Abstract
APOBEC3 proteins are potent restriction factors against retroviral infection in primates. This restriction is accompanied by hypermutations in the retroviral genome that are attributable to the cytidine deaminase activity of the APOBEC3 proteins. Studies of nucleotide sequence diversity among endogenous gammaretroviruses suggest that the evolution of endogenous retroelements could have been shaped by the mutagenic cytidine deaminase activity of APOBEC3. In mice, however, APOBEC3 appears to restrict exogenous murine retroviruses in the absence of detectable levels of deamination. AKV is an endogenous retrovirus that is involved in causing a high incidence of thymic lymphoma in AKR mice. A comparative analysis of several mouse strains revealed a relatively low level of APOBEC3 expression in AKR mice. Here we show that endogenous mouse APOBEC3 restricts AKV infection and that this restriction likely reflects polymorphisms affecting APOBEC3 abundance rather than differences in the APOBEC3 isoforms expressed. We also observe that restriction of AKV by APOBEC3 is accompanied by G→A hypermutations in the viral genome. Our findings demonstrate that APOBEC3 acts as a restriction factor in rodents affecting the strain tropism of AKV, and they provide good support for the proposal that APOBEC3-mediated hypermutation contributed to the evolution of endogenous rodent retroviral genomes.
Viruses that are restrained to infect only a specific animal species, subspecies, or strain have acquired particular features that enable them to circumvent the immune defenses of that particular host. Conversely, the natural hosts for these pathogens are alive today because they have evolved strategies to restrain the infectivities of their own pathogens. A virus with a broad host tropism will typically have evolved under selective pressure from several host factors that it will have encountered and successfully evaded. Ecotropic murine retroviruses generally have a restricted host range, due not only to the limited availability of their cellular receptor, mCAT-1 (58), but also to the various intrinsic restriction factors present in a specific host (7). Fv1 and Fv4 are the expression products of defective endogenous retroviruses that are present as germ line integrations and can interfere with and even block the infectivities of ecotropic retroviruses (6, 25).
Mouse APOBEC3 is another type of host-encoded intrinsic restriction factor that can display deoxycytidine deaminase activity on single-stranded DNA (16, 54). APOBEC3 proteins have a potent inhibitory effect on retroelements ranging from primate lentiviruses to murine retrotransposons (reviewed in reference 17). In humans and primates there are seven APOBEC3 genes, most of which have been proposed to act as restriction elements for viruses and retroelements. The most extensively characterized of the primate APOBEC3 proteins are APOBEC3F and APOBEC3G, which constitute powerful restriction factors for human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) (reviewed in reference 17). The evidence that these lentiviruses are targets for APOBEC3 action does not come just from in vitro experiments: tissue samples from HIV type 1 (HIV-1)-infected humans contain retroviral sequences exhibiting a pattern of G→A hypermutation that is characteristic of APOBEC3F/G-dependent deoxycytidine deamination (4, 12, 26, 56, 57).
In contrast to primates, mice have only a single APOBEC3 gene. This murine APOBEC3 has been shown to be able to inhibit retrotransposition of mouse MusD and intracisternal A particle elements in cotransfection assays (22, 23). However, the lack of any obvious signs of disease, developmental defect, or infertility in APOBEC3-deficient mice indicates that APOBEC3 may not play an essential role in suppressing the transposition of endogenous retroelements in laboratory mice (38, 40). With regard to exogenous retroviruses, mouse APOBEC3 has been shown to hinder the in vivo infectivity of the betaretrovirus mouse mammary tumor virus (MMTV) as well as that of the gammaretrovirus Friend murine leukemia virus (MLV) (40, 55); its activity against Moloney MLV (MoMLV), another gammaretrovirus, is apparently considerably weaker—likely reflecting the fact that MoMLV may have found ways to avoid APOBEC3-mediated restriction (14, 34, 46, 61). In none of these cases, however, does it appear that mouse APOBEC3 hypermutates the retroviral replication intermediates, suggesting that deamination is not central to its mechanism of restricting these retroviruses. Notwithstanding this failure to observe hypermutation of mouse retroviruses by mouse APOBEC3, recent studies of nucleotide sequence diversity among endogenous gammaretroviruses have suggested that the evolution of endogenous retroelements has been shaped by the mutagenic cytidine deaminase activity of APOBEC3 (28, 42). Thus, the picture which emerges is that APOBEC3 acts as one of several restriction factors of mouse retroelements, with some viruses having found ways to avoid APOBEC3-mediated restriction.
Different mouse strains exhibit different patterns of APOBEC3 expression (41, 47, 55). Thus, two major mouse APOBEC3 alleles have been identified: one encodes a protein whose sequence is similar to that of the allele expressed in C57BL/6 mice, and the other resembles that of BALB/c mice (47, 55). Two major splicing isoforms of APOBEC3, which either do or do not include exon 5, have also been detected: the relative abundance of these two isoforms differs between strains (36, 41, 47, 55). The restriction of Friend MLV and that of MMTV both appear to be dependent on the identity of the mouse strain, and it has been proposed that this reflects the polymorphism in the sequence and splicing isoforms of APOBEC3 (41, 47, 55).
In the course of our work on mouse APOBEC3, we discovered that APOBEC3 was expressed only at a low level in AKR mice. The AKR mouse strain harbors several germ line insertions of an endogenous ecotropic MLV designated AKV, which belongs to the gammaretrovirus family (5, 15, 27, 44, 45). A complex set of recombination events between AKV and nonecotropic endogenous retroviruses results in the production of leukemogenic mink cell focus-inducing viruses that are responsible for inducing a lethal form of thymic lymphoma of T-cell origin in these mice (19, 53). We were interested in determining whether the susceptibility of AKR mice to AKV infection could in part be explained by a failure of the APOBEC3 allele expressed in AKR mice to restrict this virus.
Here we show that endogenous murine APOBEC3 in C57BL/6 mice not only acts to restrict AKV infection but also hypermutates AKV replication intermediates, likely providing a powerful block to natural transmission of the virus between mouse strains. We find that the different isoforms of APOBEC3 (whether or not they include exon 5) are effective in AKV restriction and that the differential resistance of lymphocytes from different mouse strains/mutants to AKV infection correlates with the abundance of endogenous APOBEC3 mRNA. Our results indicate that APOBEC3 confers effective protection against germ line integration of retroviral pathogens in rodents, and they provide tangible support to the proposal that DNA editing by APOBEC3 may have participated in the evolution of endogenous retroviral genomes.
MATERIALS AND METHODS
Mice.
