Abstract
Babesia rossi, an intraerythrocytic protozoan, causes a severe, often life-threatening disease of domestic dogs. Dogs treated early for B. rossi infection usually recover from the disease, but dogs left untreated or treated at a later stage of infection seldom survive. Dogs infected with B. rossi have varied clinical manifestations that can be categorized as uncomplicated (with a good prognosis) or complicated (with a poor prognosis). One hundred twenty-one blood samples were collected from dogs presented to the Onderstepoort Veterinary Academic Hospital and diagnosed with babesiosis by the use of a thin blood smear. An additional 20 samples were obtained from Babesia-infected dogs from private clinics around the Onderstepoort, Johannesburg, Durban, White River, and Cape Town areas. The samples were screened by PCR targeting the Babesia rossi erythrocyte membrane antigen gene (BrEMA1) and by sequencing of the polymorphic region (i.e., region with a variable number of hexapeptide repeats). Analysis of PCR products revealed 11 different gene profiles, visualized by gel electrophoresis. Twelve distinct BrEMA1 genotypes were identified by sequencing, but the numbers of hexapeptide repeats varied from 6 to 31 (classified as genotype6 to genotype31). The genotypes were retrospectively compared to the clinical case data. The most frequently encountered B. rossi parasites were those attributed to genotype19 (36.2%), genotype28 and genotype29 (20.6% each), and genotype11 (12.7%). These genotypes were also the ones associated with the poorest prognosis. This preliminary finding suggests clinically important differences between the various B. rossi genotypes identified.
The large Babesia parasites Babesia rossi and Babesia vogeli are two of the most frequently encountered blood parasites of dogs in South Africa (3, 12, 13). Babesia rossi is the most frequently encountered species detected in dogs presented with clinical babesiosis at the outpatient clinic of the Onderstepoort Veterinary Academic Hospital (OVAH). Babesia vogeli infections are reported to be rare and less virulent than B. rossi infections (3, 18). Canine babesiosis caused by B. rossi is clinically classified as either uncomplicated or complicated. Clinical hallmarks of Babesia infections in dogs always include fever and splenomegaly. The disease is said to be uncomplicated if the clinical changes could be attributed directly to a mild or moderate anemia, with no clinical evidence of organ dysfunction or failure (9). Dogs with mild anemia are usually treated with an antibabesial drug and discharged. Dogs with moderate anemia may receive a blood transfusion and are then discharged. The survival rate in this manifestation of disease is almost 100% (11). Examples of complicated cases of the disease are those where clinical presentation is complicated by evidence of non-solid-organ failure (non-solid-organ complication [non-SOC]) characterized by severe anemia and hemoconcentration or organ dysfunction or failure (SOC). Examples of complicated disease include acute renal failure (ARF), acute respiratory distress syndrome (ARDS), cerebral involvement, coagulopathy, icterus and hepatopathy, immunity-mediated hemolytic anemia, peracute manifestations, hemoconcentration, pancreatitis, rhabdomyolysis, and shock (10, 11). These dogs all require admission to an intensive care facility, where advanced medical treatment is essential. The mortality rate in this group is around 15%, irrespective of the nature of the treatment administered (17).
The mechanisms that result in B. rossi parasites being associated with such a range of diverse clinical signs and severe disease in the host are still unknown. As suggested for Babesia canis (senso stricto), there might be genotypic differences among B. rossi strains (4) that could be associated with variable virulence. A polymorphic phosphoprotein localized on the cytoplasmic surface of B. rossi-infected red blood cells was recently characterized and named Babesia rossi erythrocyte membrane antigen 1 (BrEMA1) (B. Carcy, unpublished data). Analysis of the BrEMA1 genes of various laboratory strains of B. rossi revealed that these code for polymorphic proteins that contain various numbers of repetitive hexapeptide motifs. Although the exact function of BrEMA1 is not known, it is suspected that this gene may be unique to B. rossi parasites. Thus, since B. rossi is the most pathogenic species of the large babesias of dogs, it was hypothesized that the gene may have a role in virulence. This gene was used as a genetic marker to classify B. rossi isolates obtained from South Africa and to analyze the relationships between particular genotypes and the occurrence and severity of clinical manifestations of B. rossi-induced canine babesiosis. The molecular and biochemical characterization of BrEMA1 will be reported elsewhere by B. Carcy, University of Montpellier.
