Abstract
Highly effective glycoconjugate vaccines exist against four of the five major pathogenic groups of meningococci: A, C, W-135, and Y. An equivalent vaccine against group B meningococci (menB) has remained elusive due to the poorly immunogenic capsular polysaccharide. A promising alternative, the investigational recombinant menB (rMenB)- outer membrane vesicle (OMV) vaccine, contains fHBP, NHBA (previously GNA2132), NadA, and outer membrane vesicles (OMVs) from the New Zealand MeNZB vaccine. MenB currently accounts for 90% of meningococcal disease in England and Wales, where the multilocus sequence type (ST) 269 (ST269) clonal complex (cc269) has recently expanded to account for a third of menB cases. To assess the potential cc269 coverage of the rMenB-OMV vaccine, English and Welsh cc269 isolates from the past decade were genetically characterized with respect to fHBP, NHBA, and NadA. All of the isolates harbored fHbp and nhba alleles, while 98% of the cc269 isolates were devoid of nadA. Subvariant profiling of fHbp, nhba, and porA against STs revealed the presence of two broadly distinct and well-defined clusters of isolates, centered around ST269 and ST275, respectively. An additional molecular marker, insertion sequence IS1301, was found to be present in 100% and <2% of isolates of the respective clusters. On the basis of the genetic data, the potential rMenB-OMV coverage of cc269 in England and Wales is high (up to 100%) within both clusters. Expression studies and serum bactericidal antibody assays will serve to enhance predictions of coverage and will augment ongoing studies regarding the significance of IS1301 within the ST269 cluster.
Despite the availability of effective antibiotics, the rapid onset and progression of invasive meningococcal disease are associated with high rates of mortality (5 to 10%) and severe sequelae in survivors (28). Four of the five major pathogenic groups (pathogenic groups A, C, W-135, and Y) are now covered by highly effective glycoconjugate vaccines that are immunogenic in infants and that induce immunological memory. In addition, the prevention of acquisition and carriage by the group C vaccine has been demonstrated to promote herd immunity (3). An equivalent vaccine for group B meningococci (menB), however, has remained elusive due its poorly immunogenic (α2-8)-linked capsular polysaccharide, which is also found in fetal neural tissue (38). Outer membrane vesicle (OMV) vaccines, in which PorA constitutes the major immunodominant antigen, have proven highly effective against menB clonal epidemics (2, 4, 16, 30), and efforts to broaden their coverage are ongoing (17, 37, 39). In an alternative approach, genome mining was used to search the menB strain MC58 genome for genes for putative surface proteins, which were then characterized for their global distribution and diversity, surface expression, and immunogenicity in a process termed “reverse vaccinology” (25). Of the antigens discovered, three scored particularly well in these respects: neisserial heparin-binding antigen (NHBA; previously known as genome-derived neisserial antigen 2132 [GNA2132]), neisserial adhesin A (NadA), and factor H binding protein (fHBP; also known as GNA1870 and lipoprotein 2086) (9, 25). NadA is a pathogenicity factor involved in host cell adhesion and invasion and is reported to be present in ∼50% of isolates tested; it has a low level of representation among carriage isolates and up to 100% coverage in some hypervirulent lineages (6). All isolates possess an nhba allele. The minimum pairwise amino acid identity between subvariants is reported to be about 54%; however, these exhibit good levels of immunological cross-reactivity (14, 25). The binding of heparin by NHBA at an arginine-rich region suggests a possible role in serum resistance (Novartis Vaccines, unpublished data). fHBP is a virulence factor that specifically binds to the human complement-regulating protein factor H, thereby enhancing serum resistance (18, 27). So far, all isolates have been shown to harbor an fHbp allele, and the antigen falls into one of three major variant groups: variant 1 (also known as family B) and variants 2 and 3 (also known jointly as family A). Collectively, NadA, NHBA, and fHBP form a major constituent of the Novartis Vaccines investigational recombinant menB (rMenB)-OMV vaccine (currently in phase III trials) in which recombinant fHBP (variant 1) and NHBA are fused with two other surface proteins, GNA2091 and GNA1030, respectively (10). Also contained in the vaccine is an OMV component containing PorA subtype P1.7-2,4, which was used in the New Zealand MeNZB vaccine campaign (29). Another vaccine, developed by Wyeth Vaccine Research, comprises a single recombinant lipidated subvariant from each of the two fHBP families, families A and B (24).
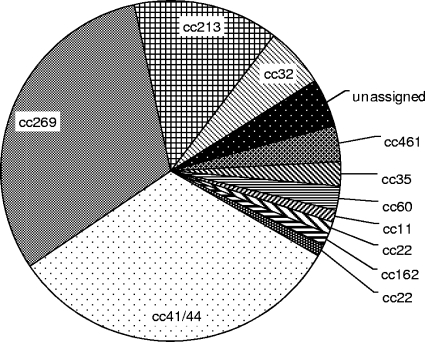
MenB currently causes over 90% of cases of invasive meningococcal disease in England and Wales, where, in recent years, the multilocus sequence type (ST) 269 (ST269) clonal complex (cc269) has expanded to account for 31% of cases of menB disease (Fig. 1). Relatively few data regarding the characterization of nadA, nhba, and fHbp within cc269 have been published, however. The aim of the present study was to determine the presence and genetic diversity of each antigen in a randomized selection of cc269 isolates from England and Wales over the past 10 years. To identify possible antigenic trends within the clonal complex, these data (along with the porA subtype) were profiled against the ST and the genomic presence of an additional genetic marker: insertion sequence IS1301. IS1301 is particularly relevant in this context, having previously been implicated in the insertional disruption of porA, nadA, and capsule expression (via siaA, the sialic acid biosynthesis gene) (6, 7, 13, 23). Under certain conditions, the insertion sequence has also been shown to enhance capsule expression, leading to increased serum resistance (34). The outcomes of this study will contribute to estimates of the level of vaccine coverage in the United Kingdom, thus informing decisions on whether to incorporate the vaccine into current vaccination schedules.
FIG. 1.
Clonal complexes of meningococcal group B isolates (n = 87) submitted to the Health Protection Agency Meningococcal Reference Unit in January 2008.
