulmonary infections in hosts who are generally immune competent (9, 14, 18).
Since C. gattii is recognized as an emerging pathogen, it is important for the clinical microbiology laboratory to accurately differentiate it from C. neoformans. A recent proficiency testing survey administered by the New York State Department of Health indicated that only 7/140 (5%) clinical laboratories participating in the event were able to correctly identify C. gattii, while the remaining 95% of laboratories surveyed misidentified the isolate as C. neoformans (15). The critique that followed this proficiency event indicated that the inability to correctly identify C. gattii was largely due to the failure to routinely use differential agar. In this report, we describe the implementation of two approaches for the routine identification of C. gattii in a high-volume clinical laboratory. The first approach uses the simple, previously reported biochemical test evaluating growth on the selective medium l-canavanine glycine bromothymol blue (CGB) agar (10, 13, 16). The agar method is low complexity and cost-effective and can be easily incorporated into the workflow of clinical laboratories of any size. We describe the ability of CGB to differentiate C. gattii from C. neoformans and other yeast encountered in our clinical practice. The second approach is a molecular method that utilizes DNA sequencing of the D2 region of the fungal 28S large ribosomal subunit to distinguish C. neoformans from C. gattii. Others have reported differentiation of C. neoformans and C. gattii by the use of sequencing of the internal transcribed spacer region (3, 6, 7), but to our knowledge, this is the first report of the use of D2 large ribosomal subunit region sequencing for the identification of C. gattii.
(This study was presented in part at the 107th General Meeting of the American Society for Microbiology, Toronto, ON, Canada, 22 to 25 May 2007.)
MATERIALS AND METHODS
Organisms.
One hundred two yeast isolates were obtained from reference sources (proficiency testing events [n = 2], American Type Culture Collection [n = 40], and fresh clinical specimens [n = 60]). Ninety-one isolates of cryptococci were tested, including C. gattii (n = 17), C. neoformans (n = 54), Cryptococcus albidus (n = 4), Cryptococcus curvatus (n = 1), Cryptococcus laurentii (n = 9), Cryptococcus luteolus (n = 1), Cryptococcus magnus (n = 1), Cryptococcus uniguttulatus (n = 2), and Cryptococcus species that are not C. neoformans (not further identified; n = 2). Eleven noncryptococcus isolates were used to evaluate the specificity of both methods and included Candida albicans (n = 1), Candida glabrata (n = 1), Candida guilliermondii (n = 1), Candida krusei (n = 1), Candida parapsilosis (n = 1), Candida tropicalis (n = 1), Trichosporon asahii (n = 1), Trichosporon loubieri (n = 1), and Trichosporon mucoides (n = 3). The isolates were obtained from a variety of specimen sources, including cerebrospinal fluid, blood, lung tissue, bronchoalveolar lavage fluid, bronchial washings, tracheal secretions, sputum, scalp swabs, sterile body fluid, and fresh tissues. A Mayo Clinic Institutional Review Board approved the use of all isolates.
Biochemical testing.
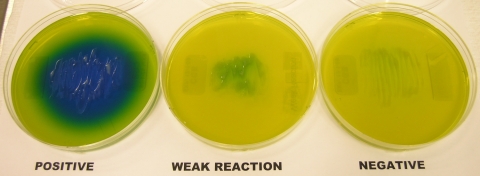
Organisms were subcultured on Sabouraud dextrose agar (Emmons modification) (Becton Dickinson [BD], Sparks, MD) or inhibitory mold agar (BD), urea agar (BD), and laboratory-prepared Niger seed agar plates. CGB agar plates were prepared as previously described (10) and were inoculated by using a loop to streak the organism down the center of the plate and then in a side-to-side motion. Various incubation temperatures were evaluated for the CGB agar reaction, which showed strong reactions for C. gattii at 30°C and at room temperature and notably weaker reactions at 37°C after 48 h (data not shown). Therefore, the CGB agar plates were incubated at room temperature for this study. Reactions on CGB agar were considered positive if a vivid cobalt blue color was produced after 48 h of incubation at room temperature and negative if the color of the CGB medium remained yellow (Fig. 1). A residual light-blue color around the inoculum was interpreted as a negative reaction.
FIG. 1.
Reactions of various cryptococci on CGB agar. The left plate depicts a positive reaction with C. gattii; the middle plate shows a weak reaction around the inoculum, which is interpreted as negative; and the right plate shows a negative reaction with C. neoformans.
