Abstract
Background
Glutamate neurotransmission plays an important role in a variety of alcohol-related phenomena, including alcohol self-administration by both animals and humans. Because the risk for alcohol dependence (AD) is genetically influenced, genes encoding glutamate receptors are candidates to contribute to the risk for AD. We examined the role of variation in the 3’ region of GRIK1, the gene that encodes the GluR5 receptor subunit of the kainic acid glutamate receptor, on risk for AD. We focused specifically on this gene because topiramate, a glutamate modulator that binds to the GluR5 subunit, has shown robust efficacy in the treatment of AD.
Methods
We genotyped 7 single nucleotide polymorphisms (SNPs) in the 3’-half of GRIK1, which includes 3 differentially spliced exons, in a sample of EA control subjects (N=507) and subjects with AD (N=1,057).
Results
We found nominally significant evidence of association to AD for 3 SNPs (rs2832407 in intron 9, rs2186305 in intron 17, and rs2832387 in the 3’UTR). Empirical p-value estimation revealed that only rs2832407 was significantly associated to phenotype (p = 0.043).
Discussion
These findings provide support for the hypothesis that variation in the 3’ portion of the gene encoding the GluR5 kainate receptor subunit contributes to the risk for AD. Further research is needed to ascertain whether this SNP is itself functional or whether the association reflects linkage disequilibrium with functional variation elsewhere in the gene and whether this SNP moderates topiramate's effects in the treatment of AD.
Keywords: psychiatric genetics, kainate receptor, GRIK1, alcohol dependence, polymorphism
Introduction
Alcohol dependence (AD) is a common psychiatric disorder. The 12-month U.S. prevalence of AD is estimated to exceed 3.8%, which equates to more than 8 million adults (Grant et al., 2004). A variety of adverse consequences are associated with AD. These include medical, social, and legal problems (Caetano and Cunradi, 1997), resulting in an estimated annual cost in the United States of nearly $185 billion (Harwood et al. 1998). Susceptibility to AD is genetically influenced, with heritability estimates ranging from 0.52 to 0.64 (Kendler 2001). Over the past two decades, considerable research has focused on understanding the specific genetic factors contributing to the risk for AD and on the development of medications to treat the disorder (Kranzler et al., in press). There has been a growing convergence between these lines of investigation, with evidence that specific neurobiological systems underlie both the risk for AD and its pharmacological treatment (Knapp et al., 2008).
Glutamatergic neurotransmission is of particular interest in relation both to AD risk and its pharmacologic treatment. Glutamate, the most abundant excitatory neurotransmitter in the central nervous system, is involved in neuroproliferative, neurotoxic, neurodegenerative, and neuromodulatory processes, and is a key component of learning and memory (Kugaya and Sanacora 2005). Glutamatergic efferents from the prefrontal cortex, amygdala, and hippocampus innervate the cell bodies of the ventral tegmental area and the nucleus accumbens shell, facilitating dopaminergic transmission in these key areas of the “reward pathway” (Tzschentke and Schmidt 2003). Alcohol modulates glutamatergic neurotransmission, producing long-lasting neurobiological changes that may contribute to the symptoms of AD. The attenuation of these glutamatergic effects reduces alcohol-induced reward and relapse-like behavior in animals (Gass and Olive 2007). Further evidence of the importance of glutamatergic neurotransmission in AD comes from alcohol treatment trials of medications that modulate glutamatergic activity, including acamprosate (Kranzler and Gage 2008) and topiramate (Johnson et al., 2003, 2007).
Several studies have examined the allelic association of a variety of genes encoding glutamate receptor subunits with AD or alcohol withdrawal phenomena (Wernicke et al. 2003, Foley et al. 2004, Rujescu et al. 2005, Tadic et al. 2005, Samochowiec et al. 2006, Schumann et al. 2008). Although none of these studies included GRIK1, the gene encoding the GluR5 kainate receptor subunit, we chose to focus on this gene because receptors containing the GluR5 subunit (as with those containing the GluR6 subunit) selectively bind topiramate (Gryder and Rogawski 2003, Kaminski et al. 2004), which could result in convergent validation of a role for this gene in AD-related phenotypes.
