Abstract
A rapid SYBR green I real-time reverse transcription-PCR (RT-PCR) assay was developed to identify pandemic influenza H1N1 virus from clinical specimens in less than 1 h. Probe real-time RT-PCR influenza A/B, H1/H3, and swNP/swHA assays were modified into the same PCR program, which allows for rapid and simultaneous typing and subtyping of influenza viruses.
A swine-origin influenza A (H1N1) virus has spread rapidly across the globe and caused a pandemic in 2009. In this study, we have developed a rapid SYBR green I real-time reverse transcription-PCR (RT-PCR) method to identify pandemic influenza H1N1 virus. We have also modified the probe real-time RT-PCR assays reported by the U.S. CDC to be more rapid. The assays were used to identify the first 10 cases of pandemic influenza H1N1 virus infection in Taiwan.
The primers used for pandemic influenza strain H1N1 detection in the SYBR green I assay were designed by the Taiwan CDC according to the sequences of strain A/California/04/2009. The primers were SWH1LF (5′-ATTACTGGACACTAGTAGAGC-3′) and SWH1LR (5′-GCATTTCTTTCCATTGCGAA-3′), and they amplify a 97-bp fragment within the hemagglutinin (HA) gene. The assay was performed using the Roche LightCycler and LightCycler RNA master SYBR green I kit. The reaction mixture consisted of 5 μl RNA template, 8.2 μl water, 7.5 μl enzyme mix, a final concentration of 500 nM of each primer, and 3.25 mM Mn(acetate)2. The reaction mixture was first incubated at 61°C for 20 min, followed by 95°C for 30 s. It was then thermal-cycled for 45 cycles as follows: 95°C for 5 s, 54°C for 10 s, and 72°C for 10 s. The modified one-step probe real-time RT-PCR (swNP and swHA) assays were carried out using the primer and probe sets published by the U.S. CDC (3). The assays were conducted using the Roche LightCycler and could be completed within 80 min. The reaction mixture consisted of 5 μl RNA template, 3.8 μl water, 7.4 μl enzyme mix, a final concentration of 1 μM of each primer, 250 nM of probe, 1.3 μl of activator, and 1 μl of enhancer. Reaction mixtures were first incubated at 63°C for 3 min, followed by 95°C for 30 s. They were then thermal-cycled for 45 cycles as follows: 95°C for 10 s, 53°C for 30 s, and 72°C for 3 s. The primer and probe sets used for routine influenza screening (influenza A/B multiplex probe assay) and subtyping (influenza A H1N1/H3N2 multiplex probe assay) were used according to the methods described in published reports (1, 2). They could be conducted in the same program as swNP and swHA assays.
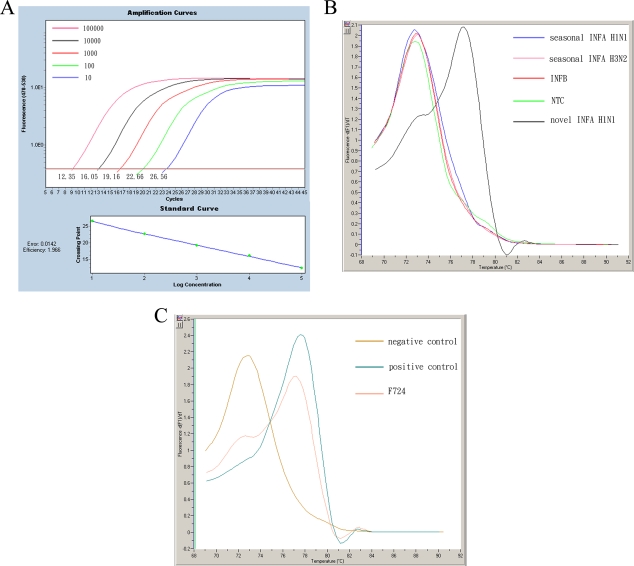
The sensitivity of the one-step SYBR green I assay was evaluated. Figure 1A shows that the detection limit of the assay was at least 10 copies of the HA gene. The specificity was tested with the other types/subtypes of influenza viruses. Melting curve analysis was used to distinguish the pandemic H1N1 virus from the others, and the results showed that only the pandemic H1N1 virus could be detected with a melting temperature (Tm) of 77.5°C. All of the nonpandemic H1N1 virus targets and the negative control (H2O) peaked at a Tm of 73°C, generated from primer dimers (Fig. 1B). The specificity of the assay was also tested with noninfluenza viruses, including parainfluenza virus types 1, 2, and 3, respiratory syncytial virus, adenovirus, rhinovirus, and herpes simplex virus. They were not detected in this assay (data not shown).
FIG. 1.