MATERIALS AND METHODS
Isolates.
The isolates used in the study (n = 168) were English and Welsh menB cc269 isolates from cases in 2000 (n = 21), 2001 (n = 22), 2005 (n = 32), 2006 (n = 41), 2007 (n = 24), and 2008 (n = 28). These constituted cc269 isolates identified by the Health Protection Agency Meningococcal Reference Unit in the corresponding year, during which multilocus sequence typing (MLST) was performed with every eighth isolate received during the whole year for 2000 and 2001 or all isolates received in January of the years 2005 to 2008. Grouping, typing, and subtyping were performed by the Meningococcal Reference Unit (11). All isolates were preserved at −80°C on Microbank cryovials containing glycerol broth (ProLab Diagnostics, Richmond Hill, Ontario, Canada). Cultures were prepared on Colombia agar with 5% (vol/vol) horse blood (Oxoid, Basingstoke, United Kingdom) and were incubated overnight at 37°C with 5% CO2.
Genomic DNA extraction.
Approximately 20 overnight colonies were suspended in 5 ml of 0.9% (wt/vol) saline with a sterile swab and were adjusted to an optical density at 650 nm of 0.1. A 1-ml aliquot of suspension was then incubated at 60°C for 70 min to ensure complete killing of the bacteria. The cells were then pelleted by centrifugation at 6,000 × g for 10 min, and the extraction was completed by using a DNeasy blood and tissue kit (Qiagen, Crawley, United Kingdom), according to the manufacturer's protocol for gram-negative bacteria. The DNA was eluted in AE buffer and was stored at 4°C.
PCR and sequencing.
PCR and sequencing of fHbp and nhba were performed according to previously published methods (14). A deletion immediately downstream of the fHbp gene in a small subset of isolates necessitated the use of an alternative reverse primer, primer gna1870R2-CGTGCCGTCGTGTCCTAG. The presence of nadA was determined by PCR (5) in which nadA-negative isolates yield an ∼400-bp product consisting of a 16-bp region (in place of nadA) and flanking regions that are common to both nadA-positive (nadA+) and nadA-negative isolates alike. The absence of nadA elsewhere in the genome was confirmed by using the internally directed primers nadaintF (5′-GTACTGACCACAGCCATCCTTGCC-3′) and nadaintR (5′-GAAGAACCGGACGAAGTGCCGAC-3′) in a PCR consisting of 33 cycles of melting at a temperature of 96°C for 30 s, annealing at a temperature of 70°C for 30 s, and extension at a temperature of 72°C for 80 s. This was performed against a comprehensive background of positive control isolates representing variants 1, 2, 3, 4, and 5 (including several alleles interrupted by IS1301 or a large 339-bp deletion [data not shown]). Sequencing of nadA was performed by a previously published protocol (5). The presence of IS1301 was determined by PCR with the internally directed primers NG742 and NG743 (34). All PCRs were performed with HotStarTaq DNA polymerase (Qiagen) with an initial activation step of 95°C for 15 min and a final step of 72°C for 7 min. The PCR products were stored at 4°C. Before the PCR products were sequenced, they were cleaned with ExoSAP-IT (USB Corporation), according to the manufacturers' instructions, and then diluted 1 in 3 in nuclease-free water. Sequencing was performed with a BigDye Terminator cycle sequencing kit (version 3.1; Applied Biosystems) in 10-μl reaction volumes, according to the manufacturer's protocol. The products were cleaned by ethanol-sodium acetate precipitation and were resuspended in 15 μl of HiDi formamide (Applied Biosystems). Sequence analysis was performed on a 3130xl sequence analyzer (Applied Biosystems). Contig assembly and manual adjustment of the bases were performed with the Sequencher program (version 4.8; Gene Codes Corporation).
Sequence alignments and phylogenetic analyses.
The nucleotide and amino acid sequences were aligned by using BioEdit software (version 6.0.8.0) (12) and the corresponding prototype sequences from the genome of fully characterized strain MC58 as a comparator. P-distance, neighbor-joining dendrograms were created by using the MEGA software package (version 4.0) (31), in which positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (by use of the pairwise deletion option). The remaining phylogenetic analyses were conducted with the eBURST program (version 3) (8), in which the minimum number of identical loci for group definition was set at ≥5.
Nucleotide sequence accession numbers.
The sequences identified in this study have been submitted to the GenBank database and may be found under the following GenBank accession numbers (the corresponding subvariant identifiers are given in parentheses): for nhba, FJ615443 (subvariant 21), FJ615442 (subvariant 21), FJ615445 (subvariant 17), FJ615446 (subvariant 24), FJ615441 (subvariant 29), FJ615447 (subvariant 31), FJ615448 (subvariant 58), FJ615449 (subvariant 90), FJ615450 (subvariant 91), FJ615451 (subvariant 92), FJ615452 (subvariant 93), and AE002098 (subvariant 3; strain MC58) (33); for nadA, FJ619642 (subvariant 2), GQ405399 (subvariant 2; truncated), and GQ405400 (subvariant 5; truncated); and for fHbp, FJ615420 (subvariant 1.4), EU541890 (subvariant 1.13), EU541896 (subvariant 1.15), EU541891 (subvariant 1.60), EU541893 (subvariant 1.61), EU541894 (subvariant 1.62), EU541895 (subvariant 1.63), FJ153806 (subvariant 1.69), FJ153807 (subvariant 1.71), EU541889 (subvariant 2.19), EU541892 (subvariant 2.19), FJ153809 (subvariant 2.68), GQ405398 (subvariant 3.31), FJ615422 (subvariant 3.59), EU541897 (subvariant 3.64), FJ615423 (subvariant 3.70), FJ615424 (subvariant 3.72), and AE002098 (subvariant 1.1; strain MC58) (33).