Kainate receptors are tetramers assembled from specific protein subunits including the GluR5, GluR6, and GluR7 subunits (Pinheiro and Mulle, 2006). Different combinations of subunits and splice variants show different regional distributions and different physiologic properties. The receptors play a key role in the regulation of neuronal excitability, including phasic and tonic modulation of excitatory and inhibitory neurotransmitter release. Although kainate receptors may play a role in the effects of alcohol, these receptors are inhibited by ethanol at concentrations higher than those required to antagonize NMDA receptors, but at levels often reached in vivo during intoxication (Dodd et al. 2000, Krystal et al. 2003).
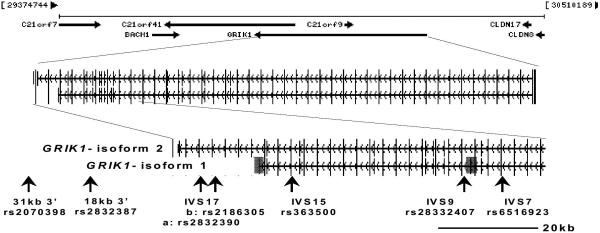
Both GluR5 and GluR6 subunit precursor RNAs undergo RNA secondary-structure-dependent adenosine-to-inosine editing, which generates a glutamine-to-arginine amino acid coding change that affects glutamate receptor function (Seeburg et al. 1998; Seeburg & Hartner 2003). GRIK1 is comprised of 18 exons spanning 403kb and maps to chromosome 21q22.11. The HapMap dataset indicates that the gene contains at least 8 haplotype blocks in Europeans. The GRIK1 transcript undergoes alternative splicing to encode two known isoforms of human GluR5. One isoform (GluR5-1d; accession number NM-000830) includes exons 1-17, while the second isoform (EAA3a; accession number U16125) includes exons 1-8, 10-16 and 18. These alternatively spliced sites reflect potential functional polymorphic variation that could influence receptor assembly and intracellular trafficking of kainate receptors. Because GRIK1 spans too great a region to tag it fully, we focused on variation in the 3’-half of the gene, including the differentially spliced exons 9, 17, and 18, which have potential functional significance for the receptor.
Materials and Methods
Subjects
Connecticut subjects
AD subjects included 372 non-Hispanic Caucasians recruited as part of ongoing studies of the genetics of AD or from treatment trials for AD at the University of Connecticut Health Center (UCHC), Farmington, CT and the VA Connecticut Healthcare Center (VA-CT), West Haven, CT. EA controls from CT (n = 507) were recruited by advertisement in the greater Hartford, CT area. Psychiatric diagnoses were made in these subjects using the Structured Clinical Interview for DSM-III-R or DSM-IV (SCID) (Spitzer and Williams 1995, First et al. 2001), or the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) (Pierucci-Lagha et al. 2005, 2007). All control subjects were screened using the SCID or the SSADDA to exclude individuals with an alcohol or drug use disorder or other major Axis I psychiatric disorder. All subjects provided written, informed consent to participate in study protocols that were approved by the institutional review boards at UCHC, Yale University School of Medicine, and/or VA-CT and were paid for their participation.
Project MATCH AD subjects
DNA and phenotypic information were obtained anonymously from 727 subjects who participated in Project MATCH, a large, multi-center trial comparing the effects of three different psychosocial interventions. The diagnosis of AD for Project MATCH subjects was made using the Computerized Diagnostic Interview for DSM-IV (Blouin et al. 1988; American Psychiatric Association 1994). Institutional review boards at UCHC, Yale, and the Medical University of South Carolina (where Project MATCH blood samples were stored prior to DNA extraction and genotyping) approved the use of coded Project MATCH samples.
Genotyping
We focused our examination on SNP markers in the region of the alternatively spliced exons 9, 17 and 18, the intron 13 region containing the complementary sequence required for RNA editing and the glutamine-to-arginine coding change in exon 13 (Barbon & Barlati 2000) and the 3’UTR (Figure 1). To provide sufficient statistical power to detect an association to phenotype, we focused on SNPs with minor allele frequencies >10%. Six tag SNPs were identified using the Tagger algorithm (de Bakker et al. 2005) incorporated in the HapMap CEPH European ancestry dataset release 21a based on the NCBI B35 assembly covering the 100kb 3’ region of GRIK1 extending from the distal portion of intron 7 to 30 kb 3’ of exon 17 (chromosome 21 position 29,800,000–29,900,000). These six Tag SNPs captured 27 of 29 known SNPs genotyped in the CEPH sample with an average R2 of 0.927. Two recombination hot spots are annotated in the HapMap dataset in this genomic interval and occur in the proximal region of intron 9, approximately 15kb distal to the alternatively spliced exon 9. We also examined rs2832407, midway between these recombination hot spots.
