Abstract
The EntericBio system uses a multiplex PCR assay for the simultaneous detection of Campylobacter spp., Salmonella enterica, Shigella spp., and Escherichia coli O157 from feces. It combines overnight broth enrichment with PCR amplification and detection by hybridization. An evaluation of this system was conducted by comparing the results obtained with the system with those obtained by routine culture, supplemented with alternative PCR detection methods. In a study of 773 samples, routine culture and the EntericBio system yielded 94.6 and 92.4% negative results, respectively. Forty-two samples had positive results by culture, and all of these were positive with the EntericBio system. This system detected an additional 17 positive samples (Campylobacter spp., n = 12; Shigella spp., n = 1; E. coli O157, n = 4), but the results for 5 samples (Campylobacter spp., n = 2; Shigella spp., n = 1; E. coli O157, n = 2) could not be confirmed. The target for Shigella spp. detected by the EntericBio system is the ipaH gene, and the molecular indication of the presence of Shigella spp. was investigated by sequence analysis, which confirmed that the ipaH gene was present in a Klebsiella pneumoniae isolate from the patient. The sensitivity, specificity, positive predictive value, and negative predictive value were 100%, 99.3%, 91.5%, and 100%, respectively. Turnaround times were significantly reduced with the EntericBio system, and a result was available between 24 and 32 h after receipt of the sample in the laboratory. In addition, the amount of laboratory waste was significantly reduced by use of this system. In summary, the EntericBio system proved convenient to use, more sensitive than the conventional culture used in this study, and highly specific; and it generated results significantly faster than routine culture for the pathogens tested.
Infectious gastroenteritis is a leading cause of morbidity and mortality worldwide (3), particularly in developing countries (2). There are also a significant economic and social costs associated with gastroenteritis (14), in addition to increased morbidity and mortality (5).
Conventional culture methods remain the norm for the isolation of bacterial enteric pathogens in clinical laboratories. A major advantage of molecular methods is the reduced time to detection (13). Faster diagnostic outputs allow earlier epidemiological investigations and infection control interventions. Furthermore, the use of molecular methods highlights that conventional methods for the isolation of Campylobacter are less sensitive than PCR (15). Molecular methods suitable for use in the detection of fecal enteric pathogens have not been routinely available to clinical laboratories, until recently. Here we describe a new multiplex PCR method for clinical diagnostic use. The method combines an overnight enrichment step with PCR and hybridization detection by use of a line blot assay for the simultaneous detection of Campylobacter spp., Salmonella enterica, Shigella spp., and Escherichia coli O157 from feces. The application of the system in a clinical laboratory is also described.
MATERIALS AND METHODS
Patient samples.
A total of 773 anonymous fecal samples were tested both by routine culture and with the EntericBio system between 16 April and 4 June 2008. All samples were collected from patients with symptoms of gastroenteritis. A history of foreign travel was noted on the specimen container.
Controls.
The following control isolates were used in the assay: Campylobacter jejuni ATCC 29428, Shigella sonnei, Salmonella enterica serovar Typhimurium, Salmonella enterica serovar Enteritidis, and E. coli O157. The non-ATCC strains had been isolated in-house; and their identities had been confirmed at the National Salmonella and Shigella Reference Laboratory, Galway, Ireland, or the National E. coli Reference Laboratory, Dublin, Ireland.
Bacterial culture.
Routine culture was performed with all samples to detect Campylobacter spp., Salmonella enterica, Shigella spp., and, when the sample was liquid, E. coli O157, as shown in Table 1.
TABLE 1.
Culture procedure used for investigation of fecal samples for the presence of Campylobacter spp., Salmonella enterica, Shigella spp., and E. coli O157
| Pathogen | Culture method | Identification method |
|---|---|---|
| Campylobacter spp. | Preston agar (Campylobacter agar base [CM689; Oxoid] with supplement SR204E); microaerophilic environment at 42°C for 48 h | Typical macroscopic and microscopic appearance; positive catalase/oxidase reactions |
| Salmonella enterica | DCA and XLDa; incubation overnight at 37°C; enrichment overnight in Selenite broth (LP0121); subculture to Harlequin medium (Hal1; LabM) | Results for suspicious colonies confirmed biochemically and serologicallyb |
| Shigella spp. | DCA and XLD and incubated overnight at 37°C | Results for suspicious colonies confirmed biochemically and serologicallyb |
| E. coli O157 | Cefixime tellurite sorbitol MacConkey agar (W11021; LIP Diagnostic Services, Galway, Ireland); incubation overnight at 37°C | Results for suspicious colonies confirmed biochemically and serologicallyb |
DCA, desoxycholate citrate agar (CM0227; Oxoid); XLD, xylose lysine desoxycholate agar (Lab 32; LabM).
