Abstract
We report the design of a single-molecule nanomotor driven by photons. The nanomotor is a DNA hairpin-structured molecule incorporated with azobenzene moieties to facilitate reversible photocontrollable switching. Upon repeated UV–vis irradiation, this nanomotor displayed 40–50% open–close conversion efficiency. This type of nanomotor displays well-regulated responses and can be operated under mild conditions with no output of waste. In contrast to multiple-component DNA nanomachines, the intramolecular interaction in this single-molecule system offers unique concentration-independent motor functionality. Moreover, the hairpin structure of the motor backbone can significantly improve the efficiency of light-to-movement energy conversion. These results suggest that azobenzene-incorporated, hairpin-structured single-molecule DNA nanomotors have promising potential for applications which require highly efficient light-driven molecular motors.
Nucleic acid (DNA) self-assembly based on spontaneous hybridization between complementary strands is an effective way to construct multidimensional nano-objects such as macromolecular architecture,1-6 biochip,7 photonic wires,8 enzyme assemblies,9 and functional DNA probes.10,11 Similar to RNA or protein-based nanostructures,12-14 DNA-based nanomachines have been developed with the ability to change their conformation upon external stimuli.15,16 Of special interest is a single DNA molecular motor developed by us a few years ago.17 The single-molecule nanomotor can switch between two conformations and perform extending–shrinking motion with stable, convenient operation and high efficiency. However, this single-DNA nanomotor requires addition and removal of fuel and waste strands for motor function.17,18 Although some artificial nanomotors can utilize alternative energy sources, including hydrolysis of the DNA backbone19 and ATP,20 applying an electromagnetic field as a convenient energy source is highly desired. As an alternative, a photon-driven single-DNA nanomotor would be of great scientific interest as well as contribute to applications in the nanosciences.
Azobenzene molecules have been extensively studied due to their reversible isomerization between planar trans form and nonplanar cis form under UV and visible light irradiation.21–23 The first light-powered molecular device using azobenzene polymer has successfully proved the concept of optomechanical energy conversion in a single device.24 Several DNA-based nanostructures incorporated with azobenzene moiety have also been constructed and investigated with photoregulated capability.25-31 A model of light driving an azobenzene-DNA molecular motor based on exchange of multiple DNA strands has been recently demonstrated.30 Although the concept of a single-molecule nanomotor was introduced in these studies, several DNA sequences were actually needed to perform motor movement. Overall, with the advent of a single-DNA nanomotor17 and the special optical properties of azobenzene well investigated,32 the development of a photoregulated single molecule nanomotor becomes feasible.
Inspired by the photoregulation capability of azobenzene to DNA hybridization, we have designed a single-molecule DNA nanomotor regulated by photons. By focusing on photonic energy driving movement, this research explores the feasibility of regulating the dehybridization (open state) and hybridization (close state) of a hairpin structure by controlling the photoresponsive azobenzene moieties integrated on the DNA bases in the hairpin's duplex stem segment. Because of its reversible extension–contraction behavior, the open–close cycle of the hairpin molecule is considered a molecular motor, and such motion can be characterized by monitoring fluorescence variation between a fluorophore and a quencher on both ends of the stem.17 Compared with previous DNA motors, in which motor cycles involve bimolecular or multimolecular interactions among several independent DNA strands, the hairpin-structured DNA motor is a single-molecule nanomotor that can facilitate the open–close cycling by virtue of its own unique structural architecture. In this single-molecule DNA nanomotor, the motor can act as a simple, yet precisely functionalized, molecular motor. Because of its simplicity and intramolecular interaction, a single-DNA nanomotor with reversible photoswitching function is expected to possess novel properties superior to those consisting of multiple-component DNA nanostructures. The nature of our single-DNA molecule motor is determined by predominant intramolecular interaction within the molecule instead of disjunctive DNA strand exchange. In this design, the photoregulation process is expected to be concentration-independent and thus ideal for developing high-density molecular motors.
Single-DNA Nanomotor Driven by Photons
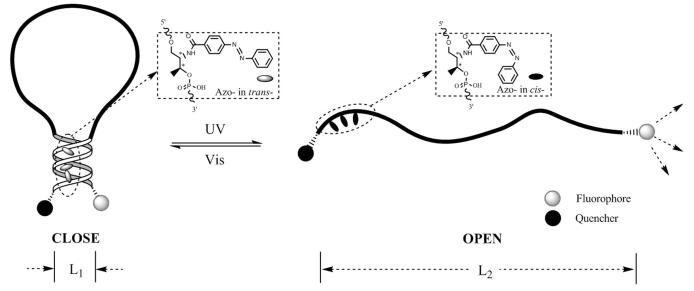
The single-molecule nanomotor was constructed as a fully functionalized DNA molecule driven by harvesting photon energy. The concept of the photoswitchable single-molecule DNA motor (PSMM) is shown in Scheme 1. The main components of PSMM include the hairpin backbone, azobenzene moiety (azobenzene phosphoramidite or Azo-) and fluorophore/quencher (F/Q) pair for signaling motor movement. Under UV and visible irradiations, the PSMM can reversibly switch conformation between open (dehybridization) and close (hybridization) states, respectively. The reversible extension (open) and contraction (close) cycle of the “arms” (forming stem duplex on close state) functions as a molecular motor because of the cycling and stretching actions by the DNA.
