Abstract
Mycobacterium ulcerans causes the devastating infectious skin disease Buruli ulcer and has a monomorphic population structure. The resolution of conventional genetic fingerprinting methods is therefore not sufficient for microepidemiological studies aiming to characterize transmission pathways. In a previous comparative genomic hybridization analysis with a microarray covering part of the M. ulcerans genome, we have found extensive insertional-deletional sequence polymorphisms among M. ulcerans isolates of diverse geographic origins that allowed us to distinguish between strains coming from different continents. Since large numbers of insertion sequences are spread over the genome of African M. ulcerans strains, we reasoned that these may drive large sequence polymorphisms in otherwise clonal local mycobacterial populations. In this study, we used a printed DNA microarray covering the whole genome of the Ghanaian M. ulcerans reference strain Agy99 for comparative genomic hybridization. The assay identified multiple regions of difference when DNA of a Japanese M. ulcerans strain was analyzed. In contrast, not a single insertional-deletional genomic variation was found within a panel of disease isolates coming from an area of Ghana where Buruli ulcer is endemic. These results indicate that, despite the expectations deduced from other mycobacterial pathogens, only analyses of single nucleotide polymorphisms will have the potential to differentiate local populations of M. ulcerans.
Buruli ulcer (BU) is a severe skin disease characterized by large ulcerative lesions, often leading to deformity and the crippling of extremities. While this neglected tropical disease occurs in more than 30 countries worldwide, it currently has its major focus in West Africa, where the prevalence is greater than that of leprosy, and incidence rates still may be underestimated (61). The social impact of BU endemicity in rural settings of Africa, where children are most affected, is dramatic. Patients often have only limited access to health care and tend to report to health care facilities at advanced stages of disease (20, 49). BU is often concentrated in areas close to stagnant or slow-moving waters. The mode of transmission is not fully understood (13, 36, 44-46, 59, 60), partly because conventional molecular fingerprinting methods have not sufficiently high resolution for microepidemiological analyses (1-4, 10, 11, 23, 24, 27, 33, 51, 53, 55-57).
Mycobacterium tuberculosis and related pathogenic mycobacteria exhibit major large sequence polymorphism (LSP)-based interstrain genomic variations (5, 9, 12, 34, 58). From these genomic insertional-deletional (InDel) markers, evolutionary scenarios were drawn for M. tuberculosis, the M. tuberculosis complex, Mycobacterium bovis, the M. bovis BCG vaccine strains, and mycolactone-producing mycobacteria related to Mycobacterium ulcerans (6-8, 25, 27, 38, 50). In recent years, proposed phylogenies were confirmed and refined by whole-genome sequence analysis identifying single nucleotide polymorphisms (SNPs) (16, 17, 21, 22). Combinations of all identified genomic polymorphisms are currently being used for studying mycobacterial evolution and transmission pathways, helping in the prevention of epidemics (34, 37).
Comparative genomic hybridization (CGH) analysis of M. ulcerans clinical isolates of diverse geographic origins also revealed extensive LSPs (47). The identified transposable element-associated InDel recombination events are indicative of progressive genome shrinking in M. ulcerans, which has emerged from the environmental mycobacterium Mycobacterium marinum by acquisition of a large virulence plasmid (52). An in-depth comparison of the recently published M. ulcerans genome of strain Agy99 to the progenitor species strain M. marinum M revealed LSPs, named M. ulcerans regions of difference (MURDs) (54). Analysis of large InDel polymorphisms allowed us to distinguish between two distinct lineages: (i) the “classical” lineage representing the most pathogenic genotypes, consisting of those that come from Africa, Australia, and Southeast Asia; and (ii) an “ancestral” lineage comprising strains from China and Japan, South America, and Mexico (29). A total of 209 copies of the insertion sequence (IS) IS2404 and 83 copies of IS2606 have been found interspersed in the genome of the M. ulcerans reference strain Agy99 from Ghana (54). Due to achievements using LSPs in other mycobacterial pathogens, we reasoned that this high abundance of transposable elements may destabilize the general architecture of the genome of African M. ulcerans strains, resulting in a rapid accumulation of LSPs in otherwise monomorphic populations of the pathogen. To assess this experimentally, we performed CGH analyses using a comprehensive whole-genome microarray. Specifically, we tried to identify LSPs within a panel of M. ulcerans strains isolated from the BU lesions of 37 patients from several districts in Ghana between the years 2002 and 2004.
MATERIALS AND METHODS
PCR product-based DNA microarray.