RESULTS
Several nomenclature systems exist for the fHBP and NHBA protein subvariants and their corresponding alleles (1, 22). NHBA subvariants have previously been described as falling into 1 of 14 main variants (1), assigned on the basis of the phylogenetic clustering of a globally representative menB panel. There is, however, no obvious subdivision of NHBA into a small number of main variants such as that seen with fHBP and NadA. The nomenclature used for the fHbp and nhba subvariants in this study follows that of the public fHBP and NHBA databases (http://www.neisseria.org), in which new allelic subvariants are assigned a sequentially allocated, numerical identifier and a preexisting or a new (sequentially allocated), numerical protein identifier (alongside the corresponding Novartis variant [variant 1, 2, or 3] and Wyeth family [family A or B], in the case of fHBP). This study pertains to both epidemiology and vaccine coverage, and as such, the allelic subvariants are described in terms of their translated protein subvariant; e.g., fHbp 1.13 refers to Novartis variant 1, Neisseria.org protein subvariant 13. The classification of NadA in this study follows a well-established scheme (1, 5, 6), in which subvariants are reported in terms of their main variant group, i.e., variant 1, 2, 3, 4, or 5. Individual protein subvariants of each of the antigens are often encoded by several alleles either within or between clonal complexes.
nadA.
Of the 168 isolates, 164 (98%) were devoid of nadA. Three of the four nadA+ isolates (isolates ST269, ST5975, and ST275 recovered in 2000, 2006, and 2000, respectively) harbored nadA-2 alleles (the latter of which was truncated owing to a single-base-pair deletion at residue 480). The fourth isolate (isolate ST6791, recovered in 2008) harbored a nadA-5 allele, apparently switched off at an internal poly(C) tract.
fHbp.
All of the isolates harbored fHbp alleles and comprised 16 subvariants in total (9 variant 1, 2 variant 2, and 5 variant 3 subvariants [Fig. 2] for 83%, 14%, and 2% of isolates, respectively). The predominant subvariant was 1.15 (64% of isolates), which appeared to decline in frequency over the period studied (from ∼70% to ∼47% of isolates per year). Subvariants 1.13 and 2.19 accounted for the majority of the remaining cc269 subvariants (15% and 14%, respectively), and collectively, these showed a trend for an increase in frequency (from ∼24% to ∼47% of isolates per year). Between them, the alleles for subvariants 1.15, 1.13, and 2.19 accounted for 93% of the cc269 isolates, while each of the remaining isolates harbored an allele for 1 of 13 minor subvariants, each of which occurred in just a single isolate. Four isolates harboring closely related variant 3 alleles had a 47-bp deletion immediately downstream of the fHbp gene, necessitating the development of an alternative reverse primer.
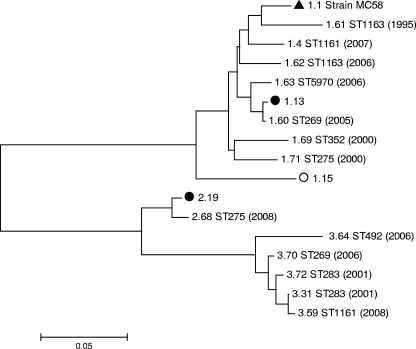
FIG. 2.
P-distance, neighbor-joining dendrogram of translated fHbp subvariants present among cc269 isolates (n = 168) collected between January 2000 and February 2008. Subvariant 1.1 (filled triangle) represents the prototype subvariant originally discovered in strain MC58 (25) and is included as a comparator; i.e., it was not present among any of the cc269 isolates. Filled and hollow circles, predominant ST275 cluster and ST269 cluster subvariants, respectively. The remaining subvariants appeared in a single isolate each (of which the corresponding ST and year [in parentheses] are included). Phylogenetic analyses were conducted with the MEGA program (version 4). All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (by use of the pairwise deletion option). There were a total of 270 positions in the final data set. The dendrogram is drawn to scale, with the sum of the branch lengths between two subvariants representing the proportion of amino acid differences between those subvariants within the pairwise alignment.
On a pairwise nucleotide alignment with the prototype strain MC58 subvariant (subvariant 1.1) as a comparator, both variant 2 alleles and seven (of nine) variant 1 alleles (including the strain MC58 variant) shared a common 15-bp deletion in a GC-rich region at residue 69. Two variant 3 alleles had 6-bp and 9-bp deletions, respectively, in the same region. A 9-bp deletion (corresponding to the amino acids KDN) was present at nucleotide residue 251 in both variant 2 and eight (of nine) variant 1 alleles (including the strain MC58 subvariant). All variant 2 and variant 3 subvariants harbored a 3-bp deletion at residue 517. A single variant 1 allele was extended by 9 bp owing to a C-to-T substitution within the stop codon. The maximum and minimum intra- and intervariant nucleotide pairwise identities of the 16 subvariants are listed in Table 1.
TABLE 1.
Maximum and minimum intra- and intervariant nucleotide pairwise identities for 16 fHbp subvariants present among cc269 isolates recovered from England and Wales between January 2000 and February 2008a
| Variant | Maximum (minimum) pairwise identity |
||
|---|---|---|---|
| 1 | 2 | 3 | |
| 1 | 0.997 (0.879) | 0.792 (0.717) | 0.770 (0.689) |
| 2 | 0.987 (0.987) | 0.896 (0.874) | |
| 3 | 0.996 (0.943) | ||
A total of 168 cc269 isolates were tested.
nhba.
All of the isolates harbored nhba alleles, representing 10 subvariants in total (Fig. 3). The predominant subvariant was 21 (66% of isolates), and the frequency of this variant showed an overall trend for a decline from ∼72% to ∼47% of isolates per year. Subvariant 17 accounted for the majority of the remaining isolates (29% overall) and showed a probable trend for an expansion in frequency over the period studied (from ∼24% to ∼50% of isolates per year). Between them, alleles for subvariants 21 and 17 accounted for 95% of cc269 isolates, while the remaining 5% of isolates harbored alleles for one of eight minor subvariants, each of which was found in just a single isolate.
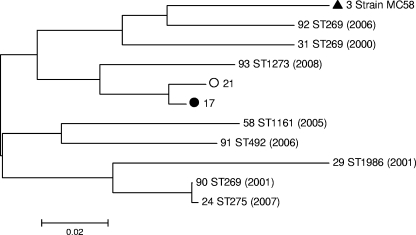
FIG. 3.
