Abstract
Arginine methylation of human immunodeficiency virus type 1 (HIV-1) Tat protein downregulates its key function in viral-gene transactivation. The fate of methylated Tat is unknown, so it is unclear whether methylated Tat is degraded or persists in the cell for additional functions. Here we show that the arginine methyltransferase PRMT6 increases Tat protein half-life by 4.7-fold. Tat stabilization depends on the catalytic activity of PRMT6 and requires arginine methylation within the Tat basic domain. In contrast, HIV-1 Rev, which is also methylated by PRMT6, is completely refractory to the stabilizing effect. Proteasome inhibition and silencing experiments demonstrated that Tat can be degraded by a REGγ-independent proteasome, against which PRMT6 appears to act to increase Tat half-life. Our data reveal a proteasome-dependent Tat degradation pathway that is inhibited by arginine methylation. The stabilizing action of PRMT6 could allow Tat to persist within the cell and the extracellular environment and thereby enable functions implicated in AIDS-related cancer, neurodegeneration, and T-cell death.
Protein arginine methylation is a posttranslational modification involved in regulating protein-protein and protein-nucleic acid interactions. It results in a plethora of effects during key cellular events, including transcriptional regulation (18, 37), T-cell activation (4), differentiation (53), cytokine signaling (42), and DNA repair (3). The protein arginine methyltransferase (PRMT) family of enzymes has at least 11 members, all of which catalytically transfer methyl groups from S-adenosyl-l-methionine to the guanidino groups of arginine (21). Either one or two methyl groups may be transferred to a single arginine residue in three distinct conformations: monomethylarginine (generated by type I and II PRMTs), asymmetric dimethylarginine (type I PRMTs), and symmetric dimethylarginine (type II PRMTs). Previously thought to be irreversible, emerging data now suggest that arginine methylation may be a reversible posttranslational modification (8).
Following infection, human immunodeficiency virus type 1 (HIV-1) reverse transcribes its RNA genome to DNA and integrates into a host cell's chromosome. Efficient gene expression from the integrated provirus requires Tat-dependent recruitment of the positive transcription elongation factor b (P-TEFb) complex, composed of cyclin T1 and CDK9 (56), as well as recruitment of chromatin-remodeling factors during transcriptional elongation (1, 40). Cofactor recruitment is enhanced by posttranslational modifications of Tat, including acetylation by PCAF, p300, and hGCN5 (13, 32); deacetylation by SIRT1 (46); and ubiquitination by MDM2 (7). It has recently been discovered that Tat transactivation function can be downmodulated by methylation of Tat arginine residues by PRMT6 (5, 58) and lysine residues by SETDB1 and SETDB2 (55). This enhancement and suppression of transactivation lend themselves to a hypothesis that posttranslational modifications regulate Tat in a “transactivation cycle” (24).
PRMT6 is a type I methyltransferase involved in regulating base excision repair (14), suppressing the HMGA1a architectural transcription factor (51), and controlling transcription by methylating histone H3 (22, 26). Arginine methylation by PRMT6 was recently shown to have a negative impact on the activities of HIV-1 Tat, Rev, and nucleocapsid proteins (5, 27, 28). PRMT6 methylates Tat at arginine residues 52 and 53 and consequently disrupts the Tat-TAR-cyclin T1 complex required for transactivation (58), presumably by inhibiting interactions between R52/R53 and the bulge region of TAR. It is therefore thought that PRMT6 is a negative regulator of Tat function, but the fate and functions, if any, of methylated Tat are unknown. Here we found that overexpression of PRMT6 led to a significant increase in Tat stability in a manner requiring PRMT6 catalytic activity. The increase in stability was due to prevention of Tat degradation by a proteasome. Our data raise the possibility that PRMT6 generates a distinct subset of stable Tat molecules able to perform functions other than Tat's established role in HIV-1 gene expression.
MATERIALS AND METHODS
Cell culture and transfections.
HeLa and HEK293T cells were cultured in RPMI 1640 medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and either 10% (vol/vol) fetal bovine serum or 10% (vol/vol) newborn bovine serum (Invitrogen Corporation). The cells were incubated at 37°C in air containing 5% CO2. Plasmid transfections were performed with Lipofectamine 2000 (Invitrogen) or FuGene 6 (Roche Diagnostics Corp.) transfection reagent. Transfections typically involved 1.5 to 2 μg of individual plasmid per 6-cm dish of cells at 80% confluence.
Plasmids.
The plasmid expressing Tat (BH10 clone; 101 amino acids) fused to the FLAG tag (pcDNA3.1/Tat-FLAG) was a gift from Monsef Benkirane, Institut de Génétique Humaine, France. The R52K R53K mutant (pcDNA3.1/Tat-R52/53K-FLAG) was derived from pcDNA3.1/Tat-FLAG by inverse PCR using primers 5′-AGAAACAGCGACGAAGACC and 5′-TCTTCTTCCTGCCATAGG. The plasmids expressing fusions between the MYC tag and wild-type PRMT6 (pMyc-PRMT6) or methyltransferase-deficient mutant PRMT6 (pMyc-PRMT6mut) were gifts from Stéphane Richard, McGill University, Canada. The mutant contains V86K and D88A amino acid mutations compared to wild-type PRMT6 (GenBank accession no. BC073866). The plasmid expressing a fusion between glutathione S-transferase (GST) and PRMT6, pGEX-6P-PRMT6, was a gift from Mark Bedford, M. D. Anderson Cancer Center, TX. The plasmid expressing a fusion between GST and GAR, pGEX-2T-GAR, was a gift from Steve Clarke, UCLA, Los Angeles, CA (19).
The plasmids expressing Tat-FLAG localization mutants were created from pcDNA3.1/Tat-FLAG either by removing the Tat basic domain by inverse PCR and ligating double-stranded oligonucleotides encoding Semliki Forest virus (SFV) capsid nuclear localization signal (NLS) P2 (17) (sense strand, 5′-AAGAAGAAAGACAAGCAAGCCGACAAGAAGAAGAAGAAA) or by inserting double-stranded oligonucleotides encoding RevNES (52) (sense strand, 5′-CCTCTTCAGCTACCACCGCTTGAGAGACTTACTCTTGAT) between the Tat second exon and the FLAG tag. The plasmids expressing hexahistidine (His6)-tagged Tat mutants were cloned from the respective pcDNA3.1 constructs into pDONR201 and then into pDEST17 using the Gateway cloning system (Invitrogen).
The HIV-1 proviral expression vector pGCH will be fully described elsewhere (L. Meredith, C. Ducloux, C. Isel, R. Marquet, and D. Harrich, unpublished data). The FLAG tag sequence was inserted at the 5′ end of the tat open reading frame via inverse PCR using primers 5′-GACGATGACAAGGAGCCAGTAGATCCTAGA and 5′-GTCCTTATAATCCATTTCTTGCTCTCCTCTG. The β-galactosidase expression plasmid pCMVβ (Clontech Laboratories) was used as a transfection control for Western blot experiments.
Western blotting.
Transfected HeLa cells were harvested 24 h posttransfection with phosphate-buffered saline containing 0.5 mM EDTA. The cells were washed in phosphate-buffered saline and lysed in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail [Roche], 1% [wt/vol] Triton X-100). The lysates were centrifuged at 1,500 × g to pellet debris, and the supernatants were collected. Total protein concentrations were determined by the Bradford method (6), and if applicable, β-galactosidase activity was measured by a chlorophenol red-β-d-galactopyranoside-based assay (16). The lysates were boiled in Laemmli sample buffer (33) and electrophoresed in a denaturating polyacrylamide gel (sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) under reducing conditions. Proteins were electroblotted to a polyvinylidene fluoride membrane (GE Healthcare) using a semidry transfer system (Bio-Rad Laboratories Inc.).