MATERIALS AND METHODS
Sample origin and grouping.
Canine blood samples (n = 121) were obtained from the outpatient clinic of OVAH. These dogs were diagnosed with canine babesiosis based on clinical signs and microscopic confirmation of infected red blood cells on a blood smear. Prior to treatment, blood samples were collected from the cephalic vein into EDTA tubes. If the presence of B. rossi was confirmed in our laboratory, clinical data for a specific dog were retrieved from patient files. A further 20 blood samples were obtained from geographically dispersed private clinics (Onderstepoort area, Johannesburg, Durban, White River, and Cape Town) from dogs diagnosed with canine babesiosis and sent to our laboratory for molecular characterization of the infection. Dogs sampled from the non-OVAH sites were not included in the clinical part of the study, since there appears to be a lack of uniformity in the criteria used by various private practices in diagnosing clinical signs. These samples were used only for establishing and identifying parasite genotype occurrences at the said geographic localities. Dogs sampled at OVAH were grouped according to their clinical presentation and outcome, as follows: group H (home), dogs that were treated with an antibabesial drug during consultation and discharged immediately; group A (admitted), dogs admitted for treatment and that survived until discharge; and group D (died), dogs that died despite treatment or that were euthanized owing to poor prognosis (3). Follow-up examinations were conducted on dogs in group H to confirm that these dogs had indeed recovered completely. Canine babesiosis cases were diagnosed and treated in a similar manner by all clinicians at the OVAH. The diagnostic criteria used for diagnoses of the various complications included in-saline positive agglutination (ISA) for immunity-mediated hemolytic anemia; respiratory distress, as evidenced by hyperpnoea or blood gas evidence of lung edema, for the diagnosis of ARDS; a rise in hematocrit above 50% or fall below 10% (identifying hemoconcentration and severe anemia, respectively); the development of any central nervous system signs that could not be attributed to any other cause (especially hypoglycemia) for the diagnosis of cerebral disease; clinical collapse for the diagnosis of shock; and the rise of blood creatinine levels above 150 μmol/liter or anuria for the diagnosis of ARF and hypoglycemia (glucose level, <3.3 mmol/liter). Based on the above criteria, dogs that were admitted with complications were then separated into two groups, i.e., those with SOC and those without SOC. Finally, dogs from both groups were separated into dogs that survived until discharge and dogs that died despite treatment or that were euthanized owing to poor prognosis.
Two additional samples, known to be positive for B. canis and B. vogeli, were obtained from Utrecht, The Netherlands, and Bloemfontein, South Africa, respectively, for the purpose of evaluating whether BrEMA1 could be detected in these species.
This project was approved by the Animal Use and Care Committee of the University of Pretoria and the Research Committee of the Faculty of Veterinary Science, University of Pretoria.
DNA extraction and PCR.
Once collected, the blood samples were sent to the Department of Veterinary Tropical Diseases, Faculty of Veterinary Science, University of Pretoria. DNA was extracted from 200 μl of each blood sample by use of a QIAamp blood and tissue extraction kit (Qiagen, Hilden, Germany) following the manufacturer's protocols. As a first step for the molecular diagnosis of Babesia species infecting each dog, a PCR was performed with reverse line blot (RLB) primers F2 (5′-GAC ACA GGG AGG TAG TGA CAA G-3′) and RLB-R2 (biotin-5′-CTA AGA ATT TCA CCT CTG ACA GT-3′). The conditions for the PCR included an initial step of 3 min at 37°C, 10 min at 94°C, and 10 cycles of 94°C (20 s), 67°C (30 s), and 72°C (30 s), with lowering of the annealing step by 2°C after every second cycle (touchdown PCR). The reaction was then followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 30 s. The PCR-amplified products, which were 460- to 540-bp fragments from the 18S rRNA gene spanning the V4 region (7, 13), were tested with RLB as previously described (13).