APOBEC3-deficent (APO3) mice were backcrossed eight times to a C57BL/6 background. These mice have the APOBEC3 gene disrupted by the insertion of a Neo selection cassette in exon 3 (38). AKR (AKR/J) mice were obtained from the Jackson Laboratory. BALB/c, C57BL/6, and NIH Swiss mice were maintained in our barrier unit. All the animal studies were performed under United Kingdom Home Office project license PPL80/2226.
Cell lines, plasmids, and cDNA cloning.
293T and NIH 3T3 cells lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. The plasmids encoding replicative AKV (AkvNBU3-EGFP has been renamed pAKV-NB-eGFP for simplicity and consistency) and MoMLV (GFP-MOV has been renamed pMOV-eGFP) have been described previously (1, 52). The plasmid carrying AKV has an internal ribosome entry site (IRES)-enhanced green fluorescent protein (eGFP) cassette inserted into the downstream U3 region, and the plasmid carrying MoMLV has the eGFP coding sequence inserted in the proline-rich region of the env gene (1, 52). Brown Norway rat APOBEC3 cDNA and the various mouse APOBEC3 cDNAs were cloned from total RNA that had been prepared from spleens using Trizol reagent (Gibco) according to the manufacturer's specifications. Ten micrograms of RNA was reverse transcribed using random hexamers (New England Biolabs) and Superscript II reverse transcriptase (Invitrogen). PCR was performed using sequence-specific primers. For rat APOBEC3, the primers were Forward1 (5′-ATGGGACCAGTGTGCCTGGGAT-3′), Reverse1 (5′-TTACAGGCCCCAGGACTCCTTG-3′), Forward2 (5′-ACTCAGATCTCGAGATGGGACCAGTGTGC-3′), and Reverse2: (5′-GCCCGCGGTACCTTACAGGCCCCAGGA-3′). For mouse APOBEC3, the primers were Forward1 (5′-ATGGGACCATTCTGTCTGGGATGC-3′), Reverse1 (5′-TCAAGACATCGGGGGTCCAAGC-3′), Forward2 (5′-ACTCAGATCTCGAGATGGGACCATTCTGT-3′), and Reverse2 (5′-GCCCGCGGTACCTCAAGACATCGGGGGTC-3′). The final PCR products were then cloned into the XhoI and KpnI sites (in boldface) of the pFLAG-C3 plasmid. Cloning of the other APOBEC3 genes has been described previously (32).
Northern blot analysis.
Total spleen RNA was resolved on a 1% agarose morpholinepropanesulfonic acid (MOPS) gel and blotted to a nylon Hybond XL (GE Healthcare) membrane according to the manufacturer's specifications. The cDNA probe was made by isolating the 579-bp XhoI-HindIII fragment from the Flag-mA3 expression vector and labeling it with [α-32P]dCTP. Hybridization was performed in ExpressHyb solution (Clontech) according to the manufacturer's instructions.
In vitro infection and encapsidation assays.
Infectious virus particles were produced by transfecting 293T cells at 70% confluence with either the pMOV-eGFP or pAKV-NB-eGFP plasmid, with or without FLAG-APOBEC3 expressing plasmids, using Genejuice transfection reagent (Novagen). The transfection ratio of proviral DNA to APOBEC3 was 5:1. The 293T cells were washed the day after transfection with phosphate-buffered saline (PBS) and were grown in RPMI 1640 culture medium for 48 h. Virus-containing supernatants were collected 72 h posttransfection and filtered through a 0.45-μm-pore-size filter. NIH 3T3 cells were then infected by spinoculation at 2,000 rpm for 2 h in the presence of 8 μg/ml of Polybrene. Target cells were monitored for eGFP fluorescence by fluorescence-activated cell sorter analysis 48 h later. APOBEC3 encapsidation assays were performed as described in reference 32.
Viral sequence analysis.
NIH 3T3 target cells were collected 48 h postinfection and treated with the restriction endonuclease DpnI to eliminate any trace of plasmid carryover from the transfection. Genomic DNA was then extracted using Puregene reagent (Gentra Systems) according to the manufacturer's specifications. DNA from MoMLV- or AKV-infected cells was amplified using the high-fidelity polymerase Pfu Turbo (Stratagene) with the following primers to amplify a 717-bp segment of the eGFP coding sequence: forward, 5′-ATGGTGAGCAAGGGCGAGGAGC-3′; reverse, 5′-CTTGTACAGCTCGTCCATGCCG-3′. The PCR products were then gel purified, cloned using the TOPO-Blunt cloning kit (Invitrogen), and sequenced using a standard M13 reverse primer. Sequences were then aligned and analyzed using Sequencher (Genecodes). Mutation databases were compiled after removal of multiply mutated sequences carrying identical sets of mutations in order to avoid repeat-counting of the same mutational events.
Ex vivo infection assays. (i) B-cell preparations.
Spleens from neonatal mouse pups 3 to 5 days of age were homogenized by enforced passage through a 70-μm-pore-size nylon cell strainer. The splenocytes were then washed in ice-cold PBS and resuspended in PBS with 1% bovine serum albumin. Anti-CD45R (B220) microbeads (Miltenyi) were added to the samples and incubated on ice for 30 min. The cells were then washed and resuspended in 500 μl of MACS Blue buffer. The B lymphocytes were purified by positive selection on MACS LS columns. B220-positive splenocytes were then washed and seeded at 250,000 per well in a round-bottom 96-well plate containing RPMI-1640 with 10% fetal bovine serum and 50 μg/ml lipopolysaccharide (Sigma).
(ii) Thymocyte preparation.
The thymuses from neonatal mouse pups 3 to 5 days of age were homogenized by enforced passage through a 70-μm-pore-size nylon cell strainer. Thymocytes were then washed in ice-cold PBS, counted, and seeded at 250,000 per well in an anti-mouse CD3-coated round-bottom 96-well plate containing RPMI 1640 with 10% fetal bovine serum and 5 ng/ml interleukin-2 (R&D Systems).
(iii) Infections.
Forty-eight hours after isolation, B cells and thymocytes were spin-infected with replicative MoMLV or AKV that had been produced in transfected 293T cells. Equal volumes (100 μl) of virus-containing supernatants were used for both viruses, since viral preparations consistently yielded similar titers (∼5 × 106 CFU/ml). The day following the infection, the cells were washed twice and cultured in fresh RPMI 1640 medium containing the appropriate growth factors for either B cells or thymocytes as indicated above (This step is necessary to ensure that there is no carryover of infectious particles from the initial infection.) This initial lymphocyte infection by replicative AKV and MoMLV resulted in more than 90% of cells expressing eGFP after 48 h. After 72 h, supernatants were collected and used to infect NIH 3T3 cells that were seeded at 104 cells per well. Target cells were monitored for eGFP fluorescence by fluorescence-activated cell sorter analysis 48 h later using a Becton Dickinson high-throughput sampler.