Scheme 1.
Scheme of PSMM with Three Azobenzene Moieties Inserted to the Stem Duplex on the Arm Labeled by Quenchera
a The structural change of DNA displayed a contraction (CLOSE) state when tethered azobenzene moiety (Azo-) takes trans conformation under visible light irradiation and an extension (OPEN) state when Azo- takes cis conformation under UV light irradiation. The L1 is the average size of hairpin structure based on the distance between F/Q pair, and L2 is the average size of extended molecules based on persistence length of a single DNA strand.
In order to demonstrate PSMM regulated by photons, a 31-base DNA was selected for a simple nanomotor. This DNA has a stable hairpin structure including a 6 base pair stem and a 19 base loop, with a Fluorescein (FAM) at the 5′ end and a Dabcyl quencher at the 3′ end: 5′ FAM-CCT AGC TCT AAA TCA CTA TGG TCG CGC TAG G-Dabcyl 3′ (underlined bases represent stem moieties). In the close state, the DNA forms a hairpin structure, and fluorescence is quenched due to the proximity of fluorescein and Dabcyl. Importantly, the fluorescence intensity is related to the distance between the F/Q pair, which indicates the close or open state of the PSMM movement. Therefore, the completely open state with fully extended structure will occur when cDNA (cDNA) is bound to the molecule with strong binding affinity, thereby disrupting the hairpin motor structure. This naturally results in maximum fluorescent intensity due to the largest separation between fluorescein and Dabcyl. We deliberately include the F/Q on the molecule with the intention of applying fluorescence variation to monitor the molecular structural changes. Hence, the difference in separation distance between the close and fully open states provides a functional “open” operational range for the molecular motor when the PSMM is driven by light irradiations in a reversibly contracting and extending movement (Scheme 1). In other words, the open state fluorescence intensity will vary between the ranges due to limitations of extension capability of a single DNA strand. In this manner, the hairpin-to-extension structure exchange can be regarded as a single molecule's motion where the push-and-pull dynamic is achieved by photons if we tether objects on the two ends of the DNA strand. Thus, a well-controlled photoswitchable DNA nanomotor can be produced based on this design.
Reversibility of the PSMM
In order to reversibly control the close–open process of the hairpin DNA, we incorporated Azo- onto the hairpin backbone to regulate the molecular structure by azobenzene isomerization upon photoirradiation. The single-molecule motor is in close state in normal conditions, since the Azo- takes the more stable trans- state, and the hairpin structure is maintained. When UV light is applied, the photons will initiate the Azo-isomerization from trans- to cis- conformation. In consequence, this conformational change destabilizes the stem duplex structure and extends the hairpin structure. This phenomenon can be regarded as a stretching movement of the motor. A subsequent visible light irradiation will rapidly isomerize the Azo- back to trans- conformation and re-form the hairpin structure. This step can be regarded as a contraction. As indicated in Scheme 1, in the open or extended state, Azo- takes on cisconformation, while, in close state, tethered Azo- takes on trans- conformation. Thus, by utilizing the hairpin-to-extension exchange, the responsive function of these molecules by otherwise ordinary target-response is converted into a reversible movement function, making a highly efficient molecular motor possible. Moreover, the whole reversible motor function can be characterized by fluorescence variation induced by structural conversion, a convenient way to monitor the movement of the motor at this stage. Accordingly, a four-step structure cycle (S1–S4) driven by photons is described in Scheme S3 in Supporting Information based on a previous study of azobenzene isomerization upon DNA hybridization. Photophysical study and experimental results on Azo-incorporated DNAs have suggested the order of these steps for a full cycle. Since a study of the kinetics needs to be undertaken to further quantitatively differentiate each step, we only focus our investigation on the conversion between hairpin structure and linear structure (S1/S2 and S3/S4) in terms of efficiency (fluorescence intensity recovery) and cycling reversibility.