Tat-FLAG was detected with mouse anti-FLAG M2 antibody (Sigma-Aldrich Inc.). Myc-PRMT6, its mutant, and Myc-Rev were detected with mouse anti-MYC clone 9E10 antibody (Millipore Corp.). Endogenous PRMT6 was detected with rabbit anti-PRMT6 antibody (LifeSpan Biosciences Inc.). HIV was detected with HIV-IG antiserum (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH). β-Tubulin was detected with mouse anti-β-tubulin clone 2-28-33 antibody (Sigma-Aldrich). His6-ubiquitin (Ub)-conjugated proteins were detected with mouse Penta-His antibody (Qiagen). Endogenous CDK9 was detected with rabbit anti-CDK9 clone C12F7 antibody (Cell Signaling Technology). Endogenous IκBα was detected with rabbit anti-phospho-IκBα (Ser32) clone 14D4 antibody (Cell Signaling). Endogenous REGγ was detected with rabbit anti-PA28γ antibody (Cell Signaling). Mouse primary antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Invitrogen), and rabbit antibodies were detected with HRP-conjugated goat anti-rabbit antibody (Invitrogen). HIV antiserum was detected with HRP-conjugated goat anti-human immunoglobulin G antiserum (Sigma-Aldrich). Bands were visualized with SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology).
RNA interference.
HeLa cells in 6-cm dishes were transfected with small interfering RNA (siRNA) against PRMT6 (target mRNA sense sequence, 5′-CAAGUGGAGCAAGACACGGACGUUU) or a percentage GC-matched negative control (Invitrogen). Typically, 300 pmol of siRNA was transfected using 60 μl/nmol of GenePorter 2 transfection reagent (Genlantis Inc.) for 24 h. The cells were then transfected again with 0.5 μg of pCMVβ and 2 μg of either pcDNA3.1/Tat-FLAG or pcDNA3.1 using 3.5 μl/μg of GenePorter 2. The dishes were supplemented with newborn bovine serum at a 10% (vol/vol) final concentration after 5 h and incubated for a further 19 h until harvest. Western blotting was performed on lysates normalized for total protein (for endogenous PRMT6 and β-tubulin expression) or normalized for β-galactosidase activity (for Tat-FLAG expression).
Microscopy.
HeLa cells growing on coverslips were transfected with pcDNA3.1/Tat-FLAG localization mutants and allowed to express for 24 h. The cells were fixed with 3% (wt/vol) paraformaldehyde, quenched with 50 mM NH4Cl, permeabilized with 0.1% (wt/vol) Triton X-100, and blocked with 10% (vol/vol) normal goat serum (Millipore Corp.). Tat-FLAG mutants were visualized with mouse anti-FLAG M2 monoclonal (Sigma-Aldrich) and fluorescein isothiocyanate-conjugated goat anti-mouse polyclonal (Invitrogen) antibodies. Nuclei were stained with 1 μM DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen) and mounted on slides with SlowFade Gold mounting medium (Invitrogen). Images were acquired with a Leica TCS SP2 confocal system (Leica Microsystems) using an oil immersion 63× objective lens. Images were uniformly postprocessed using The GIMP software (version 2.2), within which nuclei were traced using the “Threshold,” “Despeckle,” and “Edge-Detect” functions.
Recombinant proteins and in vitro methylation.
Recombinant His6-tagged wild-type Tat, Tat-R52/53K, and SFV-NLS mutant Tat were isolated from Escherichia coli as previously described (2). E. coli BL21(DE3) pLysS transformed with the GST-PRMT6 or GST-GAR plasmid was induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h before GST-PRMT6 or GST-GAR was isolated using the MagneGST protein purification system (Promega Corp.) according to the manufacturer's instructions. GST-PRMT6 was eluted in a solution containing 50 mM glutathione, 50 mM Tris-HCl (pH 8.1), 120 mM NaCl, and 2% (vol/vol) glycerol. In vitro methylation reactions between GST-PRMT6 and His6-Tat mutants or GST-GAR were performed as previously described (19), but using 50.8 kBq of [3H]AdoMet (Perkin-Elmer) per 30-μl reaction mixture incubated for 1 h. The reaction mixtures were then boiled in Laemmli sample buffer for SDS-PAGE. The gel was fixed in Coomassie G250 (Bio-Rad), incubated with Amplify fluorographic reagent (GE Healthcare), dried, and exposed to Hyperfilm MP film (GE Healthcare) at −80°C for 22 days.
Protein translation arrest.
HeLa cells cotransfected with pcDNA3.1/Tat-FLAG and either pMyc-PRMT6 or empty vector (pcDNA3.1) were treated 24 h posttransfection with medium containing 80 μg/ml cycloheximide (CHX) (Sigma-Aldrich) or vehicle (dimethyl sulfoxide [DMSO]). Cells were harvested at 0, 2, 4, and 6 h posttreatment, and Western blotting was performed on lysates equalized for total protein amounts. Band (pixel) intensities were quantified using ImageJ (version 1.39) and corrected for background levels. The mean natural log values of band intensities (from two independent experiments) were plotted against time. Tat-FLAG half-lives were determined mathematically when values were fitted to the linear-transformed exponential-decay formula: ln I = ln I0 − λt, where I is the Tat-FLAG band intensity at time t, I0 is the initial Tat-FLAG band intensity at 0 h, and λ is the decay constant. λ is equivalent to the slope of the curve, and the half-life is equal to ln 2/λ.
Ubiquitination assay and proteasome inhibition.
The ubiquitination assay has been described elsewhere (47). The His6-Ub expression plasmid was a gift from Dirk Bohmann, University of Rochester Medical Center, New York, NY. For the proteasome inhibition experiment, transfected HeLa cells were treated 24 h posttransfection with combinations of 80 μg/ml CHX, 20 μM MG-132 (Merck Chemicals Ltd.), and DMSO as the vehicle control. Cells were harvested 3 h later, and Western blotting was performed on lysates equalized for total protein.
Lentiviruses and a REGγ-silenced cell line.
To generate the lentivector expressing a synthetic microRNA (miRNA) against PSME3, the luc miRNA in pcDNA6.2-GW/EmGFP-miR-luc (Invitrogen) was first removed by inverse PCR using primers 5′-GCTGAATAGTTAACACAAGGCCTGT and 5′-AATAGCTAGCACAGCCTTCAGCAAG. An artificial REGγ pre-miRNA targeting the PSME3 mRNA sense sequence, 5′-ACAUCCAUGACCUAACUCAGA (12), was generated by annealing synthetic oligonucleotides and ligating them into pcDNA6.2-GW/EmGFP-miR-(luc−). The emerald green fluorescent protein (EmGFP)-miR-REGγ cassette was amplified by PCR using primers 5′-AGCAGTGATCAAAACCATGGTG and 5′-AGATATCTCGAGTGCGGC and was then subcloned to replace the GFP gene in pLOX-CWgfp (49) (a gift from Andreas Suhrbier, Queensland Institute of Medical Research, Australia) using BamHI and XhoI restriction enzymes. This generated the lentivector pLOX-CW/EmGFP-miR-REGγ. The negative control lentivector (pLOX-CW/EmGFP-miR-neg) was similarly derived from pcDNA6.2-GW/EmGFP-miR-neg (Invitrogen) and targets the sequence 5′-GUCUCCACGCGCAGUACAUUU.