Supplemental morphological and biochemical tests were used to help differentiate the yeasts evaluated in this study. Cornmeal agar morphology and stains (e.g., India ink) can be useful in differentiating cryptococci from noncryptococcal yeasts. In addition, urea hydrolysis was evaluated by streaking the isolate on the surface of a laboratory-prepared urea agar slant, followed by incubating at 30°C for 48 h. A positive reaction is one in which the agar turns red due to a rise in the pH of the medium containing a phenol red indicator following urea hydrolysis. Cryptococci will hydrolyze urea, while noncryptococci will fail to produce a reaction, with the notable exception of Trichosporon spp. and some Candida species. Niger seed agar was used to detect the production of melanin generated by the reaction of phenol oxidase from C. neoformans or C. gattii with caffeic acid present in the Niger seed. A positive reaction results in the production of a dark-brown pigment in the agar within 7 days of incubation at room temperature. Cryptococci other than C. neoformans or C. gattii do not produce the brown pigment on Niger seed agar.
D2 sequencing.
An approximately 300-bp portion of the D2 region of the fungal 28S ribosome was sequenced as previously described (5). Sequences were compared to a custom, laboratory-developed library populated by sequencing reference strains or conventionally identified clinical isolates. An acceptable identification was considered to be a 100% match with the library entry, provided that the sequence length obtained from the test isolate was at least 90% of the library sequence length and the quality score for the sequence was >30. Sequences for organisms that exhibited a <100% match to the laboratory-developed library sequences were also compared to sequences in a commercially available library (MicroSeq; Applied Biosystems, Foster City, CA) and the NCBI GenBank website (http://www.ncbi.nlm.nih.gov/). An in-house-developed database was selected as the primary database because commercially available libraries often have poor coverage for the cryptococci and it is not uncommon for sequences deposited in the public GenBank database to contain incorrect or incomplete entries (3, 21).
RESULTS
CGB agar.
One hundred two yeast isolates were evaluated using CGB agar, urea agar, and Niger seed agar (Table 1). By the use of CGB agar, 17/17 (100%) of C. gattii isolates had a positive reaction and turned the medium blue, while 54/54 (100%) C. neoformans isolates had a negative reaction, failing to produce a color change in the medium. A single C. neoformans isolate (1/54 [2%]) produced a weak reaction on CGB agar, indicated by a light-blue color immediately adjacent to the inoculum streak lines, and this was deemed a negative reaction (Fig. 1). Several other Cryptococcus species also produced positive reactions on CGB agar, including C. curvatus (1/1 [100%]), C. laurentii (4/9 [44%]), and C. luteolus (1/1 [100%]). Other yeasts that had positive reactions on CGB agar included C. parapsilosis (1/1 [100%]), T. asahii (1/1 [100%]) and T. mucoides (3/3 [100%]). As expected, all cryptococci (91/91 [100%]) and all the Trichosporon isolates (5/5 [100%]) were able to hydrolyze urea. Nearly all the C. gattii (16/17 [94%]) and C. neoformans (53/54 [98%]) isolates were Niger seed agar positive, while none of the noncryptococcus isolates produced melanin on Niger seed agar.
TABLE 1.
Biochemical test results
| Identification | No. of isolates tested | No. (%) of isolates positive for: |
||
|---|---|---|---|---|
| Urease production | Melanin production on birdseed agar | Glycine assimilation on CGB agar | ||
| C. gattii | 17 | 17/17 (100) | 16/17 (94) | 17/17 (100) |
| C. neoformans | 54 | 54/54 (100) | 53/54 (98) | 0/54 (0)d |
| Other cryptococcia | 20 | 20/20 (100) | 0/20 (0) | 6/20 (30) |
| Trichosporonb | 5 | 5/5 (100) | 0/5 (0) | 4/5 (80) |
| Other yeastc | 6 | Not tested | Not tested | 1/6 (17) |
C. albidus (n = 4), C. curvatus (n = 1), C. laurentii (n = 9), C. luteolus (n = 1), C. uniguttulatus (n = 2), C. magnus (n = 1), and other Cryptococcus spp. (n = 2).
T. asahii (n = 1), T. loubieri (n = 1), and T. mucoides (n = 3).
C. albicans (n = 1), C. glabrata (n = 1), C. guilliermondii (n = 1), C. krusei (n = 1), C. parapsilosis (n = 1), and C. tropicalis (n = 1).
One isolate had a weak reaction around the inoculum only.
D2 sequencing.
D2 sequencing correctly identified 76/91 (84%) of cryptococci tested with 100% sequence identity matches, using our laboratory-developed custom library (Table 2). Of note, D2 sequencing by the use of the laboratory-developed library was successful in identifying 17/17 (100%) of C. gattii isolates and 51/54 (94%) of C. neoformans isolates. The top results for all Cryptococcus species with <100% matches in the laboratory-developed database are shown in Table 3. The MicroSeq library correctly identified 48/54 isolates of C. neoformans and 2/9 isolates of C. laurentii, while C. gattii is not represented in this library.