Number of Azo- Moieties in a PSMM and the Impact on PSMM
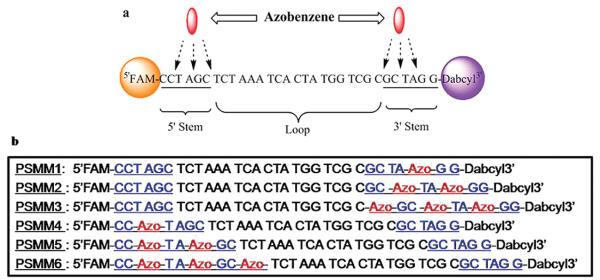
Because of the flexibility of the loop DNA sequence, the sequence in the loop strands might have little impact on stem duplex stability. However, the deliberate design of hairpin molecules with specific secondary loop structures could lead to stem modification that might have a direct impact on stem duplex stability. In our case, several Azo- moieties were incorporated to the PSMM stem in different amounts and at different positions. The six designed motors were named PSMM1 to PSMM6 (Scheme 2). We then screened these PSMMs by fluorescence changes under light irradiations and studied the photoconversion of these structures.
Scheme 2.
(a) The Available Positions for Azo-Incorporation in Stem Moiety on Each End. (b) The Sequences of the Six Types of PSMMs (PSMM 1–6)a
a PSMM1, PSMM2, and PSMM3 (or named PSMM1-3 for all three types) are hairpin structures with one to three Azo- on the 3′ end, and PSMM4, PSMM5, and PSMM6 (or named PSMM4-6 for all three types) are hairpin structures with one to three Azo- on the 5′ end. The blue bases are stem moiety; red are Azo- unit, and black bases are on loop moiety.
Since the azobenzene structure is similar to that of Dabcyl quencher, we expected quenching effect when Azo-moieties are spatially proximal to the fluorophore. Indeed, a simple fluorescence measurement revealed that PSMM 4–6 showed no fluorescence increase when cDNA was added, while PSMM 1–3 displayed the expected “on–off” fluorescence on the motor's open–close cycles. The melting curves displayed the difference of these nanomotors on fluorescence recovery in terms of stem duplex opening (Figure S1a in Supporting Information). We believe that the lack of fluorescence response in these Azo- incorporated PSMMs arises from fluorescence quenching by azobenzene (with a chemical structure similar to Dabcyl, hence a similar quenching mechanism). The fluorescence and quenching mechanism is depicted in Figure S1b in Supporting Information with possible energy transfers between fluorophore and quencher. Significant fluorescence enhancement for PSMM1–3 was observed when cDNA was added, thus confirming the selective quenching from both Dabcyl and Azo- on FAM at close distance. Because of the difficulties of monitoring structural changes by fluorescence intensity variation, we narrowed our investigation to PSMM1–3 with Azo-incorporated from the quencher end. By this selection of positions of Azo- incorporation, we can investigate the photoregulation cycle of the PSMMs by comparing fluorescence intensities from the close state to the fully open state. It may be recalled that the distance variations between the close and fully open states provides a necessary and functional “open” operational range for our molecular motors because we drive the PSMM by light in a reversible contracting and extending movement.
To accomplish this motor function with optimization, we investigated two main factors that were suspected to impact the efficiency and reversibility of our PSMM1–3 nanomotors: buffer solution and light sources (Supporting Information a, b). A buffer solution was chosen after optimizing the balance to favor both open and close states: 20 mM Tris-HCl pH 8.0, 20 mM NaCl, 2 mM MgCl2. Additionally, a 60 W table lamp with a 450 nm filter was chosen as the visible light source in all cases, and a portable 6 W UV light source (irradiated at 350 nm) was chosen for the UV light source. Reversible photoregulation was carried out by repeated irradiations at 450 and 350 nm at fixed time periods, followed by emission scans (λex = 488 nm). Although a higher power UV light source over the one selected would help to improve the trans- to cis- conversion, its use raises serious problems in terms of damaging the DNA structure and photobleaching the fluorophore, which have both occurred with these lamps after long-term irradiation.
Energy Conversion Efficiency of the PSMM
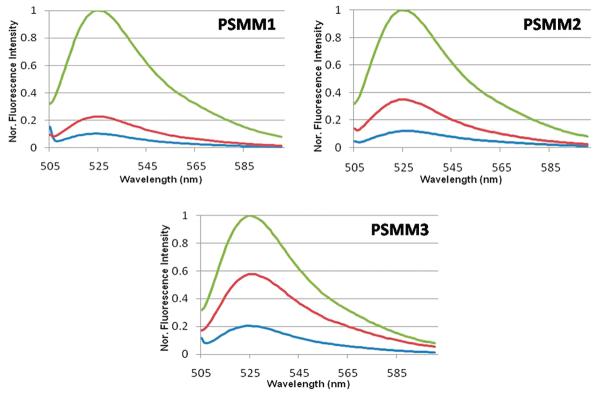
With the selected light sources and buffer conditions, PSMM1–3 were tested as described in the Experimental Section. All three nanomotors displayed different fluorescence recovery after visible irradiation followed by UV irradiation (Figure 1 and Table S1 in Supporting Information). As expected, more Azo- incorporations, such as those incorporated in PSMM3, resulted in higher fluorescence recovery, indicating that increasing of the amount of Azo- moiety can introduce a higher impact from azobenzene isomerization to hairpin structure stability. Excess amount of cDNA by 5-fold was added at the end for each type of nanomotor after photoregulation in order to compare the fluorescence intensity of all PSMMs at fully open state (induced by cDNA). By setting the fluorescence intensity when azobenzene takes the transform as a baseline (0%) and the intensity after addition of excess amount of cDNA as 100%, we could estimate the number of PSMMs in the open state for each photoregulation process. In our case, we set the fluorescence intensity on close state at 488 nm as the baseline (blue curves) and the fully open state (green curves) as 100%. We were then able to set up a fluorescence recovery parameter, recovery (%), based on the three states as defined below in order to evaluate the close–open conversion efficiency
Figure 1.