Figure 1.
SNPs examined in GRIK1 (Chr. 21q21.3)
Genotyping was performed using a closed-tube fluorescent TaqMan 5’-nuclease allelic discrimination assay and MGB-probes and primers designed using Primer Express v3.0 software [Applied Biosystems Inc. (ABI) Foster City, CA]. Fluorescence plate reads and genotype calls were made using ABI 7900 Sequence Detection Systems. Two ng of genomic DNA were PCR amplified in 384-well plates using a 5-μl reaction volume. Repeat genotyping was completed for 20% of samples with a genotype discrepancy rate of 0.2%.
To estimate genetic ancestry proportions for each subject, we used a panel of 34 short tandem repeat ancestry informative markers (AIMs), as described previously (Stein et al. 2004; Luo et al. 2005; Yang et al. 2005; Covault et al. 2008).
Data Analysis
Diagnostic groups were compared on age and the percentage of African ancestry (i.e., using a STRUCTURE-estimated ancestry proportion score) using ANOVA and on sex and the psychiatric diagnoses using a 2×2 contingency table and the χ2 test. Association between gender and SNP genotype was examined using the chi-square test and the association between age and SNP genotype was studied using ANOVA. Linkage disequilibrium (LD) plots, haplotype blocks, and tests of Hardy-Weinberg equilibrium for each group were generated using the software program Haploview v3.32 (Barrett et al. 2005).
The program STRUCTURE v2.1 (Pritchard et al. 2000; Falush et al. 2003) was used to estimate the proportion of African and European genetic ancestry for each subject based on genotype results from the 34 AIMs. Simulations used 100,000 burn-ins followed by 500,000 runs and a population parameter K=2.
Single marker analysis was conducted using logistic regressions in which the predictor included ancestry and coded genotypes for each candidate SNP. We employed an additive model, that is, we coded the three genotypes at a candidate SNP with values of –1, 0, and 1 in the logistic regression analyses.
Because seven markers were analyzed, a nominally significant result for a specific marker may not be statistically significant after accounting for the inflation of type 1 error due to multiple comparisons. Consequently, we performed 10,000 permutations to assess the effects of multiple comparisons. To generate each permuted data set, we permuted the disease status among the cases and controls and analyzed associations between the permutated trait values and seven markers individually. That is, each permuted data set retains the same number of affected and normal individuals but any potential association between disease status and genotypes is removed through the random assignment of disease status through permutation. A p-value for each of the seven markers was calculated from each permuted data set, with 10,000 permutations, resulting in a 10,000 × 7 matrix in which each row corresponded to the seven p-values from each permutation and each column represents the association between the permuted traits and a specific marker. This matrix was used to adjust the alpha level for multiple comparisons. For example, to calculate the overall p-value for the region, we first calculated the minimum p-value for the observed data, which is 0.009. Then for each permuted data set, we calculated the minimum p-value among the markers, labeling it as pmin,k. The permutation-based p-value for the whole region was estimated as the proportion of the 10,000 permutation runs whose pmin,k was <0.009. Evaluation of the empirical p-value for each marker individually, based on the permutated data sets, resulted in p-values very close to the observed p-values reported in Table 2.
Table 2.