Lentiviruses were produced by transfecting HEK293T cells with the lentivectors, pCMVΔR8.91 and pCMV-VSV-G plasmids. Filtered culture supernatants containing lentiviruses were allowed to transduce HeLa cells in the presence of 8 μg/ml hexadimethrine bromide for 48 h, and transduction was confirmed by inspecting cells with a fluorescence microscope. The cells were fluorescently sorted 2 weeks posttransduction using a MoFlo cell sorter (Beckman Coulter Inc.), thereby producing the polyclonal cell lines named HEmiR-REGγ and HEmiR-neg. REGγ silencing was confirmed by Western blotting, and expression of the miRNAs was confirmed by fluorescence microscopy, since EmGFP fluorescence correlates strongly with miRNA expression.
RESULTS
PRMT6 increases the steady-state level of Tat in the cell.
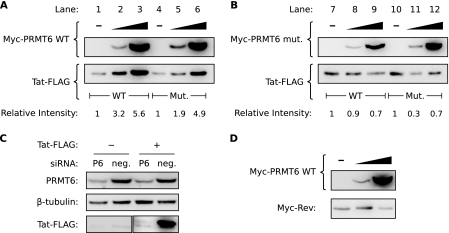
Certain PRMTs are known to alter the stabilities of their substrate proteins (10, 43). To test if PRMT6 altered the steady-state level of Tat, we cotransfected HeLa cells to overexpress PRMT6 with either wild-type Tat or a mutant of Tat in which the known PRMT6 methylation sites (arginines 52 and 53) had been mutated to lysines. Both forms of Tat were expressed as Tat-FLAG tag fusions, and PRMT6 was expressed as a Myc tag-PRMT6 fusion. A β-galactosidase expression plasmid was included in all transfections to account for variations in transfection efficiency and nonspecific effects on expression (since all plasmids express their proteins with the cytomegalovirus [CMV] promoter). When Western blotting was performed on cell lysates with equivalent β-galactosidase activities, we observed that Myc-PRMT6 induced a dose-dependent increase in the steady-state level of wild-type Tat-FLAG (Fig. 1A, lanes 1 to 3) in each of six independent experiments. The effect was also observed with mutant Tat-FLAG (lanes 4 to 6), suggesting that mutation of arginines 52 and 53 is not sufficient to block the stabilizing effect of PRMT6. To determine if the methyltransferase activity of PRMT6 is necessary for this phenomenon, we repeated the experiment with a mutant of Myc-PRMT6 (Myc-PRMT6mut) described as catalytically inactive (5, 58). Overexpression of Myc-PRMT6mut did not alter the steady-state levels of either wild-type or mutant Tat-FLAG (Fig. 1B). The data therefore show that PRMT6 can increase the steady-state level of Tat in a manner requiring PRMT6 methyltransferase activity but not requiring the reported arginine methylation sites in Tat (58).
FIG. 1.
Overexpression of catalytically active PRMT6 increases the steady-state level of Tat. (A) HeLa cells were transfected to express wild-type (WT) Tat-FLAG alone (lane 1), wild-type Tat-FLAG with increasing amounts of Myc-PRMT6 (lanes 2 and 3), mutant (Mut.) Tat-R52/53K-FLAG alone (lane 4), or mutant Tat-R52/53K-FLAG with increasing amounts of Myc-PRMT6 (lanes 5 and 6). All samples were cotransfected with a β-galactosidase expression vector, and Western blotting was performed with lysates equalized for β-galactosidase activity. The Tat-FLAG bands were quantified and are expressed relative to wild-type Tat-FLAG alone (lanes 1 to 3) or mutant Tat-R52/53K-FLAG alone (lanes 4 to 6). The blots are representative of six independent experiments. (B) Similar experiments were performed as for panel A but with a catalytically inactive version of Myc-PRMT6 (Myc-PRMT6mut). (C) HeLa cells were transfected with siRNA targeting PRMT6 (P6) or a nontargeting negative control (neg.) and then transfected 24 h later with plasmids expressing wild-type Tat-FLAG (+) or an empty-vector control (−). Cell lysates were Western blotted to confirm PRMT6 knockdown and to assess the effect on Tat steady-state levels. (D) HeLa cells transfected to express Myc-Rev alone (−) or with increasing amounts of wild-type Myc-PRMT6 were Western blotted to determine the effect on Myc-Rev steady-state levels. All samples were cotransfected with β-galactosidase vector, and the lysates were normalized for β-galactosidase activity. Panels B to D are representative of two or three independent experiments.
We next performed RNA interference to test if knockdown of endogenous PRMT6 expression can alter Tat steady-state levels. HeLa cells were initially transfected with a PRMT6 siRNA or a control siRNA and then transfected 24 h later with Tat-FLAG or empty-vector plasmids (along with β-galactosidase plasmid). When we performed Western blotting of cell lysates with equivalent β-galactosidase activities, we observed that silencing endogenous PRMT6 expression induced a marked decrease in Tat-FLAG levels compared to control siRNA (Fig. 1C). These results confirm that PRMT6 affects the steady-state level of Tat in cells.
Other HIV-1 proteins can also be methylated by PRMT6 (27, 28). We next investigated if overexpression of PRMT6 could alter the steady-state level of HIV-1 Rev. Rev shares several characteristics with Tat, in addition to being directly methylated by PRMT6, such as being translated from multiply spliced mRNA and being a small nucleocytoplasmic protein. We cotransfected HeLa cells to overexpress a Myc tag-Rev fusion protein, along with wild-type Myc-PRMT6 (the β-galactosidase control plasmid was included). Western blotting of β-galactosidase-equalized cell lysates revealed no increase in the Myc-Rev steady-state level with increasing amounts of Myc-PRMT6 (Fig. 1D). Thus, PRMT6 increases the steady-state level of Tat, but not Rev.
The basic domain of Tat is required for PRMT6 to increase Tat steady-state levels.
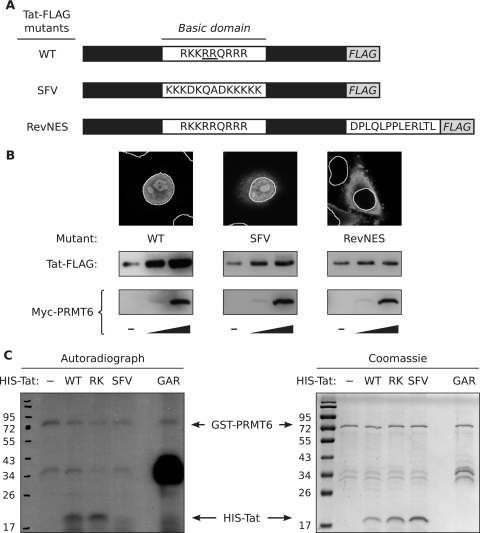
The observation that PRMT6 could increase the steady-state level of the R52/53K Tat mutant (Fig. 1A) suggested that direct Tat methylation may not be required for this effect or that PRMT6 can methylate Tat at other arginine residues. Previous in vitro investigations showed that PRMT6 methylates Tat only within its arginine-rich basic domain (5). We thus created a Tat-FLAG mutant in which the Tat basic domain, which contains both the R52-R53 sequence and an NLS, was replaced with the NLS from SFV capsid protein (17) to determine if the entire basic domain is required for PRMT6 to increase the Tat steady-state level (Fig. 2A). Of note, the SFV NLS is devoid of arginine residues. As a control, we included a Tat-FLAG mutant in which the Tat basic domain was kept intact and a nuclear export signal (NES) from HIV-1 Rev (52) was fused to the carboxyl terminus (RevNES) (Fig. 2A). Confocal microscopy of transfected HeLa cells revealed that the SFV mutant localized primarily to the nucleoplasm and nucleolus, like wild-type Tat-FLAG, but also showed some cytoplasmic accumulation (Fig. 2B). In contrast, the RevNES mutant was exclusively cytoplasmic.
