Abstract
Hepatitis C virus (HCV) is a positive-strand RNA virus replicating its genome via a negative-strand [(−)] intermediate. Little is known about replication signals residing in the 3′ end of HCV (−) RNA. Recent studies identified seven stem-loop structures (SL-I′, -IIz′, -IIy′, -IIIa′, -IIIb′, -IIIcdef′, and -IV′) in this region. In the present study, we mapped the minimal region required for RNA replication to SL-I′ and -IIz′, functionally confirmed the SL-IIz′ structure, and identified SL-IIIa′ to -IV′ as auxiliary replication elements. In addition, we show that the 5′ nontranslated region of the genome most likely does not contain cis-acting RNA structures required for RNA packaging into infectious virions.
Hepatitis C virus (HCV) is a positive-strand [(+)] RNA virus belonging to the family Flaviviridae. The viral genome has a length of about 9,600 nucleotides (nt) and codes for a polyprotein of ∼3,000 amino acids that is cleaved into at least 10 viral proteins (1). The open reading frame is flanked by two nontranslated regions (NTRs). The 5′ NTR is 341 nt in length and has at least two different functions: first, an internal ribosome entry site (IRES) directing cap-independent translation of the viral RNA (26); second, a promoter for initiation of (+) RNA synthesis (7, 14).
HCV RNA replication is believed to occur in two steps: initial synthesis of a negative-strand [(−)] RNA intermediate and subsequent synthesis of (+) RNA using the (−) RNA intermediate as a template. It has been shown that at least the 3′ NTR of (+) RNA is required for initiation of (−) RNA (5, 29, 31), and it is assumed that the 3′ end of (−) RNA is involved in (+) RNA synthesis.
Several studies have addressed the role of the 5′ NTR for RNA replication. Using exclusively genotype 1 isolates, mapping studies revealed that the first ∼120 nt of (+) RNA are sufficient for RNA replication, but high-level replication requires additional downstream RNA sequences located within the 5′ NTR (7, 14, 18, 19, 30). Moreover, intensive mutational analyses and biochemical assays established complex stem-loop (SL) structures in this region.
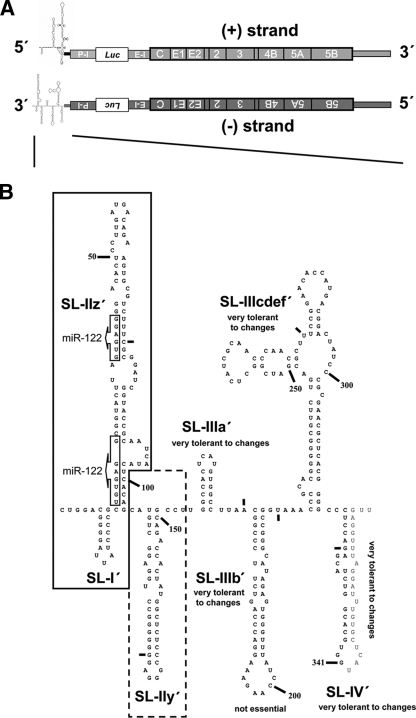
Much less is known about the structure of the 3′ end of (−) RNA. Recently, three groups independently reported structure models of this region (3, 20, 23). Distinct SL elements that are clearly different from those of the 5′ end of (+) RNA have been proposed (Fig. 1). All three studies consistently predicted five SL structures in the 3′-terminal 220 nt of (−) RNA, whereas structure predictions for the upstream sequence differ between the reports. In the study by Smith and coworkers, fewer nucleotides undergo base pairing with nucleotides complementary to sequences of the core coding region. Since core sequences are not involved in RNA replication (14; P. Friebe and R. Bartenschlager, unpublished data), we used this RNA structure prediction as a guide for our genetic analysis (Fig. 1B). Furthermore, we used the Mfold nucleic acid folding prediction algorithm (33) for design of the mutants to minimize the risk that the introduced mutations altered RNA structures in an undesired manner.
FIG. 1.