The B. rossi genetic diversity was then analyzed on genomic DNAs from samples that tested positive for B. rossi by RLB. These samples were reamplified with primers FrepBrEMA1 (5′-CCA ACA TTG ATG ATG ACA A-3′) and RrepBrEMA1 (5′-CTG CAT GTC AGC TTC ATC A-3′). These primers were designed from the B. rossi BrEMA1 gene (GenBank accession number AJ416994). They amplified an 18-nucleotide repetitive sequence whose number (16 to 31; size fragment from 375 to 645 bp) and sequence were initially shown to be variable between BrEMA1 genes from three laboratory strains (Carcy, unpublished data). To test whether the BrEMA1 gene was unique to B. rossi isolates, two additional samples, positive for B. vogeli and B. canis, were included. PCR amplification was done with 2.5 μl of extracted DNA in a final 25-μl volume containing 0.5 μl of each primer (10 μM), 4 μl of deoxynucleoside triphosphate mix (1.25 mM [each deoxynucleoside triphosphate]), 2.5 μl of 10× PCR buffer, and Taq DNA polymerase (1 U). The conditions of the PCR included in the program were 1 cycle of 5 min at 94°C, 30 cycles of amplification (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min), and 1 cycle of 7 min at 72°C.
Sequencing, phylogenetic, and statistical analyses.
PCR products (5 μl) were loaded and visualized on a 1.5% agarose gel. Samples with distinct bands were purified with a QIAquick PCR purification kit (Qiagen, Germany). The sequencing reaction was prepared with 3.2 pmol (each) of primers FrepBrEMA1 and RrepBrEMA1. Sequencing was performed in the Genetics Section of the Faculty of Veterinary Science. Sequence data for the full BrEMA1 sequences were assembled using GAP 4 of the Staden package (version 1.6.0 for Windows). Sequence alignments were manually edited using Bioedit (version 5.0.9). DNA sequences were translated into amino acid sequences, and genotyping was done according to the number of hexapeptide repeats. Sequence alignment of the amino acid sequences and phylogenetic trees were generated using GeneBee (http://www.genebee.msu.su/genebee.html) programs. The Cluster distance algorithm method was used for the construction of the phylogenetic tree (Phylip, multiline) in the phylogram format from the aligned amino acid sequences. In the Cluster algorithm, the notion of distance between groups of sequences is used for the setting of the branching order. This distance is defined as the arithmetic mean of pairwise distances between elements of the two groups (http://www.genebee.msu.su/genebee.html). The B. rossi BrAK (adenosine kinase) sequence (5) was used as an outgroup. Genotype frequencies were compared using a two-tailed binomial test. Proportions were compared using two-tailed Fisher's exact test. Analyses were done using Stata 8.2 (StataCorp, College Station, TX).
Nucleotide sequence accession numbers.
The BrEMA1 gene nucleotide sequences (designated genotype6, -11, -12, -13, -16g1, -16g2, -18, -19g1, -19g2, -22, -28, -29, and -31) have been submitted to the EMBL, GenBank, and DDBJ nucleotide sequence databases and can be retrieved under accession numbers FM164393 to FM164405.
RESULTS
Genetic analysis of B. rossi isolates.
All of the blood samples (n = 141) were positive for B. rossi in the PCR-RLB assay. These samples were further analyzed to determine the genetic heterogeneity of the BrEMA1 gene.
(i) Diversity in BrEMA1 genotypes.
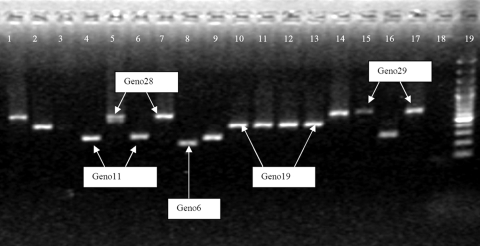
PCR amplification of the polymorphic region of the BrEMA1 gene revealed a total of 11 distinct profiles by examining the size of the amplicons which could be visualized on gel electrophoresis. The smallest amplified product was 200 bp (Fig. 1, lane 8), and the largest was ca. 645 bp (lanes 15 and 17). Sequencing results for the amplicons showed 13 different consensus amino acid sequences that ranged in size between 6 and 31 hexapeptide repeats, resulting in 12 distinct genotypes (Table 1). Genotypes with a repetitive region composed of 16 or 19 hexapeptide repeats each had two differentiable amino acid consensus sequences and were thus subdivided into two groups (designated g1 and g2). Genotype16g1 and genotype16g2 had two distinct sequences, indicating genetic heterogeneity. In contrast, genotype19g1 and genotype19g2 differed by a single amino acid, A or V, in the hexapeptide KS(A/V)ASV, suggesting a much closer relatedness. Finally, sequence analysis of genotype11 and genotype16g2 indicated that they might be related genetically. This was based on the fact that they are the only genotypes containing the motif ASPGSV and two amino acid substitutions (IP rather than VL) downstream from the repetitive sequence of BrEMA1 (Table 1).