TABLE 2.
Isolates demonstrating a 100% sequence match with library sequences
| Phenotypic identification | No. (%) of isolates correctly identified by laboratory-developed library |
|---|---|
| C. albidus | 1/4 (25) |
| C. curvatus | 0/1 (0) |
| C. gattii | 17/17 (100) |
| C. laurentii | 5/9 (56) |
| C. luteolus | 0/1 (0) |
| C. magnus | 0/1 (0) |
| C. neoformans | 51/54 (94) |
| C. uniguttulatus | 2/2 (100) |
| Cryptococcus sp. | 1/2 (50) |
TABLE 3.
D2 sequencing results for Cryptococcus sp. with <100% match in the laboratory-developed library
| Isolate | Top match in: |
||
|---|---|---|---|
| Laboratory-developed library | MicroSeq library | NCBI GenBank database | |
| C. albidus | 96% C. albidus | 93% Filobasidium uniguttulatum | 100% Ceratocystis bhutanensis |
| C. albidus | 100% Yeast unidentified | 96% C. albidus | 100% C. liquefaciens |
| C. albidus | 95% C. uniguttulatus | 100% Cryptococcus ater | 100% C. magnus |
| C. curvatus | 94% T. mucoides | 99% C. curvatus | 100% C. curvatus |
| C. laurentii | 88% C. laurentii | 90% Cryptococcus dimennae | 100% C. laurentii |
| C. laurentii | 99% C. laurentii | 95% C. laurentii | 99% Cryptococcus sp. |
| C. laurentii | 99% C. laurentii | 92% Cryptococcus heveanensis | 100% C. laurentii |
| C. laurentii | 85% Cryptococcus sp. | 91% C. dimennae | 100% Cryptococcus victoriae |
| C. luteolus | 90% C. neoformans | 90% Filobasidiella neoformans | 100% C. luteolus |
| C. magnus | 95% C. uniguttulatus | 100% C. ater | 100% C. magnus |
| C. neoformans | 99% C. neoformans | 99% Filobasidiella neoformans | 100% C. neoformans |
| C. neoformans | 99% C. neoformans | 99% Filobasidiella neoformans | 100% C. neoformans |
| C. neoformans | 99% C. neoformans | 99% Filobasidiella neoformans | 100% C. neoformans |
| Cryptococcus sp. not C. neoformans | 100% Yeast unidentified | 96% C. albidus | 100% C. liquefaciens |
DISCUSSION
The biochemical basis for the differentiation of C. neoformans and C. gattii has been elucidated by Min and Kwon-Chung (13). Cryptococcus gattii grows in the presence of l-canavanine and utilizes glycine as the sole carbon and nitrogen source, which raises the pH of the medium and causes the bromothymol blue indicator in the medium to turn the agar a blue color. Cryptococcus neoformans will not grow in the presence of canavanine and does not utilize glycine, so the CGB medium color remains unchanged. Currently available commercial methods for yeast identification, such as API 20C AUX (bioMérieux, Durham, NC), Vitek (bioMérieux), and MicroScan (Siemens, West Sacramento, CA), do not differentiate between C. neoformans and C. gattii. Serotyping is useful for differentiation of C. gattii from C. neoformans. Unfortunately, the single commercial kit previously used for serotyping of cryptococci (Crypto-Check kit; Iatron Inc., Tokyo, Japan) is no longer available, and very few laboratories are routinely serotyping cryptococci today. Recently, two groups have reported multiplex PCR and liquid array-based methods for the differentiation of C. neoformans and C. gattii (1, 11), but this technology is not routinely utilized by most clinical laboratories at this time.
Many clinical laboratories can readily differentiate cryptococci from other yeasts on the basis of their microscopic morphologies and the urease test results. Cryptococci are urease positive and most other fungi, with the notable exception of Trichosporon spp. and some Candida spp., are urease negative. Cryptococcus neoformans and C. gattii can be differentiated from other Cryptococcus species by observing the production of melanin on Niger seed agar or using the caffeic acid disk test (Thermo Fisher Scientific, Lenexa, KS). Differentiation of C. neoformans from C. gattii has traditionally been challenging but, as demonstrated by the data in Table 1, CGB agar is an effective means of differentiating these two species, with C. gattii turning the CGB agar blue and C. neoformans having no effect on the medium color. A single isolate of C. neoformans gave a weak reaction on CGB agar, but it was easily distinguished from a true positive by restriction of the blue color to the area immediately surrounding the inoculum streaks. Thirty percent (6/20) of non-C. neoformans, non-C. gattii species cryptococci tested also gave positive reactions on CGB agar, but these could easily be differentiated by a negative Niger seed agar test.