P-distance, neighbor-joining dendrogram of translated nhba subvariants present among cc269 isolates (n = 168) collected between January 2000 and February 2008. Subvariant 3 (filled triangle) represents the prototype subvariant originally discovered in strain MC58 (25) and is included as a comparator; i.e., it was not present among any of the cc269 isolates. Filled and hollow circles, predominant ST275 cluster and ST269 cluster subvariants, respectively. The remaining subvariants appeared in a single isolate each (of which the corresponding ST and year [in parentheses] are included). Phylogenetic analyses were conducted with the MEGA program (version 4). All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (by use of the pairwise deletion option). There were a total of 270 positions in the final data set. The dendrogram is drawn to scale, with the sum of the branch lengths between two subvariants representing the proportion of amino acid differences between those subvariants within the pairwise alignment.
On a pairwise nucleotide alignment, the latter third of the gene (949 bp to 1.51 kbp) was devoid of indels and was relatively well conserved (maximum and minimum pairwise identities, 0.996 and 0.956, respectively, versus 0.998 and 0.728, respectively, across the whole alignment). Upstream of this were intermittent regions of clustered substitutions, relatively conserved regions, and between three and seven indels per subvariant. Three indel hot spots were noted, in which homologous regions between subvariants harbored one of several different indels ranging from 3 to 192 bp in length.
Antigenic/genetic profiling of cc269 isolates.
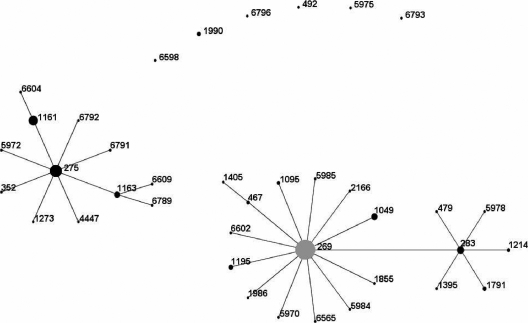
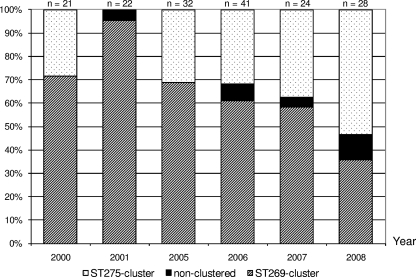
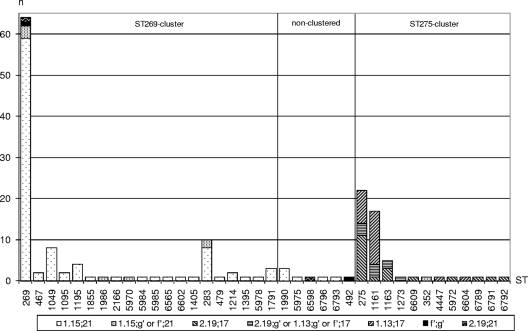
There were 38 distinct STs present among the cc269 isolates, with the ancestral ST (ST269) accounting for 38% of the isolates. Other major STs included ST275 (14% of isolates), ST1161 (10% of isolates), and ST283 (3% of isolates). The remaining 35% of isolates were distributed among 34 minor STs accounting for between one and eight isolates each. Initial ST profiling by the use of the eBURST program indicated the presence of two distinct clusters of closely related isolates centered around ST269 and the major subgroup founder, ST275, respectively. The remaining isolates were distributed among six nonclustered STs (Fig. 4). Over the period studied, the ST269 cluster exhibited a steady decline from ∼70% to ∼35% of isolates per year, while the ST275 cluster expanded from ∼30% to ∼54% of isolates per year (Fig. 5).
FIG. 4.
eBURST diagram of English and Welsh cc269 isolates (n = 168) collected between January 2000 and February 2008. The minimum number of identical loci for group definition was set at ≥5. Only single-locus variant links (i.e., those differing at a single MLST allele) are shown. The area of each circle indicates the relative frequency of each ST among the population studied. The founder ST (ST269) is shaded in gray.
FIG. 5.
ST clustering of English and Welsh cc269 isolates (n = 168) versus year of isolation.
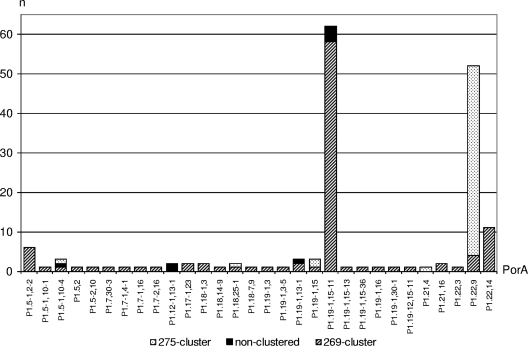
The isolates were genetically profiled with respect to the vaccine candidate antigens (including the porA subtype), the ST, and the genomic presence of IS1301. Three major profiles were identified, and these were found to segregate, for the most part, between clusters ST269 and ST275. Overall, the ST269 cluster accounted for 64% (n = 107) of cc269 isolates, and 92% (n = 98) of these harbored both fHbp subvariant 1.15 and nhba subvariant 21. Of the remaining 8% (n = 9), 7% (n = 8) harbored either of these subvariants alongside a rare/minor subvariant of the other antigen. The ST275 cluster accounted for 32% (n = 53) of cc269 isolates overall, and 92% (n = 49) of these harbored nhba subvariant 17. Of these, 41% (n = 20) also harbored fHbp subvariant 2.19, while 47% (n = 23) harbored fHbp subvariant 1.13. Collectively, fHbp subvariants 1.13 and 2.19 accounted for 87% (n = 46) of the ST275 cluster. Only a single isolate of the ST275 cluster (ST352) did not harbor at least one of these characteristic fHbp/nhba subvariants. Of the eight nonclustered isolates, six had the fHbp subvariant 1.15/nhba subvariant 21 profile associated with the ancestral ST269 cluster. Another isolate (isolate ST6598) harbored fHbp subvariant 2.19/nhba subvariant 21 and so transcended both clusters, while the remaining isolate (isolate ST492) had a minor subvariant of each antigen (Fig. 6). Insertion sequence IS1301 was present within all of the ST269 cluster isolates but only a single ST275 cluster isolate. The insertion sequence was present in seven of the eight nonclustered isolates. There were 30 different porA subtypes represented within the cc269 isolates. The ST269 cluster was relatively heterogeneous in this respect, representing 28 different subtypes. The predominant subtype among the ST269 cluster and nonclustered isolates was P1.15-1,19-11, representing 54 (n = 58) and 50% (n = 4) of isolates, respectively, while P1.15-1,19-11 was absent from the ST275 cluster. The ST275 cluster was somewhat less heterogeneous, with only five different subtypes being detected, of which 91% (n = 48) belonged to P1.22,9 (Fig. 7). The two major cc269 profiles with respect to fHbp, nhba, porA, ST, and the genomic presence of IS1301 are presented in Table 2.