GRIK1 SNP allele frequencies for self-identified EA subjects.
| SNP rs# Location | Allele1 | Controls (n=507) | CT AD (n=337) | MATCH AD (n=720) | All AD3(n=1057) | p-value2 | Haplotype Block # |
|---|---|---|---|---|---|---|---|
| rs2070398 | C | 0.783 | 0.796 | 0.805 | 0.803 | 0.201 | 1 |
| 31kb 3’ | A | 0.227 | 0.204 | 0.195 | 0.197 | ||
| rs2832387 | G | 0.667 | 0.722 | 0.712 | 0.715 | 0.012 | 1 |
| 13kb 3’ | A | 0.333 | 0.278 | 0.288 | 0.285 | ||
| rs2832390 | T | 0.752 | 0.754 | 0.783 | 0.775 | 0.172 | 2 |
| intron 17 | C | 0.248 | 0.246 | 0.217 | 0.225 | ||
| rs2186305 | A | 0.664 | 0.678 | 0.714 | 0.707 | 0.024 | 2 |
| intron 17 | G | 0.336 | 0.322 | 0.286 | 0.293 | ||
| rs363500 | G | 0.727 | 0.735 | 0.759 | 0.754 | 0.132 | -- |
| intron 15 | A | 0.273 | 0.265 | 0.241 | 0.246 | ||
| rs2832407 | A | 0.612 | 0.668 | 0.658 | 0.661 | 0.009 | -- |
| intron 9 | C | 0.388 | 0.340 | 0.341 | 0.339 | ||
| rs6516923 | A | 0.596 | 0.634 | 0.623 | 0.627 | 0.129 | -- |
| intron 7 | T | 0.404 | 0.384 | 0.376 | 0.373 |
Allele nucleotide designation refers to the chromosome plus strand sequence
Bolded p-values are nominally significant; empirical analysis supports the statistical significance of only rs2832407 when controlled for multiple comparisons.
EA Controls compared with EA Alcoholics (Connecticut and MATCH samples combined)
Results
Demographic, clinical, and ancestry characteristics of the study subjects are displayed in Table 1. The control group was significantly younger than the AD groups and included more females. To determine whether age and gender were confounders for genetic association, we examined associations between age, gender and the seven SNPs across all the cases and controls. There was no significant association found. The diagnostic groups did not differ on the proportion of African ancestry. As expected, Connecticut AD subjects had a significantly higher prevalence of lifetime diagnoses of cocaine and opioid dependence than controls. Lifetime drug dependence diagnoses were not available for the Project MATCH sample. The prevalence of antisocial personality disorder was comparable in the two AD samples and, as expected, was significantly greater than in controls.
Table 1.
Demographic and Clinical Characteristics of the Samples.
| Connecticut Samples | Project Match Sample | ||
|---|---|---|---|
| Controls (n=507) | Alcohol Dependence (n=337) | Alcohol Dependence (n=720) | |
| Age [yr (SD)] | 29.0 (9.8) | 43.0 (9.4)*** | 40.6 (11.1)*** |
| Sex (% male) | 38.1 | 70.9*** | 73.6*** |
| African Ancestry [% (SD)] | 1.9 (3.4) | 2.0 (4.3) | 1.7 (2.9) |
| Lifetime Cocaine Abuse or Dependence (%) | 0 | 40.1*** | n.a. |
| Lifetime Opioid Abuse or Dependence (%) | 0 | 16.6*** | n.a. |
| Antisocial Personality Disorder (%) | 0 | 11.1*** | 10.3*** |
p< 0.001 vs. controls
n.a., data not available
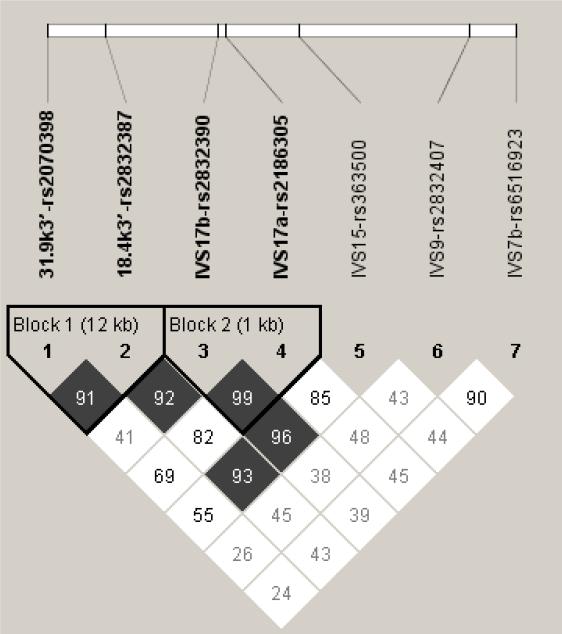
There was no evidence of deviation from Hardy-Weinberg expectations for any of the 7 SNPs examined in this study, either in subjects with AD or controls (all p's > 0.05). As shown in Figure 2, two 2-SNP haplotype blocks were identified, both of which were located in the most 3’ portion of the region examined.