FIG. 2.
PRMT6-mediated stabilization of Tat requires the Tat basic domain and nuclear localization. (A) Schematic diagrams, not drawn to scale, of wild-type Tat-FLAG (WT) and two domain substitution mutants. In SFV, the Tat basic domain, which contains an NLS and the target arginines of PRMT6-mediated methylation (underscored), was replaced with the NLS from SFV capsid protein. In RevNES, the Tat basic domain was left intact but the NES from HIV-1 Rev was fused to the carboxyl terminus of Tat. (B) (Top) Indirect immunofluorescence confocal microscopy was performed on HeLa cells expressing the Tat-FLAG mutants to determine subcellular localization. Wild-type and mutant Tat-FLAG were detected with anti-FLAG-fluorescein isothiocyanate antibodies (gray). The nuclei were stained with DAPI and are outlined with solid white lines. The micrographs are representative of 12 frames from two independent experiments. (Bottom) Western blots of HeLa cells expressing wild-type or mutant Tat-FLAG alone (−) or coexpressing increasing amounts of Myc-PRMT6 (wedges). All samples were cotransfected with a β-galactosidase expression vector, and Western blotting was performed with lysates equalized for β-galactosidase activity. All panels are representative of at least three independent experiments. (C) In vitro methylation reactions between GST-PRMT6 and His6-tagged wild-type Tat (WT), a His6-Tat-R52/53K mutant (RK), and a His6-Tat-SFV mutant (SFV), with [3H]AdoMet as a methyl donor, were analyzed by SDS-PAGE and autoradiography. GST-GAR (GAR) was used as a control methyltransferase substrate. Note that GST-PRMT6 can automethylate (19). A 3-week autoradiograph is shown on the left, and a corresponding total-protein gel, visualized with Coomassie G250, is shown on the right. Molecular masses (in kDa) are shown on the left of both images. The data are representative of two independent experiments.
HeLa cells were next cotransfected to express the Tat-FLAG mutants with increasing amounts of Myc-PRMT6 to assess the effects of the mutations on the PRMT6-mediated stabilization of Tat. In sharp contrast to wild-type Tat-FLAG, the SFV mutant showed no significant dose-dependent stabilization with increasing Myc-PRMT6 expression (Fig. 2B). The steady-state levels of the Tat-FLAG RevNES mutant similarly did not change with increasing Myc-PRMT6 despite the presence of an intact Tat basic domain (Fig. 2B). Taking the data together, we conclude that PRMT6-mediated stabilization of Tat requires the presence of an arginine-rich basic domain in Tat and concomitant nuclear localization.
To determine if the Tat-R52/53K and Tat-SFV mutants are directly methylated by PRMT6, we performed in vitro methylation experiments with recombinant GST-PRMT6 and His6-tagged Tat proteins purified from E. coli. GST-GAR was used as a control substrate for methylation. GAR is a 148-amino-acid human fibrillarin-derived peptide that contains glycine- and arginine-rich motifs robustly methylated by several arginine methyltransferases, including PRMT6 (44). Analysis of the methylation reactions showed that both wild-type Tat and the R52/53K mutant were methylated by PRMT6, whereas the SFV Tat mutant was not (Fig. 2C). The differential methylation of wild-type, R52/53K, and SFV Tat correlates with their different stabilization patterns (Fig. 1A and 2B), supporting the notion that PRMT6-mediated stabilization of Tat requires direct arginine methylation.
PRMT6 increases the steady-state level of provirus-expressed Tat.
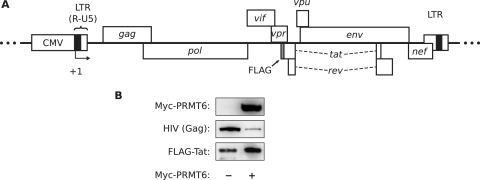
To confirm the effect of PRMT6 on Tat in the presence of other viral proteins, we coexpressed Myc-PRMT6 with the infectious proviral vector pGCH-FLAG-Tat. The vector expresses HIV-1 with a FLAG-Tat fusion in situ without requiring Tat transactivation activity, since proviral-gene expression uses a CMV promoter in place of the HIV-1 long terminal repeat promoter (Fig. 3A). HeLa cells were cotransfected with Myc-PRMT6 and pGCH-FLAG-Tat plasmids, along with the β-galactosidase plasmid to correct for effects on the CMV promoter. Western blotting of β-galactosidase-equalized cell lysates showed an increase in FLAG-Tat steady-state levels in the presence of Myc-PRMT6 compared to FLAG-Tat levels in its absence (Fig. 3B). We also observed a decrease in Gag expression in the Myc-PRMT6 sample, likely caused by downregulation of gag mRNA export as a result of previously described effects of Myc-PRMT6 on Rev function (28). The data therefore indicate that PRMT6 can increase the steady-state level of Tat when expressed by HIV-1.
FIG. 3.
Overexpression of PRMT6 increases the steady-state level of HIV-1 provirus-expressed Tat. (A) Schematic diagram of the HIV-1 provirus in the pGCH-FLAG-Tat expression vector. The promoter is a fusion between the CMV immediate-early promoter and the R and U5 regions of the HIV-1 long terminal repeat (LTR). The CMV promoter enables high-level Tat-independent proviral-gene expression. The FLAG tag sequence (gray box) was cloned onto the 5′ end of the tat open reading frame. +1, start of proviral transcription. (B) Western blot of HeLa cells expressing pGCH-FLAG-Tat alone (−) or coexpressing Myc-PRMT6 (+). Samples were cotransfected with β-galactosidase vector, and the lysates were normalized for β-galactosidase activity. HIV antiserum was used to detect provirus expression with the 55-kDa Gag band shown.
PRMT6 increases the half-life of Tat.
An increase in the steady-state level of a protein may indicate an increase in its stability. We performed translation arrest experiments with CHX to see if PRMT6 increases Tat protein stability. CHX treatment has been used to determine the half-lives of several proteins, including Tat (20, 29, 31, 41). HeLa cells transfected to express Tat-FLAG alone or with Myc-PRMT6 were treated with CHX and harvested in a time course. Western blotting was then performed on the cell lysates normalized for total protein amounts. Figure 4A shows that Tat-FLAG levels were sustained in the presence of Myc-PRMT6 but that Tat-FLAG was degraded more quickly in the absence of Myc-PRMT6. The natural log values of quantified Tat-FLAG band intensities were plotted against time to calculate protein half-lives (Fig. 4B). The half-life of Tat-FLAG alone was 2.6 h, in fair agreement with the results of Gargano and colleagues (20). In the presence of Myc-PRMT6, the half-life of Tat-FLAG increased 4.7-fold to 12.1 h. Protein-radiolabeling pulse-chase experiments confirmed these results and determined that CHX did not artificially affect Tat-FLAG stability (data not shown). Overall, these results demonstrate that PRMT6 increases Tat steady-state levels by increasing Tat protein stability.
FIG. 4.
