Abstract
Yeast retrotransposons form intracellular particles within which replication occurs. Because fungal nuclear membranes do not break down during mitosis, similar to retroviruses infecting nondividing cells, the cDNA produced must be translocated through nuclear pore complexes. The Saccharomyces cerevisiae long terminal repeat retrotransposon Ty3 assembles its Gag3 and Gag3-Pol3 precursor polyproteins into viruslike particles in association with perinuclear P-body foci. These perinuclear clusters of Ty3 viruslike particles localized to sites of clustered nuclear pore complexes (NPCs) in a nup120Δ mutant, indicating that Ty3 particles and NPCs interact physically. The NPC channels are lined with nucleoporins (Nups) with extended FG (Phe-Gly) motif repeat domains, further classified as FG, FxFG, or GLFG repeat types. These domains mediate partitioning of proteins between the cytoplasm and the nucleus. Here we have systematically examined the requirements for FG repeat domains in Ty3 nuclear transport. The GLFG domains interacted in vitro with virus-like particle Gag3, and this interaction was disrupted by mutations in the amino-terminal domain of Gag3, which is predicted to lie on the external surface of the particles. Accordingly, Ty3 transposition was decreased in strains with the GLFG repeats deleted. The spacer-nucleocapsid domain of Gag3, which is predicted to be internal to the particle, interacted with GLFG repeats and nucleocapsid localized to the nucleus. We conclude that Ty3 particle docking on nuclear pores is facilitated by interactions between Gag3 and GLFG Nups and that nuclear entry of the preintegration complex is further promoted by nuclear localization signals within the nucleocapsid and integrase.
Fungal long terminal repeat (LTR) retrotransposons assemble into cytoplasmic viruslike particles (VLPs) and undergo proteolytic maturation followed by reverse transcription of the RNA genome into cDNA. Because the Saccharomyces cerevisiae nuclear envelope (NE) does not dissociate during mitosis (reviewed in reference 14) and LTR retrotransposon particles are likely too large to transit the nuclear pore (41), VLPs probably undergo remodeling or uncoating as well as active translocation through the nuclear pore. Despite identification of redundant nuclear localization signals (NLS) in both retrotransposon and retrovirus proteins, the actual mechanism of nuclear entry remains obscure. In this study, we examined the role of major capsid structural proteins and the FG nucleoporins (Nups), which line the pore channel, in retrotransposition of the S. cerevisiae LTR retrotransposon Ty3.
Nuclear pore complexes (NPCs) are highly conserved structures, with apparent eightfold rotational symmetry perpendicular to the NE plane and a central aqueous channel. Cytoplasmic filaments and a nuclear basket extend from the NPC faces. Modeling studies have recently concluded that the yeast NPC is composed of 456 subunits, assembled from multiple copies of approximately 30 different proteins termed nucleoporins (Nups) (1). Nups are described as asymmetric if they are strongly biased toward the cytoplasmic or nucleoplasmic face of the pore and symmetric if more centrally distributed. NPC constituents are generally classified as pore membrane, structural, and phenylalanine-glycine (FG) repeat proteins (reviewed in references 45, 54, 56, and 57). Pore membrane proteins anchor the peripheral NPC structures in the pore formed by fusion of the inner and outer nuclear membranes. Structural Nups form inner and outer rings which support nuclear pore architecture and participate in pore biogenesis. In their native state, the domains formed from FG repeats flanked by more polar amino acid spacer sequences are predicted to be unfolded and of potential extended topologies (12, 34). S. cerevisiae encodes 11 FG Nups with such FG domains. This results in approximately 176 unfolded FG domains (1). The FG Nups can be further subdivided according to the predominant specific type of FG repeat in the respective domain. In addition, the FG Nups localize differentially within the NPC. The cytoplasmic face of the NPC is enriched for two FG Nups, Nup42 and Nup159. The nucleoplasmic face is enriched for three Nups with FxFG (x is any amino acid) repeats, Nup1, Nup2, and Nup60. Nsp1, which is located centrally inside the pore and distributed symmetrically, has both FG and FxFG repeats. Nups with GLFG (L = leucine) repeats (Nup49, Nup57, Nup100, Nup116, and Nup145) are collectively located more centrally (46).