The results for all suspicious isolates were confirmed by reference laboratories.
Molecular detection with EntericBio system.
The CE-marked EntericBio system (Serosep Ltd., Limerick, Ireland) was used to test all samples according to the manufacturer's instructions. The method uses a combination of enrichment and then DNA extraction, PCR amplification, and detection of the results by means of hybridization and color development.
Enrichment of the fecal samples was done by inoculation of 1.0 g of a fecal specimen into EntericBio enrichment broth, which equated with the fill line on the broth tube. The broth container was capped and mixed by inversion at least five times before it was incubated for between 16 and 24 h at 37°C.
Following overnight incubation, the enriched broths were thoroughly mixed by inversion after removal from the incubator. The broth was allowed to settle for 5 min at room temperature to allow large particles to settle. To perform DNA extraction, a volume of 200 μl of the supernatant from the enriched fecal samples was transferred to an appropriately labeled EntericBio extract tube (which contains a two-phase separation system). Each extract tube was mixed by inversion approximately 20 times and sonicated for 30 min with a Transsonic digital sonicator (S-TP 680DH), according to the manufacturer's instructions. The remainder of the enriched EntericBio broth was placed in a freezer at −80°C for storage. These broths were thawed, mixed, and subcultured in cases in which conventional culture did not yield a positive result when the EntericBio system PCR result was positive.
PCR amplification was then carried out with lyophilized PCR strips, which were removed from the kit, labeled, and placed on a PCR rack. A volume of 14 μl of molecular biology-grade (MBG) water was pipetted into each PCR tube. Ten microliters of the lower phase of the DNA extract solution was transferred from the sonicated EntericBio extract tube to the prelabeled PCR tube. A volume of 1 μl of uracil DNA glycosylase (UNG; 111025; Bioron, Germany) was also added to each PCR tube. A negative control that consisted of 24 μl of MBG water and 1 μl of UNG in a PCR tube was run with each batch of tests. A positive control that comprised 5 μl of the positive control and 20 μl of MBG water was also run.
The PCR tubes were capped firmly, and a touchdown PCR program was run. A pre-PCR incubation at 21°C for 10 min was followed by a two-cycle step of 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s. This was followed by single cycles with the same step times but with incrementally decreasing annealing temperatures of 64, 63, 62, and 61°C. The program continued with a further 29 cycles at an annealing temperature of 60°C, which were followed by a final extension step of 72°C for 10 min. After amplification, the PCR tubes were removed from the thermocycler and the samples were denatured by adding 25 μl of denaturation solution to each tube, which was then incubated at room temperature for 10 min.
Hybridization and color development were performed on the EntericBio AutoProcessor tray (Tecan) by using a substrate solution, a wash solution, distilled water, and detection and hybridization solutions.
The hybridization strips were prepared by using the forceps provided with the kit to remove the hybridization line blot strips from their storage tube, and the strips were placed at the top end of individual channels in the single-use plastic AutoProcessor tray. The entire solution from the PCR tube was transferred to the bottom end of individual channels, taking care not to place the solution onto the strip, as the pH would destroy the probes. A volume of 500 μl of hybridization solution was added to the top end of each channel, and the tray was agitated manually to mix both solutions. The tray was placed securely on the AutoProcessor tray, and the hybridization process was initiated. When this process finished after 3 h, the results were interpreted. A positive internal control line signified that an individual test was valid. The test results were interpreted on the basis of the absence or the presence of a line blot at each of the four locations denoting the absence or the presence of Campylobacter spp., Shigella spp., E. coli O157, and Salmonella enterica in the original sample (see Fig. 1, in which it may be seen that for N, the single hybridization line occurs for the internal control and all other targets are negative and in which for P, hybridization lines are detectable for all four pathogens and the internal control).
FIG. 1.