A shotgun clone library of M. ulcerans strain Agy99 DNA, comprising 1,536 Escherichia coli plasmids (pCDNA2.1; Invitrogen, Basel, Switzerland) was used, with each plasmid containing an M. ulcerans fragment of ∼2.3 to 2.7 kb (54). Plasmids were selected so that the contigs covered the entire genome at least once, with particular regard to all coding sequences (CDSs), whereas sequence redundancies were excluded. Given a genome size of 5.8 Mb and after exclusion of redundant fragments, this set of plasmid inserts represents a theoretical coverage of >75% of the M. ulcerans genome. Plasmid DNA was prepared by using a Biomek 2000 Workstation (Beckman Coulter, Krefeld, Germany) and dissolved at a concentration of 150 ng/μl in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). For all plasmids, PCR was performed using primers pCDNA2_F (5′-TAAAACGACGGCCAGTGAAT-3′) and pCDNA2_R (5′-CGAGCTCGGATCCACTAGTAA-3′), with binding in the multiple cloning sites outside of the inserted regions. PCR products were checked by gel electrophoresis using 1% agarose gels, purified using a MinElute 96 ultracentrifugation PCR purification kit (Qiagen, Hombrechtikon, Switzerland), and dissolved at a concentration of 400 ng in 3× SSC. The resulting 1,520 PCR products were loaded on a nanoliter dispenser printhead in a 96-well format (Topspot, Biofluidix, Freiburg, Germany). A piezo-actuator-driven pressure pulse was applied for spotting of 1-nl droplets onto glass slides coated with poly-l-lysine (Superfrost Plus, Menzel, Braunschweig, Germany). The spots had an average diameter of 150 μm, with a distance of 500 μm to each other, and were arranged in “standard packing” onto the arrays for establishing and evaluating hybridization conditions (Fig. 1). In the final microarray, arranging spots in “orange packing” allowed for a microarray layout of 1,520 spots in triplicates on one glass slide. The microarray slides were incubated at 4°C overnight and rehydrated under 60% humidity for 1 h in an incubator, and DNA was fixed by exposure to UV light at 650 mJ in a Stratalinker 2400 (Strategene, La Jolla, CA). Controls spotted included 3× SSC buffer only, a β-lactamase gene fragment labeled with either Cy3 (spotting control) or biotin (colorization control), PCR products comprising IS2404 and IS2606 stretches (hybridization controls), and PCR products from the unrelated species Neisseria meningitidis and Plasmodium falciparum (nonhybridization controls).
FIG. 1.
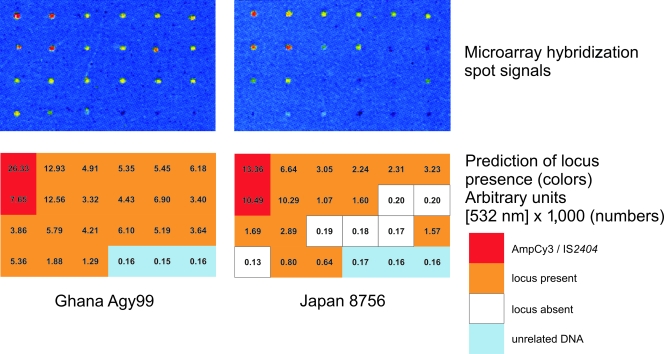
Setup of comparative genomic hybridization conditions. PCR products sized between 0.6 and 2.3 kb in length were selected to establish spotting and hybridization conditions with the required sensitivity and reliability. PCR products comprising a β-lactamase fragment linked with Cy3 or DNA stretches of IS2404 and IS2606 present in all M. ulcerans strains (red) and DNA regions known to be present in both M. ulcerans strains Ghana 970483 and Japan 8756 (orange) served as positive controls. PCR products of DNA from unrelated species (N. meningitidis and P. falciparum; light blue) and regions known to be absent from strain Japan 8756 only (white) were used as negative controls.
Bacterial strains.
M. ulcerans strains were isolated from lesions of BU patients as described earlier (62). Isolates used for microarray hybridization were as follows: Agy99; Japan 8756; strains NM01/03, NM14/02, NM18/02, NM21/02, NM23/02, NM27/02, NM28/02, NM32/02, NM33/02, NM34/02, NM40/02, NM41/02, NM43/02, NM44/02, NM46/02, NM47/02, NM49/02, NM50/02, NM51/02, NM52/02, NM54/02, NM56/02, NM59/02, NM60/02, NM61/02, NM62/02, NM68/02, NM69/02, NM72/03, NM89/03, and NM90/03 from the Greater Accra region, Ghana; strains NM22/02, NM37/02, and NM38/02 from the Ashanti region, Ghana; and strains NM42/02 and NM53/02 from the Eastern Region, Ghana (for a distribution map see reference 24). DNA from clinical isolates M. marinum M (ATCC BAA-535 from LGC Standards; http://www.lgcstandards-atcc.org/ATCCCulturesandProducts/tabid/979/Default.aspx), N. meningitidis N3221, and P. falciparum 3D7 were used as controls.
DNA extraction and biotinylation of M. ulcerans genomic DNA fragments.