Figure 2.
Haploview LD plot of GRIK1 3’region markers: LD plot from Haploview 3.32 for EA subjects (507 control subjects and 1057 subjects with AD). Pairwise SNP |D’| values (x100) of linkage are shown together with 2 haplotype blocks identified using the four-gamete rule. Darkened blocks indicate SNP pairs without evidence of extensive recombination (i.e. 4-gamete rule for haplotype block characterization with at least one 2-SNP haplotype having a frequency < 0.02).
Three markers (rs2832407 in intron 9, rs2186305 in intron 17, and rs2832387 in the 3’UTR) showed nominally significant allelic association to AD in this sample (Table 2). Accounting for multiple comparisons, we compared the minimum p-value among the seven markers (0.009 for rs2832407) to the empirical distribution of the minimum p-value among the seven markers from the 10,000 permuted samples. A total of 425 permuted samples had the minimum p-value less than 0.009, suggesting that there is statistically significant evidence (an empirical p-value of 0.0425) when the minimum p-value is considered for multiple comparison adjustments.
Discussion
These findings provide support for the hypothesis that variation in the gene encoding the GluR5 glutamate receptor subunit contributes to the risk for AD. Three GRIK1 SNPs (rs2186305, rs2832407, and rs2832387) showed nominally significant evidence of association in allelewise analyses. Correction for multiple testing showed that only one of the SNPs (rs2832407) was significantly associated to AD.
Using the UCSC Genome Browser, the HapMap site, and a number of software programs, (e.g. SNPStudio), we found that the region that includes rs2832407 has very low conservation across species and it is far from intron-exon boundaries. Despite evidence that it is not directly involved in splicing, LD patterns can be complex and many SNPs from recent genomewide association studies do not have apparent functional consequences. Consequently, this finding does not exclude the possibility that this SNP is functionally relevant to the AD phenotype and further investigation is needed to ascertain whether it has a direct effect on GluR5 kainate receptor function. An alternate explanation is that the association reflects linkage disequilibrium with functional variation elsewhere in the gene.
Of the glutamate receptors, the NMDA type has received the most research attention in relation to AD (Gass and Olive 2008). Consistent with that literature, a recent survey of 10 glutamate system genes showed a moderate association of AD (i.e., an odds ratio of 2.18) with markers in GRIN2A, which encodes NR2A, the N-methyl-D-aspartate receptor subunit 2A (Schumann et al. 2008). The effect was initially demonstrated in one sample and replicated in an independent sample. That study did not, however, examine any of the genes encoding kainate receptor subunits. Kainate receptors are important modulators of interneuron excitability in the hippocampus. Carta et al. (2003) showed that ethanol potently inhibits this effect at plasma concentrations associated with the ingestion of 1-2 alcoholic drinks, an effect that appeared to be specific for kainate receptors and physiologically relevant. The authors argued that the effect may partially explain some of the paradoxical excitatory actions of alcohol, which are perceived as positive by some individuals and could contribute to the development of alcoholism. The only other kainate receptor gene that has been studied as a candidate for alcohol-related disorders is GRIK3, which encodes the GluR7 subunit. Preuss et al. (2006) found an association in a German case-control sample of a functional Ser310Ala polymorphism with a history of delirium tremens. That finding, however, was not replicated in Polish family-based or case-control samples, in which delirium tremens and other alcohol-related phenotypes, including AD, were examined (Samochowiec et al. 2006).
In addition to providing evidence of the contribution of GRIK1 to the risk of AD, the present findings may have relevance for the pharmacogenetic analysis of topiramate in the treatment of AD. The use of the intron 9 SNP to characterize alcohol-dependent subjects in treatment trials of topiramate may be warranted based on the findings reported here and would be feasible, given the relatively high prevalence of this allele. GRIK1 was recently shown by Uhl et al. (2008) to be among the genes that were nominally associated with successful attempts to quit smoking in pooled genomewide association analyses in at least two of three independent treatment samples, supporting its potential utility as a predictor of treatment response in addictive disorders.