The NPC imposes a diffusion barrier to macromolecules greater than ∼30 kDa (17, 41). Recent measurements of both the effects of asymmetric FG domain absence and symmetric FG domain interactions suggest that members of the symmetric GLFG domain subclass may interact to form a centrally located meshwork, whereas a subset of noninteracting asymmetric FxFG Nups might be required for fully normal gating function (16, 42). Proteins larger than the diffusion limit are translocated into the nucleus via transport receptors, which predominantly belong to the karyopherin β (Kapβ; importin β) family of proteins (43). Kapβ transporters interact with the transport substrate NLS and with FG Nups. Collectively, the FG Nups are required for facilitated diffusion of macromolecules through the NPC. The reticulum of FG repeats undergoes cycles of association and dissociation with Kapβ transporters or cargos (17, 43, 52). Interactions with nuclear-localized Ran-GTP trigger release of translocated cargo, thus conferring directionality. S. cerevisiae has 14 Kapβ transporters and 1 Kapα, Kap60, which acts as a substrate binding adapter for Kapβ Kap95. The Kap60/95 complex imports proteins containing the classic monobasic cNLS, and other karyopherins interact preferentially with other NLS types, which have been defined to various extents (43). Interestingly, there are distinct requirements for subsets of FG domains (51, 54). None of the 11 individual FG domains are essential for viability. Thus, the functions of specific FG domains have been examined using yeast strains harboring multiple in-frame deletions of the respective FG-encoding sequences (42, 51, 54, 63). Two other Nups, Nup53 and Nup59, are not amenable to this analysis, as they have few FG repeats and these are not tightly clustered. Surprisingly, cells lacking all five asymmetric FG domains (cytoplasmic FG and nucleoplasmic FxFG) are viable (51). However, all the GLFG domains cannot be simultaneously deleted, indicating that these perform nonredundant functions (51, 56). Most intriguing, deletion of certain FG domains attenuates the transport rate of specific karyopherins, indicating that the roles of specific FG Nups are not redundant and there are transport pathway preferences (51, 54).
Evidence from both retroviruses and retrotransposons suggests that multiple subdomains potentially contribute to passage of particle components through the NPC. Human immunodeficiency virus type 1 (HIV-1), which infects nondividing cells, has multiple components, including the matrix (MA), integrase (IN), Vpr, and cDNA flap with NLS activity that is expected to mediate interaction with Kaps. Surprisingly, these signals are dispensable for HIV-1 infection of nondividing cells (reviewed in references 13 and 53). In contrast to HIV-1, Moloney murine leukemia virus (Mo-MLV) does not infect nondividing cells. HIV-1 containing substitutions of MoMLV MA and capsid (CA) for the corresponding domains of HIV-1 or mutated in the CA domain was also blocked for infection of nondividing cells, indicating that this property is conferred by CA (59, 60). HIV-1 CA does not contain a known NLS motif, and the precise mechanism of nuclear entry remains unclear. Rous sarcoma virus Gag (10) and foamy virus Gag (48, 62) have NLS activity. However, this activity has not been shown to mediate infection in nondividing cells, so the relationship with preintegration complex trafficking, if any, is also unknown.
Similar to retroviruses that infect nondividing cells, a subset of retrotransposon proteins must transit into the nucleus together with the cDNA. The major Ty3 structural polyprotein, Gag3, is processed into CA, spacer (SP), and nucleocapsid (NC) during Ty3 VLP maturation. The amino-terminal domain (NTD) of CA is hypothesized to be similar to retroviral CA in forming the outer surface of the particle (32). POL3 encodes protease (PR), reverse transcriptase, and IN. Immature Ty3 VLPs range in size from 44 to 53 nm (29). Thus, given the ∼39-nm maximal transport capacity of the NPC (41), remodeling of the immature particle, perhaps in association with maturation, must occur prior to nuclear entry. Similar to HIV-1 IN, Ty3 IN contains an NLS competent to mediate nuclear localization of a heterologous protein (35). Ectopically expressed IN of the distantly related Ty1 also displays NLS activity (26, 38), and nuclear localization of Ty1 IN was recently shown to be Kap60/Kap95 dependent (36). However, based on similarities of proteins and particle structure with retroviruses, it is doubtful that these retrotransposon IN NLS motifs are exposed in the immature VLPs. In the case of another metavirus element, Tf1 in Schizosaccharomyces pombe, multiple independent subdomains of Gag displayed NLS activity (11).