Fluorescence spectra of PSMM1–3 (λex = 488 nm) from top to bottom after irradiation under a 6 W UV lamp (350 nm) and a 60 W desktop lamp with 450 nm filter at 25 °C. All other conditions are the same. The blue curve is pure DNA in buffer solution (100 nM); green line is with five times of cDNA; and red line is UV/vis irradiation. Buffer: 20 mM Tris buffer pH 8.0, Na+: 20 mM, Mg2+:2 mM, [PSMM3] = 100 nM, [cDNA] = 500 nM.
IUV is the fluorescence intensity of DNA solution after UV irradiation, I0 is the fluorescence intensity after visible irradiation, It is the fluorescence intensity of DNA solution after adding extra cDNA. The higher the recovery value, the higher the amount of molecules that will be found in open state driven by photonic energy, and the better the efficiency of the conversion from close to open state. Since the reversible open to close conversion step is very fast for each of our PSMMs, we focus the recovery percentage on close to open conversion and discuss the motor efficiency accordingly.
We used the recovery (%) to compare the efficiency of energy conversion from photonic energy to molecular motion for PSMM1–3 under the same conditions. We found approximately 14.2% of recovery from trans- to cis- for PSMM1, 26.3% for PSMM2, and 54.7% for PSMM3. This result supported our assumption that multiple azobenzene incorporation will introduce higher impact to hairpin structure stability or photoregulation capability. The improvement of this recovery (%) is, however, not proportional to the number of Azo- moiety, although it does gradually increase as the number of Azo- increases. Specifically, the triple Azonanomotor (PSMM3) displayed a much higher open-to-close ratio than PSMM1 and PSMM2, which supports this argument. Furthermore, the result is consistent with a previous study of the relationship between Azo- moiety and duplex association/dissociation conversion. Because the energy barrier of azobenzene isomerization from trans- to cis- is higher than cis- to trans-, the trans- to cis- conversion requires a long UV light irradiation time to drive this conversion. Our results demonstrate that recovery (%) can reach to about 60% (±3.0%) after 20 min of UV irradiation before photobleaching of FAM fluorophore appears to become a serious problem. Thus, for realistic usage, which balances input and output energy, a UV irradiation time from 2 to 10 min is satisfactory for the PSMM running in numerous cycles without losing apparent functionality and efficiency.
Theoretical calculation of the extension and contraction forces is based on free energy and extending capability of single- and double-stranded DNA. We estimated the force based on Gibbs free energy and the distance variation from close and open structures. The L1 and L2 are approximately 10.2 and 2.2 nm and give the estimated values of two forces of 1.5 and 3.1 pN (the average forces based on our irradiation timeline), respectively (see Supporting Information g). Noticeably, the single-stranded DNA has the nature of forming random coil in solution instead of an extended structure. Here, we estimated the sizes by free energy and persistent length when applied. The actual size variation will determine the potential motor strength. The total input photon energy on extending the nanomotor can be calculated by UV lamp power (0.197 mW) and irradiation time (5 min) with Einput = Ps = 5.91 × 10−2 J. On the basis of our previously calculated extension force (1.5 pN) and distance (8 nm), each nanomotor has the extension work of 1.2 × 10−20 J (woutput = Fs). We can regard the extension work as the output mechanical energy. Therefore, the total output work Woutput for each type of nanomotor under our conditions can be calculated by Woutput = [(extended molecule)%] × [total molecule number (100 nM × 120 μL × NA = 7.22 × 1012] × [woutput]: PSMM1 (14.2%), 1.23 × 10−8 J; PSMM2 (26.3%), 2.28 × 10−8 J; and PSMM3(54.7%), 4.74 × 10−8 J. The energy conversion efficiencies (Einput/Woutput) are 2.09 × 10−7, 3.85 × 10−7, and 8.02 × 10−7, respectively.