(A) Schematic presentation of the JFH1-derived reporter virus genome used throughout this study in the (+) and (−) orientations. Secondary structures of 5′ end of (+) RNA and 3′ end of (−) RNA are indicated. The poliovirus IRES (P-I) is positioned upstream of the firefly luciferase (Luc) reporter gene, and it is separated from the HCV 5′ NTR by a 61-nt-long spacer element (not indicated). Translation of the HCV coding region is mediated by the EMCV IRES (E-I). (B) Predicted RNA secondary structure of the 3′ end of JFH1 (−) RNA according to reference 23. The nomenclature of SL structures is given. Nucleotides are counted from the 3′ to 5′ direction; small bars indicate 25-nt intervals. Nucleotides downstream of position 341 are absent in the construct (see the supplemental material) and given only to highlight SL-IV′. Box arrows indicate complementary sequences of seed sequences for miRNA-122 binding in the 5′ NTR of (+) RNA. Minimal sequence required for RNA replication is framed; sequences that significantly contribute to RNA replication are framed with a dashed box.
Introduction of mutations into the 3′-terminal sequences of (−) RNA will most likely interfere with IRES activity residing in the complementary sequences of the 5′ NTR. For this reason, and to determine whether earlier results obtained with genotype 1 isolates hold true for the widely used genotype 2a isolate JFH1 that supports production of infectious virus (27), we generated a reporter virus genome in which IRES activity of the HCV 5′ NTR is not required (Fig. 1A). Instead, the start codon at nt position 341 was removed and the 5′ NTR was fused via a 61-nt-long random spacer to the IRES of the poliovirus (P-I) directing translation of the firefly luciferase reporter gene. Translation of the HCV open reading frame is directed by an engineered encephalomyocarditis virus IRES (E-I), and, therefore, the HCV 5′ NTR of (+) RNA only acts as a “promoter” for RNA synthesis. The nucleotide sequence of this bicistronic reporter virus genome, designated pFK341-Sp-PI-luc-EI-core-3′JFH1, which was used for insertion of all mutations in this study, is provided in the supplemental material. An analogous subgenomic JFH1 replicon that replicates with kinetics comparable to those of the genomic replicon used here has been described earlier (2).
The fact that the borders of SL-I′ and SL-IIz′ fit the minimal sequence required for RNA replication prompted us to investigate whether SL-I′ and SL-IIz′ (nt 1 to 104) constitute the minimal sequence at the 3′ end of (−) RNA required for RNA replication. A mutant in which nt 1 to 106 (corresponding to SL-I′ and SL-IIz′) were directly fused to P-I was generated, and replication competence was tested in transient assays as described elsewhere (6). As shown in Fig. 2B, replication of this mutant was severely impaired as compared to that of the wild type (wt), but was still about 10-fold above the background as determined with the NS5B active site polymerase mutant GND. Thus, SL-I′ and SL-IIz′ are sufficient for RNA replication, but the profound reduction of mutant nt 106 argues that additional sequences are required (Fig. 2B).
FIG. 2.
Mutations affecting SL-I′ and SL-II′ in the 3′ end of (−) RNA and their impact on HCV RNA replication. (A) Detailed presentation of the mutants targeting the three 3′-terminal elements of (−) RNA. The wt structures are boxed with black lines; corresponding mutants are displayed next to each wt structure. Nucleotide substitutions are indicated in red, and their positions are highlighted with red lines; changes in the predicted RNA structure are indicated by dislocated sequences. The name of the mutant is given above each depicted RNA structure. Complementary sequences corresponding to seed sequences of miRNA-122 binding sites in the 5′ NTR of (+) RNA are boxed. Replication competence is summarized below each mutant. (B) Transient replication assays of mutants specified below each bar. In vitro transcripts were transfected into Huh7-Lunet cells (15) that were harvested 4, 24, 48, and 72 h posttransfection. RNA replication was scored by luciferase assay after normalization to the 4-h value reflecting transfection efficiency (15). Shown is the normalized 48-h luciferase value as a percentage of the normalized wt activity. We have shown earlier that the 48-h value reflects RNA replication competence best (6). GND, inactive replicon with a single amino acid substitution destroying the active site of the NS5B RNA polymerase. Mean values from three independent experiments are shown.
By using a genotype 1b replicon system, Luo and coworkers recently reported that preservation of the stem of SL-I′, but not the loop sequence, is required for RNA replication (18). To investigate the role of SL-I′ in RNA replication in more detail, we first determined whether results obtained with genotype 1 replicons are valid for genotype 2 as well. Two JFH1-derived mutants were constructed in which either the stem sequence, but not the structure, or the loop sequence was altered (Fig. 2A [SL-I′-AU or SL-I′-loop, respectively]). Mutant SL-I′-loop replicated to the wt level, as described for genotype 1b by Luo and colleagues (18). However, mutant SL-I′-AU was completely blocked in RNA replication, in contrast to what is described for genotype 1b (18), arguing that either the requirements for RNA replication differ between the genotypes or differences in experimental conditions used in the study by Luo and by us account for the discrepant results.