FIG. 1.
Field polymorphism of B. rossi strains in South Africa, as evidenced by PCR amplification of the BrEMA1 repeat region with primers FrepBrEMA1 and RrepBrEMA1. PCR products were loaded on a 1.5% agarose gel. PCR profiles of the most encountered genotypes (11, 19, 28, and 29) and that of genotype6 are indicated on the picture as follows: lanes 15 and 17 represent genotype29; lanes 1, 5, 7, and 14 represent genotype28; lanes 2, 10, 11, 12, and 13 represent genotype19; lanes 4, 6, 9, and 16 represent genotype11; lane 8 represents genotype6, and lane 19 is the 100-bp marker.
TABLE 1.
Consensus amino acid sequence of 13 BrEMA1 genotypesa
| Genotype | Sample origin (no. of samples sequenced) | Consensus amino acid sequenceb |
|---|---|---|
| Genotype6 | OVAH (1) | NIDDDKASV KSADSL KSAGSA KSVASV RSADSV ESAGSA KSVASV RSADADVLHDTHLDEADMQ 64 |
| Genotype11 | OVAH (14), Onderstepoort (4) | NIDDDKASV KSAGSV (RSADSV ASPGSV KSAASV)2 RSADSV ESVASV RSADSV ESVASV RSADADIPHDTHLDEADMQ 94 |
| Genotype12 | OVAH (1) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA KSVASV (RSADSV ESAGSA KSVASV)2 RSADADVLHDTHLDEADMQ 100 |
| Genotype13 | Onderstepoort (1) | NIDDDKASV KSAASL KSADSA KSVASV RSADSV ESAGSA KSAGSV KSAASV RSADSV ESAASA KSAASV RSADSV ESAGSA KSAASV RSADADVLHDTHLDEADMQ 106 |
| Genotype16 | ||
| Genotype16g1 (repeated region from GenBank sequence AJ416994) | Johannesburg (1), OVAH (1) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA KSAASV KSAASV RSADSV ESAASA KSAASV (RSADSV ESAGSA KSVASV)2 RSADADVLHDTHLDEADMQ 124 |
| Genotype16g2 | Durban (1), White River (4) | NIDDDKASV KSAGSV (KSAASV RSADSV ASPGSV)4 KSAASV RSADSV ESVASV RSADADIPHDTHLDEADMQ 124 |
| Genotype18 | OVAH (1), Onderstepoort (1) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA (KSVASV RSADSV ESAASA)3 KSVASV RSADSV ESAGSA KSVASV RSADADVLHDTHLDEADMQ 136 |
| Genotype19 | ||
| Genotype19g1 (repeated region from GenBank sequence AJ416997) | OVAH (30), Onderstepoort (1) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA KSAGSV KSAASV RSADSV ESAASA KSAASV RSADSV ESAGSA KSVASV RSADSV ESAGSA KSA*ASV RSADSV ESAGSA KSVASV RSADADVLHDTHLDEADMQ 142 |
| Genotype19g2 | OVAH (18), Onderstepoort (1), White River (1) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA KSAGSV KSAASV RSADSV ESAASA KSAASV RSADSV ESAGSA KSVASV RSADSV ESAGSA KSV*ASV RSADSV ESAGSA KSVASV RSADADVLHDTHLDEADMQ 142 |
| Genotype22 | Onderstepoort (1) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA KSAASV KSAASV RSADSV ESAASA KSAASV RSADSV ESAGSA KSAASV RSADSV ESAGSA KSVASV RSADSV ESAGSA KSAASV RSADSV ESAGSA KSVASV RSADADVLHDTHLDEADMQ 160 |
| Genotype28 | OVAH (27), Onderstepoort (3) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA KSAGSV KSAASV RSADSV ESAASA KSAASV RSADSV ESAGSA (KSVASV RSADSV ESAGSA)5 KSVASV RSADADVLHDTHLDEADMQ 196 |
| Genotype29 | OVAH (28) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA KSAGSV KSAGSV KSAASV RSADSV ESAASA KSAASV RSADSV ESAGSA (KSVASV RSADSV ESAGSA)5 KSVASV RSADADVLHDTHLDEADMQ 202 |
| Genotype31 (repeated region from GenBank sequence AJ416996) | Cape Town (1) | NIDDDKASV KSAASL KSADSA KSVASV RSPDSV ESAGSA KSAGSV (KSAASV RSADSV ESAGSA)2 (KSVASV RSADSV ESAGSA)6 KSVASV RSADADVLHDTHLDEADMQ 214 |
Sequencing of the repetitive region of the gene was performed on 141 samples from dogs diagnosed with B. rossi infections.