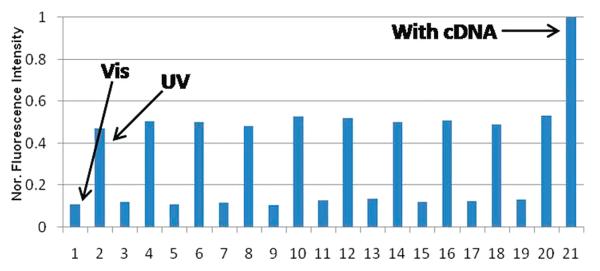
Figure 2 shows the reversibility of the PSMM3 nanomotor for ten rounds of close-open cycles. For each cycle, 1 min visible irradiation and 3 min of UV irradiation were applied. Compared to an approximate 40% decrease in cycling recovery for the previous DNA-fueled nanomachines under more favorable conditions (higher temperatures, more Azo- ratio), the cycling of PSMM3 maintains its recovery consistency and has no tendency to decrease after ten cycles. Additionally, all the cycles were performed at room temperature (25 °C) where the close state is more favored. As such, we can predict that conversion efficiency of our motor can be further improved at higher temperatures (close to Tm) as the stem is close to the transition point from duplex to single-strand state. This result demonstrates that the DNA hairpin nanomotor possesses high close-open conversion efficiency and molecular stability under current operating conditions. Repeated nanomotor cycles displayed no obvious decomposition of the motor (up to 20 cycles; data not shown). Overall, these results demonstrate a long-lasting molecular motor with high conversion efficiency using a clean energy input.
Figure 2.
Cycling of close-open from Vis/UV irradiations at 25 °C by repeated visible and UV irradiations. Vis (450 nm), 1 min; UV (350 nm), 3 min. Fluorescence intensities at the maximum emission (525 nm) were recorded immediately after each irradiation. Buffer, 20 mM Tris buffer pH 8.0; Na+, 20 mM; Mg2+, 2 mM; [PSMM3] = 100 nM, [cDNA] = 500 nM.
A comparably high fluorescence recovery seems very interesting from the perspective of energy application since it is related to energy conversion efficiency. That is, under the same operating conditions for similar molecular nanomotors labeled with F/Q, a higher fluorescence variation illustrates that higher ratios of molecules are driven from one state to another state. This comparison is applicable to hairpin structures as well as linear DNA strands as long as we normalize the conditions and set up correlated parameters. In this case, we can use the previously defined recovery percentage as the indicator of energy conversion efficiency. Under these conditions, a higher recovery percentage means that comparable DNA motor systems can convert more absorbed energy to drive the structure changes. This efficiency can therefore be related to the capability of fulfilling motorlike function. Although it is possible that different molecular structures might absorb different amounts of photon energy under the same conditions, we can still count it as part of the overall energy conversion capability for specific molecules.
In order to compare this conversion efficiency between our hairpin single-molecule nanomotors and previously investigated models, we designed a series linear DNAs, as well as another hairpin structure, using melting temperature (Tm) as the correlated parameter for comparison. The details of the rationale underlying this assumption are discussed in Supporting Information. The definition of Tm for short DNA duplex can be expressed by
ΔH° and ΔS° are the melting parameters; R is the ideal gas constant; CDNA is the molar concentration. For DNA sequences with the same concentrations, CDNA is the same, while ΔS° varies slightly. Therefore, Tm is approximately proportional to ΔH°, which is the energy absorbed by the DNA molecule to dissociate duplex structures. Therefore, Tm value can be regarded as the capability of absorbing sufficient energy to dissociate DNA duplex structures. For DNA nanomotors involved in duplex dissociation, we can always use Tm as the standard by which to evaluate the structure conversion or motor operation capability by absorbing external energy. For different duplex structures, such as hairpin and linear duplex with the same Tm value, they are expected to display the same recovery percentage under the same photoregulation conditions.
Linear DNA Probe Comparison
Two groups of linear DNAs bearing Azo- and Dabcyl and their cDNA bearing FAM have been synthesized (L12-1, L12-2 and L12-3/L12-cDNA; L10-1 and L10-2/L10-cDNA, Supporting Information, Scheme S4). The sequences of linear DNAs are designed in a manner similar to the truncation of the hairpin DNA from the 3′ end and have a Tm comparable to that of PSMM1–3 in order to compare the fluorescence recovery and quality of motor function (Supporting Information, Table S1). For L12-1 to L12-3, all three linear DNAs have almost the same Tm (53.7.2–55.2 °C) as PSMMs (55–57 °C). Therefore, they should be able to dissociate the duplex structure with the same recovery as PSMMs under the same conditions. As we mentioned above, it is possible that the energy absorption capabilities between heat and light are different for hairpin structure and linear structures. If that is the case, then our hairpin-structured motor has the advantage over linear DNAs of absorbing higher light energy that can be converted to molecular movement. L10-1 and L10-2 were intentionally selected with much lower Tm (47.5–48.6 °C) than PSMMs where they are expected to display higher recovery.