Genomic DNA was extracted from bacterial pellets using an optimized method for mycobacterial DNA preparation (30). Bacterial pellets of about 20 mg (wet weight) were heat inactivated for 1 h at 95°C, followed by cell wall disruption and digestion. DNA was extracted from the supernatants by phenol-chloroform (Fluka, Buchs, Switzerland) extraction and subjected to ethanol precipitation as described previously (30). DNA was measured by the optical density at 260 nm using a NanoDrop 1000 spectrophotometer (Thermo Fisher, Waltham, MA). Digestion and biotinylation of genomic DNA were optimized to yield high-quality labeling as follows: 1 μg of genomic DNA was digested with 8 U of Sau3AI (New England Biolabs, Hitchin, United Kingdom) in a volume of 60 μl, incubated for 75 min at 37°C, and ethanol precipitated, followed by measurement of the DNA concentration. Of the digested DNA, 250 ng was biotinylated using a BioPrime DNA Labeling System (Invitrogen, Basel, Switzerland) for 7 h at 37°C, ethanol precipitated, and eluted in water. DNA concentration measurement showed average yields of 25 μg of biotinylated DNA.
Hybridization and colorization of microarray slides.
Hybridization was done under identical conditions for all strains. Nine nanograms of biotinylated DNA was mixed with 16.5 μg of human Cot-1 DNA (Roche Applied Science, Indianapolis, IN) and 90 μg of yeast (Saccharomyces cerevisiae) tRNA (Gibco/BRL, Gaithersburg, MD). The hybridization mix was concentrated with a Speed Vac Concentrator System (Eppendorf, Basel, Switzerland), resuspended in 20 μl of hybridization buffer (30% formamide, 5× SSC, 0.1% sodium dodecyl sulfate, 100 ng/μl double-stranded herring sperm DNA), denatured for 2 min at 95°C, and applied to the preheated (5 min at 95°C) microarray slide. Following hybridization at 60°C for 15 h, slides were washed once with 2× SSC-0.1% sodium dodecyl sulfate for 5 min at 60°C, three times with 0.1× SSC for 5 min at room temperature, and twice with 0.01× SSC for 10 min at room temperature. The colorization step was performed with 2 ml of staining solution containing 50% caseine (Roche Applied Science, Basel, Switzerland), 1× maleic acid buffer (Roche Applied Science, Basel, Switzerland), and 2 μl of streptavidin Cy3 fluorolink (Amersham, Piscataway, NJ) for 30 min at room temperature under slow agitation, followed by additional washing steps (twice with 0.1× SSC for 5 min, twice with 0.01× SSC for 5 min, and once with water) before slides were dried by centrifugation at 900 × g for 1 min.
Microarray scanning and data evaluation.
Images of the microarrays were retrieved using the laser scanner GenePix 4100 A (Axon Instruments Inc., Foster City, CA) with an excitation wavelength of 532 nm, an emission wavelength of 570 nm, and standardized measurement parameters (laser power of 100%; photomultiplier tube gain of 600). The resulting images were analyzed using GenePix Pro, version 6.0, software (Axon Instruments Inc., Foster City, CA) for background subtraction and calculation of spot intensities (fluorescence signal intensity units). A total of 170 spots with insufficient amounts of DNA spotted were excluded, as were 7 spots for which the coefficient of variation in Agy99 was >30% between the replicates. Control strains (Agy99 and M. marinum M) were hybridized in duplicates, yielding six values per spot (each), and test strains yielded three values per spot. For each spot, the background signal was subtracted, and mean values were determined. To equalize values across hybridization experiments, the values were normalized for the fluorescence signal of a spotted control DNA on each array. In order to determine outlier fluorescence signals that are much lower than signals in the reference strain or absent from the reference strain, we calculated fractions for each spot as follows: number of fluorescence units of Agy99/number of fluorescence units of the test strain. Thus, spots with signal intensities similar to the reference DNA gave values close to 1, whereas spots with no or weak fluorescence signals in the test strain resulted in high fluorescence intensity ratios. These outlier signal ratios suggested absent genomic regions compared to the Agy99 genome. Raw data were sorted and visualized as outlined (see the legend of Fig. 2) using the data computational program Spotfire (Spotfire DecisionSite FunctionalGenomics; Tibco Spotfire, Göteborg, Sweden).
FIG. 2.
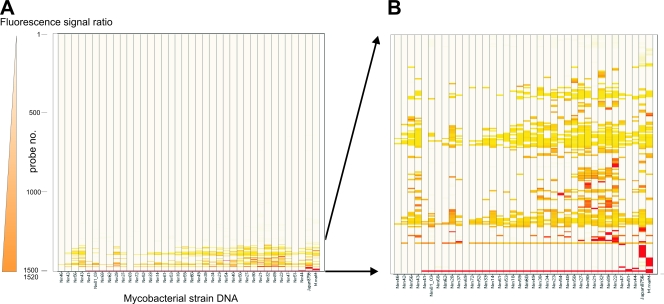
Heat map representation of microarray hits. In the heat map, each row represents the fluorescence signal ratio (reference strain Agy99/test strain) of one of the 1,520 immobilized insert probes, and each column represents a mycobacterial strain DNA sample. Outlier ratios of normalized fluorescence signals were sorted using an algorithm combining both the ratio value (color ranges from light yellow for low values to dark red for high values) and the number of affected strains (Spotfire DecisionSite). Panel A shows all 1,520 spots, and panel B shows a zoomed-in version of the 75 hit-generating spots across the 37 patient isolates and control strains. M.marM, M. marinum M.