FIG. 6.
fHbp nhba profile (translated) versus ST in English/Welsh cc269 isolates (n = 168) collected between January 2000 and February 2008. STs are grouped into their respective clusters, as indicated by eBURST analysis. g′, a minor subvariant of nhba; f′, a minor subvariant of fHbp.
FIG. 7.
porA subtype versus ST clustering among cc269 isolates (n = 168).
TABLE 2.
Major fHbp, nhba, porA, and IS1301 profile versus ST cluster for 160 cc269 isolates
| Cluster | Major antigenic subvariant (proportion of cluster) |
Presence of IS1301a | ||
|---|---|---|---|---|
| fHbp | nhba | porA subtype | ||
| 269 centered | 1.15 (94) | 21 (96) | P1.19-1,15-11 (54) | 100 (107/107) |
| 275 centered | 1.13 (47) or 2.19 (40) | 17 (92) | P1.22,9 (91) | 2 (1/53) |
Data are presented as percent (number of isolates in which IS1301 was present/total number of isolates tested).
The four nadA+ cc269 isolates were distributed evenly between the clusters. Two (isolates ST6791 and ST275, respectively) belonged to the ST275 cluster and, with the exception of a minor fHbp variant (subvariant 1.71) in the latter isolate, had an antigenic/genetic profile to match. The remaining two isolates were of the ST269 cluster (isolate ST269) and nonclustered ST5975, respectively. Other than the lack of IS1301 in the ST5975 isolate, these conformed to the typical ST269 cluster profile and shared a common porA subtype, P1.5-1,10-4.
DISCUSSION
The poorly immunogenic capsular polysaccharide and the considerable diversity among menB immunodominant surface epitopes have hindered the pursuit of a universal vaccine against meningococcal disease (26, 38). OMV vaccines have proven to be effective against clonal epidemics but, thus far, offer little cross-protection on a broader scale (26, 32). A number of relatively well-conserved surface proteins, fHBP, NadA, and NHBA, have aroused considerable interest as prospective components of broadly cross-protective meningococcal vaccines (9, 10). This study aimed to characterize the presence and genetic diversity of fHbp, nadA, and nhba in recently expanded English and Welsh cc269 isolates. Further profiling of the antigenic data for ST, the porA subtype, and the presence of IS1301 was used to highlight underlying antigenic/genetic trends within the clonal complex.
The frequency of nadA among isolates has previously been reported to be approximately 50% (6). In this study, 4 of 168 cc269 isolates (2%) were found to harbor nadA alleles. The ST6791 isolate in the ST275 cluster harbored an allele for nadA-5 which shares significant homology with the poorly cross-reactive, carriage-associated allele nadA-4. The allele was apparently switched off at an internal poly(C) tract (unique to nadA-5); however, Western blot analyses with similar nadA-5+ cultures have shown that the protein is present, indicating the simultaneous presence of both on and off genes within the culture (data not presented). Previously, nadA-5 has been described only in cc213 isolates (1), suggesting a possible source from which this isolate (or a recent predecessor) may have acquired the allele via the horizontal route. All of the remaining three isolates (isolates ST5975, ST269, and ST275) harbored nadA-2 alleles, but in the ST275 isolate, the nadA-2 allele was truncated due to a frameshift mutation. Apart from their atypical nadA+ status, these isolates are interesting since they transcend both the ST269 and ST275 clusters and the nonclustered ST5975 ST. The fact that they harbor the same variant suggests that these isolates may descend from a common nadA+ ancestor. This seems possible in the case of isolates ST269 and ST5975, which are identical in terms of fHbp and nhba and which share a relatively atypical porA P1.5-1,10-4. The respective cases, however, were separated by ∼6 years (2000 and 2006, respectively) and were geographically remote (southeast England and Devon, respectively), thus excluding any clear epidemiological links. An independent horizontal transfer event appears to be likely in the case of the remaining ST275 cluster isolate, however, since a common nadA+ ancestor would have had to have preceded the divergence of the two clusters, both of which are similarly devoid of nadA. Despite its role as a pathogenicity factor, therefore, it seems that cc269 has not yet adapted so that it may incorporate nadA to any large degree.
In accordance with the findings of previous studies (14, 20), all of the isolates harbored fHbp and nhba. The ST clustering of 95% of the cc269 isolates (all years) into two distinct groups (centered around ST269 and ST275, respectively) was greatly reflected by the genetic profiles generated for fHbp, nhba, and porA. Among the predominant antigenic subvariants described for these antigens, there was minimal crossover between clusters ST269 and ST275. The most discriminatory marker between the two clusters was the genomic presence of IS1301, which occurred in all of the ST269 cluster isolates but in just a single ST275 cluster isolate. With the exception of isolates ST492 and ST6598, the nonclustered isolates conformed to the typical ancestral ST269 cluster profiles. Isolate ST6598 was largely similar to the isolates in the ST269 cluster in harboring IS1301 and nhba subvariant 21, but it also harbored fHbp subvariant 2.19, characteristic of the ST275 cluster. Isolate ST492 harbored minor subvariants of both fHbp and nhba and was devoid of IS1301. A wider eBURST analysis of the whole Neisseria MLST database indicates that ST492 is relatively distant from ST269 (having diverged via two subgroup founder STs) and that both ST492 and ST6598 diverged via a common subgroup founder (isolate ST1354). Interestingly, both isolate ST492 and isolate ST6598 were unique among the isolates in sharing the porA P1.12,13-1 subtype. The wider eBURST analysis also identified ST492 as the founder of a considerable subgroup. Given the antigenic profile of isolate ST492 in this study, it is conceivable that this subgroup may form another largely distinct cluster, similar to the case for the ST275 cluster. The fact that no single-locus variants of ST492 were identified among the English and Welsh isolates may be an indicator that this subgroup has emerged elsewhere and later been imported into the region. Other subgroup founders worthy of surveillance include ST283, ST1161, and ST1163.