RESULTS
APOBEC3 mRNA levels differ between mouse strains.
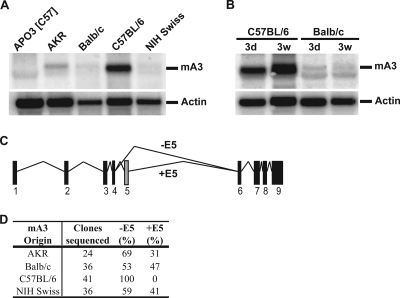
The APOBEC3 family appears to be rapidly evolving, with the sequences of APOBEC3G, for example, providing strong evidence of positive selection during primate evolution (48, 60). However, although there are multiple APOBEC3 genes in primates, as well as in several other mammals, there is only a single copy of the APOBEC3 gene in mouse. We were therefore interested in analyzing the polymorphism of mouse APOBEC3. Splenic RNA prepared from various strains of laboratory mice was analyzed by Northern blotting using a cDNA probe complementary to APOBEC3 exons 1 to 4. We found that splenic APOBEC3 expression levels differed dramatically among the selected mouse strains, with C57BL/6 mice expressing 6- to 12-fold more APOBEC3 mRNA than any other mouse strain tested (Fig. 1A). This difference was evident in both adult and neonatal mice (Fig. 1B). In order to find out whether this difference in expression levels was accompanied by differences in the coding sequence, we sequenced APOBEC3 cDNAs from each of the mouse strains following PCR amplification and cloning. The APOBEC3 cDNA sequences obtained from BALB/c, AKR, and NIH Swiss mice were all identical except for the presence of a 99-bp insertion corresponding to exon 5 in roughly half the clones sequenced for each mouse (Fig. 1C and D). In contrast, C57BL/6 mice expressed a different APOBEC3 allele, exhibiting 15 amino acid differences (6 of which are conservative) from the BALB/c and AKR alleles and lacking exon 5 in all of the clones that we sequenced (Fig. 1D) (41, 47, 55).
FIG. 1.
Endogenous APOBEC3 mRNA expression in mice. (A) Total RNA (10 μg) was prepared from the spleens of various strains of mice (3 weeks old). (Top) Northern blots were probed with a fragment of the mouse APOBEC3 (mA3) cDNA encompassing exons 1 to 4. (Bottom) An actin cDNA probe served as a loading control. (B) Comparison of APOBEC3 expression in mice 3 days (3d) or 3 weeks (3w) old. (C) Pictogram showing the alternative splicing of exon 5 of mouse APOBEC3 mRNA. (D) Representation of the proportions of mA3 mRNA splicing variants in various mouse strains. mA3 was amplified from splenic cDNA using flanking primers to the coding sequence and was cloned into the FLAG-C3 plasmid. Exon 5-negative (−E5) and exon 5-positive (+E5) clones are represented as percentages of the total number of clones sequenced.
Transfected mouse APOBEC3 restricts AKV in vitro.
We were struck by the fact that, in contrast to C57BL/6 mice, AKR mice (in which the AKV MLV is an endogenous ecotropic virus) and NIH Swiss mice (in which AKV has been shown to be able to establish a productive infection [3]) express only low levels of APOBEC3. We therefore wondered whether mouse APOBEC3 might in fact serve to restrict AKV infection. To test this, we used a recombinant replicative AKV provirus in which the region in the capsid gene that is responsible for sensitivity to Fv1 restriction was replaced with the corresponding region in MoMLV, thereby allowing for infection of murine cells carrying either the Fv1n or the Fv1b allele (1). Both the MoMLV and AKV proviruses that we used throughout our experiments express the fluorescent reporter eGFP, which enables direct measurement of infectivity by flow cytometry (1, 52).
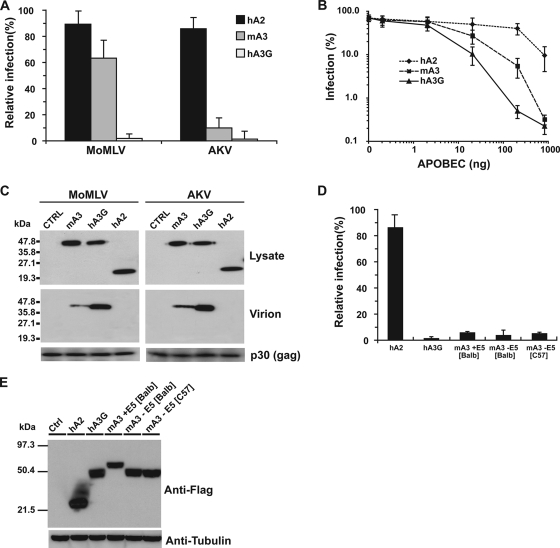
To test whether the C57BL/6 APOBEC3 allele could restrict AKV infection in vitro, AKV particles produced in the presence or absence of cotransfected mouse APOBEC3 in 293T cells were used to infect NIH 3T3 cells, and viral infection was assessed by monitoring eGFP expression. The results in Fig. 2A show that APOBEC3 from C57BL/6 mice efficiently restricts AKV infectivity but has only a marginal effect on MoMLV in the same assay. Although this inhibition is potent, titration experiments reveal it to be slightly weaker than that achieved with human APOBEC3G (Fig. 2B). We also tested mouse APOBEC3 on a recombinant AKV that did not contain the eGFP coding sequence, using an anti-Env antibody to monitor infection. This virus was restricted to the same degree as AKV containing the eGFP sequence, thus excluding the possibility that eGFP itself could facilitate the restriction of the virus (data not shown).
FIG. 2.