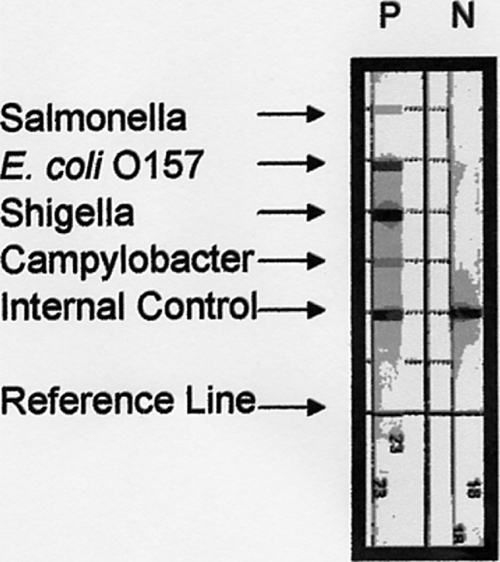
EntericBio line blot hybridization results for a positive control (P) and a negative control (N).
Molecular confirmation of results.
In cases in which the molecular results were not confirmed by conventional culture (this also included in every case subculture of a sample from the incubated EntericBio broth), alternative molecular confirmation methods were used.
For Shigella spp., the PCR method of Aranda et al. (1) was used to detect the ipaH gene. However, in the single case in which culture for Shigella spp. remained negative, a subculture of the EntericBio broth was made on MacConkey agar (MAST DM140D), which was incubated overnight at 37°C. Fermentative and nonfermentative colonies were cultured for purity onto further MacConkey agar plates. Following overnight incubation, DNA was extracted from each colony type by a standard boiling method, and this DNA was tested for the presence of the ipaH gene. The colony in which the ipaH gene was located was subcultured for purity and retested for the presence of the gene, and a species identification was conducted by using the API 20E system (BioMérieux, France).
For Campylobacter spp., the PCR method of Linton et al. (8) was used. This method detects the hippuricase gene of C. jejuni, and the method of Maher et al. (9) was used to detect a genus-specific product for the samples for which positive results were not obtained by using the method of Linton et al. (8).
For E. coli O157, the method of Müller et al. (10) was used to investigate the samples for the presence of the verotoxin 1 (VT1) and/or VT2 gene among the specimens found to be positive with the EntericBio system. Furthermore, each of the fecal samples for which positive results were derived was sent for confirmatory testing to the National E. coli Reference Laboratory.
RESULTS
The results of tests with 773 clinical samples are shown in Table 2.
TABLE 2.
Comparison of results of routine bacterial culture with those of EntericBio system for detection of Salmonella enterica, Shigella spp., Campylobacter spp., and E. coli O157
| Organism | No. (%) of samples with positive results by use of the followinga: |
|
|---|---|---|
| Routine culture | EntericBio system | |
| Salmonella enterica | 4 (0.5) | 4 (0.5) |
| Shigella spp. | 0 | 1 (0.1) |
| Campylobacter spp. | 30 (3.9) | 42 (5.4) |
| E. coli O157b | 8 (1.0) | 12 (1.6) |
| Total | 42 (5.4) | 59 (7.6) |
A total of 731 (94.6%) samples were negative by routine culture and 714 (92.4%) samples were negative with the EntericBio system.
A total of 10 positive results were generated by the National E. coli Reference Laboratory by using PCR, and subculture from EntericBio broth yielded an additional 2 positive culture results compared with the number of positive results obtained by the routine culture methods for the same 10 samples.
All positive results by conventional culture were matched by positive results with the EntericBio system. However, 17 additional positive results were generated by the EntericBio system. A single Shigella ipaH gene target was detected by the EntericBio system, but Shigella spp. was not detected by routine culture or by retrospective culture of the EntericBio broth sample for that patient. The ipaH gene was, however, also detected by the method of Aranda et al. (1), and this target was located in Klebsiella pneumoniae, isolated from the sample from that patient. Sequence analysis showed >98% similarity of that sequence with published sequences of ipaH gene amplicons from Shigella spp. for short segments flanking the central section of the amplicon which corresponded to sequences of K. pneumoniae (data not shown). Ten of the 12 additional positive results for Campylobacter spp. generated by the EntericBio system gave positive results by the hippuricase gene-specific PCR, indicating a species identification of C. jejuni in each case. The remaining two positive results remained unconfirmed by the methods of either Linton et al. (8) or Maher et al. (9). When the 12 samples with positive results for E. coli O157 obtained with the EntericBio system were tested by the National E. coli Reference Laboratory, 10 samples with positive results for E. coli O157 were generated; 9 of those samples with positive results were found to be positive for VT2. The EntericBio system results for these remaining two samples were not confirmed either by the method of Müller et al. (10) or by the National E. coli Reference Laboratory.