Overexpression of PRMT6 increases the half-life of Tat. (A) HeLa cells expressing Tat-FLAG with or without Myc-PRMT6 were treated with CHX and harvested at 0, 2, 4, and 6 h posttreatment. Western blotting was performed on the total-protein-equalized lysates. The β-tubulin levels show equivalent sample loadings. (B) The Tat-FLAG band intensities in panel A were quantified, and their natural log values were plotted as a function of time. Values for Tat-FLAG with Myc-PRMT6 are indicated by black boxes and values for Tat-FLAG alone by white boxes. The calculated Tat-FLAG half-lives are shown. The data points represent the means and standard deviations of two independent experiments.
PRMT6 prevents degradation of Tat by a proteasome.
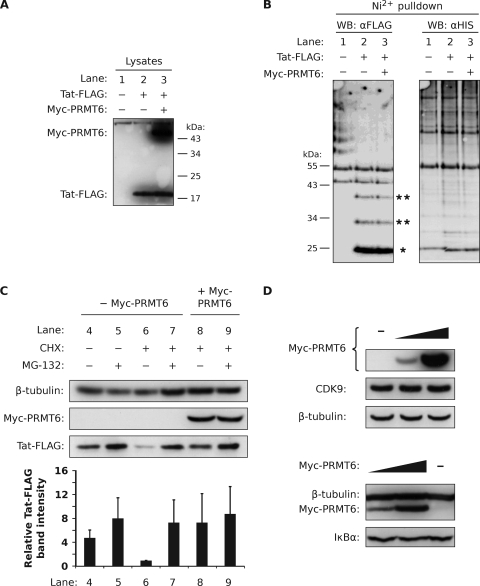
Protein stability is often regulated by the ubiquitin-proteasome system, in which the polyubiquitination of a protein marks the protein for degradation by a proteasome. We assessed if PRMT6 increases Tat stability by suppressing Tat polyubiquitination in a standard ubiquitination assay. HEK293T cells (chosen for their ability to express transfected proteins at very high levels) were cotransfected with Tat-FLAG and Myc-PRMT6 plasmids, along with a plasmid expressing His6-Ub. Cell lysates with equivalent Tat-FLAG protein levels (Fig. 5A) were loaded onto nickel-nitrilotriacetic acid agarose beads in order to capture proteins ubiquitinated with the His6-Ub moieties. Western blotting was performed using the anti-FLAG antibody to detect ubiquitinated Tat.
FIG. 5.
Tat is not polyubiquitinated but is degraded by a proteasome. (A) HEK293T cells were transfected with a plasmid expressing His6-Ub alone (lane 1), with Tat-FLAG (lane 2), or with Tat-FLAG and Myc-PRMT6 plasmids (lane 3). Nickel-coated nitrilotriacetic acid-agarose beads were added to lysates equalized for Tat-FLAG protein levels to pull down ubiquitinated conjugates via the His6 tag. (B) Bead eluates were subjected to Western blotting (WB) using the anti-FLAG (αFLAG) antibody (left blot) and the anti-His6 (αHIS) antibody (right blot). The locations of monoubiquitinated Tat-FLAG (single asterisk) and multiply ubiquitinated Tat-FLAG (double asterisks) are indicated. (C) HeLa cells expressing Tat-FLAG with (+) or without (−) Myc-PRMT6 were treated for 3 h with combinations of CHX, the proteasome inhibitor MG-132, or vehicle (DMSO). The cell lysates were then analyzed by Western blotting. The Tat-FLAG bands were quantified and are expressed relative to lane 6, where columns represent mean band intensities and error bars indicate standard deviations. (D) HeLa cells expressing increasing amounts of Myc-PRMT6 were Western blotted for endogenous CDK9 expression (upper blots) and endogenous IκBα expression (lower blots). The β-tubulin levels show equivalent sample loadings. All panels are representative of two or three independent experiments.
Figure 5B, lane 2, demonstrates that only three ubiquitinated forms of Tat were pulled down. Monoubiquitinated Tat-FLAG was the major form detected, as previously observed by Brès and coworkers (7). The two additional forms, presumably di- and triubiquitinated Tat-FLAG, were observed at lower intensities. Importantly, no other Ub concatemers were present; the two higher bands in lane 2 are nonspecific, as seen in lane 1 (His6-Ub only). Coexpression of Myc-PRMT6 did not alter the Tat ubiquitination pattern (Fig. 5B, lane 3). When we probed the blot with anti-His6 antibody, we confirmed that ubiquitinated proteins were being efficiently captured (Fig. 5B, anti-His6 Western blot). The same results were observed when HeLa cells were used, and Tat-FLAG was not pulled down in the absence of His6-Ub (data not shown). We infer from the data that Tat is not polyubiquitinated but is probably multiply ubiquitinated with only a few Ub monomers.
Tat may still be subject to proteasomal degradation (20) despite its lack of polyubiquitination. To test this hypothesis, HeLa cells expressing Tat-FLAG were treated with the proteasome inhibitor MG-132. Treatment for 3 h resulted in a slight increase (1.7-fold) of Tat-FLAG signal compared to vehicle-treated controls as assessed by Western blotting (Fig. 5C, lanes 4 and 5), consistent with other studies (7, 20, 30). However, constitutive plasmid expression may mask any significant rescue of Tat-FLAG by the proteasome inhibitor. Thus, we repeated the experiment but treated the cells with CHX in order to stop translation and to create a finite pool of Tat-FLAG in the cell. Compared to vehicle controls, CHX greatly reduced the Tat-FLAG signal after the 3-h treatment period (4.8-fold) (Fig. 5C, lane 6), confirming efficient translation arrest. Significantly, simultaneous treatment of cells with CHX and MG-132 (lane 7) rescued Tat-FLAG protein levels compared to treatment with CHX alone (a 7.3-fold increase compared to lane 6), clearly demonstrating that proteasome activity is required for Tat-FLAG degradation. Coexpression of Myc-PRMT6 in CHX-arrested cells also rescued Tat-FLAG levels (7.4-fold rescue; lane 8 versus lane 6). Furthermore, no additive increase of Tat-FLAG levels was observed in CHX-arrested cells when Myc-PRMT6 coexpression and MG-132 treatment were combined (compare lanes 7, 8, and 9). This may indicate that Myc-PRMT6 and MG-132 share a common pathway to stabilize Tat-FLAG. PRMT6 therefore prevents Tat degradation by a proteasome-dependent pathway.
Myc-PRMT6 may prevent Tat-FLAG degradation by directly inhibiting proteasome activity. To test this possibility, we transfected HeLa cells with Myc-PRMT6 plasmid and assessed endogenous CDK9 levels by Western blotting. CDK9 is known to be degraded by a proteasome presumably located in the nucleus, since CDK9 degradation requires binding to the exclusively nuclear cyclin T1 (31, 45). Western blotting of total-protein-normalized samples revealed no change in CDK9 steady-state levels with increasing amounts of Myc-PRMT6 (Fig. 5D, upper blots). We similarly observed no change in the levels of IκBα, a protein degraded by a cytoplasmic proteasome (48), in Myc-PRMT6-transfected cells (Fig. 5D, lower blots). The data therefore suggest that PRMT6 does not directly inhibit proteasome activity.
REGγ is not involved in the PRMT6-sensitive Tat degradation pathway.