Genome-wide analysis of the host factors important for Ty3 transposition (6, 24) underscored the importance of NPC components for transposition. The Nup84 subcomplex (Nup84, Nup85, Nup120, Nup133, Nup145C, Seh1, and Sec13) lies in the outer ring of the pore and participates in nuclear pore biogenesis (1). Mutants with genes deleted that encode members of the Nup84 subcomplex, NUP84, NUP133, and NUP120, were identified with elevated transposition phenotypes in the screen of the knockout collection (24), suggesting that the Nup84 subcomplex restricts Ty3 nuclear entry. On the other hand, deletion of various FG-Nups resulted in variable changes in transposition levels. For example, a nup100Δ strain and a strain with truncated NUP159 had elevated transposition, but a nup59Δ strain and a strain with truncated NUP116 exhibited decreased transposition.
The present study was undertaken to elucidate the role of Nups in Ty3 nuclear entry. Cells expressing Ty3 accumulate P-body and Ty3 components into one or two perinuclear foci (8), which we show here are physically associated with pores. A collection of yeast strains with minimal subsets of FG Nup domains (51, 54) was used to test for roles of FG domain subtypes in Ty3 transposition. Gag3 interacted directly with GLFG repeats, and deletion of these Nup domains decreased transposition. We hypothesize that Ty3 particles dock on the NPC via interactions between Gag3 and GLFG Nups; uncoating and nuclear entry are promoted by interactions between Kaps and internal particle components, including NC and IN.
MATERIALS AND METHODS
Yeast and bacterial strains and culture conditions.
Yeast and bacterial culture methods were as previously described except where noted (3, 5). Bacterial strain DH5α [F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ−] (Invitrogen, Carlsbad, CA) was used for plasmid preparations, and Rosetta [F− ompT hsdSB(rB− mB−) gal dcm (DE3) pRARE (Camr)] (Novagen) was used for recombinant protein expression and purification. Genotypes of the yeast strains used in this study are detailed in Table S1 of the supplemental material. Yeast strain BY4741 (his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Open Biosystems, Huntsville, AL) was used in fluorescence experiments localizing Gag3 and its subdomains. Derivatives of BY4741 used to examine VLP localization and NC nuclear entry were nup120Δ, nup133Δ, kap108Δ, kap114Δ, kap120Δ, kap122Δ, kap123Δ, kap142/msn5 Δ, and los1Δ (Open Biosystems). Strain yTM443 (trp1-H3 ura3-52 his3-200 ade2-101 lys2-1 leu1-12 can1-100 bar1::hisG GAL+ ΔTy3) (37) was used to prepare extracts for affinity capture assays. Strains SWY 2283 (ura3-1 his3-11,15 leu2-3,112 lys2) and SWY2284 (ura3-1 his3-11,15 leu2-3,112 trp1-1) are the strains from which the ΔFG mutants were derived; they were used for all comparisons to the ΔFG mutants (51). Cells with prototrophic markers were selected by growth on synthetic dextrose (SD; 0.67% yeast nitrogen base, 2% dextrose), synthetic galactose (SG; 0.67% yeast nitrogen base, 2% galactose), or synthetic raffinose (SR; 0.67% yeast nitrogen base, 1% raffinose, 2% [vol/vol] glycerol, 2% [vol/vol] lactic acid) medium lacking the amino acid provided for by the prototrophic marker. To induce Ty3 expression for Southern blot analysis and to obtain cell extracts for affinity capture assays, cells transformed with corresponding plasmids were grown in SR to late logarithmic phase, diluted in SR to an optical density at 600 nm of 0.35, and induced for 24 h by addition of galactose to a final concentration of 2%.
Plasmids.
Plasmids carrying recombinant Nups were as follows: glutathione-S-transferase (GST) vector pGEX-2TK (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), with GST-NUP59 (full length), and GST-NUP159 (codons 441 to 881) (gift of M. Rexach, University of California, Santa Cruz) (2); pGEX-2T vector with GST-NUP116 (codons 161 to 730) (GST-GLFGNup116), pGEX-3X vector with GST-NUP100 (codons 2 to 610) (GST-GLFGNup100) (50), pGEX-3X vector with GST-NSP1 FxFG (codons 167 to 600) (GST-FxFGNsp1), and GST-NUP42 (full-length) GST-FGNup42. Wild-type (wt) Ty3 and mutant derivatives cloned under the control of the GAL10 upstream activating sequence (UAS) were expressed from a high-copy, URA3-marked shuttle vector, pDLC201, to obtain protein for affinity capture assays (19, 30, 32). The pET29a plasmid (Novagen, Gibbstown, NJ) with Gag3 and its subdomains cloned between NdeI and XhoI sites was used to express Ty3 protein in Escherichia coli for affinity capture assays. For direct fluorescence microscopy, plasmid pSSL2010 (35) was modified to allow expression of Gag3 and its subdomains fused to a tandem repeat of the green fluorescence protein (2×GFP). Ty3 marked with an antisense insertion of HIS3 (Ty3-HIS3) was expressed under the control of the GAL10 UAS from a low-copy-number plasmid (pNB2361) in order to measure transposition. Plasmids are summarized in Table S2 of the supplemental material.