To prove the predicted structure of SL-IIz′ in a functional manner and to dissect the individual regions of this element for RNA replication, a panel of mutants was constructed (Fig. 2A). In mutant SL-IIz′-25-38, SL-IIz′-77-100, or SL-IIz′-38-45, the lower or upper stem was disrupted, whereas in mutants SL-IIz′-25-38 + 77-100 and SL-IIz′-38-45 + 70-77, the stem was restored, but in an inverted orientation. The role of the loop and bulge sequences was probed with mutants SL-IIz′-56-59, SL-IIz′-78-82, and SL-IIz′-91-97 (Fig. 2A). Disruption of either the lower or middle part of SL-IIz′ inhibited RNA replication, whereas restoration of the stem also restored RNA replication, albeit to about 10-fold-lower levels than those of the wt (Fig. 2B). These data thus corroborate structure prediction of SL-IIz′.
Reduced replication of double-stem mutants (SL-IIz′-25-38 + 77-100 and SL-IIz′-38-45 + 70-77) compared to the wt could be due to less favorable stem formation or to an impact of the primary sequence: e.g., the loss of microRNA (miRNA)-122 binding sites (Fig. 2A) (10, 11). Nevertheless, the replication competence of the two mutants clearly demonstrates that miRNA-122 binding-sites are not essential for genotype 2 RNA replication, which is in contrast to what was found with genotype 1 replicons (10, 11). Replication of mutants SL-IIz′-56-59, SL-IIz′-78-82, and SL-IIz′-91-97 was very similar to that of the wt, arguing that the primary sequence of the loop and bulges in SL-IIz′ play a minor role or no role in RNA replication (Fig. 2B).
Having functionally confirmed the requirements of SL-I′ and SL-IIz′ in the 3′ end of (−) RNA for RNA replication, we next probed the structure of SL-IIy′ in the same way. For this purpose, mutants SL-IIy′-108-121 and SL-IIy′-138-150, in which the stem was disrupted, and mutant SL-IIy′-108-121 + 138-150, in which the stem was restored, were constructed. Mutant SL-IIy′-127-130 had an altered loop sequence (Fig. 2A). The two mutations disrupting the stem reduced RNA replication about 100-fold (Fig. 2B). No significant restoration of RNA replication was found with double mutant SL-IIy′-108-121 + 138-150. We therefore conclude that either this mutation did not restore the stem structure or the primary RNA sequence rather than the stem structure is required for RNA replication. In support of the structure of SL-IIy′, replication of mutant SL-IIy′-127-130 was not affected (Fig. 2B), arguing that the primary sequence of the loop is less important for RNA replication. These findings suggest that SL-IIy′ plays a crucial role for RNA replication.
To further dissect nucleotide sequence requirements of SL-IIIa′ and SL-IIIb′, we deleted either the loop or the complete SL of SL-IIIa′ or SL-IIIb′ or deleted both SLs (Fig. 3A [mutants SL-IIIa′-Del 162-165, SL-IIIa′-Del 156-170, SL-IIIb′-Del 192-205, SL-IIIb′-Del 178-220 and SL-IIIa′+b′-Del IIIa′+b′, respectively]). As shown in Fig. 3B, the two loop deletion mutants replicated to the wt level, whereas the three other mutants were slightly impaired, replicating on the average about twofold less efficiently than the wt.
FIG. 3.
Mutations in SL-III′ and SL-IV′ and their impact on HCV RNA replication. (A) Detailed overview of constructed mutations. Nucleotide substitutions are highlighted with red letters, and their positions are emphasized with red lines; deletions are highlighted with blue letters and blue lines. Mutations predicted to disrupt proposed RNA elements are indicated by dislocation of the altered sequence. The wt structures are boxed with black lines; mutations affecting the same RNA element are given next to the corresponding wt structure. Names of mutants are given at the top of each panel, and replication competence is summarized below each mutant. (B) Replication competence of mutants that are specified below each bar. Mean values of three independent experiments are shown. For further details, see the legend to Fig. 2.
The role of SL-IIIcdef′ for RNA replication was probed with three different mutations specified in the lower panel of Fig. 3A: one deletion mutation affecting the lower stem (SL-IIIcdef′-Del 229-238), one deletion affecting a bulge (SL-IIIcdef′-Del 265-271), and one nucleotide substitution disrupting the upper stem (SL-IIIcdef′-306-311). Finally, even though SL-IV′ is not formed in our construct, which lacks core coding sequences, we probed the sequence of “SL-IV'” by introducing nucleotide substitutions (SL-IV′-325-330). All mutations had only minor effect on RNA replication, reducing replication about twofold compared to the wt. These results suggest that most of SL-III′ and SL-IV′ have only a minor contribution to RNA replication.