*, difference in a single conservative amino acid.
The two positive B. vogeli and B. canis samples could not be amplified with the BrEMA1 primers and were therefore negative for the BrEMA1 gene.
(ii) Phylogenetic relationships of BrEMA1 sequences.
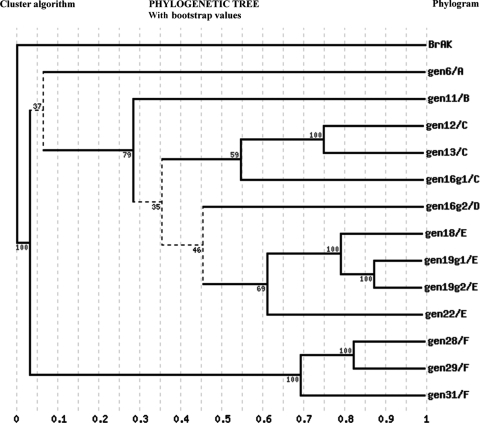
Based on the 13 BrEMA1 genotype sequences available to date, it follows that the genotypes could belong to six distinct monophyletic groups, labeled A to F in Fig. 2. The low bootstrap values obtained at certain nodes, especially those of <50, strongly suggest that the phylogenetic analysis of BrEMA1 genotypes presented in our study is incomplete (Fig. 2).
FIG. 2.
Cluster algorithm tree showing the phylogenetic relationships between various genotypes based on the BrEMA1 gene sequences. The genotypes appear to belong to six monophyletic groups, labeled A to F. BrAK (adenosine kinase gene from B. rossi; GenBank accession number AJ223322) was used as an outgroup.
(iii) Occurrence of BrEMA1 genotypes among all samples.
The total analysis of the 141 blood samples revealed 12 different BrEMA1 genotypes that clustered in six monophyletic groups (Fig. 2). The most abundant genotype was genotype19, followed by genotype28, genotype29, and genotype11 (Fig. 3). The remaining genotypes represented, at most, 10% of the cases.
FIG. 3.
Occurrence of B. rossi BrEMA1 genotypes among all samples (141 dogs).
Occurrence of BrEMA1 genotypes among OVAH samples.
The results revealed that there were four frequently occurring genotypes (19, 28, 29, and 11) among clinical cases, with genotype19 being the most frequently encountered genotype (Table 2). The four representative genotypes occurred with complicated clinical presentations, but the frequency of genotype19 was significantly greater over all cases (P < 0.001). Genotype19 was significantly more present in SOC patients than in non-SOC patients (P = 0.02; Fisher's exact test). Furthermore, the odds of genotype19 occurring in fatal cases were 3.71 times greater than those for surviving cases.
TABLE 2.