In our laboratory, we have incorporated the routine inoculation of CGB agar for all suspected isolates of C. neoformans into our yeast identification algorithm. Inoculation of the plate requires <1 min of technologist time, and the plate is incubated simultaneously with the urea slant and the Niger seed agar plate. The CGB agar 48-h results provide differentiation between C. neoformans and C. gattii with no noticeable increase in technologist hands-on time, at a cost of only $0.55 per plate. Since implementing the use of CGB agar in July 2007, we have identified a single C. gattii isolate from a clinical specimen of a patient from the eastern coast of the United States. Since C. gattii has not been reported yet in the upper Midwest, the paucity of C. gattii isolates in our clinical practice is probably not surprising. However, given C. gattii's increasing profile worldwide as a pathogen and its documented spread to nontraditional areas of endemicity, we believe CGB agar is an inexpensive and convenient way to screen for emergence in our patient population.
D2 large ribosomal subunit DNA sequencing can be used for identification of fungi following growth in pure culture (5). The results can be available within 24 h after growth of the organism, but this methodology is technically challenging, can be expensive, and is generally done only in reference laboratories or larger clinical laboratories. Accurate identification of the organism is heavily dependent on the entries in the library that is being searched, and many commercial sequence libraries, such as the MicroSeq D2 library, do not currently include C. gattii. While we are routinely using CGB agar in our yeast identification algorithm, we have found utility for D2 sequencing to identify selected isolates that demonstrate unusual reactions on traditional agar media. In this study, two isolates, one C. neoformans and one C. gattii, were Niger seed agar negative and therefore would have traditionally been identified as “Cryptococcus spp., not C. neoformans” but these isolates were able to be identified using D2 DNA sequencing. Interestingly, each of these isolates did produce the expected reaction on CGB agar. As another example, the single isolate of C. neoformans that produced a weak reaction around the inoculum on CGB agar was confirmed to be C. neoformans by DNA sequencing.
D2 sequencing identified all 91 Cryptococcus sp. isolates to the correct genus level and 76/91 (84%) isolates to the correct species level, using a 100% sequence match criterion. Several of the Cryptococcus species tested were not represented in our laboratory-developed library and therefore gave lower-than-acceptable sequence matches. It is important for the diagnostic laboratory to know which database is being searched and what the criteria are for an acceptable match. For example, in Table 3, an isolate was found to be a 96% match to C. albidus in our laboratory-developed library, a 93% match to Filobasidium uniguttulatum, a teleomorph of C. uniguttulatus, in the commercially available MicroSeq library, and a 100% match to Ceratocystis bhutanensis in the NCBI GenBank database. These sequencing results should not be reported to the clinician unless they meet the match criteria discussed previously, including the critical review of published sequences in GenBank for C. bhutanensis.
In summary, CGB agar provides a simple, low-cost, and efficient method to routinely differentiate C. neoformans from C. gattii in the clinical laboratory. It can easily be incorporated into the laboratory screening algorithm, which already likely includes urea and Niger seed agars, and results can be available within 48 h. On the other hand, D2 DNA sequencing can identify most cryptococci seen in the clinical laboratory setting to the species level within 24 h after growth in pure culture, and it can be used to differentiate isolates that give weak reactions on CGB agar or that have aberrant Niger seed agar results. Sequencing is becoming more cost-effective and less labor-intensive, and the turnaround time required for results continues to decrease. Given the recent emergence of C. gattii as a human pathogen, it is important for laboratories to be able to distinguish this organism from closely related species, especially C. neoformans. Both CGB agar and D2 DNA sequencing can provide definitive identification and discrimination of C. neoformans and C. gattii.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Bovers, M., M. R. Diaz, F. Hagen, L. Spanjaard, B. Duim, C. E. Visser, H. L. Hoogveld, J. Scharringa, I. M. Hoepelman, J. W. Fell, and T. Boekhout. 