Verification of microarray data.
Spots that indicated the presence of a deletion compared to strain Agy99 were analyzed for verification. First, the spotted genome locus was identified by sequencing of the PCR products spotted on the respective positions using primers pCDNA_F and pCDNA_R. Second, the defined region was identified on the Agy99 genome sequence (BuruList server; http://genopole.pasteur.fr/Mulc/BuruList.html), and primers complementary to these regions were designed that allowed scanning of the respective DNA stretch in steps of a maximum of 1 kb. Using these primer sets, a test PCR and sequencing were performed for each strain detected by the microarray data including control strains at conditions that allowed for identification of deletions as small as 50 bp, thus verifying the spot signals as true- or false-positive hits.
DNA fragment amplification and sequencing.
PCR was performed using the FirePol 10× BD buffer and 0.5 μl of FirePol Taq polymerase (Solis BioDyne, Tartu, Estonia) with 5 ng of genomic DNA, 0.6 μM (each) forward and reverse primer, 1.7 mM MgCl2, and a 0.3 mM concentration of each deoxynucleoside triphosphate in a total volume of 30 μl. PCRs were run in a GeneAmp PCR System 9700 PCR machine. The thermal profile for PCR amplification of E. coli plasmids and M. ulcerans genomic DNA included an initial denaturation step of 95 to 98°C for 3 min, followed by 32 cycles of 95°C for 20 s, annealing at 58 to 65°C for 20 s, and elongation at 72°C for 30 s up to 2 min. The PCRs were finalized by an extension step at 72°C for 10 min. PCR products were analyzed on 1% agarose gels by gel electrophoresis using ethidium bromide staining and an AlphaImager illuminator (Alpha Innotech, San Leandro, CA). Primers (Sigma-Aldrich, Steinheim, Germany) were designed using Primer3 software, version 0.4.0 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Sequencing was performed using a Big Dye kit and an ABI Prism 310 genetic sequence analyzer (Perkin-Elmer, Waltham, MA).
RESULTS
In order to detect new LSPs in M. ulcerans by CGH, we developed a PCR product-based printed DNA microarray covering a large proportion of the genome of M. ulcerans strain Agy99. Test hybridizations were performed with spotted DNA derived from regions of difference (RDs) identified in previous studies (29, 47). Using DNA of strains differing in these target sequences, we established spotting and hybridization conditions that yielded the predicted hybridization signals in a reliable and reproducible manner (Fig. 1). CGH was performed with genomic DNA of 37 Ghanaian M. ulcerans isolates, one Japanese M. ulcerans strain representing the ancestral lineage, and the M. marinum strain M (Fig. 2). Seventy-five of the spotted PCR products (Fig. 2) yielded a total of 321 outlier signals that were >100-fold lower in intensity than the corresponding signal of the reference strain Agy99. Thirty-five of these were >1,000-fold lower.
None of the negative control spots showed a significant outlier signal. Instead, probes containing IS2404/IS2606 fragments showed an outlier ratio in M. marinum, as expected due to its lack of either IS element (ISE). For M. marinum M, other outlier signals >1,000-fold lower than the reference were found that corresponded either to non-ISEs not present in the M. marinum M genome or to regions in the M. ulcerans plasmid pMUM001, which is also not present in M. marinum M. In addition, outlier signals of 100-fold intensity difference yielded two regions from within ISEs and three regions in close vicinity to either a MURD or to genome inversions between the sequences of M. marinum M and M. ulcerans Agy99, which likely results in steric hindrance of DNA-DNA hybridization. These results confirm the reliability established in the above test hybridizations.
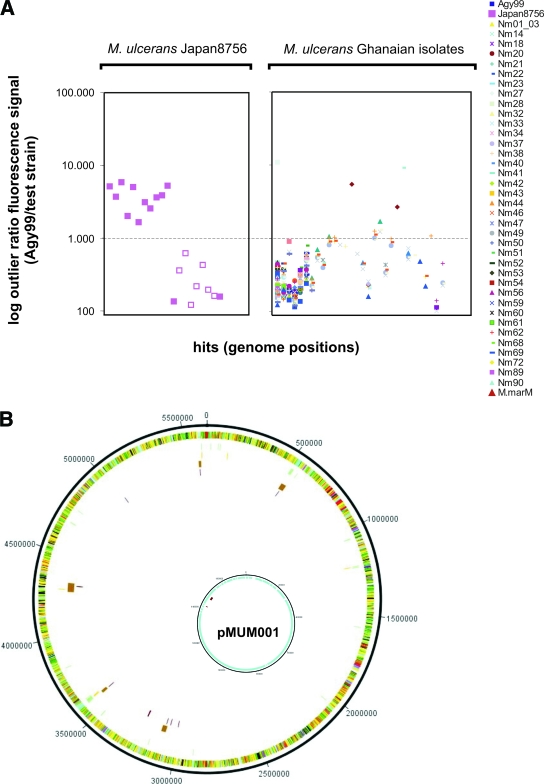
CGH analysis of the M. ulcerans strain Japan 8756 yielded 11 outlier signals that were >1,000-fold lower. All of these hits were associated with deletions in the corresponding genomic loci (Fig. 3). Thereby, all previously described deletions of the Asian haplotype in the RDs 9/15, 11, and 12 (28) were traced, and five additional deletions were newly identified (Fig. 3B). PCR analyses showed that all five of these deletions are larger than 2 kb. Moreover, hits relating to the same RDs but derived from different spotted probes demonstrated the reliability of the microarray-based CGH technology. Of the spotted probes associated with outlier signals with only a 130- to 160-fold reduction in signal intensity, one was related to a new RD. Furthermore, one spotted sequence directly adjacent to MURD 87 (and thus prone to reduced hybridization) yielded outlier signals with both M. ulcerans Japan 8756 and M. marinum M DNA. The newly identified deletions in the Japanese strains affected two IS transposition genes and CDSs of the following functional categories: PE/PPE family (three CDSs), conserved hypotheticals (eight CDSs), regulatory proteins (one CDS), cell wall/cell processes (three CDSs), and intermediary metabolism/respiration (six CDSs), the latter including a cytochrome P450 and dehydrogenase cluster.