Restriction of MoMLV and AKV by APOBEC3 proteins. (A) The infectivities of MoMLV or AKV particles that were produced by cotransfecting subconfluent 293T cells with 1 μg of either pMOV-eGFP or pAKV-NB-eGFP proviral DNA together with 0.2 μg of one of the FLAG-APOBEC3-expressing vectors were determined by transferring the virus-containing supernatants to NIH 3T3 cells at 36 h posttransfection and monitoring the percentages of cells displaying eGFP fluorescence after a further 48 h. The results are displayed as the level of infection relative to that obtained with cotransfection of the empty vector. Error bars represent the standard errors of the means for six independent experiments (as in panel B). hA2, human APOBEC2; mA3, mouse APOBEC3; hA3G, human APOBEC3G. (B) AKV infectivity, monitored by eGFP fluorescence of infected NIH 3T3 cells as a function of the amount of an APOBEC-expressing plasmid cotransfected with 1 μg of pAKV-NB-eGFP proviral DNA into subconfluent 293T cells during viral production. (C) Packaging of mouse APOBEC3 into AKV and MoMLV virions. Subconfluent 293T cells were cotransfected with 1 μg of either pMOV-eGFP or pAKV-NB-eGFP proviral DNA and 0.2 μg of each of the FLAG-APOBEC3-expressing vectors. After 36 h, virus was collected from the supernatants by ultracentrifugation. Western blot analysis with a horseradish peroxidase-conjugated anti-FLAG antibody was performed on transfected-cell lysates (top) or on virions pelleted from the supernatants (center). (Bottom) Virion encapsidation immunoblots were stripped and reprobed with an anti-p30 (Gag) antibody. CTRL, control. (D) Effects of different mouse APOBEC3 isoforms on AKV infectivity. Assays were performed and results presented as in panel A. +E5, with exon 5; −E5, without exon 5. (E) The expression of the various FLAG-tagged APOBEC proteins in the transfected 293T cells used in panel D was assayed by Western blot analysis using a horseradish peroxidase-conjugated anti-FLAG antibody on transfected-cell lysates. The immunoblot was then stripped and reprobed with an anti-tubulin antibody.
Aside from some instances where APOBEC3 proteins have been reported to mediate an antiviral effect when expressed in target cells, restriction has generally been associated with encapsidation of APOBEC3 into viral particles (18, 36). It has been an object of significant debate whether or not the resistance of MoMLV to restriction by mouse APOBEC3 could be attributable to a defect in APOBEC3 packaging (2, 21, 29). Recent reports, however, support the claim that mouse APOBEC3 is incorporated into MoMLV particles (albeit at levels lower than or equivalent to that of human APOBEC3G), and thus poor encapsidation has been proposed to be an unlikely explanation for the resistance of MoMLV to restriction by mouse APOBEC3 (14, 46, 61). In agreement with this, in our assays we also see clearly that mouse APOBEC3 is present in purified viral particles of both MoMLV and AKV (Fig. 2C). We therefore believe that differences in protein encapsidation are unlikely to explain why APOBEC3 can potently restrict AKV but not MoMLV.
AKV is similarly restricted in vitro by different mouse APOBEC3 isoforms.
In order to discover whether the restriction of AKV by mouse APOBEC3 was peculiar to the C57BL/6 allele, we compared the infectivities of AKV virions produced in the presence of cotransfected APOBEC3 cDNAs that derived from the C57BL/6 allele (lacking exon 5) or from the AKR or BALB/c allele (with or without exon 5). All the APOBEC3 isoforms tested were well expressed in these assays, and all inhibited AKV infectivity similarly (Fig. 2D and E). Thus, with respect to AKV restriction, the different isoforms of mouse APOBEC3 tested appear to exhibit broadly similar antiviral potencies at similar expression levels.
Transfected mouse APOBEC3 hypermutates AKV.
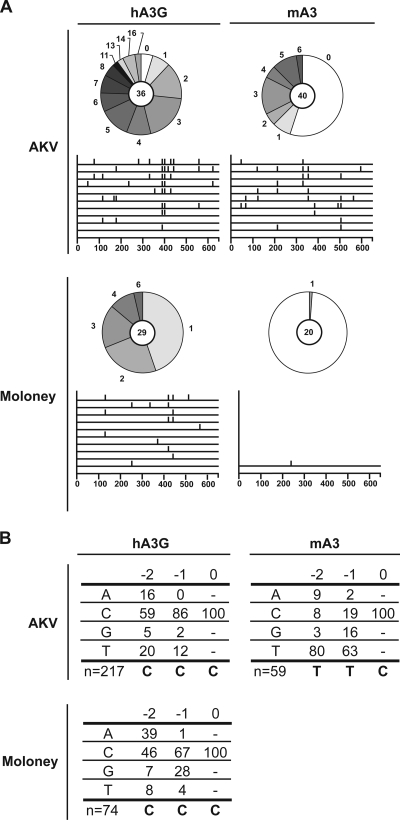
Although it has been shown that mouse APOBEC3 can restrict several murine retroviruses, such as Friend MLV and MMTV (40, 47, 55), mutations caused by the catalytic activity of mouse APOBEC3 have generally been undetectable, aside from one report showing low-frequency hyperediting of Friend MLV DNA by mouse APOBEC3 using a highly sensitive PCR approach (43). Whether these exogenous retroviruses have evolved a specific way to avoid being deaminated by mouse APOBEC3 is as yet unclear. To determine if AKV DNA is a target for APOBEC3-mediated deamination, the eGFP coding sequence from the proviral DNA was amplified by PCR from cells infected with AKV particles produced in the presence of various APOBEC proteins; then the PCR product was cloned and sequenced. This region was chosen to allow a direct comparison of hypermutated sequences and DNA deamination specificities between AKV and MoMLV using the same PCR conditions and primers. In contrast to what was observed with MoMLV, a large number of AKV clones contained G-to-A transition mutations on the plus-strand proviral DNA when AKV was produced in the presence of mouse APOBEC3 (Fig. 3A and Table 1). The target DNA sequence specificity for the mutations (Fig. 3B) was clearly indicative of the catalytic activity of mouse APOBEC3 on the minus-strand DNA, since the deoxycytidine mutations occurred in the context of two preceding thymidines, according well with the deamination context consensus previously identified for mouse APOBEC3 (9, 28, 43). Sequencing of a corresponding fragment of the env gene from the proviral DNAs of both viruses revealed similar results for AKV and no detectable mutations attributable to mouse APOBEC3 for MoMLV (data not shown).
FIG. 3.
APOBEC3-mediated hypermutation of MoMLV and AKV. (A) Pie charts depict the proportions of sequences with the indicated numbers of mutations. The total number of clones sequenced is given at the center of each pie. Below each pie, a line drawing depicts the distribution of G→A mutations along the eGFP gene in the first 10 mutated sequences analyzed in each data set. hA3G, human APOBEC3G; mA3, mouse APOBEC3. (B) Local sequence preference for deamination by hA3G or mA3, computed with respect to the deaminated cytidine (position zero) on the viral minus strand. “n” indicates the total number of mutations analyzed.
TABLE 1.