The fluorescence spectra of all the linear DNAs under the same conditions as PSMM are displayed in Supporting Information, Figure S2, and the recovery values are summarized in Supporting Information, Table S1. Under the same conditions, the recovery is from 14.2 to 54.7% for PSMM motors compared to 2.9–11.5% for L12 DNAs and 6.4–13.8% for L10 DNAs. These findings indicated that neither linear DNAs (L12s) with similar thermal response nor those with thermal response at lower temperature (L10) have the same response to light energy as PSMMs. If we assume these linear DNAs can do the same work as PSMM (actually less than PSMM due to smaller size changes), the energy conversion efficiencies based on recovery percentage values for L12 DNAs (L12-1, L12-2, L12-3) are 4.26 × 10−8, 8.38 × 10−8, and 1.69 × 10−7 and for L10 DNAs (L10-1, L10-2) are 9.26 × 10−8 and 2.03 × 10−7. As a result, linear DNAs cannot produce a comparable recovery by absorbing light energy from the same input light energy and display less photon to mechanical energy conversion efficiency. The energy conversion efficiency of PSMM3 is 4.76 times over L12-3 and 3.96 times over L10-2. Moreover, our hairpin structure has the same or fewer Azo- moieties tethered on the backbone than linear DNAs which theoretically absorb fewer photons per molecule, yet display a higher recovery percentage. These results strongly validate the expectation that the PSMMs have much higher energy conversion efficiency than linear DNA structures and that this directly results from the nanomotor structure. Additionally, our highly efficient nanomotors are operated under mild conditions of room temperature, illustrating that improvement of recovery can be further achieved by simply increasing the operational temperature. Although we did not systematically examine the recovery for PSMMs at temperatures closer to their Tm, higher conversion recoveries were observed when we increased the operational temperature.
As we discussed before, the main factor contributing to the high conversion efficiency is the hairpin structure of the motor and the intramolecular interaction arising from the single-stranded DNA. To investigate the influence of hairpin structure alone, we synthesized another Azo- incorporated hairpin molecule with a substitution of polyT loop from PSMM loop moiety. This hairpin structure also displays high response to photon energy, although it is lower than PSMM3 (Supporting Information h and Figure S3). This result suggests that carefully designed Azo- nanomotors with hairpin structure are intended to have high response to light energy.
Molecular interaction is also believed to play a key role in the significance of PSMMs. First, for linear DNA nanomotor systems, at least two pieces of DNA are needed to trigger a motor movement. Therefore, the hybridization process is based on an intermolecular interaction, and the concentration of each DNA strand contributes to the overall hybridization rate and hence the motor efficiency. For PSMMs, there is only one DNA strand that functions as a single-molecule motor. Thus, the hybridization within the PSMMs takes place by intramolecular interaction. We compared the photoregulation of PSMM3 and ten base linear DNAs (L10-1) for their photocontrollability (Supporting Information, Table S2) at different concentrations. The constant distance of paired hybridizing moieties within a single hairpin-structured molecule produces a type of concentration-independent nanomotor that maintains a high conversion efficiency (from 54.7 to 44.7% on recovery), while on the other hand the linear L10–1 displays a significant concentration-dependence effect (6.3–0.7%). Therefore, the hairpin nanomotors are expected to overcome the low efficiency problem experienced by linear DNA nanomotors, especially in a situation where high density DNA motor packing is required.
In conclusion, we have designed a photoswitchable single-molecule DNA nanomotor. It is the first fully reversible single-molecule DNA nanomachine driven by photons without any additional DNA strands as fuel. The incorporation of photosensitive Azo- moiety successfully produces a reversible photoregulated DNA nanomotor. This clean, photon-fueled nanomotor holds promise for applications that require the conversion of photonic energy into other forms of energy, such as mechanical movement. In addition, photoregulated DNA nanomotors can be easily manipulated and reproduced, as our design is simple and easy in terms of synthesis and operational strategy. Furthermore, PSMM is a novel single-molecule nanomotor system, which in contrast to previous linear DNA nanomotors or hairpin nanostructures does not require several DNA strands for operation. Thus, the intramolecular interaction that occurs in this system circumvents the complexity introduced by intermolecular reactions that occurred in previous DNA nanomotors. Compared to concentration-dependent linear DNAs, the concentration-independence of our PSMM allows high efficiency at any concentrations and situations that would otherwise be difficult to achieve as a result of interference by multimolecular interactions. Continued investigation of the relationship between hairpin structure and energy conversion efficiency will further elucidate the mechanisms underlying the energy conversion process. Meanwhile, the coupling of nanomotors to useful applications for force production on the nanoscale remains a significant challenge.