These findings raise several interesting points, the first of which relates to the way in which we view cc269. It is clear that, at least in England and Wales, cc269 consists of two broadly distinct groups (described here as clusters) and that this distinction spans both housekeeping genes and surface antigens and includes the genomic presence of the mobile genetic element IS1301. Unsurprisingly, the distinction between these clusters is mainly blurred by the porA subtype, yet even this major immunodominant antigen displays a remarkable tendency toward particular subtypes that differ between the two clusters. By definition, the two clusters are from the same clonal complex; however, for practical purposes it may be prudent to view them as distinct. For example, the ancestral ST269 cluster is apparently in decline, while the ST275 cluster and nonclustered isolates are expanding (frequencies, 35, 54, and 11% of cc269 isolates in 2008, respectively). It is clearly misleading, therefore, to view the group solely in terms of ancestral traits, which the title “cc269” may infer. It is also worth considering that several STs with four or more MLST alleles in common with ST275 (but less than four alleles in common with the founder ST269) are currently unassigned to a clonal complex (http://www.neisseria.org) and, by definition, are excluded from cc269. It seems to be reasonable to assume that these constitute an important arm of the ST275 cluster, yet they may often go unrepresented in seemingly appropriate studies (including the present study).
The second point raised by these findings, directly related to the first, is the possibility that relatively distinct antigenic profiles might result in similarly distinct vaccine coverage. It is likely that the NadA component of the rMenB-OMV vaccine would have a limited impact against both, largely nadA-deficient clusters. The coverage afforded by the vaccine's PorA P1.7-2,4 component is also likely to be limited, since only two of the isolates (isolate ST5978, recovered in 2006, and isolate ST1163, recovered in 2007) were found to belong to the corresponding subtypes (subtypes P1.7-2,16 and P1.21,4, respectively). At best, the heterogeneity of PorA among these isolates would be likely, however, to limit the coverage of any single PorA vaccine component to a maximum of ∼46%; i.e., the PorA P1.19-1,15-11 component would potentially cover ∼64% of the ST269 cluster and ∼4% of the ST275 cluster (those isolates harboring either or both variable epitopes, P1.19-1 and P1.15-11). The coverage afforded by the vaccine's NHBA and fHBP components ought to be somewhat less susceptible to such diversity, owing to their high inter- and intravariant cross-reactivities, respectively (9, 20, 36). Any major disparity in coverage between the two clusters would likely result from the relatively poor intervariant cross-reactivity exhibited by fHBP (9, 20). The fHBP variant 1 component of the rMenB-OMV vaccine would theoretically cover approximately 83% of the cc269 isolates, i.e., those harboring variant 1 fHbp alleles (71% in January 2008). Of the remaining 17% of the non-variant 1 isolates, not covered by this component, at least 14% (21% in January 2008) belonged to the ST275 cluster; i.e., they harbored fHbp subvariant 2.19. Nonetheless, the high residual proportion of fHbp variant 1 among the cc269 isolates as a whole favors the use of a variant 1 fHBP vaccine component. All of the isolates harbored nhba, giving this broadly immunologically cross-reactive component a theoretical coverage of 100%. A minor subvariant of at least one of fHbp or nhba was represented by 11% of the cc269 isolates, and a minor subvariant of both occurred in two (1%) of the isolates. Consideration of these is also important in terms of vaccine coverage, although the fact that they occurred in just a single isolate each suggests that many of these unusual subvariants likely lack longevity within the clonal complex. That most of the minor subvariants had not previously been submitted to the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) or the Neisseria fHBP database (http://www.neisseria.org) may indicate horizontal transfer from carriage isolates, with which large-scale studies of these antigens are still to be performed/reported.
Collectively, fHbp subvariants 1.15 and 1.13 accounted for 79% of the isolates, while nhba subvariant 21 and subvariant 17 accounted for ∼95% of the isolates. These high theoretical projections of coverage are, therefore, heavily reliant on there being sufficient cross-reactivity between the vaccine subvariants and these four highly representative subvariants. Previous studies relating to the intra- and intervariant cross-reactivities of fHBP and NHBA, respectively, however, are very encouraging (25, 36). The remaining OMV-related antigens and the GNA2091 and GNA1030 components may also serve to enhance the vaccine's coverage; however, the epidemiology of these antigens is beyond the scope of this study. The diversity among menB as a whole precludes the use of just a single antigen in a vaccine that is intended to provide broad cross-protection, a situation that is mirrored, on a smaller scale, by cc269. A vaccine based on just a single antigen may also be susceptible to the emergence of escape mutants (10).
The apparently universal presence of IS1301 in the ST269 cluster and its effective absence from the ST275 cluster are clearly significant findings. The insertion sequence has been implicated in the phase variation of several genes and operons, including porA, nadA, and the capsule locus (6, 7, 13, 23). Under certain conditions the insertion of IS1301 into the capsule locus can result in capsular upregulation and enhanced serum resistance (34). Despite the potential benefits that IS1301 may confer, the frequency of the ST269 cluster harboring the insertion sequence has been waning over the past decade. Work is currently ongoing to understand the significance of the insertion sequence within the clonal complex and the wider implications concerning, e.g., pathogenesis and potential effects on vaccine efficacy.
Despite distinct clustering and the overall lack of nadA and porA P1.7-2,4, the potential coverage of cc269 by the rMenB-OMV vaccine (due to fHBP and NHBA) is encouraging. As well as antigenic cross-reactivity, however, potential coverage is likely to be dependent on phenotypic expression levels meeting a threshold (15, 21). Further work in this area will therefore be to assess the levels of expression of each antigen in all of the isolates. Representatives of each antigenic/expression profile may then be selected for serum bactericidal antibody assays (the accepted surrogate of protection for menB) that will compare antibody-mediated complement killing by pre- and postvaccination serum samples from clinical trials. This will provide a reliable estimate of vaccine coverage among this important component of menB disease in the United Kingdom, thereby informing decisions on whether the vaccine should be incorporated into current schedules while informing future vaccine design.