DISCUSSION
Conventional culture has been the “gold standard” method for the detection of bacterial enteric pathogens. The advantages of these methods include identification to the species or serovar level, the facilitation of outbreak management, and the generation of an antimicrobial susceptibility profile. A disadvantage of the use of culture is the time taken to generate a result, positive or negative, from a fecal sample. A further disadvantage is the occurrence in some cases of viable but nonculturable C. jejuni isolates (11), which makes the recovery of the organism challenging when culture techniques are used. In this comparative study of conventional culture techniques and a multiplex PCR method for the detection of Campylobacter spp., Salmonella enterica, Shigella spp., and E. coli O157, the minimum time taken to confirm a positive result for these pathogens by conventional culture ranged from 48 h for Campylobacter spp. to 96 h for Salmonella enterica. The time taken to generate a negative report was 48 h, or within the next 2 working days following receipt of the sample. For the EntericBio system, the time taken to generate a report, positive or negative, was between 24 and 32 h, or within the next working day following receipt of the sample in the laboratory. The advantages of an early laboratory report include the early diagnosis of infection, which allows the timely reporting of food-borne diseases and the prevention of additional cases in outbreak settings (3).
As shown in Table 2, the overall positivity rate for this prospective study was 5.4% by conventional culture techniques and 7.6% by the method with the EntericBio system. For the detection of Salmonella, culture and the EntericBio system generated identical results. In the case of Shigella, detection by the EntericBio system, which detects the ipaH gene, for which the results by culture and with the EntericBio system for a single sample did not match, yielded a positive result. By using the primer set of Aranda et al. (1), the target was located in Klebsiella pneumoniae, which showed 100% similarity with the ipaH target primers, following sequencing and BLAST analysis, to accessions of Shigella spp. and which contained an internal sequence with >98% similarity to accessions of K. pneumoniae. The sample from which this result was obtained was from a patient with gastroenteritis on return from travel in India. The sample did not yield any further pathogens, either bacterial or parasitic. This finding may serve to highlight the transmissibility of virulence factors such as the plasmid-borne ipaH gene among members of the Enterobacteriaceae family and suggests the possibility that an effective means of detection of a pathogen may originate from first identifying the presence of a virulence gene and then identifying the organism carrying it.
For Campylobacter spp., a total of 30 culture isolations were made from the 773 samples, all of which were detected with the EntericBio system. However, the EntericBio system detected an additional 12 samples with positive results for Campylobacter, and the isolates in 10 of these samples were confirmed to be C. jejuni by the PCR method of Linton et al. (8). The PCR method of Maher et al. (9) was applied to the remaining two samples with unconfirmed positive results for Campylobacter spp. by use of the EntericBio system, but the results for these samples also remained unconfirmed by that method. These results were thus considered to be false positive. It should be noted that these additional 12 samples with positive results were also subcultured from the EntericBio broths onto Preston agar (incubated at 37 and 42°C) but did not result in any positive culture results. The finding that the currently used culture-based methods may miss a substantial proportion of Campylobacter infections has been noted previously (6). It probably reflects the detection of Campylobacter cells in metabolic states that are less amenable to culture on selective media, whether they be damaged, viable but nonculturable, or dead (7). The clinical significance of nonculturable Campylobacter isolates in human infection remains poorly understood and warrants future investigation. When the results of the current study were compared to the findings of an Irish study by Maher et al. (9), however, some differences were found. The earlier study compared conventional culture to the detection of Campylobacter spp. directly from feces by the use of a combination of primer/probe sets with colorimetric membrane-based detection. The study also showed that molecular detection was more sensitive than culture; but the DNA of a variety of species was detected, including the DNA of Campylobacter concisus, Campylobacter curvus, and Campylobacter gracilis, in addition to the DNA of C. jejuni. Those findings are in contrast to the findings of a 2007 study performed in the United Kingdom by Wilson and Aitchison (16), in which the use of filtration and culture methods in combination yielded only C. jejuni. In the current study, only C. jejuni was identified among the nonculturable Campylobacter spp., although the molecular method was designed by using a gene idiosyncratic for Campylobacter spp., according to the manufacturer. Further studies are needed to establish whether the overnight enrichment step used in this method facilitates the growth of all species. Nevertheless, the detection of an additional 25% Campylobacter-positive results compared to the number of Campylobacter-positive results obtained by routine culture points to the greater sensitivity of the EntericBio system than routine culture, at least for the detection of C. jejuni.