Transposition assays.
Mutant and parent strains were transformed with plasmid pNB2361 carrying Ty3-HIS3, and His+ Ura+ transformants were selected on SD −Ura −His medium. Four independent transformants of each type were patched onto the same selective medium. After 2 days at 24°C, cells were replica plated to SG −Ura −His at 24°C to induce Ty3 transposition and after 3 days to YPD to allow plasmid loss. After 1 day on YPD, cells were replica plated to SD −His supplemented with 5′ fluoro-orotic acid (2 mg/ml) in order to select for His+ cells that had acquired a genomic Ty3-HIS3 and lost the URA3-marked donor plasmid. Cells that had undergone transposition formed papillations on selective medium, and the phenotype was scored (+ to ++++, or − for no papillations).
Southern blot analysis of Ty3 cDNA.
Mutant and parent strains were transformed with Ty3 expression plasmid pDLC201. Transformants were grown in SR −Ura, and Ty3 expression was induced by growth in SG −Ura for 24 h at 24°C. DNA was extracted and Southern blot analysis of Ty3 cDNA was performed (32).
Fluorescence and electron microscopy.
For microscopy cells were grown in appropriate dropout medium to an optical density at 600 nm of 0.2 to 0.35, induced by addition of galactose to a final concentration of 2%, and grown at 24°C. Cells and slides were prepared as described previously (40). For immunofluorescence cells were incubated with anti-CA rabbit polyclonal immunoglobulin G (IgG; diluted 1:1,000) (37) and anti-Nsp1 mouse monoclonal IgG (diluted 1:500; EnCor Biotechnology, Inc., Gainesville, FL) overnight in a moist chamber at 24°C. Rabbit IgG was detected with Alexa 488-conjugated goat anti-rabbit IgG (2 mg/ml; Sigma-Aldrich, Inc., St. Louis, MO) in phosphate-buffered saline (PBS)-bovine serum albumin (20 mg/ml) at a 1:1,000 dilution. Mouse IgG was detected with Alexa 568-conjugated goat anti-mouse IgG (2 mg/ml; Sigma-Aldrich, Inc.) in PBS-bovine serum albumin at a 1:1,000 dilution. Cells were visualized using a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss Inc.) as described previously (8). Electron microscopy (EM) was performed as described previously (32).
Preparation of recombinant proteins for affinity capture assays.
Induction and purification of N-terminal GST Nup fusions from E. coli were performed essentially as described previously (2). After purification on glutathione-coated Sepharose beads (Sigma-Aldrich, Inc., St. Louis, MO), eluted proteins were dialyzed into lysis buffer [20 μM HEPES, pH 6.8, 150 μM KOAc, 250 μM sorbitol, 2 μM Mg(OAc)2] using Spectra/Por membrane (molecular weight cutoff, 6 to 8,000; Spectrum Laboratories, Rancho Dominguez, CA). Concentrations of purified proteins for binding assays were determined by the method of Bradford (9).
Affinity capture assays.
For affinity capture from total yeast extracts, cultures of yTM443 transformed with pDLC201 and its mutant derivatives were induced for Ty3 expression for 24 h, and cell extracts were prepared by breaking cells with glass beads in binding buffer [20 μM HEPES, pH 6.8, 150 μM KOAc, 250 μM sorbitol, 2 μM Mg(OAc)2] (2). One hundred micrograms of total cell extract was incubated with GST-Nup protein bound to glutathione-coated Sepharose beads. For affinity capture from E. coli extracts, Gag3 and its subdomains fused at the C-terminal end to His6-tagged proteins (Gag3 or Gag3 subdomains-His6, etc.) were expressed in E. coli under the T7 promoter. Gag3-His6 was expressed at 4°C to increase protein solubility, whereas His6-tagged Gag3 subdomains were expressed at 24°C and IN-His6 was expressed at 30°C. Cell extracts were prepared by sonication in the same lysis buffer as used for yeast cells. Relative amounts of His6-tagged protein in each extract were estimated by immunoblot analysis (see below) using dilutions of the total cell extracts. Approximately equal amounts of each protein were incubated with GST-Nup protein bound to glutathione beads. Affinity capture assays were performed as described previously (2).