Experimental Section. Chemicals and Reagents
The chemicals for synthesis of phosphoramidite monomer were purchased from Aldrich Chemical, Inc. The materials for DNA synthesis, including CPG columns and reagents for DNA modification and coupling, were purchased from Glen Research Co. All the chemicals were used without further purification, except as otherwise explained. In this case, a 31 base DNA probe, which fits most of our requirements for a simple and highly responsive nanodevice, was selected. These DNA probes are called photoswitchable single molecular DNA motor (PSMM). The PSMMs were synthesized by a DNA synthesizer from 3′ end to 5′ end, starting with CPG column labeled with Dabcyl quencher (Dabcyl CPG) and with FAM fluorophore coupled on 5′. The target cDNA for the PSMMs is a partially complementary strand with the sequence complementary to the PSMMs from 3′ end throughout the entire loop. The linear DNAs were synthesized with the same Dabcyl CPG on azobenzene-incorporated sequences, and general bases were CPG-coupled with FAM on 5′ end for the complementary sequences.
Instruments
The purification of chemical compounds was performed by glass column for the silica gel chromatography and identified by thin layer chromatography (TLC) plate (silica gel 60F254; Merck) and NMR spectrometer (Mercury 300). An ABI3400 DNA/RNA synthesizer (Applied Bio-systems) was used for all the DNA-related synthesis. The purifications were carried out on a ProStar HPLC system equipped with gradient unit (Varian) with 18 column (Econosil, 5U, 250 × 4.6 mm) (Alltech Associates). The characterizations of all DNAs on concentration were performed with a Cary Bio-300UV spectrometer (Varian) by calculating the absorbance of DNA at 260 nm. The melting properties were studied with MyiQ single-color RT-PCR system (Bio-Rad). A Fluorolog-Tau-3 Spectrofluorometer with a temperature controller (Jobin Yvon) was used for all steady-state fluorescence measurements. The samples were loaded with quartz cell for fluorescent spectrum (Starna Cells, Inc.). The UV light source was a portable 6 W UV-A fluorescent lamp (FL6BL-A; Toshiba), and the visible light source was a general table lamp with a 60 W lamp and optical filters (Asahi Technoglass).
Photoregulation of Photoswitchable Single Molecular DNA Motor (PSMM)
The phosphoramidite monomer was synthesized from protocol of Asanuma et al. with minor modifications (Supporting Information c). The synthesis and purification of PSMMs and linear sequences followed usual procedures, except as otherwise mentioned (Supporting Information d). The concentration of each DNA was calculated by the absorption at 260 nm by UV spectrometer. The melting curves were measured with a PCR machine with the same buffer solution for all the measurements (Supporting Information f). For comparison, PSMMs were diluted to equal concentration for the entire experiment at room temperature (25 °C) using the same buffer solution. For the photoregulation of PSMMs, irradiations by UV light at 350 nm and visible light at 450 nm were applied for all experiments, although the irradiation times varied, as described. Fluorescence intensity was recorded under excitation at 488 nm. In our protocol, once the temperature is set at 25 °C and stabilized, the quartz cell with samples is maintained at the set temperature for at least 5 min before each test. The steps for UV/vis irradiation measurements are listed below.
The sample is diluted and added in quartz cell.
The quartz cell is set in a holder and maintained in place for 5 min.
Irradiation is performed with visible light at 450 nm for 1 min.
Fluorescence spectrum is measured (excited at 488 nm).
Irradiation is performed with UV lamp at 350 nm for 5 min.
Fluorescence spectrum is again measured (excited at 488 nm).
Steps 1–6 are repeated to confirm the reversibility of the photoregulation.
The intensity changes are compared after several rounds with a 5-fold addition of cDNA. For optimization experiments, the UV light varied from 1–20 min. An efficiency of around 50% for PSMM3 was obtained at 10 min UV irradiation time. Longer periods can improve the efficiency slightly, but prolonged irradiation under current portable UV lamp over 30 min tends to result in photobleaching the fluorophore and decomposing the structure (data not shown). For the reversibility test, the UV irradiation time was set at 3 min for all the tests, except as otherwise noted.
Photoregulation of Linear DNAs
The 12 and 10 bases FAM DNAs were designed and synthesized as described in Supporting Information e. They were first diluted in the same buffer solution as the hairpin molecules with a final concentration of 100 nM and set for 5 min until stable. An emission scan was performed with 488 nm excitation. Azobenzene-incorporated DNA (1.5-fold of each) was then added to the cell and stabilized for 5 min until the two strands were fully hybridized, followed by a second emission scan. The reversible irradiations with UV–vis light were obtained by repeating steps 3–7 established for photoregulation of PSMMs.