FIG. 3.
Identification and validation of deletions. (A) Microarray hits with outlier ratios (reference strain Agy99/test strain) of normalized fluorescence signals higher than 100 (accounting for low signal/absence of a genomic region) are indicated as single symbols, as indicated on the figure. Each column represents one genome position that yielded a microarray hit. Hybridization was done with M. ulcerans Japan 8756 (filled symbols, confirmed absent genomic loci; open symbols, confirmed present genomic loci) and 37 Ghanaian isolates (all loci confirmed present). M.marM, M. marinum M. (B) Mapping of identified deletions to regions known to be absent. The circular view depicts the M. ulcerans Agy99 genome (outer circle), positions of non-ISEs not present in M. marinum M (http://www.sanger.ac.uk/Projects/M_marinum/) larger than 500 bp (second circle from outside) with their confirmation of absence by the microarray (third circle from outside), and deletions formerly found for the Asian haplotype (fourth circle from outside), with newly identified deletions in strain Japan 9756 (third circle from outside and innermost circle). Inset: regions absent from M. marinum M that corresponded to the M. ulcerans plasmid pMUM001.
CGH with DNA of 37 disease isolates from Ghana resulted in 235 hits with signals >100-fold lower than the reference. These were associated with 40 of the 1,520 printed PCR products (Fig. 3A). Only 15 of the outlier signals were >1,000-fold weaker than the corresponding Agy99 signal. When the genome regions of the 37 Ghanaian patient isolates, corresponding to the 40 implicated spotted probes, were scanned by PCR for the presence of deletions, no LSP was found.
DISCUSSION
A thorough knowledge of the epidemiology of a pathogen is of great importance for public health interventions and the identification of targets for diagnostic tests and drug and vaccine development (17). Determining genetic variability is a prerequisite for genomic fingerprinting and microepidemiological analyses. In various mycobacterial species, LSPs represent robust and accurate markers for mapping of chromosomal imbalances. In M. tuberculosis (15, 32, 58), Mycobacterium africanum (40), M. bovis (39), the vaccine strain M. bovis BCG (6, 19), and in other mycobacteria (18, 35, 42, 43, 48), most known LSPs have been identified using microarray technologies. Previous identification of RDs in a collection of M. ulcerans patient isolates originating from different regions worldwide where BU is endemic suggested that a whole-genome microarray might lead to the identification of LSPs also at a regional level (47).
In this study, we have used the largest collection thus far of regional strains, isolated between 2002 and 2004 from patients living in different districts of Ghana, including the Greater Accra, the Ashanti, and the Eastern regions. It was thought that such a panel of strains would show LSPs in a screening of the whole genome. Based on the genomic versatility discovered in other mycobacterial species (5, 9, 12, 34, 58), it was commonly assumed that LSPs would provide the key source of diversity, with genetic discriminatory power for microepidemiological purposes. However, no such variation was found across the investigated collection.
Suitability of the microarray assay was evidenced by confirmation of known LSPs, as well as by identification of new RDs in comparison to M. marinum M and M. ulcerans Japan 9756. Still, absence of PCR-reconfirmed hits in the CGH experiments with the Ghanaian M. ulcerans isolates does not strictly prove the absence of any LSPs. Some genomic regions might not be covered by the library; others might have been lost either because Sau3AI restriction sites were possibly overrepresented in the respective sites or because amplification or spotting was incomplete and deletions of <500 bp are not necessarily identified with the microarray setup used. However, the theoretical coverage, the confirmation of already known deletions, and the identification of new LSPs in M. marinum and an M. ulcerans strain belonging to the ancestral lineage indicate that the number of undetected deletions is expected to be small using this assay.