Occurrence of B. rossi BrEMA1 genotypes identified from blood samples and clinical outcomes of dogs presented at OVAH
| Genotype | No. of cases |
Total % of cases | % Complicated cases | % SOC cases | % Fatal cases | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Complicated (admitted) |
Home | Total | ||||||||
| SOC |
Non-SOC |
|||||||||
| Died | Survived | Died | Survived | |||||||
| Genotype11 | 1 | 2 | 8 | 3 | 14 | 11.6 | 12.8 | 10.3 | 5.0 | |
| Genotype19g1 (GenBank accession no. AJ416997) | 8 | 3 | 11 | 8 | 30 | 24.8 | 25.6 | 37.9 | 40.0 | |
| Genotype19g2 | 5 | 2 | 6 | 5 | 18 | 14.9 | 15.1 | 24.1 | 25.0 | |
| Genotype28 | 4 | 2 | 15 | 6 | 27 | 22.3 | 23.3 | 20.6 | 20.0 | |
| Genotype29 | 1 | 1 | 1 | 14 | 11 | 28 | 23.1 | 20.9 | 6.9 | 10.0 |
| Genotype6 | 1 | 1 | 3.0 | 2.3 | 0.0 | 0.0 | ||||
| Genotype12 | 1 | 1 | ||||||||
| Genotype13 | 0 | |||||||||
| Genotype18 | 1 | 1 | ||||||||
| Genotype16g1 (GenBank accession no. AJ416994) | 1 | 1 | ||||||||
| Total | 19 | 10 | 1 | 56 | 35 | 121 | ||||
Association between BrEMA1 genotypes and clinical signs.
The frequencies of clinical signs and associated genotypes among the 29 dogs diagnosed with SOC and among the 19 dogs that died from SOC-induced death are shown in Table 3. There was no association of a specific genotype with any set of clinical signs. Although not statistically significant, it was observed that the odds of genotype19 occurring in ARDS patients were 2.64 times greater than those for non-ARDS patients.
TABLE 3.
Clinical signs from complicated cases with SOC and their associated BrEMA1 genotypesa
| Sample | Genotype | Outcome | Complication |
|---|---|---|---|
| ES15 | 11 | S | ARDS |
| BC214* | 11 | S | ARDS |
| ES80 | 19g1 | S | ARDS |
| ES90 | 19g1 | S | ARDS |
| ES46 | 19g2 | S | ARDS |
| BC232* | 19g1 | D | ARDS |
| BC289 | 19g1 | D | ARDS |
| BC298* | 19g1 | D | ARDS |
| ES78 | 19g1 | D | ARDS |
| BC238 | 19g1 | D | ARDS |
| BC228* | 19g1 | D | ARDS |
| BC265 | 19g2 | D | ARDS |
| ES53 | 19g2 | D | ARDS |
| ES1 | 28 | S | ARDS |
| BC302* (h) | 28 | D | ARDS |
| BC303* | 29 | D | ARDS |
| BC232* | 19g1 | D | Shock |
| BC298* | 19g1 | D | Shock |
| BC228* | 19g1 | D | Shock |
| ES10 | 19g2 | D | Shock |
| ES76 | 19g2 | D | Shock |
| BC138 | 19g2 | S | Shock |
| BC302* (h) | 28 | D | Shock |
| BC251* (h) | 28 | D | Shock |
| BC303* | 29 | D | Shock |
| ES19 | 11 | D | ARF |
| ES84* | 19g1 | D | ARF |
| BC288 (h) | 28 | S | ARF |
| BC251* (h) | 28 | D | ARF |
| BC303* | 29 | D | ARF |
| BC268* | 28 | D | Icterus and hypoglycemia |
| BC168* | 28 | D | Icterus and hypoglycemia |
| BC228* | 19g1 | D | Neurological signs |
| ES84* | 19g1 | D | Pancreatitis |
| BC214* | 11 | S | Splenomegaly |
| BC211* | 19g1 | S | Icterus and ISA |
| BC217* | 29 | S | Icterus and ISA |
| ES63** | 29 | D | Hemoconcentration |
*, sample associated with two distinct clinical signs; h, sample associated with hemolysis; **, sample associated with non-SOC; S, survived; D, died.
DISCUSSION
It should be noted that relatively large numbers of disease phenotypes and parasite genotypes were studied in a relatively small population. This means that associations between disease phenotype and parasite genotype, although interesting, require a much larger prospective study to elucidate them further. It should also be noted that a nonbiased approach of sampling asymptomatic dogs might reveal other genotypes not described in the current study.