Packaging of the viral RNA genome into the nucleocapsid may require distinct RNA structures, which are often located near the 5′ end of the RNA genome or at some other position within the viral genome (8, 12, 17, 28). In the case of HCV, little is known about possible RNA signals required for packaging. The fact that subgenomic replicons composed of only the NTRs and the NS3-to-NS5B coding region can be encapsidated demonstrates that the region coding from the core to NS2 does not contain an essential packaging signal (24). However, since core was found to interact with the 5′ NTR of (+) RNA (4, 13, 16, 21, 22, 25, 32), we analyzed whether an RNA packaging signal may reside there by using the panel of mutants depicted in Fig. 4A. Design of the mutants was based on a homology model deduced from the secondary structure of the 5′ NTR of (+) RNA of genotype 1 (9). To analyze the impact of these mutations on viral RNA packaging, we transfected Huh7.5 cells with these reporter genomes. Filtered supernatant harvested 48 h after transfection was used to infect naive Huh7.5 cells, and 72 h later, cells were lysed and luciferase activity was measured (Fig. 4B). As a negative control, we used a reporter virus lacking the glycoproteins E1 and E2 (Del E1/E2) (Fig. 4B). This RNA replicates to wt levels but does not produce infectious virus and therefore determines the background of our infection assay (27). Figure 4B summarizes the replication competence of the mutants and their competence for infectious virus production (upper and lower panels, respectively). All replication-competent RNAs supported production of infectious virus, and in all cases, a direct correlation between efficiency of RNA replication and virus production was found. We therefore conclude that the RNA structures in the 5′ NTR of (+) RNA most likely are not required for RNA packaging into infectious virus particles or that signals required for RNA packaging directly overlap with signals essential for RNA replication.
FIG. 4.
Mutations in the 5′ NTR of (+) RNA and their impact on production of infectious HCV particles. (A) Secondary structure of the 5′ NTR of JFH1 (+) wt RNA is framed (9). Nucleotide positions are indicated by small bars in 25-nt intervals; the start codon is circled. Nucleotides in blue indicate deletions, and their positions are further highlighted by blue lines; nucleotide substitutions are highlighted in red and with red lines. Possible rearrangements of RNA secondary structures induced by mutations are not considered. (B) Results of transient RNA replication assays are shown in the upper graph. Experimental conditions were the same as those described in the legend to Fig. 2, with the exception that Huh7.5 cells were used. Infectivity assays of the various mutants are shown in the lower panel. Forty-eight hours posttransfection, culture supernatants were harvested and used to infect naïve Huh7.5 cells. These were harvested 72 h after inoculation, and luciferase activities in cell lysates were determined. Values are normalized to the wt, which was set to 100%. Names of the constructs are displayed between both panels. Mutant nt 296 contains only the first 296 nt of the 5′ NTR and served as a reference; mutant Del E1/E2 is derived from the bicistronic wt genome, but lacks the envelope glycoproteins (27). This RNA replicates to wt levels, but does not release infectious particles and therefore was used to define background of the infectivity assay. Mean values of three independent experiments are shown.
As summarized in Fig. 1B, we describe a genetic analysis of the 3′ end of JFH1 (−) RNA with respect to its role in RNA replication and thus complement structure predictions generated for genotype 1 isolates in a functional manner. In addition, we provide evidence that the 5′ NTR of (+) RNA most likely does not contain an RNA packaging signal. Based on these and earlier results, it is likely that RNA packaging signals either reside in the region encoding the replicase (NS3 to NS5B) or genome packaging is mediated by specific interaction between viral (RNA binding) proteins.
Supplementary Material
Acknowledgments
We are grateful to Ulrike Herian for excellent technical assistance, Volker Lohmann for stimulating discussions, and Sandra Bühler for editorial assistance. We thank Takaji Wakita for the gift of the JFH-1 isolate and Charles Rice for Huh7.5 cells.
This work was supported by grants of the Deutsche Forschungsgemeinschaft (BA1505/2-2 and Sonderforschungsbereich 638, Teilprojekt A5 [both to R.B.]).