In contrast, our data suggest that in spite of the presence of numerous transposable ISs in the genome of African M. ulcerans strains, genetic changes leading to LSPs appear too rare to provide a basis for genetic fingerprinting of local M. ulcerans populations. We conclude that on a regional geographic scale, differentiation within these largely monomorphic local clones might be possible through identification of SNPs. First evidence for a SNP-based genetic discrimination between and within African countries has already been provided (31).
The M. ulcerans classical lineage, responsible for the vast majority of BU cases worldwide, is thought to have undergone massive genome reduction while adapting to a relatively stable ecological niche (14, 28, 29, 47, 54). The nature of some of the genomic changes speaks for an adaptation to an environment which is screened by immune effector mechanisms (14, 26, 28, 41). Genetic monomorphism indicates that this lineage has been forged through a bottleneck situation (29, 54, 63). The genomic stability demonstrated here of this successful clone may be indicative of decreased selective pressure, rendering further genome shrinkage unnecessary. The natural habitat and transmission pathways of M. ulcerans remain unknown, and it is not clear whether long-term persistence in ulcerative wounds of mammalian hosts is contributing significantly to the survival of the species. Development of a SNP-based rather than an LSP-based genetic fingerprinting method may help answer these key questions.
Acknowledgments
This research activity was part of the Stop Buruli initiative funded by the UBS Optimus Foundation, Switzerland.
We thank Simona Rondini for initial work and Martin Naegeli, Kerensa McElroy, Adriana Ille, and Michel Tessier for technical assistance.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Ablordey, A., P. A. Fonteyne, P. Stragier, P. Vandamme, and F. Portaels. 2007. Identification of a new variable number tandem repeat locus in Mycobacterium ulcerans for potential strain discrimination among African isolates. Clin. Microbiol. Infect. 13:734-736. [DOI] [PubMed] [Google Scholar]
- 2.Ablordey, A., M. Hilty, P. Stragier, J. Swings, and F. Portaels. 2005. Comparative nucleotide sequence analysis of polymorphic variable-number tandem-repeat Loci in Mycobacterium ulcerans. J. Clin. Microbiol. 43:5281-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ablordey, A., R. Kotlowski, J. Swings, and F. Portaels. 2005. PCR amplification with primers based on IS2404 and GC-rich repeated sequence reveals polymorphism in Mycobacterium ulcerans. J. Clin. Microbiol. 43:448-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ablordey, A., J. Swings, C. Hubans, K. Chemlal, C. Locht, F. Portaels, and P. Supply. 2005. Multilocus variable-number tandem repeat typing of Mycobacterium ulcerans. J. Clin. Microbiol. 43:1546-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alland, D., D. W. Lacher, M. H. Hazbon, A. S. Motiwala, W. Qi, R. D. Fleischmann, and T. S. Whittam. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 45:39-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 7.Brosch, R., S. V. Gordon, T. Garnier, K. Eiglmeier, W. Frigui, P. Valenti, S. S. Dos, S. Duthoy, C. Lacroix, C. Garcia-Pelayo, J. K. Inwald, P. Golby, J. N. Garcia, R. G. Hewinson, M. A. Behr, M. A. Quail, C. Churcher, B. G. Barrell, J. Parkhill, and S. T. Cole. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. USA 104:5596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosch, R., A. S. Pym, S. V. Gordon, and S. T. Cole. 2001. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 9:452-458. [DOI] [PubMed] [Google Scholar]
- 10.Chemlal, K., K. De Ridder, P. A. Fonteyne, W. M. Meyers, J. Swings, and F. Portaels. 2001. The use of IS2404 restriction fragment length polymorphisms suggests the diversity of Mycobacterium ulcerans from different geographical areas. Am. J. Trop. Med. Hyg. 64:270-273. [DOI] [PubMed] [Google Scholar]
- 11.Chemlal, K., G. Huys, P. A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 39:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, S. T. 2002. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology 148:2919-2928. [DOI] [PubMed] [Google Scholar]
- 13.Debacker, M., F. Portaels, J. Aguiar, C. Steunou, C. Zinsou, W. Meyers, and M. Dramaix. 2006. Risk factors for Buruli ulcer, Benin. Emerg. Infect. Dis. 12:1325-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demangel, C., T. P. Stinear, and S. T. Cole. 2009. Buruli ulcer: reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat. Rev. Microbiol. 7:50-60. [DOI] [PubMed] [Google Scholar]
- 15.Diaz, R., N. Siddiqi, and E. J. Rubin. 2006. Detecting genetic variability among different Mycobacterium tuberculosis strains using DNA microarrays technology. Tuberculosis 86:314-318. [DOI] [PubMed] [Google Scholar]
