Abstract
Matrix-assisted laser desorption ionization-time of flight mass spectrometry has emerged as a rapid, cost-effective alternative for bacterial species identification. Identifying 60 blind-coded nonfermenting bacteria samples, this international study (using eight laboratories) achieved 98.75% interlaboratory reproducibility. Only 6 of the 480 samples were misidentified due to interchanges (4 samples) or contamination (1 sample) or not identified because of insufficient signal intensity (1 sample).
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has emerged as a fast and cost-effective alternative for bacterial species identification in microbiology. On the basis of mass analysis of the protein composition of a bacterial cell, which is assumed to be characteristic for each bacterial species, it is possible to determine the species within few minutes, starting from whole cells, cell lysates, or crude bacterial extracts (2, 3, 5, 6). The proof of principle of MALDI-TOF MS for bacterial species identification was shown a decade ago (2, 5, 6); however, due to low reproducibility, it has not been widely adopted in clinical microbiology. We have recently shown that use of a larger mass range for detection (2,000 to 20,000 Da), dedicated analysis software for spectral pattern matching, and a high-quality reference database of spectra generated from quality-controlled culture collection strains resulted in accurate species identifications, with high intralaboratory reproducibility (7). For interlaboratory reproducibility, there are only very limited data available (8, 10). We therefore evaluated the interlaboratory reproducibility for MALDI-TOF MS-based species identification in a multicenter study, applying the above-described MALDI-TOF MS improvements.
(This study was presented in part at the 19th European Congress of Clinical Microbiology and Infectious Diseases [ECCMID] in Helsinki, Finland, 16 to 19 May 2009.)
Sixty blind-coded samples were shipped worldwide by mail to eight laboratories with access to Bruker MALDI-TOF MS platforms and personnel trained in MALDI-TOF MS-based species identification (Centre de Ressources Biologiques de l′Institut Pasteur [CRBIP], Department of Microbiology, Institut Pasteur, Paris, France; Department of Microbiology and Molecular Cell Biology, Center for Biomedical Proteomics, Eastern Virginia Medical School, Norfolk, VA; Labor Limbach, Heidelberg, Germany; Research Institute of Physical-Chemical Medicine, Moscow, Russia; Bruker Daltonik, GmbH, Bremen, Germany; Institut de Bactériologie, Strasbourg, France; Microbiology Unit, Bambino Gesù Children's Hospital, Health Care and Research Institute, Rome, Italy; and Molecular Infectious Diseases Laboratory, Vanderbilt University Hospital, Nashville, TN). Samples 001 to 030 of the 60 samples included pure cultures of different nonfermenting bacteria, either culture collection strains from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) and Laboratorium voor Microbiologie, Universiteit Gent (LMG, Gent, Belgium), or strains isolated at the Institute of Hygiene, Münster, Germany, during routine diagnostic efforts. They contained single strains of Alcaligenes faecalis subsp. faecalis (DSMZ 30030), Brevundimonas andropogonis (DSMZ 9511), Brevundimonas aurantiaca (DSMZ 4731), Burkholderia caribensis (DSMZ 13236), Brevundimonas diminuta (DSMZ 7234), Brevundimonas intermedia (DSMZ 4732), Brevundimonas vesicularis (DSMZ 7226), Comamonas nitrativorans (DSMZ 13191), Comamonas testosteroni (DSMZ 50244), Flavobacterium johnsoniae (DSMZ 2064), Inquilinus limosus (DSMZ 16000), Sphingobacterium mizutaii (DSMZ 11724), Pseudomonas beteli (LMGZ 978), Pseudomonas boreopolis (LMGZ 979), Pseudomonas extremorientalis (DSMZ 15824), 13 Pseudomonas aeruginosa strains (DSMZ 50071 and 12 clinical isolates), and 2 Stenotrophomonas maltophilia strains (clinical isolates). The 16 culture collection strains listed above were among the 248 strains used for constructing a nonfermenter reference database (7). All species designations were unambiguously confirmed using partial 16S rRNA gene sequencing as described elsewhere (7). Samples 031 to 060 contained preprocessed cell extracts from the first 30 strains as described recently (7). Accompanying the samples, each participating laboratory received a “sample cultivation and preparation guide” and a “result reporting guide” to facilitate and standardize data generation and interpretation. Briefly, the laboratories were asked to streak out samples 001 to 030 onto blood agar plates (irrespective of the vender) and to incubate them for 48 h at 30°C in an ambient atmosphere. Using a single colony, extraction for MALDI-TOF MS analysis was initiated. For preprocessed samples 031 to 060, the guide included instructions for matrix preparation and MALDI-TOF MS analysis (7). For spectral calibration, the Bruker bacterial test standard (Escherichia coli lysate) was used during the measuring step. All laboratories used the MALDI Biotyper 2.0 software package (Bruker Daltonik, GmbH, Bremen, Germany) and the MALDI Biotyper database, containing spectra of more than 2,800 microorganisms (including the 248 nonfermenter species) as reference data. The software generates a list of probable species identifications ranked by the log(score) value, which reflects the peak matches as well as intensities and results in values between 0 and 3 (0 to 100% pattern match). After comparison of an unknown spectrum with all reference spectra of the database, the log(score)s are ranked. Values of ≥2.0 were required for secure identification at the species level and values between <2 and ≥1.7 for secure identification at the genus level. Results based on log(score) values of <1.7 were rated as not identifiable. These thresholds were empirically determined based on the whole MALDI Biotyper database contents.