The present study has a number of strengths. First, it is based on large, well-characterized samples of AD cases and screened controls. Second, the gene studied encodes a protein that may have important implications for understanding both the etiology and pharmacological treatment of AD. Third, we examined multiple SNPs in GRIK1, which were chosen based on potential functional effects related either to a glutamine-to-arginine amino acid coding change that affects glutamate receptor function (Seeburg et al. 1998; Seeburg & Hartner 2003) or to potential alternative splice sites resulting in isoforms that could influence receptor assembly and intracellular trafficking of receptors that contain the GluR5 subunit. Although the study is limited by the fact that the control sample was drawn from Connecticut, while the majority of cases were recruited from sites around the United States, analysis of ancestry informative markers revealed no difference in ancestry proportion as a function of diagnostic group. Similarly, although the age and sex distributions differed by diagnostic group, these differences were not correlated with genotype for the seven SNPs examined. Thus, these potential confounders did not appear to influence the findings. Based on these findings, the association of variation in GRIK1 to AD reported here warrants further research, including replication studies in EA and other populations, examination of the functional effects of the observed variation, and consideration of a potential pharmacogenetic effect of this variation in AD subjects.
Acknowledgments
We are grateful to investigators from the multi-center study, Project MATCH, and the National Institute on Alcohol Abuse and Alcoholism for providing access to clinical data and blood samples from that study. This study was supported by NIH grants P50 AA03510, M01 RR06192, R01 AA11330, R01 AA015606, K24 AA13736, and K24 DA15105.
References
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. 4th Edition American Psychiatric Press; Washington, DC: 2004. [Google Scholar]
- Angehagen M, Ronnback L, Hansson E, Ben-Menachem E. Topiramate reduces AMPA-induced Ca(2+) transients and inhibits GluR1 subunit phosphorylation in astrocytes from primary cultures. J Neurochem. 2005;94:1124–1130. doi: 10.1111/j.1471-4159.2005.03259.x. [DOI] [PubMed] [Google Scholar]
- Barbon A, Barlati S. Genomic organization, proposed alternative splicing mechanisms, and RNA editing structure of GRIK1. Cytogenet Cell Genet. 2000;88:236–9. doi: 10.1159/000015558. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Blouin AG, Perez EL, Blouin JH. Computerized administration of the Diagnostic Interview Schedule. Psychiatry Res. 1988;23:335–344. doi: 10.1016/0165-1781(88)90024-8. [DOI] [PubMed] [Google Scholar]
- Caetano R, Cunradi C. Alcohol dependence: A public health perspective. Addiction. 1997;97:633–645. doi: 10.1046/j.1360-0443.2002.00184.x. [DOI] [PubMed] [Google Scholar]
- Carta M, Ariwodola OJ, Weiner JL, Valenzuela CF. Alcohol potently inhibits the kainate receptor-dependent excitatory drive of hippocampal interneurons. Proc Natl Acad Sci USA. 2003;100:6813–6818. doi: 10.1073/pnas.1137276100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5’Region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Dodd PR, Beckmann AM, Davidson MS, Wilce PA. Glutamate-mediated transmission, alcohol, and alcoholism. Neurochem Int. 2000;37:509–533. doi: 10.1016/s0197-0186(00)00061-9. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First M, Spitzer R, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition With Psychotic Screen (SCID-I/P W/ PSY SCREEN) New York Biometrics Research. New York State Psychiatric Institute; 2001. [Google Scholar]
- Foley PF, Loh EW, Innes DJ, Williams SM, Tannenberg AE, Harper CG, Dodd PR. Association studies of neurotransmitter gene polymorphisms in alcoholic Caucasians. Ann N Y Acad Sci. 2004;1025:39–46. doi: 10.1196/annals.1316.005. [DOI] [PubMed] [Google Scholar]