In an attempt to characterize the Tat degradation pathway, we investigated whether REGγ (also known as PA28γ and PSME3) is required for Tat degradation. Heptameric REGγ forms a regulatory cap to facilitate polyubiquitin-independent protein degradation by the REGγ proteasome in the nucleus (9, 38, 39), thereby making REGγ a candidate cofactor for Tat instability. We created a REGγ-silenced polyclonal cell line by transducing HeLa cells with lentivirus containing a CMV promoter-driven expression cassette comprised of the EmGFP gene and a synthetic miRNA targeting REGγ (or a nontargeting negative control miRNA). EmGFP and the synthetic miRNA are expressed as a single transcript, thus tightly associating fluorescence with REGγ silencing. Highly fluorescent integrants were isolated by fluorescence-activated cell sorting to establish the polyclonal cell lines. The cell lines were termed HEmiR-REGγ and HEmiR-neg, for cells expressing the REGγ miRNA and negative control miRNA, respectively.
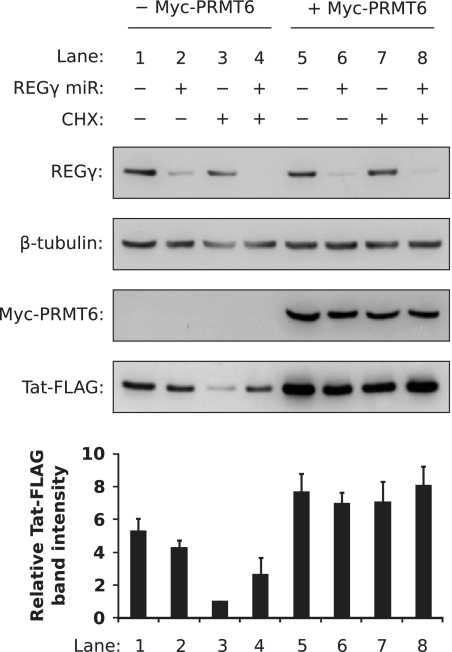
Using these cell lines, we determined if silencing REGγ alters the stabilizing effect of PRMT6 on Tat. The cell lines were transfected to express Tat-FLAG alone or Tat-FLAG with Myc-PRMT6 (along with β-galactosidase for transfection control) and treated 24 h posttransfection with CHX or vehicle for 3 h. Western blotting of β-galactosidase-normalized lysates revealed that CHX treatment decreased Tat-FLAG steady-state levels in control cells by 5.3-fold compared to vehicle treatment (Fig. 6, lanes 1 and 3) but decreased Tat-FLAG levels by only 1.6-fold in REGγ-silenced cells (lanes 2 and 4). This suggests that REGγ may play a role in Tat degradation, since depletion of Tat-FLAG in translation-arrested cells was slower when REGγ was silenced. In cells coexpressing Myc-PRMT6, in contrast, CHX treatment had minimal effects on Tat-FLAG levels in control cells (a 1.1-fold difference compared to vehicle) (Fig. 6, lanes 5 and 7) and REGγ-silenced cells (a 1.2-fold difference) (Fig. 6, lanes 6 and 8). This indicates that Myc-PRMT6 equally increased Tat-FLAG levels in both translation-arrested cell lines. Furthermore, Myc-PRMT6 increased the steady-state levels of Tat-FLAG by 3.1-fold in the REGγ-silenced cell line (Fig. 6, lanes 4 and 8). Taken together, the data imply that Myc-PRMT6 prevents Tat degradation from a proteasomal pathway that does not involve REGγ.
FIG. 6.
Silencing REGγ expression does not affect the ability of PRMT6 to stabilize Tat. HeLa-derived cell lines stably expressing miRNA against PSME3 (REGγ miR +) or a nontargeting negative control miRNA (REGγ miR −) were transfected to express Tat-FLAG with or without Myc-PRMT6. All samples were cotransfected with β-galactosidase vector. The cells were treated with CHX (CHX +) or vehicle (CHX −) for 3 h before Western blotting was performed on β-galactosidase-equalized lysates (for Myc-PRMT6 and Tat-FLAG blots) or total-protein-equalized lysates (for REGγ and β-tubulin blots). The Tat-FLAG bands were quantified and are expressed relative to lane 3, where columns represent mean band intensities and error bars indicate standard deviations. The data are representative of three independent experiments.
DISCUSSION
Methylation of HIV-1 Tat at arginine (5, 58) and lysine (55) residues diminishes Tat function in transactivation. In contrast, the other known posttranslational modifications of Tat (including acetylation, deacetylation, ubiquitination, and phosphorylation [24]) enhance transactivation. The precise role of Tat methylation is currently unclear; it may mark Tat for degradation and thus represent a form of innate cellular immunity (58), or it may act to counterbalance stimulated transcription and thereby function in a Tat transactivation cycle (24, 55). We provide evidence in this study that PRMT6, an arginine methyltransferase that directly methylates HIV-1 Tat, does not promote Tat degradation but instead increases Tat stability within cells by 4.7-fold (Fig. 4). Stabilization requires the catalytic activity of PRMT6 (Fig. 1B), the Tat basic domain, and Tat nuclear localization (Fig. 2B and C). We observed a lack of polyubiquitination of Tat (Fig. 5B) but determined that both PRMT6 and the proteasome inhibitor MG-132 rescue Tat steady-state levels in translation-arrested cells in a nonadditive manner (Fig. 5C), suggesting that PRMT6 inhibits Tat degradation by a proteasomal pathway. The pathway, however, does not require the polyubiquitin-independent REGγ proteasome, since silencing PSME3 expression did not affect the ability of PRMT6 to stabilize Tat (Fig. 6). We therefore suggest that arginine methylation by PRMT6 stabilizes Tat by preventing its degradation by a proteasome.
Protein methylation has been documented to influence the stabilities of several proteins. Lysine methylation by SETD7, for example, increases the stability of the tumor suppressor p53 (11). In contrast, arginine methylation of NCOA3 (also called p/CIP and SRC-3) by CARM1 promotes the degradation of NCOA3 (43), presumably by the REGγ proteasome (39). Another arginine methyltransferase, PRMT3, increases the stability of the 40S ribosomal protein S2 by suppressing ribosomal protein S2 ubiquitination in a manner independent of PRMT3 methyltransferase activity (10). Our study provides one of the first pieces of evidence that catalytically active arginine methyltransferase activity can increase protein stability. PRMT6-mediated stabilization of Tat appears to require direct methylation. Mutating the reported arginine methylation sites in Tat, however, does not abrogate the effect (Fig. 1A), nor does it prevent the in vitro methylation of Tat (Fig. 2C), in agreement with previous observations (58). PRMT6 may thus stabilize Tat by methylating arginines other than R52 and R53. This hypothesis is supported by a Tat mutant, Tat-SFV, whose arginine-rich basic domain was replaced with an NLS from SFV (Fig. 2A). Tat-SFV was unresponsive to overexpression of PRMT6 (Fig. 2B) and, importantly, was not methylated by PRMT6 in vitro (Fig. 2C). Since PRMT6 methylates Tat only within the basic domain (5), these results suggest that methylation of Tat basic-domain arginines is likely to be a prerequisite for stabilization.