Editing of MoMLV and AKV by APOBEC3 proteinsa
| Virus and APOBEC proteinb | No. of sequences | No. of sequences mutated | Total no. of mutations | No. of G-to-A mutations | G-to-A mutation rate (mutations/kb) |
|---|---|---|---|---|---|
| MoMLV | |||||
| hA2 | 24 | 2 | 2 | 0 | |
| hA3G | 29 | 29 | 78 | 74 | 3.6 |
| mA3 | 20 | 2 | 3 | 1 | 0.1 |
| rA3 | 24 | 11 | 19 | 17 | 1.0 |
| AKV | |||||
| hA2 | 22 | 1 | 3 | 0 | |
| hA3G | 36 | 34 | 224 | 217 | 8.4 |
| mA3 | 40 | 22 | 62 | 59 | 2.1 |
| C57BL/6 (wt) | 64 | 17 | 20 | 16 | 0.3 |
| C57BL/6 (APO3) | 18 | 2 | 2 | 1c | 0.1 |
Genomic DNA was extracted from target cells infected by either MoMLV or AKV. A 717-bp segment of integrated viral DNA was amplified by PCR and cloned. Independent clones were sequenced, and mutations computed on the plus-strand DNA were analyzed.
hA2, human APOBEC2; hA3G, human APOBEC3G; mA3, mouse APOBEC3; rA3, rat APOBEC3. C57BL/6 (wt), editing of AKV by endogenous APOBEC3 following passage on splenocytes isolated from C57BL/6 (wt) mice. C57BL/6 (APO3), editing of AKV following passage on splenocytes isolated from mice with a targeted APOBEC3 gene disruption (APO3) of C57BL/6 background.
This sole G-to-A mutation occurred in a GGC context as read on the minus-strand viral DNA.
Endogenous mouse APOBEC3 restricts and hypermutates AKV.
The data therefore show that mouse APOBEC3, regardless of strain-specific polymorphisms and splicing isoforms, can perform as a potent restriction factor (and hypermutator) for AKV, as judged by in vitro assays. If mouse APOBEC3 is indeed also a natural intrinsic restriction factor for AKV, then the infectivity of the recombinant NB-tropic AKV should be restricted in mouse lymphocytes that express sufficient levels of endogenous APOBEC3.
Although the leukemogenic mink cell focus-inducing viruses of AKV origin that are produced in AKR mice have acquired long-terminal-repeat enhancer sequences conferring T-cell specificity, the enhancer sequence of AKV has been shown to promote predominantly B-cell specificity (33). For this reason, we isolated CD45R (B220)-positive B cells from the neonatal spleens of various mouse strains, including C57BL/6 mice carrying a targeted APOBEC3 gene disruption (APO3) (38). After activation with lipopolysaccharide, the cells were infected with MoMLV or AKV, and culture supernatants were collected after 3 days and used to infect NIH 3T3 cells. Relative infectivity was calculated by comparing the percentages of NIH 3T3 cells infected by viruses produced in lymphocytes from each of the mouse strains. The percentage of NIH 3T3 cells infected by viruses from C57BL/6 mice was set as the reference value.
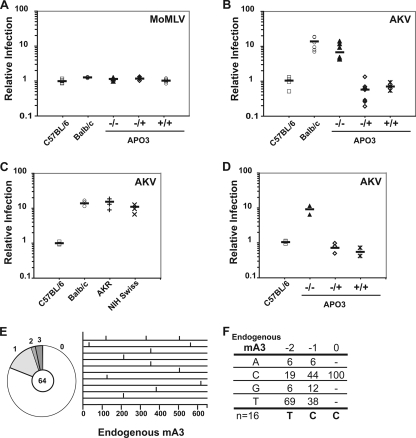
MoMLV particles produced from infected primary mouse splenocytes showed similar levels of infectivity when assayed on NIH 3T3 cells regardless of the strain of mouse from which the splenocytes were obtained. Thus, APOBEC3 expression had no apparent effect on MoMLV infectivity in these assays (Fig. 4A). On the other hand, AKV infectivity was greatly affected by the expression of APOBEC3 when the virus was produced from the splenocytes of C57BL/6 mice (Fig. 4B and C). Endogenous APOBEC3 mRNA expression levels clearly predict the outcome of the infection. Infection assays with BALB/c, NIH Swiss, and AKR mice all produced results similar to those seen with APOBEC3-deficient C57BL/6 mice (Fig. 4B and C). The results therefore show that APOBEC3 in C57BL/6 mice is indeed a restriction factor for AKV but not MoMLV infection, and they indicate that the higher levels of AKV infection achieved in AKR, NIH Swiss, and BALB/c mice correlate with lower levels of in vivo APOBEC3 expression (Fig. 4B and C). We were also curious to determine whether APOBEC3 could restrict AKV infection in thymocytes as well as in B cells. Ex vivo infection assays with AKV were performed on thymocytes isolated from APOBEC3-deficient neonatal mice of C57BL/6 genetic background (Fig. 4D). Endogenous APOBEC3 in thymocytes restricted AKV in a manner similar to that in B cells (Fig. 4D).
FIG. 4.
Restriction of AKV by endogenous mouse APOBEC3. (A, B, and C) Purified mouse splenocytes were infected with replicative MoMLV (A) or AKV (B and C) that had been produced by plasmid transfection into 293T cells. At 72 h postinfection, virus-containing culture supernatants were harvested from the cultured splenocytes and used to infect NIH 3T3 cells. Infection levels were determined by assessing the percentages of eGFP-positive cells 48 h later. Relative infection levels were established by setting the average percentage of eGFP-positive cells from the C57BL/6 mouse cohort to 1. Each point represents the mean of five independent infectivity measurements from a mouse splenocyte preparation. (D) Purified thymocytes from APOBEC3-deficient C57BL/6 mice were infected with AKV, and restriction of the virus was analyzed as for panel B. Each point represents the mean of four independent infectivity measurements from a mouse thymocyte preparation. (E) The pie chart depicts the proportions of sequences with the indicated numbers of mutations. The total number of clones sequenced is given at the center of the pie. A line drawing depicts the distribution of G→A mutations along the eGFP gene in the first 10 mutated sequences analyzed. mA3, mouse APOBEC3. (F) Local sequence preference for deamination by endogenous mouse APOBEC3, computed with respect to the deaminated cytidine (position zero) on the viral minus strand. “n” indicates the total number of mutations analyzed.
Sequencing of the integrated proviral AKV DNA from the infected target cells revealed the presence of mutations, almost all of which could be ascribed to APOBEC3, since they were largely G-to-A mutations occurring on the retroviral plus strand in a cytidine deamination context typical of mouse APOBEC3 (Table 1; Fig. 4E and F). In contrast, sequence analysis of retroviruses passaged through lymphocytes of APOBEC3-deficient mice revealed no mutations that were likely to have arisen through cytidine deamination by any other member of the APOBEC family (Table 1).