Acknowledgments
We thank Lynn Newbould, Andrew Birtles, Jayne Blake, Tony Carr, and Stefanie Gilchrist for their contributions to MLST and the PorA subtyping of the isolates used in the study.
This work was sponsored by Novartis Vaccines.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Bambini, S., A. Muzzi, P. Olcen, R. Rappuoli, M. Pizza, and M. Comanducci. 2009. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 27:2794-2803. [DOI] [PubMed] [Google Scholar]
- 2.Bjune, G., E. A. Høiby, J. K. Grønnesby, O. Arnesen, J. H. Fredriksen, A. Halstensen, E. Holten, A. K. Lindbak, H. Nøkleby, E. Rosenqvist, L. K. Solberg, O. Closs, J. Eng, L. O. Froholm, A. Lystad, L. S. Bakketeig, and B. Hareide. 1991. Effect of outer membrane vesicle vaccine against group B meningococcal disease in Norway. Lancet 338:1093-1096. [DOI] [PubMed] [Google Scholar]
- 3.Borrow, R., and E. Miller. 2006. Long term protection in children with meningococcal C conjugation vaccination: lessons learned. Expert Rev. Vaccines 6:851-857. [DOI] [PubMed] [Google Scholar]
- 4.Boslego, J., J. Garcia, C. Cruz, W. Zollinger, B. Brandt, S. Ruiz, M. Martinez, J. Arthur, P. Underwood, W. Silva, E. Moran, W. Hankins, J. Gilly, J. Mays, et al. 1995. Efficacy, safety, and immunogenicity of a meningococcal group B (15:P1.3) outer membrane protein vaccine in Iquique, Chile. Vaccine 13:821-829. [DOI] [PubMed] [Google Scholar]
- 5.Comanducci, M., S. Bambini, D. A. Caugant, M. Mora, B. Brunelli, B. Capecchi, L. Ciucchi, R. Rappuoli, and M. Pizza. 2004. NadA diversity and carriage in Neisseria meningitidis. Infect. Immun. 72:4217-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Aricò, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elias, J., and U. Vogel. 2007. IS1301 fingerprint analysis of Neisseria meningitidis strains belonging to the ET-15 clone. J. Clin. Microbiol. 45:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher, L. D., L. Bernfield, V. Barniak, J. E. Farley, A. Howell, M. Knauf, P. Ooi, R. P. Smith, P. Weise, M. Wetherell, X. Xie, R. Zagursky, Y. Zhang, and G. W. Zlotnick. 2004. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect. Immun. 72:2088-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giuliani, M. M., J. Adu-Bobie, M. Comanducci, B. Aricò, S. Savino, L. Santini, B. Brunelli, S. Bambini, A. Biolchi, B. Capecchi, E. Cartocci, L. Ciucchi, F. Di Marcello, F. Ferlicca, B. Galli, E. Luzzi, V. Masignani, D. Serruto, D. Veggi, M. Contorni, M. Morandi, A. Bartalesi, V. Cinotti, D. Mannucci, F. Titta, E. Ovidi, J. A. Welsch, D. Granoff, R. Rappuoli, and M. Pizza. 2006. A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. USA 103:10834-10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray, S. J., C. L. Trotter, M. E. Ramsay, M. Guiver, A. J. Fox, R. Borrow, R. H. Mallard, and E. B. Kaczmarski. 2006. Epidemiology of meningococcal disease in England and Wales 1993/94 to 2003/04: contribution and experiences of the Meningococcal Reference Unit. J. Med. Microbiol. 55:887-896. [DOI] [PubMed] [Google Scholar]
- 12.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 41:95-98. [Google Scholar]
- 13.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobsson, S., S. Thulin, P. Mölling, M. Unemo, M. Comanducci, R. Rappuoli, and P. Olcén. 2006. Sequence constancies and variations in genes encoding three new meningococcal vaccine candidate antigens. Vaccine 24:2161-2168. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, H. Q., S. L. Harris, C. Tan, A. Scott, K. Alexander, K. Mason, I. DaSilva, M. Mack, X. J. Zhao, S. Guttmann, D. Liu, P. Wong, L. McNeil, M. Pride, A. Arora, D. Zhu, S. K. Hoiseth, T. L. Mininni, M. Hagen, T. Jones, K. U. Jansen, and G. W. Zlotnick. 2008. Prediction of broad vaccine coverage for a bivalent rLP2086 based vaccine which elicits serum bactericidal activity against a diverse collection of serogroup B meningococci, abstr. O41. Abstr. 16th Int. Pathogenic Neisseria Conf.