2007. Identification of genotypically diverse Cryptococcus neoformans and Cryptococcus gattii isolates by Luminex xMAP technology. J. Clin. Microbiol. 45:1874-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrnes, E. J., III, R. J. Bildfell, S. A. Frank, T. G. Mitchell, K. A. Marr, and J. Heitman. 2009. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J. Infect. Dis. 199:1081-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciardo, D. E., G. Schar, E. C. Bottger, M. Altwegg, and P. P. Bosshard. 2006. Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J. Clin. Microbiol. 44:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colom, M. F., S. Frases, C. Ferrer, A. Jover, M. Andreu, S. Reus, M. Sanchez, and J. M. Torres-Rodriguez. 2005. First case of human cryptococcosis due to Cryptococcus neoformans var. gattii in Spain. J. Clin. Microbiol. 43:3548-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall, L., S. Wohlfiel, and G. D. Roberts. 2003. Experience with the MicroSeq D2 large-subunit ribosomal DNA sequencing kit for identification of commonly encountered, clinically important yeast species. J. Clin. Microbiol. 41:5099-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaocharoen, S., B. Wang, K. M. Tsui, L. Trilles, F. Kong, and W. Meyer. 2008. Hyperbranched rolling circle amplification as a rapid and sensitive method for species identification within the Cryptococcus species complex. Electrophoresis 29:3183-3191. [DOI] [PubMed] [Google Scholar]
- 7.Katsu, M., S. Kidd, A. Ando, M. L. Moretti-Branchini, Y. Mikami, K. Nishimura, and W. Meyer. 2004. The internal transcribed spacers and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans species complex. FEMS Yeast Res. 4:377-388. [DOI] [PubMed] [Google Scholar]
- 8.Kidd, S. E., P. J. Bach, A. O. Hingston, S. Mak, Y. Chow, L. MacDougall, J. W. Kronstad, and K. H. Bartlett. 2007. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg. Infect. Dis. 13:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon-Chung, K. J., and J. E. Bennett. 1992. Cryptococcosis. In K. J. Kwon-Chung and J. E. Bennett (ed.), Medical mycology. Lea & Febiger, Philadelphia, PA.
- 10.Kwon-Chung, K. J., I. Polacheck, and J. E. Bennett. 1982. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J. Clin. Microbiol. 15:535-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leal, A. L., J. Faganello, M. C. Bassanesi, and M. H. Vainstein. 2008. Cryptococcus species identification by multiplex PCR. Med. Mycol. 46:377-383. [DOI] [PubMed] [Google Scholar]
- 12.MacDougall, L., S. E. Kidd, E. Galanis, S. Mak, M. J. Leslie, P. R. Cieslak, J. W. Kronstad, M. G. Morshed, and K. H. Bartlett. 2007. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg. Infect. Dis. 13:42-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min, K. H., and K. J. Kwon-Chung. 1986. The biochemical basis for the distinction between the two Cryptococcus neoformans varieties with CGB medium. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 261:471-480. [DOI] [PubMed] [Google Scholar]
- 14.Morgan, J., K. M. McCarthy, S. Gould, K. Fan, B. Arthington-Skaggs, N. Iqbal, K. Stamey, R. A. Hajjeh, and M. E. Brandt. 2006. Cryptococcus gattii infection: characteristics and epidemiology of cases identified in a South African province with high HIV seroprevalence, 2002-2004. Clin. Infect. Dis. 43:1077-1080. [DOI] [PubMed] [Google Scholar]
- 15.New York State Department of Health. 2005. Mycology critique, Mycology Proficiency Testing Program, May 2005. New York State Department of Health, Albany, NY.
- 16.Nishikawa, M. M., O. D. Sant'Anna, M. S. Lazera, and B. Wanke. 1996. Use of d-proline assimilation and CGB medium for screening Brazilian Cryptococcus neoformans isolates. J. Med. Vet. Mycol. 34:365-366. [PubMed] [Google Scholar]
- 17.Olivares, L. R., K. M. Martinez, R. M. Cruz, M. A. Rivera, W. Meyer, R. A. Espinosa, R. L. Martinez, and G. M. Santos. 2009. Genotyping of Mexican Cryptococcus neoformans and C. gattii isolates by PCR-fingerprinting. Med. Mycol. 20:1-9. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 18.Sharp, S. E. 2009. Cryptococcus gattii. Clin. Microbiol. Newsl. 31:84-86. [Google Scholar]
- 19.Soares, B. M., D. A. Santos, L. M. Kohler, G. da Costa Cesar, I. R. de Carvalho, M. dos Anjos Martins, and P. S. Cisalpino. 2008. Cerebral infection caused by Cryptococcus gattii: a case report and antifungal susceptibility testing. Rev. Iberoam. Micol. 25:242-245. [PubMed] [Google Scholar]
- 20.Trilles, L., B. Fernandez-Torres, S. Lazera Mdos, B. Wanke, and J. Guarro. 2004. In vitro antifungal susceptibility of Cryptococcus gattii. J. Clin. Microbiol. 42:4815-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]