Immunoblot analysis.
Elution fractions from affinity capture assays using yeast lysates were analyzed with a polyclonal anti-GST IgG (Sigma-Aldrich, Inc.) diluted 1:20,000, anti-CA IgG diluted 1:20,000, and anti-IN IgG diluted in 1:5,000 in 1× PBS-0.1% Tween 20-2.5% milk. Elution fractions from affinity capture of His6-tagged proteins in bacterial lysates were analyzed with antipolyhistidine IgG (Sigma-Aldrich, Inc.) diluted 1:1,000. Subsequent incubations with secondary antibodies and detection were as described previously (32).
RESULTS
The current study was undertaken in order to examine whether Gag3, the primary component of the Ty3 VLP, has a role in nuclear entry of a Ty3 integration-competent species.
Ty3 VLPs are physically associated with nuclear pore complexes.
Mutations which disrupt the Nup84 NPC subcomplex, such as deletion of genes encoding subunits Nup120 and Nup133, inhibit NPC biogenesis and result in clusters of NPCs in localized NE regions (22, 33, 49). Since the majority of NPC are included in these clusters, if Ty3 VLPs were physically associated with NPC, this would result in clear colocalization of Ty3 VLPs and NPC in mutant strains. Such colocalization with NPC clusters is a common test for distinguishing between NPC versus NE association. Therefore the localization of Ty3 VLP clusters and NPC clusters in nup120Δ mutant cells was examined. Wild-type and nup120Δ mutant cells were transformed with high-copy-number Ty3 expression plasmid pDLC201, induced for Ty3 expression for 2, 4, and 6 h, and subjected to immunofluorescence microscopy using anti-CA IgG to visualize Ty3 and anti-Nsp1 IgG to visualize NPCs (Fig. 1A). In wt cells the NPCs were observed as red fluorescence (Nsp1 signal) ringing the 4′,6-diamidino-2-phenylindole-stained DNA. In contrast, in mutant cells, the Nsp1 signal showed bright, enlarged foci representing the collection of NPCs at the nuclear periphery, as is characteristic of clustered NPCs (22). At early times after induction, small Ty3 clusters observed as green fluorescence were scattered in wt and nup120Δ cells. However, by 6 h, large Ty3 clusters were observed in the proximity of the nucleus in a majority of cells. In nup120Δ cells, 92% of these clusters were adjacent to or overlapping NPC clusters. Ty3 particle clusters observed by EM also localized adjacent to nuclear pores in wt cells (Fig. 1B). A pattern of Ty3 particles coinciding with NPC foci, similar to that of NPC foci in nup120Δ, was observed in the nup133Δ strain at 6 h postinduction (data not shown). The association between Ty3 clusters and NPC in these mutants argues that Ty3 VLPs interact physically with the NPC during the time that complete cDNA products can first be detected. In spite of the association of wt Ty3 VLPs with NPCs, wt Ty3 components are not readily observed within the nuclei of wt cells expressing Ty3 (unpublished data).
FIG. 1.
Ty3 VLPs localize in the proximity of nuclear pores. (A) Ty3 VLP clusters colocalize or overlap with pore clusters in the nup120Δ mutant. wt and nup120Δ cells were transformed with pDLC201 Ty3 expression plasmid, grown in selective medium, and induced for Ty3 expression for 2, 4, or 6 h. Ty3 was visualized using anti-CA IgG and goat anti-rabbit IgG conjugated to Alexa 488 (green). Nuclear pores were visualized using anti-Nsp1 IgG and goat anti-mouse IgG conjugated to Alexa 568 (red). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). White arrows indicate specific examples of colocalized Ty3 and Nsp1 clusters. Bar (lowest right panel), 5 μm. Ty3 and nuclear pore cluster localization assessment was performed in 116 cells, among which 92 cells had one Ty3 cluster, 17 cells had two Ty3 clusters, and 7 cells had three Ty3 clusters, for the total of 147 clusters. Of these, 135 clusters (92%) were adjacent to or overlapped with the pore clusters. (B) EM of perinuclear VLP cluster in wt cells. C, cytoplasm; N, nucleus; the arrow points to a nuclear pore. Bar, 100 nm.