- 16.Kelly, C., R. Arnold, Y. Galloway, and J. O'Hallahan. 2007. A prospective study of the effectiveness of the New Zealand meningococcal B vaccine. Am. J. Epidemiol. 166:817-823. [DOI] [PubMed] [Google Scholar]
- 17.Koeberling, O., S. Giuntini, A. Seubert, and D. M. Granoff. 2009. Meningococcal outer membrane vesicle vaccines derived from mutant strains engineered to express factor H binding proteins from antigenic variant groups 1 and 2. Clin. Vaccine Immunol. 16:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madico, G., J. A. Welsch, L. A. Lewis, A. McNaughton, D. H. Perlman, C. E. Costello, J. Ngampasutadol, U. Vogel, D. M. Granoff, and S. Ram. 2006. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol. 177:501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Masignani, V., M. Comanducci, M. M. Giuliani, S. Bambini, J. Adu-Bobie, B. Arico, B. Brunelli, A. Pieri, L. Santini, S. Savino, D. Serruto, D. Litt, S. Kroll, J. A. Welsch, D. M. Granoff, R. Rappuoli, and M. Pizza. 2003. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J. Exp. Med. 197:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNeil, L. K., E. Murphy, X. J. Zhao, S. Guttmann, S. L. Harris, A. A. Scott, C. Tan, M. Mack, I. DaSilva, K. Alexander, K. Mason, H. Q. Jiang, D. Zhu, T. L. Mininni, G. W. Zlotnick, S. K. Hoiseth, T. R. Jones, M. W. Pride, K. U. Jansen, and A. S. Anderson. 2009. Detection of LP2086 on the cell surface of Neisseria meningitidis and its accessibility in the presence of serogroup B capsular polysaccharide. Vaccine 27:3417-3421. [DOI] [PubMed] [Google Scholar]
- 22.Murphy, E., L. Andrew, K. L. Lee, D. A. Dilts, L. Nunez, P. S. Fink, K. Ambrose, R. Borrow, J. Findlow, M. K. Taha, A. E. Deghmane, P. Kriz, M. Musilek, J. Kalmusova, D. A. Caugant, T. Alvestad, L. W. Mayer, C. T. Sacchi, X. Wang, D. Martin, A. von Gottberg, M. du Plessis, K. P. Klugman, A. S. Anderson, K. U. Jansen, G. W. Zlotnick, and S. K. Hoiseth. 2009. Sequence diversity of the factor h binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J. Infect. Dis. 200:379-389. [DOI] [PubMed] [Google Scholar]
- 23.Newcombe, J., K. Cartwright, S. Dyer, and J. McFadden. 1998. Naturally occurring insertional inactivation of the porA gene of Neisseria meningitidis by integration of IS1301. Mol. Microbiol. 30:453-454. [DOI] [PubMed] [Google Scholar]
- 24.Pillai, S., A. Howell, K. Alexander, B. E. Bentley, H. Q. Jiang, K. Ambrose, D. Zhu, and G. Zlotnick. 2005. Outer membrane protein (OMP) based vaccine for Neisseria meningitidis serogroup B. Vaccine 23:2206-2209. [DOI] [PubMed] [Google Scholar]
- 25.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Aricò, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 26.Rosenqvist, E., E. A. Høiby, E. Wedege, K. Bryn, J. Kolberg, A. Klem, E. Rønnild, G. Bjune, and H. Nøkleby. 1995. Human antibody responses to meningococcal outer membrane antigens after three doses of the Norwegian group B meningococcal vaccine. Infect. Immun. 63:4642-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider, M. C., R. M. Exley, H. Chan, I. Feavers, Y. H. Kang, R. B. Sim, and C. M. Tang. 2006. Functional significance of factor H binding to Neisseria meningitidis. J. Immunol. 176:7566-7575. [DOI] [PubMed] [Google Scholar]
- 28.Segal, S., and A. J. Pollard. 2005. Vaccines against bacterial meningitis. Br. Med. Bull. 72:65-81. [DOI] [PubMed] [Google Scholar]
- 29.Sexton, K., D. Lennon, P. Oster, S. Crengle, D. Martin, K. Mulholland, T. Percival, S. Reid, J. Stewart, and J. O'Hallahan. 2004. The New Zealand Meningococcal Vaccine Strategy: a tailor-made vaccine to combat a devastating epidemic. N. Z. Med. J. 117:U1015. [PubMed] [Google Scholar]
- 30.Sierra, G. V., H. C. Campa, N. M. Varcacel, I. L. Garcia, P. L. Izquierdo, P. F. Sotolongo, G. V. Casanueva, C. O. Rico, C. R. Rodriguez, and M. H. Terry. 1991. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann. 14:195-207. [PubMed] [Google Scholar]
- 31.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 32.Tappero, J. W., R. Lagos, A. M. Ballesteros, B. Plikaytis, D. Williams, J. Dykes, L. L. Gheesling, G. M. Carlone, E. A. Høiby, J. Holst, H. Nøkleby, E. Rosenqvist, G. Sierra, C. Campa, F. Sotolongo, J. Vega, J. Garcia, P. Herrera, J. T. Poolman, and B. A. Perkins. 1999. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: a randomized controlled trial in Chile. JAMA 281:1520-1527. [DOI] [PubMed] [Google Scholar]
- 33.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 34.Uria, M. J., Q. Zhang, Y. Li, A. Chan, R. M. Exley, B. Gollan, H. Chan, I. Feavers, A. Yarwood, R. Abad, R. Borrow, R. A. Fleck, B. Mulloy, J. A. Vazquez, and C. M. Tang. 2008. A generic mechanism in Neisseria meningitidis for enhanced resistance against bactericidal antibodies. J. Exp. Med. 205:1423-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Welsch, J. A., G. R. Moe, R. Rossi, J. Adu-Bobie, R. Rappuoli, and D. M. Granoff. 2003. Antibody to genome-derived neisserial antigen 2132, a Neisseria meningitidis candidate vaccine, confers protection against bacteremia in the absence of complement-mediated bactericidal activity. J. Infect. Dis. 188:1730-1740. [DOI] [PubMed] [Google Scholar]
- 37.Weynants, V., P. Denoël, N. Devos, D. Janssens, C. Feron, K. Goraj, P. Momin, D. Monnom, C. Tans, A. Vandercammen, F. Wauters, and J. T. Poolman. 2009. Genetically modified L3,7 and L2 lipooligosaccharides from Neisseria meningitidis serogroup B confer a broad cross-bactericidal response. Infect. Immun. 77:2084-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyle, F. A., M. S. Artenstein, B. L. Brandt, E. C. Tramont, D. L. Kasper, P. L. Altieri, S. L. Berman, and J. P. Lowenthal. 1972. Immunologic response of man to group B meningococcal polysaccharide vaccines. J. Infect. Dis. 126:514-521. [DOI] [PubMed] [Google Scholar]
- 39.Zollinger, W. D., M. Donets, B. L. Brandt, B. Ionin, E. E. Moran, D. Schmiel, V. Pinto, M. Fisseha, J. Labrie, R. Marques, and P. Keiser. 2008. Multivalent group B meningococcal vaccine based on native outer membrane vesicles (NOMV) has potential for providing safe, broadly protective immunity, abstr. O35. Abstr. 16th Int. Pathogenic Neisseria Conf.