This study identified the existence of 12 B. rossi genotypes, with the most common being genotype19. This genotype was also associated with large numbers of dogs that were admitted at OVAH suffering from B. rossi-induced canine babesiosis. Furthermore, our data have shown that B. rossi genotype19 is the most common genotype occurring among SOC and fatal cases. Since genotype19 appears to be more virulent, this might suggest an association between this genotype and severe clinical cases of canine babesiosis.
Our data analysis appears to suggest that SOC can be a precursor to a fatal outcome. It has been reported that once SOC appear in infected dogs, the mortality rate is usually around 15% (17). In agreement with published data, we estimated from our results that once a dog is infected with B. rossi, its chance of developing SOC would be ca. 24%, with a 16.5% chance of dying from these complications.
Phylogenetic analysis of the BrEMA1 sequences identified six monophyletic groups. The low bootstrap values (<50) obtained for certain nodes of the BrEMA1 phylogenetic tree imply that the diversity in B. rossi genotypes in the field is higher than that described in this report. More B. rossi-positive samples will need to be collected from geographically dispersed areas to elucidate a more comprehensive picture. These data, however, indicate that genotypes 19, 28, 29, and 11 are the most frequently occurring among the 12 identified genotypes. These four genotypes belong to three monophyletic groups (i.e., genotype19, genotype28/29, and genotype11), in agreement with their separation into three distinct groups of genotypes on the basis of their frequency and virulence. For comparison, genotype19 appears to be highly frequent and virulent, genotypes 28 and 29 appear to be moderate in frequency and virulence, and the frequency and virulence of genotype11 appear to be low.
Our analysis also indicates that the BrEMA1 gene might be a good genetic marker for investigating B. rossi virulent genotypes in areas where babesiosis is endemic, especially since the less pathogenic species B. canis and B. vogeli were found not to have the BrEMA1 gene. This finding is likened to related studies through which specific genes have been found to be unique to virulent protozoal species. For example, the virulence of Plasmodium falciparum and Babesia bovis is influenced by their ability to adhere to capillary endothelium, a phenomenon known as sequestration (1, 14, 15). Sequestration has been linked to proteins of both P. falciparum and B. bovis expressed on the surfaces of infected red blood cells, which seem to alter the adhesive properties of these cells (6, 15). Preliminary evidence has suggested that the expression of parasite-derived antigens (with adhesive properties) on the extracellular surface of B. rossi-infected red blood cells may be responsible for their ability to sequester in vivo and to aggregate in vitro (16). This has not been proven, however. Furthermore, there is no evidence related to the involvement of BrEMA1 in sequestration. It has been demonstrated, though, that both adhesive and mechanical properties of P. falciparum- or B. bovis-infected red blood cells play a pivotal role in their ability to sequester parasites and, consequently, in the virulence of these parasites (6, 8). If virulence is indeed directly related to cytoadhesion, as suggested by the B. bovis model (1, 2), then more work should be undertaken to correlate the relationship between the virulent BrEMA1 genotypes and cytoadhesion or to identify auxiliary genes that may play a role in this phenomenon.
Conclusion.
The significant suggestion of this work is that different parasite genotypes may cause differing host responses to infection (i.e., there could be a relationship between parasite genotypes and disease pathogenesis). Changing disease outcome through treatment depends heavily on understanding disease pathogenesis. In this sense, these preliminary data may eventually prove clinically significant.
Acknowledgments
We sincerely thank M. Böhm and E. Scheepers for offering clinical records. We also thank Peter Thompson for statistical analysis.
This work was funded by the Thuthuka NRF fund and an institutional collaboration agreement (95401) between the Institute of Tropical Medicine, Antwerp, Belgium, and the Department of Veterinary Tropical Diseases, University of Pretoria (V042/05).