MoMLV is restricted by rat APOBEC3.
These results suggest that an interaction between a retrovirus and the host APOBEC3 may contribute to the narrow species/strain tropism of AKV, somewhat analogously to that inferred previously for HIV-1/SIV in primates (11, 35, 42, 49, 60). We therefore wondered whether MoMLV, although able to evade restriction by mouse APOBEC3, might nevertheless be susceptible to APOBEC3 from another rodent species, such as the rat. This would add further weight to the argument that APOBEC3 restriction might indeed be one factor affecting retroviral host tropism in rodents.
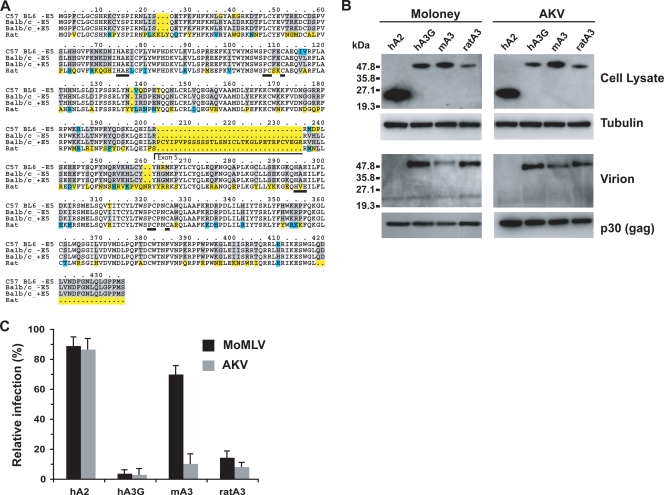
Rat APOBEC3 cDNA was cloned from the spleen of a Brown Norway rat. Sequence analysis revealed that the APOBEC3 proteins from mouse and rat share strong sequence similarities except for a few conservative amino acid substitutions (Fig. 5A). None of the rat APOBEC3 cDNAs we cloned contained a mouse-like exon 5 sequence. However, a region can be found in the genomic sequence of the rat gene that would share 24 of the 33 amino acid bases of mouse exon 5, if it were to be expressed. Rat APOBEC3, similarly to mouse APOBEC3, is evidently incorporated into MoMLV and AKV virions (Fig. 5B). Infection assays performed on NIH 3T3 cells with MoMLV and AKV particles produced in the presence of the various APOBEC3 proteins revealed that MoMLV and AKV were both strongly restricted by human APOBEC3G and by rat APOBEC3, but MoMLV was only marginally affected by mouse APOBEC3 (Fig. 5C). Analysis of integrated proviral DNA that had been PCR amplified from infected NIH 3T3 cells revealed that the restriction by rat APOBEC3 was accompanied by hypermutation (Table 1).
FIG. 5.
Restriction of MoMLV and AKV by rat APOBEC3. (A) Amino acid alignment of rat and mouse APOBEC3 proteins. Yellow boxes indicate major amino acid differences; blue boxes indicate conservative amino acid changes. Residues making up exon 5 and the zinc coordination motif of the first and second protein domains are underlined. (B) Encapsidation of rat APOBEC3 by MoMLV and AKV. FLAG-tagged APOBEC3 proteins were detected in transfected cell lysates or in virions pelleted from culture supernatants by Western blot analysis using a horseradish peroxidase-conjugated anti-FLAG antibody. The lysate immunoblots were stripped and reprobed with an anti-tubulin antibody, whereas the virion blots were reprobed with an anti-p30 (Gag) antibody. hA2, human APOBEC2; hA3G, human APOBEC3G; mA3, mouse APOBEC3; ratA3, rat APOBEC3. (C) The infectivities of MoMLV or AKV particles that were produced by cotransfecting subconfluent 293T cells with 1 μg of either pMOV-eGFP or pAKV-NB-eGFP proviral DNA together with 0.2 μg of either the rat, mouse, or human FLAG-APOBEC3-expressing vector (or the human APOBEC2 control) were determined by transferring the virus-containing supernatants to NIH 3T3 cells at 36 h posttransfection and monitoring the percentages of cells displaying eGFP fluorescence after a further 48 h.
DISCUSSION
The first evidence that APOBEC3 proteins could function as viral restriction factors came with the demonstration that forced expression of human APOBEC3G could restore an ability to restrict Vif-deficient HIV-1 to an APOBEC3F/G-deficient human T-cell line (50). The resistance of Vif-proficient HIV-1 virions to APOBEC3G restriction was then explained by the ability of Vif to direct APOBEC3G degradation (20, 37, 51, 59). Strong support for the proposal that HIV-1 is actually targeted by APOBEC3G and APOBEC3F during natural infection is provided by the fact that hypermutated HIV-1 sequences carrying the characteristic and idiosyncratic hallmarks of APOBEC3G/F deoxycytidine deamination are readily detected in human patients (4, 12, 26, 56, 57).
Following these early discoveries with human APOBEC3G, several APOBEC proteins were shown, by use of transfection assays, to be able to inhibit the replication of different viruses or transposable elements (17). Some of these observations likely reflect restriction pathways that operate naturally in vivo, although considerable caution must obviously be exercised in inferring a physiological restriction role based simply on in vitro experiments: it is readily imaginable that forced expression of a nucleic acid-binding protein such as APOBEC3 could lead to a nonphysiological inhibition of infectivity.
Here we have studied mouse APOBEC3, and we provide evidence not only that it can restrict and mutate AKV in transfected cells, but also that the endogenous APOBEC3 present in primary mouse B cells and thymocytes restricts AKV infection and hypermutates the AKV genome. Strikingly, this resistance is evident in cells from C57BL/6 mice but not from other mouse strains, such as BALB/c, NIH Swiss, or AKR (the natural host of AKV). Resistance to infection by AKV, in contrast to resistance to infection by MMTV or Friend MLV (40, 41, 47, 55), is not attributable to the specific isoform of murine APOBEC3 mRNA expressed but rather to low APOBEC3 abundance. This and the presence of hypermutations could indicate that AKV is restricted by mouse APOBEC3 by a mechanism different from that used for restriction of Friend MLV and MMTV. Thus, whereas some viruses might exploit the polymorphic difference in patterns of APOBEC3 expression, AKV may simply exploit the differences in expression levels. The molecular basis of this difference in expression levels remains to be identified. In agreement with Okeoma and colleagues, who recently identified a number of polymorphisms in the various alleles of APOBEC3 (41), it may be that the promoter of the APOBEC3 gene is functionally polymorphic. Or perhaps endogenous APOBEC3 levels are kept low through the action of some factor functioning in trans (e.g., possibly the product of an endogenous retroviral genome).