Nup GLFG repeat domains play a role in Ty3 transposition.
Since Ty3 clusters appeared physically associated with NPCs in the nup120Δ and nup133Δ mutant cells, we next asked whether functional interactions between Ty3 proteins and specific NPC components could be identified. Previous genome-wide mutant screens for genes which affect transposition (6, 24) implicated several FG-type Nups. However, as discussed above, FG Nups have redundant functions, so that a single deletion may not be sufficient to uncover function in Ty3 nuclear entry. We therefore undertook a qualitative assessment of the effect of internal in-frame deletions of sequences encoding specific types of FG repeat domains, both singly and in combination. Nup mutant strains were transformed with the low-copy-number plasmid pNB2361 carrying Ty3-HIS3, and cells were assayed for transposition (see the summary of results in Table 1). Of the nine single GLFG or FxFG Nup deletion mutants tested, five showed no difference from wt in Ty3 transposition efficiency. Although nup116ΔGLFG showed wt levels of transposition, a truncation mutation which eliminated most of the coding domain decreased Ty3 transposition (6), indicating that there may be Ty3-related functions of the non-GLFG repeat portion of this Nup. In contrast, Ty3 transposition was slightly decreased in nup49ΔGLFG and nup145ΔGLFG mutants and slightly increased for nup57ΔGLFG and nup100ΔGLFG mutants (Table 1).
TABLE 1.
Transposition rates and cDNA production
| Strain group | Strain | MAT type | Descriptiona | Transposition at 24°Cb | cDNAc |
|---|---|---|---|---|---|
| Wild type | SWY2283 | a | wt | +++ | 1.0 |
| SWY2284 | α | wt | +++ | 1.0 | |
| Single FxF(G) and GLFG deletion mutants | SWY2801 | α | nup1ΔFxFG | +++ | ND |
| SWY2731 | a | nup2ΔFxFG | +++ | 0.4 | |
| SWY2772 | a | nup60ΔFxF | +++ | ND | |
| SWY2922 | a | nsp1ΔFGΔFxFG | +++ | 1.0 | |
| SWY2825 | a | nup49ΔGLFG | ++ | 0.8 | |
| SWY2869 | a | nup145ΔGLFG | ++ | 0.4 | |
| SWY2754 | a | nup57ΔGLFG | ++++ | 0.6 | |
| SWY2764 | a | nup100ΔGLFG | ++++ | 0.5 | |
| SWY2789 | a | nup116ΔGLFG | +++ | 4.2/1.7** | |
| Double GLFG deletion mutants | SWY2882 | a | nup49ΔGLFG nup57ΔGLFG | ++ | 1.2 |
| SWY2916 | a | nup116ΔGLFG nup145ΔGLFG | + | 1.6/2.5** | |
| SWY2819 | a | nup116ΔGLFG nup57ΔGLFG | +++* | 1.7/2.2** | |
| SWY2841 | a | nup116ΔGLFG nup49ΔGLFG | + | 1.7 | |
| SWY2973 | a | nup100ΔGLFG nup145ΔGLFG | ++ | 0.5 | |
| SWY2785 | a | nup100ΔGLFG nup57ΔGLFG | +++ | 0.5 | |
| SWY2835 | a | nup100ΔGLFG nup49ΔGLFG | ++ | ND | |
| SWY2924 | a | nup145ΔGLFG nup57ΔGLFG | ++ | 0.7 | |
| Triple symmetric deletion mutants | SWY2968 | a | nup100ΔGLFG nup145ΔGLFG nup49ΔGLFG | − | 5.3*** |
| SWY2951 | a | nup100ΔGLFG nup145ΔGLFG nup57ΔGLFG | + | 1.3 | |
| SWY2953 | α | nup100ΔGLFG nup145ΔGLFG nup57ΔGLFG | + | 1.3 | |
| SWY3012 | a | nup100ΔGLFG nup57ΔGLFG nsp1ΔFGΔFxFG | + | 1.5 | |
| SWY2980 | a | nup100ΔGLFG nup145ΔGLFG nsp1ΔFGΔFxFG | ++ | 1.8 | |
| Asymmetric mutants | SWY2844 | a | ΔC | +++ | 0.5 |
| SWY2852 | α | ΔC nsp1ΔFG | ++++ | 1.8*** | |
| SWY2897 | α | ΔN | ++ | 0.4 | |
| SWY2905 | α | ΔN nsp1ΔFxFG | +++ | 0.7/1.2** | |
| SWY3062 | a | ΔNΔC nsp1ΔFGΔFxFG | ++ | 1.26 | |
| SWY3063 | α | ΔNΔC nsp1ΔFGΔFxFG | + | 0.9*** | |
| SWY2971 | α | ΔNΔC | ++ | 0.2 | |
| Asymmetric + GLFG deletions | SWY3462 | a | ΔNΔC nup145ΔGLFG | ++ | 0.3 |
| SWY3410 | α | ΔNΔC nup57ΔGLFG | +++ | 0.7 | |