- 16.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagneux, S., and P. M. Small. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7:328-337. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Pelayo, M. C., K. C. Caimi, J. K. Inwald, J. Hinds, F. Bigi, M. I. Romano, D. van Soolingen, R. G. Hewinson, A. Cataldi, and S. V. Gordon. 2004. Microarray analysis of Mycobacterium microti reveals deletion of genes encoding PE-PPE proteins and ESAT-6 family antigens. Tuberculosis (Edinb.) 84:159-166. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 20.Grietens, K. P., A. U. Boock, H. Peeters, S. Hausmann-Muela, E. Toomer, and J. M. Ribera. 2008. “It is me who endures but my family that suffers”: social isolation as a consequence of the household cost burden of Buruli ulcer free of charge hospital treatment. PLoS Negl. Trop. Dis. 2:e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutacker, M. M., B. Mathema, H. Soini, E. Shashkina, B. N. Kreiswirth, E. A. Graviss, and J. M. Musser. 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J. Infect. Dis. 193:121-128. [DOI] [PubMed] [Google Scholar]
- 22.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilty, M., M. Kaser, J. Zinsstag, T. Stinear, and G. Pluschke. 2007. Analysis of the Mycobacterium ulcerans genome sequence reveals new loci for variable number tandem repeats (VNTR) typing. Microbiology 153:1483-1487. [DOI] [PubMed] [Google Scholar]
- 24.Hilty, M., D. Yeboah-Manu, D. Boakye, E. Mensah-Quainoo, S. Rondini, E. Schelling, D. Ofori-Adjei, F. Portaels, J. Zinsstag, and G. Pluschke. 2006. Genetic diversity in Mycobacterium ulcerans isolates from Ghana revealed by a newly identified locus containing a variable number of tandem repeats. J. Bacteriol. 188:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber, C. A., M. T. Ruf, G. Pluschke, and M. Kaser. 2008. Independent loss of immunogenic proteins in Mycobacterium ulcerans suggests immune evasion. Clin. Vaccine Immunol. 15:598-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaser, M., J. Hauser, P. Small, and G. Pluschke. 2009. Large sequence polymorphisms unveil the phylogenetic relationship of environmental and pathogenic mycobacteria related to Mycobacterium ulcerans. Appl. Environ. Microbiol. 75:5667-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaser, M., and G. Pluschke. 2008. Differential gene repertoire in Mycobacterium ulcerans identifies candidate genes for patho-adaptation. PLoS Negl. Trop. Dis. 2:e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaser, M., S. Rondini, M. Naegeli, T. Stinear, F. Portaels, U. Certa, and G. Pluschke. 2007. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evol. Biol. 7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaser, M., M. T. Ruf, J. Hauser, L. Marsollier, and G. Pluschke. 2009. Optimized method for preparation of DNA from pathogenic and environmental mycobacteria. Appl. Environ. Microbiol. 75:414-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Käser, M., J. Hauser, and G. Pluschke. 2009. Single nucleotide polymorphisms on the road to strain differentiation in Mycobacterium ulcerans. J. Clin. Microbiol. 47:3647-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavender, C. J., T. P. Stinear, P. D. Johnson, J. Azuolas, M. E. Benbow, J. R. Wallace, and J. A. Fyfe. 2008. Evaluation of VNTR typing for the identification of Mycobacterium ulcerans in environmental samples from Victoria, Australia. FEMS Microbiol. Lett. 287:250-255. [DOI] [PubMed] [Google Scholar]
- 34.Malik, A. N., and P. Godfrey-Faussett. 2005. Effects of genetic variability of Mycobacterium tuberculosis strains on the presentation of disease. Lancet Infect. Dis. 5:174-183. [DOI] [PubMed] [Google Scholar]
- 35.Marsh, I. B., J. P. Bannantine, M. L. Paustian, M. L. Tizard, V. Kapur, and R. J. Whittington. 2006. Genomic comparison of Mycobacterium avium subsp. paratuberculosis sheep and cattle strains by microarray hybridization. J. Bacteriol. 188:2290-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marsollier, L., R. Robert, J. Aubry, J. P. Saint Andre, H. Kouakou, P. Legras, A. L. Manceau, C. Mahaza, and B. Carbonnelle. 2002. Aquatic insects as a vector for Mycobacterium ulcerans. Appl. Environ. Microbiol. 68:4623-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathema, B., N. E. Kurepina, P. J. Bifani, and B. N. Kreiswirth. 2006. Molecular epidemiology of tuberculosis: current insights. Clin. Microbiol. Rev. 19:658-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 39.Mostowy, S., J. Inwald, S. Gordon, C. Martin, R. Warren, K. Kremer, D. Cousins, and M. A. Behr. 2005. Revisiting the evolution of Mycobacterium bovis. J. Bacteriol. 187:6386-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mostowy, S., A. Onipede, S. Gagneux, S. Niemann, K. Kremer, E. P. Desmond, M. Kato-Maeda, and M. Behr. 2004. Genomic analysis distinguishes Mycobacterium africanum. J. Clin. Microbiol. 42:3594-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mve-Obiang, A., R. E. Lee, F. Portaels, and P. L. Small. 