Although AKV and MoMLV share strong sequence homology and exhibit similar capacities to encapsidate APOBEC3 in their viral particles, the outcomes of infection by these two retroviruses are very different: MoMLV but not AKV escapes restriction and deamination by mouse APOBEC3 under our experimental conditions. The mechanism by which MoMLV avoids APOBEC3 restriction is unknown, but it could result from a strategy that the virus has specifically tailored for mouse APOBEC3, since MoMLV is sensitive to both the rat and human orthologs of the enzyme. We have initiated experiments that take advantage of the strong homology between AKV and MoMLV to try to identify regions responsible for these important differences in sensitivity to mouse APOBEC3. The answer so far appears to be complex and requires further investigation. The different patterns of AKV and MoMLV restriction by rat and mouse APOBEC3 proteins certainly reveal the importance of small differences in either host or viral sequences; this is a typical instance of selective pressure where resistant strains of a virus are selected in the presence of a potent antiviral agent. It is also interesting that MoMLV but not AKV has evolved a strategy for evading restriction by mouse APOBEC3. Perhaps AKV has experienced less selective pressure to evolve such a strategy—possibly because it is an N-tropic virus and/or is confined to a subset of hosts naturally low in APOBEC3 expression. Indeed, it will be interesting to measure APOBEC3 levels and sensitivity to AKV in feral mice, given that wild mice often appear to lack Fv1 restriction (24, 30).
It is striking that restriction of AKV by mouse APOBEC3 (both in vitro and ex vivo) is accompanied by G→A hypermutation. This contrasts with the situation reported for the in vivo restriction of Friend MLV and MMTV by mouse APOBEC3, where no hypermutation was observed (40, 41, 43, 47, 55). However, in two recent reports, researchers have analyzed sequences of endogenous gammaretroviruses in mice and primates to look for evidence of APOBEC3-mediated deamination in their evolution (28, 42). Both groups found strong evidence of a role for G→A hypermutation in the evolution and inactivation of these elements. The site preference for the mutations identified by these groups in the inactivated endogenous retroelements of C57BL/6 mice accords well with the site preference for mouse APOBEC3-mediated hypermutation of AKV identified in this work.
The finding that rat APOBEC3 can act on both AKV and MoMLV in vitro could suggest that in the wild, restriction by APOBEC3 may contribute to protecting rats from infection by certain mouse retroviruses. However, it is not clear whether these retroviruses can productively infect rats in the wild and cause tumors or whether they are possibly restricted for reasons other than the effect of APOBEC3 (13, 31). If such is the case, these murine retroviruses may not have been under pressure to evolve evasion mechanisms against rat APOBEC3. It is also unknown whether APOBEC3 is polymorphic in rats and whether MoMLV restriction is exclusive to the allele expressed in the Brown Norway rat. It will clearly be interesting to ascertain whether the sequences of rat retroelements bear the imprint of rat APOBEC3-like hypermutation.
Whether deamination is an essential component of the physiological process of AKV restriction in vivo remains to be established. Indeed, analogous questions have been raised regarding the in vivo restriction of HIV-1 by APOBEC3G/3F in humans (8, 10, 39). However, the availability of this mouse model in which endogenous mouse APOBEC3 mediates both the restriction and the hypermutation of AKV should enable the issue to be addressed by analysis of restriction in mice bearing suitable targeted modifications of their endogenous APOBEC3 gene. It will also be interesting to determine whether increasing APOBEC3 expression in AKR mice (possibly by serially back-crossing in a C57BL/6 APOBEC3 allele) will be sufficient to inhibit AKV-induced leukemogenesis, and if so, whether this depends on the catalytic site of APOBEC3 proteins. Thus, we anticipate that the identification of AKV as an endogenous target for restriction and hypermutation by mouse APOBEC3 in such a model should facilitate investigations into the physiological pathways of APOBEC3-mediated restriction of viral infection and pathogenesis.
Acknowledgments
We are especially grateful to Finn Skou Pedersen and Lars Aagaard for helpful discussions. We also thank F. S. Pedersen for providing the various AKV vectors and Barbara Schnierle for providing the pMOV-eGFP vector. Special thanks go to Silvestro Conticello for programming a PEARL script that facilitates mutation and target sequence analysis.
K.K. is supported by studentships from the Cambridge European Trust and the German Academic Exchange Service (DAAD). M.-A.L. was supported by a fellowship from the Canadian Institutes of Health Research and by an MRC Career Development Fellowship.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Aagaard, L., J. G. Mikkelsen, S. Warming, M. Duch, and F. S. Pedersen. 2002. Fv1-like restriction of N-tropic replication-competent murine leukaemia viruses in mCAT-1-expressing human cells. J. Gen. Virol. 83:439-442. [DOI] [PubMed] [Google Scholar]
- 2.Abudu, A., A. Takaori-Kondo, T. Izumi, K. Shirakawa, M. Kobayashi, A. Sasada, K. Fukunaga, and T. Uchiyama. 2006. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 16:1565-1570. [DOI] [PubMed] [Google Scholar]
- 3.Bachrach, E., M. Pelegrin, M. Piechaczyk, F. S. Pedersen, and M. Duch. 2002. Efficient gene transfer into spleen cells of newborn mice by a replication-competent retroviral vector. Virology 293:328-334. [DOI] [PubMed] [Google Scholar]
- 4.Beale, R. C., S. K. Petersen-Mahrt, I. N. Watt, R. S. Harris, C. Rada, and M. S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585-596. [DOI] [PubMed] [Google Scholar]
- 5.Bedigian, H. G., N. G. Copeland, N. A. Jenkins, K. Salvatore, and S. Rodick. 1983. Emv-13 (Akv-3): a noninducible endogenous ecotropic provirus of AKR/J. mice. J. Virol. 46:490-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 7.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 8.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 10.Bishop, K. N., M. Verma, E. Y. Kim, S. M. Wolinsky, and M. H. Malim. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borman, A. M., C. Quillent, P. Charneau, C. Dauguet, and F. Clavel. 1995. Human immunodeficiency virus type 1 Vif− mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J. Virol. 69:2058-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brightman, B. K., M. Okimoto, V. Kulkarni, J. K. Lander, and H. Fan. 1998. Differential behavior of the Mo + PyF101 enhancer variant of Moloney murine leukemia virus in rats and mice. Virology 242:60-67. [DOI] [PubMed] [Google Scholar]