- Gass JT, Olive MF. Glutamatergic substrates of drug addiction and alcoholism. Biochem Pharmacol. 2008;75:218–265. doi: 10.1016/j.bcp.2007.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991-1992 and 2001-2002. Drug Alcohol Depend. 2004;74:223–234. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Gryder DS, Rogawski MA. Selective antagonism of GluR5 kainate-receptor-mediated synaptic currents by topiramate in rat basolateral amygdala neurons. J Neurosci. 2003;23:7069–7074. doi: 10.1523/JNEUROSCI.23-18-07069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood H, Fountain D, Livermore G. Report prepared for the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Department of Health and Human Services. National Institute of Health Publication No. 98-4327. National Institutes of Health.; Rockville, MD: 1998. The Economic Costs of Alcohol and Drug Abuse in the United States 1992. http://www.nida.nih.gov/EconomicCosts/Index.html. [Google Scholar]
- Johnson BA. Progress in the development of topiramate for treating alcohol dependence: from a hypothesis to a proof-of-concept study. Alcohol Clin Exp Res. 2004;28:1137–1144. doi: 10.1097/01.alc.0000134533.96915.08. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Ait-Daoud N, Bowden CL, DiClemente CC, Roache JD, Lawson K, Javors MA, Ma JZ. Oral topiramate for treatment of alcohol dependence: a randomised controlled trial. Lancet. 2003;361(9370):1677–1685. doi: 10.1016/S0140-6736(03)13370-3. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Rosenthal N, Capece JA, Wiegand F, Mao L, Beyers K, McKay A, Ait-Daoud N, Anton RF, Ciraulo DA, Kranzler HR, Mann K, O'Malley SS, Swift RM, Topiramate for Alcoholism Advisory Board. Topiramate for Alcoholism Study Group Topiramate for treating alcohol dependence: a randomized controlled trial. JAMA. 2007;298:1641–1651. doi: 10.1001/jama.298.14.1641. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J. CHRM2 gene predisposes to alcohol dependence, drug dependence, and affective disorders: results from an extended case-control structured association study. Hum Mol Genet. 2005;14:2421–2434. doi: 10.1093/hmg/ddi244. [DOI] [PubMed] [Google Scholar]
- Kaminski RM, Banerjee M, Rogawski MA. Topiramate selectively protects against seizures induced by ATPA, a GluR5 kainate receptor agonist. Neuropharmacology. 2004;46:1097–1104. doi: 10.1016/j.neuropharm.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Kendler KS. Twin studies of psychiatric illness: an update. Arch Gen Psychiatry. 2001;58:1005–14. doi: 10.1001/archpsyc.58.11.1005. [DOI] [PubMed] [Google Scholar]
- Knapp CM, Ciraulo DA, Kranzler HR. Neurobiology of alcohol. In: Galanter M, Kleber HD, editors. Textbook of Substance Abuse Treatment. 4th Edition American Psychiatric Press, Inc.; Washington, DC: 2008. [Google Scholar]
- Kranzler HR, Ciraulo D, Jaffe JH. Medications for use in alcohol rehabilitation. In: Ries R, Fiellin D, Miller S, Saitz R, editors. Principles of Addiction Medicine. 4th Edition. Lippincott Williams & Wilkins; Philadelphia: in press. [Google Scholar]
- Kranzler HR, Gage A. Acamprosate efficacy in alcohol-dependent patients: Reanalysis of results from 3 pivotal trials. Am J Addict. 2008;17:70–76. doi: 10.1080/10550490701756120. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Petrakis IL, Mason G, Trevisan L, D'Souza DC. N-methyl-D-aspartate glutamate receptors and alcoholism: reward, dependence, treatment, and vulnerability. Pharmacol Ther. 2003;99:79–94. doi: 10.1016/s0163-7258(03)00054-8. [DOI] [PubMed] [Google Scholar]
- Kugaya A, Sanacora G. Beyond monoamines: glutamatergic function in mood disorders. CNS Spectr. 2005;10:808–819. doi: 10.1017/s1092852900010403. [DOI] [PubMed] [Google Scholar]
- McDonald RL, Rogawski MA. Cellular effects of antiepileptic drugs. In: Pedley TA, editor. Epilepsy: A Comprehensive Textbook. 2nd Ed. Lippincott, Williams, & Wilkins; Philadelphia: 2006. [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells J, Pearson D, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment of Drug Dependence and Alcoholism (SSADDA). Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells J, Farrer L, Kranzler HR. Reliability of DSM-IV Diagnostic Criteria Using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA). Drug Alcohol Depend. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro P, Mulle C. Kainate receptors. Cell Tissue Res. 2006;326:457–482. doi: 10.1007/s00441-006-0265-6. [DOI] [PubMed] [Google Scholar]