Footnotes
Published ahead of print on 9 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 2.Binder, M., D. Quinkert, O. Bochkarova, R. Klein, N. Kezmic, R. Bartenschlager, and V. Lohmann. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J. Virol. 81:5270-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutkiewicz, M., A. Swiatkowska, M. Figlerowicz, and J. Ciesiolka. 2008. Structural domains of the 3′-terminal sequence of the hepatitis C virus replicative strand. Biochemistry 47:12197-12207. [DOI] [PubMed] [Google Scholar]
- 4.Fan, Z., Q. R. Yang, J. S. Twu, and A. H. Sherker. 1999. Specific in vitro association between the hepatitis C viral genome and core protein. J. Med. Virol. 59:131-134. [PubMed] [Google Scholar]
- 5.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friebe, P., J. Boudet, J.-P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolova, E., I. Frolov, and S. Schlesinger. 1997. Packaging signals in alphaviruses. J. Virol. 71.1:248-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda, M., M. R. Beard, L.-H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jopling, C. L., S. Schutz, and P. Sarnow. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jopling, C. L., M. Yi, A. M. Lancaster, S. M. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577-1581. [DOI] [PubMed] [Google Scholar]
- 12.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, M., Y. Ha, and H. J. Park. 2006. Structural requirements for assembly and homotypic interactions of the hepatitis C virus core protein. Virus Res. 122:137-143. [DOI] [PubMed] [Google Scholar]
- 14.Kim, Y. K., C. S. Kim, S. H. Lee, and S. K. Jang. 2002. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 290:105-112. [DOI] [PubMed] [Google Scholar]
- 15.Koutsoudakis, G., E. Herrmann, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, D., S. T. Takyar, W. B. Lott, and E. J. Gowans. 2003. Amino acids 1-20 of the hepatitis C virus (HCV) core protein specifically inhibit HCV IRES-dependent translation in HepG2 cells, and inhibit both HCV IRES- and cap-dependent translation in HuH7 and CV-1 cells. J. Gen. Virol. 84:815-825. [DOI] [PubMed] [Google Scholar]
- 17.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157:125-152. [DOI] [PubMed] [Google Scholar]
- 18.Luo, G., S. Xin, and Z. Cai. 2003. Role of the 5′-proximal stem-loop structure of the 5′ untranslated region in replication and translation of hepatitis C virus RNA. J. Virol. 77:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reusken, C. B., T. J. Dalebout, P. Eerligh, P. J. Bredenbeek, and W. J. Spaan. 2003. Analysis of hepatitis C virus/classical swine fever virus chimeric 5′NTRs: sequences within the hepatitis C virus IRES are required for viral RNA replication. J. Gen. Virol. 84:1761-1769. [DOI] [PubMed] [Google Scholar]
- 20.Schuster, C., C. Isel, I. Imbert, C. Ehresmann, R. Marquet, and M. P. Kieny. 2002. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J. Virol. 76:8058-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimoike, T., C. Koyama, K. Murakami, R. Suzuki, Y. Matsuura, T. Miyamura, and T. Suzuki. 2006. Down-regulation of the internal ribosome entry site (IRES)-mediated translation of the hepatitis C virus: critical role of binding of the stem-loop IIId domain of IRES and the viral core protein. Virology 345:434-445. [DOI] [PubMed] [Google Scholar]
- 22.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, R. M., C. M. Walton, C. H. Wu, and G. Y. Wu. 2002. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J. Virol. 76:9563-9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinmann, E., C. Brohm, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2008. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J. Virol. 82:7034-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka, Y., T. Shimoike, K. Ishii, R. Suzuki, T. Suzuki, H. Ushijima, Y. Matsuura, and T. Miyamura. 2000. Selective binding of hepatitis C virus core protein to synthetic oligonucleotides corresponding to the 5′ untranslated region of the viral genome. Virology 270:229-236. [DOI] [PubMed] [Google Scholar]
- 26.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss, B., H. Nitschko, I. Ghattas, R. Wright, and S. Schlesinger. 1989. Evidence for specificity in the encapsidation of Sindbis virus RNAs. J. Virol. 63:5310-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagi, M., M. St. Clair, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye, L., K. A. Timani, L. Ye, L. Kong, X. Yang, Q. Liao, and J. Wu. 2005. Two cis-acting elements in negative RNA strand of hepatitis C virus involved in synthesis of positive RNA strand in vitro. Acta Virol. 49:83-90. [PubMed] [Google Scholar]
- 31.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology 293:141-150. [DOI] [PubMed] [Google Scholar]
- 33.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.