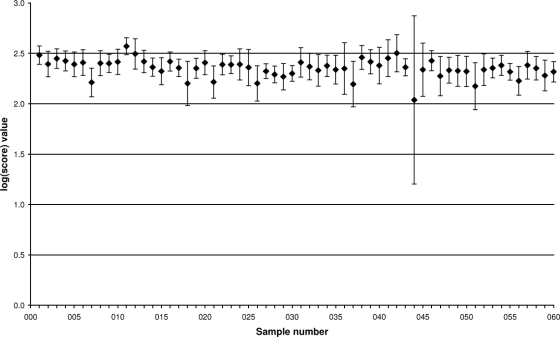
Each of the eight participating laboratories received 60 blind-coded samples for MALDI-TOF MS-based species identification. The aggregated results for each laboratory and the machines used are shown in Table 1. Of the total 480 samples, 474 (98.75%) were correctly identified at the species level by using the highest log(score) value for the identified species after MALDI-TOF MS spectral comparisons. Five of the remaining six samples were misidentified, and one sample did not result in any valid species designation, due to low signal intensity. Overall, six of the eight laboratories identified all 60 samples correctly (Table 1). Four hundred sixty-seven of the 480 samples (97.29%) with log(score) values of ≥2 (mean, 2.353; standard deviation, 0.146) were identified, indicating a probable secure species identification level (Table 1). Twelve of the remaining 13 samples showed log(score) values between 1.7 and 2, which correlated with at least a secure identification at the genus level; only sample 044, which was not identified, due to low signal intensity, had a log(score) value of 0. The 12 samples were distributed among four laboratories; no pattern of an especially problematic sample was discerned. The five misidentified samples showed log(score) values of ≥2. Figure 1 displays the mean log(score) value for each sample and its standard deviation. Of the 60 samples investigated, only sample 044 showed a significantly higher standard deviation and lower mean value due to the failure in laboratory B. There was no significant difference between the mean log(score) values of cultured samples versus those of preprocessed samples as determined by t test statistics (P = 0.20).
TABLE 1.
Aggregated log(score) values and final species identification results for each of the eight participating laboratories (not specified) for 60 blind-coded samples (n = 480 in total) containing nonfermenting bacteria either as pure culture or as preprocessed cell extract
| Laboratory | MALDI-TOF MS instrument (purchase yr) | No. (%) of log(score) values |
No. (%) of samples |
|||
|---|---|---|---|---|---|---|
| ≥2 | <2-1.7 | <1.7 | Correctly identified | Mis- or nonidentified | ||
| A | Microflex LT (2007) | 60 (100) | 0 (0) | 0 (0) | 60 (100) | 0 (0) |
| B | Autoflex LT (2002) | 53 (88.33) | 6 (10.0) | 1 (1.67) | 58 (96.67) | 2a (3.33) |
| C | Microflex LT (2007) | 60 (100) | 0 (0) | 0 (0) | 60 (100) | 0 (0) |
| D | Ultraflex III (2007) | 59 (98.33) | 1 (1.67) | 0 (0) | 56 (93.33) | 4b (6.67) |
| E | Microflex LT (2007) | 58 (96.67) | 2 (3.33) | 0 (0) | 60 (100) | 0 (0) |
| F | Microflex LRF (2005) | 57 (95.0) | 3 (5.0) | 0 (0) | 60 (100) | 0 (0) |
| G | Biflex (1999) | 60 (100) | 0 (0) | 0 (0) | 60 (100) | 0 (0) |
| H | Microflex LT (2009) | 60 (100) | 0 (0) | 0 (0) | 60 (100) | 0 (0) |
| Total | 467 (97.29) | 12 (2.5) | 1 (0.21) | 474 (98.75) | 6 (1.25) | |
One of the two samples yielded a log(score) value of ≥2, and the other sample had a log(score) value of <1.7.
All four samples yielded log(score) values of ≥2.
FIG. 1.
Mean log(score) values and standard deviations for all 60 blind-coded samples identified using MALDI-TOF MS, calculated from the results for all eight participating laboratories.