| SWY3930 | a | ΔNΔC nup57ΔGLFG | +++ | 1.4 | |
| SWY3043 | a | ΔNΔC nup100ΔGLFG | +++ | 0.3 | |
| Slow-growing strains | SWY2965 | a | nup145ΔGLFG nup49ΔGLFG | ND | 0.5 |
| SWY2871 | a | nup100ΔGLFG nup49ΔGLFG nup57ΔGLFG | ND | 1.2 | |
| SWY3008 | a | nup100ΔGLFG nup49ΔGLFG nsp1ΔFGΔFxFG | ND | No cDNA | |
| SWY3603 | α | ΔNΔC nup116ΔGLFG | ND | No cDNA |
ΔC, Nup42ΔFG Nup159ΔFG; ΔN, Nup1ΔFxFG Nup2ΔFxFG Nup60ΔFxF.
Cells that had undergone transposition formed papillations on selective medium, and the phenotype was scored for three to four transformants for each mutant (+ to ++++; − indicates no papillations). *, results shown are for three of four transformants tested (reduced for one of four).
For cDNA analysis, two transformants were tested unless stated otherwise. The cDNA-to-plasmid ratio was quantified using Quantity One software (Bio-Rad, Hercules, CA) and was normalized to the ratio for the wt, which was designated as 1. One of the two transformants is shown if the difference between levels of cDNA compared to wt was less than 0.2. **, results shown are for two transformants with a cDNA difference of more than 0.2; ***, only one transformant was tested; ND, not determined.
Each of the three FG Nup repeat classes (FG, FxFG, and GLFG) is represented by multiple members, so that even deficiencies in a single member contributing to transposition would not necessarily affect transposition at a detectable level if there were functional redundancy among FG Nups of the same repeat type. Transposition was therefore assayed in mutants containing combinations of FG domain deletions. Despite the collection of VLPs at the NPC, deletion of FG repeats in Nups on the cytoplasmic NPC face (ΔC) did not have a significant effect on transposition. Deletion of all three of the FxF(G) repeats on the nucleoplasmic NPC side (ΔN) resulted in only a modest decrease in transposition. We observed reduced Ty3 transposition in the ΔNΔC mutant, which lacks all asymmetric FG domains.
Nsp1, a central channel Nup, is unique among the FG Nups in that it contains repeats of both the FG and FxFG classes. Mutant nsp1ΔFGΔFxFG had a wt transposition frequency, but combining nsp1ΔFxFG with ΔN actually suppressed the negative effect of deleting the nuclear basket FxFG repeat domains. Combining the nsp1ΔFG mutation with ΔC deletions increased transposition. This result suggested that FxFG Nup domains are not absolutely essential for wt levels of transposition and that there is complex interplay between the central channel and nuclear basket FG Nups.
We have previously used the ΔNΔC mutant as a platform for investigating transport through NPCs with a limiting number of FG repeats (54). To this end, we assayed Ty3 transposition in mutants lacking all asymmetric FG domains and also lacking one symmetric FG domain. Combining nup57ΔGLFG or nup100ΔGLFG, each of which had slightly elevated transposition, with ΔNΔC resulted in wt levels of transposition. Combining nup145ΔGLFG with ΔNΔC did not further reduce transposition beyond the reduction observed for the individual mutations. Transposition was considerably reduced upon simultaneous deletion of all FG and FxFG repeats (ΔNΔC nsp1ΔFGΔFxFG), including nsp1ΔFG and nsp1ΔFxFG, but the specificity of this effect was not clear because these mutants grow very slowly (reference 51 and unpublished data).