Rescue of Tat by PRMT6 appears to be through prevention of Tat degradation from a proteasomal pathway (Fig. 5C). PRMT6 does not perturb general proteasome function, since the stabilities of endogenous CDK9 and IκBα, nuclear and cytoplasmic proteins, respectively, that are degraded by proteasomes (31, 48), were not altered by PRMT6 overexpression (Fig. 5D). An unanswered question is against which class of proteasome does PRMT6 prevent Tat degradation. Coexpression of Myc-PRMT6 did not alter Tat-FLAG's nuclear localization (data not shown), indicating that the proteasome must be nuclear. Gargano and colleagues speculated that the nuclear REGγ proteasome may be involved in Tat degradation (20). While our results may support this hypothesis to a limited extent (Fig. 6, lanes 3 and 4), PRMT6 appears to block Tat degradation from a more dominant proteasomal pathway. Lassot and coworkers demonstrated that Tat specifically disassembles another nuclear proteasome, the 26S multicatalytic proteinase, by recruiting the PAAF1 proteasome disassembly factor during transactivation (36). They showed that Tat subsequently recruits the liberated 19S regulatory particle to enhance HIV-1 gene expression (36). Whether 26S promotes Tat degradation outside of transactivation is unknown, and as such, it remains a candidate proteasome from which PRMT6 protects Tat. Finally, the possibility that the PA200 nuclear proteasome (54) is somehow involved in regulating Tat stability cannot be excluded.
Our study presents an apparent paradox with the work of Xie and colleagues (58), indicating that PRMT6 can be both a positive and a negative cofactor of HIV-1 Tat. This dilemma may be resolved by considering that Tat plays a spectrum of roles during HIV-1 replication and AIDS pathogenesis (25), many of which require Tat to be present in the cell cytoplasm (23, 50, 57) or extracellular milieu (15, 34, 35). The fate of Tat after it becomes methylated by PRMT6 and can no longer participate in transactivation is unknown, but our data show that methylated Tat is unlikely to be degraded efficiently. We propose a model in which methylated Tat, by way of its increased stability, becomes the source pool for cytoplasmic and extracellular Tat. PRMT6 would enable Tat to bypass or escape the transactivation cycle, thereby making Tat available for other functions. In this model, arginine methylation acts as a molecular switch of Tat function, and PRMT6 is therefore a positive regulator of Tat pathogenesis.
Acknowledgments
This work was supported by grants from the Australian National Health and Medical Research Council (NHMRC) (no. 389826) and the Australian Centre for HIV and Hepatitis Virology, awarded to D.H., and an Australian Postgraduate Award to H.S.
We thank David Warrilow for helpful discussions.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Agbottah, E., L. Deng, L. O. Dannenberg, A. Pumfery, and F. Kashanchi. 2006. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology. 3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apolloni, A., L. W. Meredith, A. Suhrbier, R. Kiernan, and D. Harrich. 2007. The HIV-1 Tat protein stimulates reverse transcription in vitro. Curr. HIV Res. 5:473-483. [DOI] [PubMed] [Google Scholar]
- 3.Bedford, M. T., and S. Richard. 2005. Arginine methylation: an emerging regulator of protein function. Mol. Cell 18:263-272. [DOI] [PubMed] [Google Scholar]
- 4.Blanchet, F., A. Cardona, F. A. Letimier, M. S. Hershfield, and O. Acuto. 2005. CD28 costimulatory signal induces protein arginine methylation in T cells. J. Exp. Med. 202:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulanger, M. C., C. Liang, R. S. Russell, R. Lin, M. T. Bedford, M. A. Wainberg, and S. Richard. 2005. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79:124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brès, V., R. E. Kiernan, L. K. Linares, C. Chable-Bessia, O. Plechakova, C. Tréand, S. Emiliani, J. M. Peloponese, K. T. Jeang, O. Coux, M. Scheffner, and M. Benkirane. 2003. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat. Cell Biol. 5:754-761. [DOI] [PubMed] [Google Scholar]
- 8.Chang, B., Y. Chen, Y. Zhao, and R. K. Bruick. 2007. JMJD6 is a histone arginine demethylase. Science 318:444-447. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., L. F. Barton, Y. Chi, B. E. Clurman, and J. M. Roberts. 2007. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGγ proteasome. Mol. Cell 26:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi, S., C. R. Jung, J. Y. Kim, and D. S. Im. 2008. PRMT3 inhibits ubiquitination of ribosomal protein S2 and together forms an active enzyme complex. Biochim. Biophys. Acta 1780:1062-1069. [DOI] [PubMed] [Google Scholar]
- 11.Chuikov, S., J. K. Kurash, J. R. Wilson, B. Xiao, N. Justin, G. S. Ivanov, K. McKinney, P. Tempst, C. Prives, S. J. Gamblin, N. A. Barlev, and D. Reinberg. 2004. Regulation of p53 activity through lysine methylation. Nature 432:353-360. [DOI] [PubMed] [Google Scholar]
- 12.Cioce, M., S. Boulon, A. G. Matera, and A. I. Lamond. 2006. UV-induced fragmentation of Cajal bodies. J. Cell Biol. 175:401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Col, E., C. Caron, D. Seigneurin-Berny, J. Gracia, A. Favier, and S. Khochbin. 2001. The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J. Biol. Chem. 276:28179-28184. [DOI] [PubMed] [Google Scholar]
- 14.El-Andaloussi, N., T. Valovka, M. Toueille, R. Steinacher, F. Focke, P. Gehrig, M. Covic, P. O. Hassa, P. Schar, U. Hubscher, and M. O. Hottiger. 2006. Arginine methylation regulates DNA polymerase beta. Mol. Cell 22:51-62. [DOI] [PubMed] [Google Scholar]
- 15.Ensoli, B., G. Barillari, S. Z. Salahuddin, R. C. Gallo, and F. Wong-Staal. 1990. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature 345:84-86. [DOI] [PubMed] [Google Scholar]
- 16.Eustice, D. C., P. A. Feldman, A. M. Colberg-Poley, R. M. Buckery, and R. H. Neubauer. 1991. A sensitive method for the detection of beta-galactosidase in transfected mammalian cells. BioTechniques 11:739-740. [PubMed] [Google Scholar]
- 17.Favre, D., E. Studer, and M. R. Michel. 1994. Two nucleolar targeting signals present in the N-terminal part of Semliki Forest virus capsid protein. Arch. Virol. 137:149-155. [DOI] [PubMed] [Google Scholar]
- 18.Fingerman, I. M., and S. D. Briggs. 2004. p53-mediated transcriptional activation: from test tube to cell. Cell 117:690-691. [DOI] [PubMed] [Google Scholar]
- 19.Frankel, A., N. Yadav, J. Lee, T. L. Branscombe, S. Clarke, and M. T. Bedford. 2002. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277:3537-3543. [DOI] [PubMed] [Google Scholar]
- 20.Gargano, B., M. Fiorillo, S. Amente, B. Majello, and L. Lania. 2008. p14ARF is capable of promoting HIV-1 tat degradation. Cell Cycle 7:1433-1439. [DOI] [PubMed] [Google Scholar]
- 21.Gary, J. D., and S. Clarke. 1998. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 61:65-131. [DOI] [PubMed] [Google Scholar]
- 22.Guccione, E., C. Bassi, F. Casadio, F. Martinato, M. Cesaroni, H. Schuchlautz, B. Luscher, and B. Amati. 2007. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449:933-937. [DOI] [PubMed] [Google Scholar]
- 23.Harrich, D., C. Ulich, L. F. García-Martínez, and R. B. Gaynor. 1997. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 16:1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hetzer, C., W. Dormeyer, M. Schnolzer, and M. Ott. 2005. Decoding Tat: the biology of HIV Tat posttranslational modifications. Microbes Infect. 7:1364-1369. [DOI] [PubMed] [Google Scholar]