Besides the lack of comprehensive reference databases for spectral comparisons and of sophisticated software tools for data interpretation, the broad use of MALDI-TOF MS for species identification was hampered in the past by the limited reproducibility (9). Dedicated software tools are now available, along with comprehensive databases for some genera (e.g., anaerobic [4] or nonfermenting [7] bacteria), and intralaboratory reproducibility has been proven (7); however, interlaboratory reproducibility remained unclear. Therefore, we present here for the first time a large international multicenter study, using 60 blind-coded nonfermenting bacterial samples, showing a high interlaboratory reproducibility, with 98.75% correct species identifications (Table 1). There was no significant difference in achieved log(score) values between cultured and preprocessed samples, indicating that both methods were equally reproducible. In contrast to previous studies (9), the Bruker system yields high reproducibility if a minimum standard is followed, as recommended in the “sample cultivation and preparation guide.” Although all six mis- or nonidentified samples were preprocessed samples, only in sample 044 of laboratory B was a failure due to low signal intensity noted. For the remaining misidentified samples, sample interchange (four samples) or contamination with skin flora (one sample; Staphylococcus epidermidis) was a highly likely reason for misidentification. Not only were samples correctly identified at a high rate, but reliability was high: 97.29% of all log(score) values were >2, the threshold for a secure species identification. It was even possible to correctly identify 12 of the 13 samples with log(score) values of <2. This level of reproducibility is usually achievable only with DNA sequence-based methods (1). Moreover, sending preprocessed (inactivated and therefore noninfectious) samples greatly facilitated the exchange of specimens.
In summary, this study demonstrated that MALDI-TOF MS has become a highly reproducible alternative platform for partial 16S rRNA gene sequencing for the identification of bacterial species in the microbiology laboratory. Whereas MALDI-TOF MS has a higher discriminatory power than 16S rRNA gene PCR, the latter is more sensitive, but neither of them can adequately resolve mixed bacterial samples.
Acknowledgments
This study was supported by OPBG Grant Ricerca Corrente 200702P002153.
We thank Isabell Ramminger and Ursula Keckevoet (Münster, Germany); Leopoldo Dimiziani (Rome, Italy); Ulrike Wild and Anke Veldenzer (Heidelberg, Germany); Beatrix Wegmann (Bremen, Germany); Haijing Li, Criziel Quinn, and Beth Mutai (Nashville, TN); and Gongyi Shi and Sam Fu (Fremont, CA) for skillful technical assistance.
M. Kostrzewa and T. Maier have declared a potential conflict of interest. They are both employees of Bruker Daltonik, GmbH, the company that produces the MALDI-TOF MS instruments and the software mentioned in the manuscript. All other authors have declared that no competing interests exist.
Footnotes
Published ahead of print on 23 September 2009.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. W. Friedrich, D. Harmsen, A. Holmes, X. Huijsdens, A. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claydon, M. A., S. N. Davey, V. Edwards-Jones, and D. B. Gordon. 1996. The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 14:1584-1586. [DOI] [PubMed] [Google Scholar]
- 3.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20:157-171. [DOI] [PubMed] [Google Scholar]
- 4.Grosse-Herrenthey, A., T. Maier, F. Gessler, R. Schaumann, H. Bohnel, M. Kostrzewa, and M. Kruger. 2008. Challenging the problem of clostridial identification with matrix-assisted laser desorption and ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe 14:242-249. [DOI] [PubMed] [Google Scholar]
- 5.Holland, R. D., J. G. Wilkes, F. Rafii, J. B. Sutherland, C. C. Persons, K. J. Voorhees, and J. O. J. Lay. 1996. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:1227-1232. [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurthy, T., P. L. Ross, and U. Rajamani. 1996. Detection of pathogenic and non-pathogenic bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:883-888. [DOI] [PubMed] [Google Scholar]
- 7.Mellmann, A., J. Cloud, T. Maier, U. Keckevoet, I. Ramminger, P. Iwen, J. Dunn, G. Hall, D. Wilson, P. Lasala, M. Kostrzewa, and D. Harmsen. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker, J., A. J. Fox, V. Edwards-Jones, and D. B. Gordon. 2002. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J. Microbiol. Methods 48:117-126. [DOI] [PubMed] [Google Scholar]
- 9.Wang, Z., L. Russon, L. Li, D. C. Roser, and S. R. Long. 1998. Investigation of spectral reproducibility in direct analysis of bacteria proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 12:456-464. [DOI] [PubMed] [Google Scholar]
- 10.Wunschel, S. C., K. H. Jarman, C. E. Petersen, N. B. Valentine, K. L. Wahl, D. Schauki, J. Jackman, C. P. Nelson, and E. White V. 2005. Bacterial analysis by MALDI-TOF mass spectrometry: an inter-laboratory comparison. J. Am. Soc. Mass Spectrom. 16:456-462. [DOI] [PubMed] [Google Scholar]