2003. Heterogeneity of mycolactones produced by clinical isolates of Mycobacterium ulcerans: implications for virulence. Infect. Immun. 71:774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paustian, M. L., V. Kapur, and J. P. Bannantine. 2005. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J. Bacteriol. 187:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paustian, M. L., X. Zhu, S. Sreevatsan, S. Robbe-Austerman, V. Kapur, and J. P. Bannantine. 2008. Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species. BMC Genomics 9:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Portaels, F., P. Elsen, A. Guimaraes-Peres, P. A. Fonteyne, and W. M. Meyers. 1999. Insects in the transmission of Mycobacterium ulcerans infection. Lancet 353:986. [DOI] [PubMed] [Google Scholar]
- 45.Pouillot, R., G. Matias, C. M. Wondje, F. Portaels, N. Valin, F. Ngos, A. Njikap, L. Marsollier, A. Fontanet, and S. Eyangoh. 2007. Risk factors for Buruli ulcer: a case control study in Cameroon. PLoS Negl. Trop. Dis. 1:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quek, T. Y., E. Athan, M. J. Henry, J. A. Pasco, J. Redden-Hoare, A. Hughes, and P. D. Johnson. 2007. Risk factors for Mycobacterium ulcerans infection, southeastern Australia. Emerg. Infect. Dis. 13:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rondini, S., M. Käser, T. Stinear, M. Tessier, C. Mangold, G. Dernick, M. Naegeli, F. Portaels, U. Certa, and G. Pluschke. 2007. Ongoing genome reduction in Mycobacterium ulcerans. Emerg. Infect. Dis. 13:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semret, M., G. Zhai, S. Mostowy, C. Cleto, D. Alexander, G. Cangelosi, D. Cousins, D. M. Collins, D. van Soolingen, and M. A. Behr. 2004. Extensive genomic polymorphism within Mycobacterium avium. J. Bacteriol. 186:6332-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sizaire, V., F. Nackers, E. Comte, and F. Portaels. 2006. Mycobacterium ulcerans infection: control, diagnosis, and treatment. Lancet Infect. Dis. 6:288-296. [DOI] [PubMed] [Google Scholar]
- 50.Smith, N. H., S. V. Gordon, R. Rua-Domenech, R. S. Clifton-Hadley, and R. G. Hewinson. 2006. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat. Rev. Microbiol. 4:670-681. [DOI] [PubMed] [Google Scholar]
- 51.Stinear, T., J. K. Davies, G. A. Jenkin, F. Portaels, B. C. Ross, F. Oppedisano, M. Purcell, J. A. Hayman, and P. D. Johnson. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stinear, T. P., H. Hong, W. Frigui, M. J. Pryor, R. Brosch, T. Garnier, P. F. Leadlay, and S. T. Cole. 2005. Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J. Bacteriol. 187:1668-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stinear, T. P., G. A. Jenkin, P. D. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stinear, T. P., T. Seemann, S. Pidot, W. Frigui, G. Reysset, T. Garnier, G. Meurice, D. Simon, C. Bouchier, L. Ma, M. Tichit, J. L. Porter, J. Ryan, P. D. Johnson, J. K. Davies, G. A. Jenkin, P. L. Small, L. M. Jones, F. Tekaia, F. Laval, M. Daffe, J. Parkhill, and S. T. Cole. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stragier, P., A. Ablordey, L. M. Bayonne, Y. L. Lugor, I. S. Sindani, P. Suykerbuyk, H. Wabinga, W. M. Meyers, and F. Portaels. 2006. Heterogeneity among Mycobacterium ulcerans isolates from Africa. Emerg. Infect. Dis. 12:844-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stragier, P., A. Ablordey, L. Durnez, and F. Portaels. 2007. VNTR analysis differentiates Mycobacterium ulcerans and IS2404 positive mycobacteria. Syst. Appl. Microbiol. 30:525-530. [DOI] [PubMed] [Google Scholar]
- 57.Stragier, P., A. Ablordey, W. M. Meyers, and F. Portaels. 2005. Genotyping Mycobacterium ulcerans and Mycobacterium marinum by using mycobacterial interspersed repetitive units. J. Bacteriol. 187:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner, T., M. E. Benbow, T. O. Brenden, J. Qi, and R. C. Johnson. 2008. Buruli ulcer disease prevalence in Benin, West Africa: associations with land use/cover and the identification of disease clusters. Int. J. Health Geogr. 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wansbrough-Jones, M., and R. Phillips. 2006. Buruli ulcer: emerging from obscurity. Lancet 367:1849-1858. [DOI] [PubMed] [Google Scholar]
- 61.World Health Organization. 2008. Buruli ulcer: progress report, 2004-2008. Wkly. Epidemiol. Rec. 83:145-156. [PubMed] [Google Scholar]
- 62.Yeboah-Manu, D., T. Bodmer, E. Mensah-Quainoo, S. Owusu, D. Ofori-Adjei, and G. Pluschke. 2004. Evaluation of decontamination methods and growth media for primary isolation of Mycobacterium ulcerans from surgical specimens. J. Clin. Microbiol. 42:5875-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yip, M. J., J. L. Porter, J. A. Fyfe, C. J. Lavender, F. Portaels, M. Rhodes, H. Kator, A. Colorni, G. A. Jenkin, and T. Stinear. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J. Bacteriol. 189:2021-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]