- Preuss UW, Zill P, Koller G, Bondy B, Hesselbrock V, Soyka M. Ionotropic glutamate receptor gene GRIK3 SER310ALA functional polymorphism is related to delirium tremens in alcoholics. Pharmacogenomics J. 2006;6:34–41. doi: 10.1038/sj.tpj.6500343. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujescu D, Soyka M, Dahmen N, Preuss U, Hartmann AM, Giegling I, Koller G, Bondy B, Moller HJ, Szegedi A. GRIN1 locus may modify the susceptibility to seizures during alcohol withdrawal. Am J Med Genet B Neuropsychiatr Genet. 2005;133:85–87. doi: 10.1002/ajmg.b.30112. [DOI] [PubMed] [Google Scholar]
- Samochowiec J, Grzywacz A, Kucharska-Mazur J, Samochowiec A, Horodnicki J, Pelka-Wysiecka J, Syrek S. Family-based and case-control association studies of glutamate receptor GRIK3 Ser310Ala polymorphism in Polish patients and families with alcohol dependence. Neurosci Lett. 2006;396:159–162. doi: 10.1016/j.neulet.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Schumann G, Johann M, Frank J, Preuss U, Dahmen N, Laucht M, Rietschel M, Rujescu D, Lourdusamy A, Clarke TK, Krause K, Dyer A, Depner M, Wellek S, Treutlein J, Szegedi A, Giegling I, Cichon S, Blomeyer D, Heinz A, Heath S, Lathrop M, Wodarz N, Soyka M, Spanagel R, Mann K. Systematic analysis of glutamatergic neurotransmission genes in alcohol dependence and adolescent risky drinking behavior. Arch Gen Psychiatry. 2008;65:826–838. doi: 10.1001/archpsyc.65.7.826. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Hartner J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr Opin Neurobiol. 2003;13:279–83. doi: 10.1016/s0959-4388(03)00062-x. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Higuchi M, Sprengel R. RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res Brain Res Rev. 1998;26:217–29. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Williams JBW. Structured Clinical Interview for DSM-III-R Patient Version. Biometric Research Department, New York State Psychiatric Institute; New York: 1995. [Google Scholar]
- Skradski S, White HS. Topiramate blocks kainate-evoked cobalt influx into cultured neurons. Epilepsia. 2000;41(Suppl 1):S45–47. doi: 10.1111/j.1528-1157.2000.tb02171.x. [DOI] [PubMed] [Google Scholar]
- Stein MB, Schork NJ, Gelernter J. A polymorphism of the β1-adrenergic receptor is associated with shyness and low extraversion. Biol Psychiatry. 2004;56:217–224. doi: 10.1016/j.biopsych.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Tadic A, Dahmen N, Szegedi A, Rujescu D, Giegling I, Koller G, Anghelescu I, Fehr C, Klawe C, Preuss UW, Sander T, Toliat MR, Singer P, Bondy B, Soyka M. Polymorphisms in the NMDA subunit 2B are not associated with alcohol dependence and alcohol withdrawal-induced seizures and delirium tremens. Eur Arch Psychiatry Clin Neurosci. 2005;255:129–135. doi: 10.1007/s00406-004-0545-7. [DOI] [PubMed] [Google Scholar]
- Tzschentke TM, Schmidt WJ. Glutamatergic mechanisms in addiction. Mol Psychiatry. 2003;8:373–382. doi: 10.1038/sj.mp.4001269. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, David SP, Niaura R, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernicke C, Samochowiec J, Schmidt LG, Winterer G, Smolka M, Kucharska-Mazur J, Horodnicki J, Gallinat J, Rommelspacher H. Polymorphisms in the N-methyl-D-aspartate receptor 1 and 2B subunits are associated with alcoholism-related traits. Biol Psychiatry. 2003;54:922–928. doi: 10.1016/s0006-3223(03)00072-6. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: Characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol. 2005;28:302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- Yu Z, Schaid DJ. Sequential haplotype scan methods for association analysis. Genet Epidemiol. 2007;31:553–64. doi: 10.1002/gepi.20228. [DOI] [PubMed] [Google Scholar]