Finally, the isolation of E. coli O157 by routine culture without enrichment yielded eight samples with positive results, which were augmented by an additional two samples with positive results both by subculture of the EntericBio broth onto sorbitol MacConkey agar and by the National E. coli Reference Laboratory by the use of immunomagnetic concentration techniques in combination with molecular detection. Nine of these samples contained isolates that were VT2 positive; the 10th sample contained an isolate that was nonverotoxigenic. The remaining two samples with positive results by use of the EntericBio system remained unconfirmed either as E. coli O157 or as verotoxigenic strains by the National E. coli Reference Laboratory and by the additional molecular testing which we undertook to detect VT1 and VT2, and so these results were interpreted to be false positive.
An expanded gold standard was used to calculate the sensitivity, specificity, positive predictive value, and negative predictive value of the method with the EntericBio system compared with the results of the combination of culture and alternative molecular techniques. Those calculations showed that the sensitivity was 100%, the specificity was 99.3%, the positive predictive value was 91.5%, and the negative predictive value was 100%.
The detection of E. coli O157 is clinically more highly significant when the organism is demonstrated to be verotoxigenic (4). In this study, the two unconfirmed E. coli O157 isolates did not give positive signals for VT1 or VT2, thereby lessening the impact of a false-positive result in these cases. It could be argued that the EntericBio system might benefit more from a facility for the detection of VT1 and VT2 rather than E. coli O157 alone, as many serotypes of E. coli have been identified to be verotoxigenic (12). Future modifications of the system should address this issue.
The detection of E. coli O157, Salmonella spp., and Shigella spp. by the EntericBio system required a subsequent culture analysis of samples positive for these organisms. In each case, the identification of the pathogen to the serovar or serotype level was vital for epidemiological purposes. This necessitated a delay in the generation of a report of at least 48 h after a positive result was indicated by the EntericBio system. However, the low rate of positive results (7.6%) by the EntericBio system meant that the results for more than 90% of the fecal samples negative for the four pathogens detected by the assay, even during a peak seasonal period (between April and June), as in this study, can be reported with confidence at least 1 working day earlier than when routine culture is used.
A comparison of the amounts of waste generated by culture and by use of the EntericBio system was also made. Approximately 3,000 agar plates were generated by culture, and the total waste for autoclaving amounted to approximately 306 liters, which contrasted sharply with the 36 liters generated by the EntericBio system. Most of the waste generated by the latter was recyclable packaging.
In summary, the EntericBio system proved convenient to use, more sensitive than the conventional culture method used in this study, and highly specific; and it generated results significantly faster than routine culture for the pathogens tested. At the moment, the method is more expensive than conventional culture. However, this is compensated for in the laboratory by not having to perform immunomagnetic separation for E. coli O157 and by the potential savings in the numbers of personnel needed once the method is established in the laboratory. The use of an enrichment broth suitable for the enrichment of all four pathogens necessitates the use of a category 3 containment facility, however, which may not be available in all laboratories. Finally, the simultaneous detection of the four most common bacterial enteric pathogens by one test and with a turnaround time significantly reduced compared with that of culture is likely to be the major advantage offered by this system for clinical diagnostic laboratories. The clinical impact of the decreased turnaround time means that bacterial diarrhea is more promptly ruled out by the use of the EntericBio system than by the use of conventional culture. This reduces pressure on infection control resources and, in particular, in cases of sporadic diarrhea, helps reduce the requirement for scarce isolation rooms. In addition, the earlier availability of results is helpful in the management of outbreaks which are largely community based.