- 25.Huigen, M. C., W. Kamp, and H. S. Nottet. 2004. Multiple effects of HIV-1 trans-activator protein on the pathogenesis of HIV-1 infection. Eur. J. Clin. Investig. 34:57-66. [DOI] [PubMed] [Google Scholar]
- 26.Hyllus, D., C. Stein, K. Schnabel, E. Schiltz, A. Imhof, Y. Dou, J. Hsieh, and U. M. Bauer. 2007. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21:3369-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Invernizzi, C. F., B. Xie, F. A. Frankel, M. Feldhammer, B. B. Roy, S. Richard, and M. A. Wainberg. 2007. Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. AIDS 21:795-805. [DOI] [PubMed] [Google Scholar]
- 28.Invernizzi, C. F., B. Xie, S. Richard, and M. A. Wainberg. 2006. PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA. Retrovirology 3:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumi, T., A. Takaori-Kondo, K. Shirakawa, H. Higashitsuji, K. Itoh, K. Io, M. Matsui, K. Iwai, H. Kondoh, T. Sato, M. Tomonaga, S. Ikeda, H. Akari, Y. Koyanagi, J. Fujita, and T. Uchiyama. 2009. MDM2 is a novel E3 ligase for HIV-1 Vif. Retrovirology 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jadlowsky, J. K., M. Nojima, A. Schulte, M. Geyer, T. Okamoto, and K. Fujinaga. 2008. Dominant negative mutant cyclin T1 proteins inhibit HIV transcription by specifically degrading Tat. Retrovirology 5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiernan, R. E., S. Emiliani, K. Nakayama, A. Castro, J. C. Labbe, T. Lorca, K. I. Nakayama, and M. Benkirane. 2001. Interaction between cyclin T1 and SCFSKP2 targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 21:7956-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiernan, R. E., C. Vanhulle, L. Schiltz, E. Adam, H. Xiao, F. Maudoux, C. Calomme, A. Burny, Y. Nakatani, K. T. Jeang, M. Benkirane, and C. Van Lint. 1999. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 18:6106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King, J., and U. K. Laemmli. 1971. Polypeptides of the tail fibres of bacteriophage T4. J. Mol. Biol. 62:465-477. [DOI] [PubMed] [Google Scholar]
- 34.King, J. E., E. A. Eugenin, C. M. Buckner, and J. W. Berman. 2006. HIV tat and neurotoxicity. Microbes Infect. 8:1347-1357. [DOI] [PubMed] [Google Scholar]
- 35.Lafrenie, R. M., L. M. Wahl, J. S. Epstein, I. K. Hewlett, K. M. Yamada, and S. Dhawan. 1996. HIV-1-Tat protein promotes chemotaxis and invasive behavior by monocytes. J. Immunol. 157:974-977. [PubMed] [Google Scholar]
- 36.Lassot, I., D. Latreille, E. Rousset, M. Sourisseau, L. K. Linares, C. Chable-Bessia, O. Coux, M. Benkirane, and R. E. Kiernan. 2007. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol. Cell 25:369-383. [DOI] [PubMed] [Google Scholar]
- 37.Lee, D. Y., C. Teyssier, B. D. Strahl, and M. R. Stallcup. 2005. Role of protein methylation in regulation of transcription. Endocrine Rev. 26:147-170. [DOI] [PubMed] [Google Scholar]
- 38.Li, X., L. Amazit, W. Long, D. M. Lonard, J. J. Monaco, and B. W. O'Malley. 2007. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGγ-proteasome pathway. Mol. Cell 26:831-842. [DOI] [PubMed] [Google Scholar]
- 39.Li, X., D. M. Lonard, S. Y. Jung, A. Malovannaya, Q. Feng, J. Qin, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2006. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGγ proteasome. Cell 124:381-392. [DOI] [PubMed] [Google Scholar]
- 40.Marcello, A., M. Lusic, G. Pegoraro, V. Pellegrini, F. Beltram, and M. Giacca. 2004. Nuclear organization and the control of HIV-1 transcription. Gene 326:1-11. [DOI] [PubMed] [Google Scholar]
- 41.Marshall, R. M., D. Salerno, J. Garriga, and X. Grana. 2005. Cyclin T1 expression is regulated by multiple signaling pathways and mechanisms during activation of human peripheral blood lymphocytes. J. Immunol. 175:6402-6411. [DOI] [PubMed] [Google Scholar]
- 42.Mowen, K. A., B. T. Schurter, J. W. Fathman, M. David, and L. H. Glimcher. 2004. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell 15:559-571. [DOI] [PubMed] [Google Scholar]
- 43.Naeem, H., D. Cheng, Q. Zhao, C. Underhill, M. Tini, M. T. Bedford, and J. Torchia. 2007. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 27:120-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Najbauer, J., B. A. Johnson, A. L. Young, and D. W. Aswad. 1993. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J. Biol. Chem. 268:10501-10509. [PubMed] [Google Scholar]
- 45.Napolitano, G., P. Licciardo, R. Carbone, B. Majello, and L. Lania. 2002. CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. J. Cell Physiol. 192:209-215. [DOI] [PubMed] [Google Scholar]
- 46.Pagans, S., A. Pedal, B. J. North, K. Kaehlcke, B. L. Marshall, A. Dorr, C. Hetzer-Egger, P. Henklein, R. Frye, M. W. McBurney, H. Hruby, M. Jung, E. Verdin, and M. Ott. 2005. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 3:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng, J., D. Schwartz, J. E. Elias, C. C. Thoreen, D. Cheng, G. Marsischky, J. Roelofs, D. Finley, and S. P. Gygi. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21:921-926. [DOI] [PubMed] [Google Scholar]
- 48.Roff, M., J. Thompson, M. S. Rodriguez, J. M. Jacque, F. Baleux, F. Arenzana-Seisdedos, and R. T. Hay. 1996. Role of IκBα ubiquitination in signal-induced activation of NFκB in vivo. J. Biol. Chem. 271:7844-7850. [DOI] [PubMed] [Google Scholar]
- 49.Salmon, P., J. Oberholzer, T. Occhiodoro, P. Morel, J. Lou, and D. Trono. 2000. Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol. Ther. 2:404-414. [DOI] [PubMed] [Google Scholar]
- 50.Seeger, M., K. Ferrell, R. Frank, and W. Dubiel. 1997. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J. Biol. Chem. 272:8145-8148. [DOI] [PubMed] [Google Scholar]
- 51.Sgarra, R., J. Lee, M. A. Tessari, S. Altamura, B. Spolaore, V. Giancotti, M. T. Bedford, and G. Manfioletti. 2006. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J. Biol. Chem. 281:3764-3772. [DOI] [PubMed] [Google Scholar]
- 52.Stauber, R. H., and G. N. Pavlakis. 1998. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology 252:126-136. [DOI] [PubMed] [Google Scholar]
- 53.Torres-Padilla, M. E., D. E. Parfitt, T. Kouzarides, and M. Zernicka-Goetz. 2007. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445:214-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ustrell, V., L. Hoffman, G. Pratt, and M. Rechsteiner. 2002. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21:3516-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Duyne, R., R. Easley, W. Wu, R. Berro, C. Pedati, Z. Klase, K. Kehn-Hall, E. K. Flynn, D. E. Symer, and F. Kashanchi. 2008. Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 57.Xiao, H., C. Neuveut, M. Benkirane, and K. T. Jeang. 1998. Interaction of the second coding exon of Tat with human EF-1 delta delineates a mechanism for HIV-1-mediated shut-off of host mRNA translation. Biochem. Biophys. Res. Commun. 244:384-389. [DOI] [PubMed] [Google Scholar]
- 58.Xie, B., C. F. Invernizzi, S. Richard, and M. A. Wainberg. 2007. Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat Interactions with both cyclin T1 and the Tat transactivation region. J. Virol. 81:4226-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]