- 14.Browne, E. P., and D. R. Littman. 2008. Species-specific restriction of APOBEC3-mediated hypermutation. J. Virol. 82:1305-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chattopadhyay, S. K., W. P. Rowe, N. M. Teich, and D. R. Lowy. 1975. Definitive evidence that the murine C-type virus inducing locus Akv-1 is viral genetic material. Proc. Natl. Acad. Sci. USA 72:906-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chelico, L., P. Pham, P. Calabrese, and M. F. Goodman. 2006. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13:392-399. [DOI] [PubMed] [Google Scholar]
- 17.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317-353. [DOI] [PubMed] [Google Scholar]
- 18.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 19.Cloyd, M. W., J. W. Hartley, and W. P. Rowe. 1980. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J. Exp. Med. 151:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 21.Doehle, B. P., A. Schafer, H. L. Wiegand, H. P. Bogerd, and B. R. Cullen. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79:8201-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 23.Esnault, C., J. Millet, O. Schwartz, and T. Heidmann. 2006. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 34:1522-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartley, J. W., and W. P. Rowe. 1975. Clonal cell lines from a feral mouse embryo which lack host-range restrictions for murine leukemia viruses. Virology 65:128-134. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda, H., and T. Odaka. 1983. Cellular expression of murine leukemia virus gp70-related antigen on thymocytes of uninfected mice correlates with Fv-4 gene-controlled resistance to Friend leukemia virus infection. Virology 128:127-139. [DOI] [PubMed] [Google Scholar]
- 26.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins, N. A., N. G. Copeland, B. A. Taylor, and B. K. Lee. 1982. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J. Virol. 43:26-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jern, P., J. P. Stoye, and J. M. Coffin. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 3:2014-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi, M., A. Takaori-Kondo, K. Shindo, A. Abudu, K. Fukunaga, and T. Uchiyama. 2004. APOBEC3G targets specific virus species. J. Virol. 78:8238-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak, C. A. 1985. Analysis of wild-derived mice for Fv-1 and Fv-2 murine leukemia virus restriction loci: a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J. Virol. 55:281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubo, Y., A. Ishimoto, T. Ono, H. Yoshii, C. Tominaga, C. Mitani, H. Amanuma, and N. Yamamoto. 2004. Determinant for the inhibition of ecotropic murine leukemia virus infection by N-linked glycosylation of the rat receptor. Virology 330:82-91. [DOI] [PubMed] [Google Scholar]
- 32.Langlois, M. A., R. C. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovmand, J., A. B. Sorensen, J. Schmidt, M. Ostergaard, A. Luz, and F. S. Pedersen. 1998. B-cell lymphoma induction by AKV murine leukemia viruses harboring one or both copies of the tandem repeat in the U3 enhancer. J. Virol. 72:5745-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Low, A., C. M. Okeoma, N. Lovsin, M. de Las Heras, T. H. Taylor, B. M. Peterlin, S. R. Ross, and H. Fan. 2009. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology 385:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 36.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 37.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 38.Mikl, M. C., I. N. Watt, M. Lu, W. Reik, S. L. Davies, M. S. Neuberger, and C. Rada. 2005. Mice deficient in APOBEC2 and APOBEC3. Mol. Cell. Biol. 25:7270-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 40.Okeoma, C. M., N. Lovsin, B. M. Peterlin, and S. R. Ross. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445:927-930. [DOI] [PubMed] [Google Scholar]
- 41.Okeoma, C. M., J. Petersen, and S. R. Ross. 2009. Expression of murine APOBEC3 alleles in different mouse strains and their effect on mouse mammary tumor virus infection. J. Virol. 83:3029-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Caballero, D., S. J. Soll, and P. D. Bieniasz. 2008. Evidence for restriction of ancient primate gammaretroviruses by APOBEC3 but not TRIM5α proteins. PLoS Pathog. 4:e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petit, V., D. Guetard, M. Renard, A. Keriel, M. Sitbon, S. Wain-Hobson, and J. P. Vartanian. 2009. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J. Mol. Biol. 385:65-78. [DOI] [PubMed] [Google Scholar]
- 44.Rowe, W. P., J. W. Hartley, and T. Bremner. 1972. Genetic mapping of a murine leukemia virus-inducing locus of AKR mice. Science 178:860-862. [DOI] [PubMed] [Google Scholar]
- 45.Rowe, W. P., and C. A. Kozak. 1980. Germ-line reinsertions of AKR murine leukemia virus genomes in Akv-1 congenic mice. Proc. Natl. Acad. Sci. USA 77:4871-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rulli, S. J., Jr., J. Mirro, S. A. Hill, P. Lloyd, R. J. Gorelick, J. M. Coffin, D. Derse, and A. Rein. 2008. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J. Virol. 82:6566-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santiago, M. L., M. Montano, R. Benitez, R. J. Messer, W. Yonemoto, B. Chesebro, K. J. Hasenkrug, and W. C. Greene. 2008. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science 321:1343-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schröfelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 51.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 52.Sliva, K., O. Erlwein, A. Bittner, and B. S. Schnierle. 2004. Murine leukemia virus (MLV) replication monitored with fluorescent proteins. Virol. J. 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoye, J. P., C. Moroni, and J. M. Coffin. 1991. Virological events leading to spontaneous AKR thymomas. J. Virol. 65:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suspène, R., P. Sommer, M. Henry, S. Ferris, D. Guetard, S. Pochet, A. Chester, N. Navaratnam, S. Wain-Hobson, and J. P. Vartanian. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 32:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeda, E., S. Tsuji-Kawahara, M. Sakamoto, M. A. Langlois, M. S. Neuberger, C. Rada, and M. Miyazawa. 2008. Mouse APOBEC3 restricts Friend leukemia virus infection and pathogenesis in vivo. J. Virol. 82:10998-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vartanian, J. P., M. Henry, and S. Wain-Hobson. 2002. Sustained G→A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 strain Vau group O genome. J. Gen. Virol. 83:801-805. [DOI] [PubMed] [Google Scholar]
- 57.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, H., M. P. Kavanaugh, R. A. North, and D. Kabat. 1991. Cell-surface receptor for ecotropic murine retroviruses is a basic amino-acid transporter. Nature 352:729-731. [DOI] [PubMed] [Google Scholar]
- 59.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, J., and D. M. Webb. 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 13:1785-1791. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, L., X. Li, J. Ma, L. Yu, J. Jiang, and S. Cen. 2008. The incorporation of APOBEC3 proteins into murine leukemia viruses. Virology 378:69-78. [DOI] [PubMed] [Google Scholar]