Acknowledgments
We gratefully acknowledge Tim Buckley, laboratory manager at the Microbiology Department of Cork University Hospital, for facilitating this work. We thank M. Pierre Douarre at the Department of Biological Sciences at the Cork Institute of Technology for his confirmatory work with a subset of the results for Campylobacter spp. and the National E. coli Reference Laboratory for its assistance with the investigation of putative positive results for E. coli O157 by use of the EntericBio system. Our gratitude also goes to our colleagues for their support of this project.
We thank the health service executive of Cork University Hospital for bearing the cost of the validation of the EntericBio system prior to its incorporation for routine use in the department.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Aranda, K. R., S. H. Fabbricotti, U. Fagundes-Neto, and I. C. Scaletsky. 2007. Single multiplex assay to identify simultaneously enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and Shiga-toxin-producing Escherichia coli strains in Brazilian children. FEMS Microbiol. Lett. 267:145-150. [DOI] [PubMed] [Google Scholar]
- 2.Britton, R. A., and J. Veraslovic. 2008. Probiotics and gastrointestinal infections. Interdiscip. Perspect. Infect. Dis. 2008:290769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2001. Updated guidelines for evaluating public health surveillance systems: recommendations from the guidelines working group. MMWR Recommend. Rep. 50(RR-13):1-35. [PubMed] [Google Scholar]
- 4.Foley, B., and P. McKeown. 2001. Epidemiology of verotoxigenic E. coli O157 in Ireland, 2001. NDSC Annual Report. National Disease Surveillance Centre, Dublin, Ireland.
- 5.Hall, G., M. D. Kirk, N. Becker, J. E. Gregory, L. Unicomb, G. Millard, R. Stafford, K. Lalor, and the OzFoodNet Working Group. 2005. Estimating foodborne gastroenteritis, Australia. Emerg. Infect. Dis. 11:1257-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen, A., K. Stark, J. Kunkel, E. Schreier, R. Ignatius, O. Liesenfeld, D. Werber, U. B. Göbel, M. Zeitz, and T. Schneider. 2008. Aetiology of community-acquired, acute gastroenteritis in hospitalised adults: a prospective cohort study. BMC Infect. Dis. 8:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawson, A. J., J. M. Logan, G. L. O'Neill, M. Desai, and J. Stanley. 1999. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37:3860-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linton, D., A. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher, M., C. Finnegan, E. Collins, B. Ward, C. Carroll, and M. Cormican. 2003. Evaluation of culture methods and a DNA probe-based PCR assay for detection of Campylobacter spp. in clinical specimens of feces. J. Clin. Microbiol. 41:2980-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller, D., P. Hagedorn, S. Brast, G. Heusipp, M. Bielaszewska, A. W. Friedrich, H. Karch, and M. A. Schmidt. 2006. Rapid identification and differentiation of clinical isolates of enteropathogenic Escherichia coli (EPEC), atypical EPEC, and Shiga-toxin-producing Escherichia. coli by a one-step multiplex PCR method. J. Clin. Microbiol. 44:2626-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nogva, H. K., A. Bergh, A. Holck, and K. Rudi. 2000. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pradel, N., Y. Bertin, C. Martin, and V. Livrelli. 2008. Molecular analysis of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients and dairy samples in France. Appl. Environ. Microbiol. 74:2118-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuurman, T., R. F. de Boer, E. van Zanten, K. R. van Slochteren, H. R. Scheper, B. G. Dijk-Alberts, A. V. Möller, and A. M. Kooistra-Smid. 2007. Feasibility of a molecular screening method for detection of Salmonella enterica and Campylobacter jejuni in a routine community-based clinical microbiology laboratory. J. Clin. Microbiol. 45:3692-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van den Brandhof, W. E., G. A. De Wit, M. A. de Wit, and Y. T. van Duynhoven. 2004. Costs of gastroenteritis in The Netherlands. Epidemiol. Infect. 132:211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanniasinkam, T., J. A. Lanser, and M. D. Barton. 1999. PCR for the detection of Campylobacter spp. in clinical specimens. Lett. Appl. Microbiol. 28:52-56. [DOI] [PubMed] [Google Scholar]
- 16.Wilson, G., and L. B. Aitchison. 2007. The use of a combined enrichment-filtration technique for the isolation of Campylobacter spp. from clinical samples. Clin. Microbiol. Infect. 13:643-644. [DOI] [PubMed] [Google Scholar]