Supplementary Material
Acknowledgment
We acknowledge the support by the NIH NIGMS GM 066137 and NIH U54NS058185 and the NSF grants.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Seeman NC. J. Biomol. Struct. Dyn. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 2.Seeman NC. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 3.Chen JH, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 4.Zhang YW, Seeman NC. J. Am. Chem. Soc. 1994;116:1661–1669. [Google Scholar]
- 5.Shih WM, Quispe JD, Joyce GFA. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 6.Goodman RP, Schaap AT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 7.Robinson BH, Seeman NC. Protein Eng. 1987;1:295–300. doi: 10.1093/protein/1.4.295. [DOI] [PubMed] [Google Scholar]
- 8.Heilemann M, Tinnefeld P, Mosteiro SG, Parajo GM, Van Hulst FN, Sauer M. J. Am. Chem. Soc. 2004;126:6514–6515. doi: 10.1021/ja049351u. [DOI] [PubMed] [Google Scholar]
- 9.Niemeyer CM, Koehler J, Wuerdermann C. ChemBioChem. 2002;3:242–245. doi: 10.1002/1439-7633(20020301)3:2/3<242::AID-CBIC242>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 10.Yang X, Vologodskii AV, Liu B, Kemper B, Seeman NC. Biopolymers. 1998;45:69–83. doi: 10.1002/(SICI)1097-0282(199801)45:1<69::AID-BIP6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 11.Shin JS, Pierce NA. J. Am. Chem. Soc. 2004;126:10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- 12.Higashi-Fujime S, Ishikawa R, Iwasawa H, Kagami O, Kurimoto E, Kohama K, Hozumi T. FEBS Lett. 1995;375:151–154. doi: 10.1016/0014-5793(95)01208-v. [DOI] [PubMed] [Google Scholar]
- 13.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech RT, Doudna AJ. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 14.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. Annu. Rev. Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 15.Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 16.Yan H, Zhang X, Shen Z, Seeman NC. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 17.Li JW, Tan WH. Nano Lett. 2002;2:315–318. [Google Scholar]
- 18.Turberfield AJ, Mitchell JC, Yurke B, Mills AP, Jr., Blakey MI, Simmel FC. Phys. Rev. Lett. 2003;90:118102–118109. doi: 10.1103/PhysRevLett.90.118102. [DOI] [PubMed] [Google Scholar]
- 19.Tian Y, He Y, Peng Y, Mao C. Angew. Chem., Int. Ed. 2005;44:4355–4358. doi: 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- 20.Yin P, Yan H, Daniell XG, Turberfield AJ, Reif JH. Angew. Chem., Int. Ed. 2004;43:4906–4911. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- 21.Sudesh G, Neckers DC. Chem. Rev. 1989;89:1915–1925. [Google Scholar]
- 22.Berg RH, Hvilsted S, Ramanujam PS. Nature. 1996;383:505–508. [Google Scholar]
- 23.Guang Diau EW. J. Phys. Chem. A. 2004;108:950–956. [Google Scholar]
- 24.Hugel T, Holland NB, Cattani A, Moroder L, Seitz M, Gaub HE. Science. 2002;296:1103–1106. doi: 10.1126/science.1069856. [DOI] [PubMed] [Google Scholar]
- 25.Asanuma H, Takarada T, Yoshida T, Liang X, Komiyama M. Angew. Chem., Int. Ed. 2001;40:2671–2673. [PubMed] [Google Scholar]
- 26.Liang X, Asanuma H, Komiyama M. Tetrahedron Lett. 2001;42:6723–6725. [Google Scholar]
- 27.Asanuma H, Liang X, Yoshida T, Komiyama M. ChemBioChem. 2001;2:39–44. doi: 10.1002/1439-7633(20010105)2:1<39::AID-CBIC39>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Liang X, Asanuma H, Komiyama M. J. Am. Chem. Soc. 2002;124:1877–1883. doi: 10.1021/ja011988f. [DOI] [PubMed] [Google Scholar]
- 29.Asanuma H, Matsunaga D, Komiyama M. Nucleic Acids Symp. Ser. 2005;49:35–36. doi: 10.1093/nass/49.1.35. [DOI] [PubMed] [Google Scholar]
- 30.Liang X, Nishioka H, Takenaka N, Asanuma H. ChemBioChem. 2008;9:702–705. doi: 10.1002/cbic.200700649. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Yaegashi S, Asanuma H, Hagiya M. Lect. Notes Comput. Sci. 2006;3892:336–346. [Google Scholar]; Kim YM, Phillips AJ, Liu HP, Kang HZ, Tan WH. PNAS. 2009;106:6489–6494. doi: 10.1073/pnas.0812402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths J. Chem. Soc. Rev. 1972;1:481–493. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.