Sensitivity and specificity of the one-step SYBR green I real-time RT-PCR assay. (A) HA plasmids from strain A/California/07/2009 were serially diluted from 101 to 105 copies and submitted to the assay. Positive signals are obtained from all the diluted plasmids, with each CT value indicated. The linear response of the assay is shown by the standard curve in the bottom panel. (B) Various viral RNAs of human seasonal influenza A (H1N1), influenza A (H3N2), and influenza B viruses were submitted to the assay. Fluorescent signals with a Tm of 77.5°C are observed only from the pandemic influenza A (H1N1) virus, and the signals from the other viruses are the same as those of the negative control (H2O), with a Tm of 73°C. (C) Melting curve analysis of the first case of H1N1 infection in Taiwan (F724). Fluorescent signals from the positive control (A/Califorina/07/2009) and the negative control (H2O) are also indicated.
On 20 May 2009, an individual tested positive for strain H1N1 by the influenza A/B multiplex assay at a threshold cycle (CT) value of 32.9, but virus could not be detected by the H1N1/H3N2 multiplex assay. In the one-step SYBR green I assay, the melting curve analysis showed a fluorescent peak with a Tm of 77.5°C (Fig. 1C). In this case, we performed DNA sequencing of the PCR product amplified in the SYBR green I assay, and the result showed perfect sequence homology with strain A/California/07/2009. In the swNP and swHA assays, there were fluorescent signals with CT values of 33.5 and 37.4, respectively. The first case of pandemic H1N1 virus infection in Taiwan was thus confirmed.
From 20 to 27 May 2009, there were nine further cases of pandemic H1N1 infection, including the first domestic case, a friend of the fourth individual who was infected in Taiwan. The melting curve analysis also showed melting curves, with a Tm of 77.5°C, and the CT values of these nine specimens determined by influenza A/B multiplex, swNP, and swHA assays are shown in Table 1.
TABLE 1.
First 10 cases of novel H1N1 identified in Taiwan from 20 to 27 May 2009
| Case no. | Date infection confirmed | PCR product with Tm of 77.5°Ca |
CT of the specimen determined by indicated probe assay |
||
|---|---|---|---|---|---|
| FluA/B | swNP | swHA | |||
| 1 | May 20 | + | 32.9 | 33.5 | 37.4 |
| 2 | May 21 | + | 29.0 | 29.0 | 30.8 |
| 3 | May 21 | + | 33.2 | 33.9 | 36.2 |
| 4 | May 22 | + | 32.0 | 32.5 | 35.4 |
| 5 | May 22 | + | 33.3 | 31.8 | 34.5 |
| 6 | May 22 | + | 28.3 | 27.6 | 28.8 |
| 7 | May 24 | + | 34.9 | 33.6 | 37.3 |
| 8 | May 24 | + | 32.5 | 33.8 | 36.9 |
| 9 | May 24 | + | 32.3 | 32.8 | 34.2 |
| 10 | May 27 | + | 30.2 | 29.3 | 29.1 |
+, positive result as determined by SYBR green I assay.
For the detection of influenza virus, the real-time RT-PCR assay exhibits high levels of sensitivity and specificity and allows influenza infection to be confirmed rapidly. Therefore, this assay has become a good laboratory diagnostic method for influenza virus. In our laboratory, we have developed a rapid SYBR green I real-time RT-PCR method to identify pandemic H1N1 virus. Probe is not required for this assay, and it is faster (finished within 50 min) and less expensive than the probe real-time RT-PCR assays. We also modified the multiplex real-time RT-PCR influenza A/B, influenza A H1N1/H3N2, and swNP/swHA assays into the same PCR program, which allows the rapid and simultaneous typing and subtyping of influenza viruses.
Acknowledgments
We are indebted to Tim J. Harrison at the Department of Medicine, Royal Free and University College Medical School, for critical reading of the manuscript.
This study was supported by grants from the National Science Council (National Research Program for Genome Medicine; 98-0324-01-F-20) and the Centers for Disease Control, Department of Health, Taiwan, Republic of China.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Suwannakarn, K., S. Payungporn, T. Chieochansin, R. Samransamruajkit, A. Amonsin, T. Songserm, A. Chaisingh, P. Chamnanpood, S. Chutinimitkul, A. Theamboonlers, and Y. Poovorawan. 2008. Typing (A/B) and subtyping (H1/H3/H5) of influenza A viruses by multiplex real-time RT-PCR assays. J. Virol. Methods 152:25-31. [DOI] [PubMed] [Google Scholar]
- 2.Ward, C. L., M. H. Dempsey, C. J. Ring, R. E. Kempson, L. Zhang, D. Gor, B. W. Snowden, and M. Tisdale. 2004. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J. Clin. Virol. 29:179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. 2009. CDC protocol of real-time RT-PCR for swine influenza A (H1N1). W. H. O. Collaborating Center for Influenza, Centers for Disease Control, Atlanta, GA.