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Allred, D. R., and B. Al-Khedery. 2004. Antigenic variation and cytoadhesion in Babesia bovis and Plasmodium falciparum: different logics achieve the same goal. Mol. Biochem. Parasitol. 13:27-35. [DOI] [PubMed] [Google Scholar]
- 2.Allred, D. R., and B. Al-Khedery. 2006. Antigenic variation as an exploitable weakness of babesial parasites. Vet. Parasitol. 138:50-60. [DOI] [PubMed] [Google Scholar]
- 3.Böhm, M., A. L. Leisewitz, P. N. Thompson, and J. P. Schoeman. 2006. Capillary and venous Babesia canis rossi parasitaemias and their association with outcome of infection and circulatory compromise. Vet. Parasitol. 141:18-29. [DOI] [PubMed] [Google Scholar]
- 4.Bourdoiseau, G. 2006. Canine babesiosis in France. Vet. Parasitol. 138:118-125. [DOI] [PubMed] [Google Scholar]
- 5.Carret, C., F. Walas, B. Carcy, N. Grande, E. Precigout, K. Moubri, T. P. Schetters, and A. Gorenflot. 1999. Babesia canis canis, Babesia canis vogeli, Babesia canis rossi: differentiation of the three subspecies by a restriction fragment length polymorphism analysis on amplified small subunit ribosomal RNA genes. J. Eukaryot. Microbiol. 46:298-303. [DOI] [PubMed] [Google Scholar]
- 6.Cooke, B. M., N. Mohandas, and R. L. Coppel. 2001. The malaria-infected red blood cell: structural and functional changes. Adv. Parasitol. 50:1-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubbels, J. M., A. P. de Vos, M. van der Weide, J. Viseras, L. M. Schouls, E. de Vries, and F. Jongejan. 1999. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J. Clin. Microbiol. 37:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutchings, C. L., A. Li, K. M. Fernandez, T. Fletcher, L. A. Jackson, J. B. Molloy, W. K. Jorgensen, C. T. Lim, and B. M. Cooke. 2007. New insights into the altered adhesive and mechanical properties of red blood cells parasitized by Babesia bovis. Mol. Microbiol. 65:1092-1105. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson, L. S., and I. A. Clark. 1994. The pathophysiology of canine babesiosis: new approaches to an old puzzle. J. S. Afr. Vet. Assoc. 65:134-145. [PubMed] [Google Scholar]
- 10.Jacobson, L. S., and R. G. Lobetti. 1996. Rhabdomyolysis as a complication of canine babesiosis. J. Small Anim. Pract. 37:286-291. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson, L. S. 2006. The South African form of severe and complicated canine babesiosis: clinical advances 1994-2004. Vet. Parasitol. 138:126-139. [DOI] [PubMed] [Google Scholar]
- 12.Matjila, P. T., A. L. Leisewitz, F. Jongejan, and B. L. Penzhorn. 2008. Molecular detection of tick-borne protozoal and ehrlichial infections in domestic dogs in South Africa. Vet. Parasitol. 155:152-157. [DOI] [PubMed] [Google Scholar]
- 13.Matjila, P. T., B. L. Penzhorn, C. P. Bekker, A. M. Nijhof, and F. Jongejan. 2004. Confirmation of occurrence of Babesia canis vogeli in domestic dogs in South Africa. Vet. Parasitol. 122:119-125. [DOI] [PubMed] [Google Scholar]
- 14.Miller, L. H., D. I. Baruch, K. Marsh, and O. K. Doumbo. 2002. The pathogenic basis of malaria. Nature 415:673-679. [DOI] [PubMed] [Google Scholar]
- 15.O'Connor, R. M., J. A. Long, and D. R. Allred. 1999. Cytoadherence of Babesia bovis-infected erythrocytes to bovine brain capillary endothelial cells provides an in vitro model for sequestration. Infect. Immunol. 67:3921-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schetters, T. P., and W. M. Eling. 1999. Can Babesia infections be used as a model for cerebral malaria? Parasitol. Today 15:492-497. [DOI] [PubMed] [Google Scholar]
- 17.Shakespeare, A. S. 1995. The incidence of canine babesiosis amongst sick dogs presented to the Onderstepoort Veterinary Academic Hospital. J. S. Afr. Vet. Assoc. 66:247-250. [PubMed] [Google Scholar]
- 18.Uilenberg, G., F. F. Franssen, N. M. Perie, and A. A. Spanjer. 1989. Three groups of Babesia canis distinguished and a proposal for nomenclature. Vet. Q. 11:33-40. [DOI] [PubMed] [Google Scholar]