Mutants harboring single deletions of the GLFG repeat domains were the only class of single deletions that reduced Ty3 transposition. As noted above, decreased Ty3 transposition was observed with the nup145ΔGLFG and nup49ΔGLFG single-deletion mutants. The cumulative effect of multiple GLFG domain deletions was investigated using viable combinations of nup116ΔGLFG, nup100ΔGLFG, nup145ΔGLFG, nup49ΔGLFG, and nup57ΔGLFG. Since nup116ΔGLFG in combination with more than one of the other GLFG domain deletions is inviable (51), triple-deletion combinations with nup116ΔGLFG could not be studied. A number of viable strains carrying multiple deletions that included deletion of one or more of the other GLFG domains exhibited slow growth and could not be assayed reliably for transposition (see Table 1).
Although the nup116ΔGLFG mutant alone showed no defect, addition of nup116ΔGLFG to the nup145ΔGLFG mutant or to the nup49ΔGLFG mutant exacerbated the decrease in transposition caused by nup145ΔGLFG or nup49ΔGLFG alone (Table 1). Different combinations of the deletions of three GLFG domains at a time (or two GLFG domains and Nsp1 FG and FxFG domains, which phenocopy triple GLFG mutants) resulted in severe transposition reduction (Table 1). The similar transport phenotypes for all triple symmetric deletion mutants was not unexpected, as their other transport defects are indistinguishable (51).
We observed reduced Ty3 transposition in the ΔNΔC mutant, which lacks all asymmetric FG domains. We have previously used the ΔNΔC mutant as a platform for investigating transport through NPCs with a limiting number of FG repeats. To this end, we assayed Ty3 transposition in mutants lacking all asymmetric FG domains and also lacking one symmetric FG domain. Combining nup57ΔGLFG or nup100ΔGLFG, each of which had slightly elevated transposition, with ΔNΔC resulted in wt levels of transposition. Combining nup145ΔGLFG with ΔNΔC did not further reduce transposition beyond the reduction observed for the individual mutations. In general, GLFG domain deletions decreased transposition frequency in proportion to the number of GLFG domain deletions. These results argue that the GLFG Nups are not redundant with the asymmetric Nups but rather play a special role in Ty3 transposition. These results also suggest that a subset of nucleoplasmic FxFG Nups is required for efficient transposition.
GLFG repeat domains play a postreplication role in transposition.
Although deletion of GLFG and FxF(G) domains decreased transposition, it is possible that this decrease resulted from an indirect effect on a function required for Ty3 transposition rather than directly affecting nuclear entry of Ty3 VLPs or PICs. Such an indirect effect could block transposition at any stage of the replication cycle, whereas a disruption of Ty3 nuclear entry would necessarily occur late, for example, subsequent to cDNA production. Nup mutants with decreased transposition were therefore examined for cDNA production. Mutants were transformed with pDLC201, which carries Ty3 under the control of the GAL10 UAS, and induced to express Ty3 by growth in galactose as a carbon source for 24 h. Ty3 cDNA production was assayed by Southern blot analysis (Table 1). Two mutants (ΔNΔC nup116ΔGLFG and nup100ΔGLFG nup49ΔGLFG nsp1ΔFGΔFxFG), which grew too poorly to assay for transposition, did not produce detectable cDNA. Remarkably, triple symmetric FG domain deletion combinations (nup100ΔGLFG nup145ΔGLFG nup57ΔGLFG, nup100ΔGLFG nup57ΔGLFG nsp1ΔFGΔFxFG, and nup100ΔGLFG nup145ΔGLFG nsp1ΔFGΔFxFG) and double GLFG domain deletions (nup57ΔGLFG nup49ΔGLFG, nup116ΔGLFG nup145ΔGLFG, nup116ΔGLFG nup57ΔGLFG, and nup116ΔGLFG nup49ΔGLFG) had similar or slightly higher than wt amounts of cDNA and reduced transposition (Table 1). These GLFG mutants therefore affected Ty3 at a postreplication stage of transposition. The strain with a single GLFG deletion (nup116ΔGLFG), which had wt transposition, displayed elevated amounts of cDNA. On the other hand, deletion of the nuclear FxF(G) domains decreased the amount of cDNA, and so that analysis did not provide support for a late-acting effect.
Ty3 VLPs associate with GLFG repeat domains.
If Ty3 VLPs interact with Nups, this would likely be mediated by Gag3, the major structural protein of the VLP (18). In order to test this hypothesis, we utilized affinity capture assays with recombinant GST-Nup fusions as bait for Ty3 from total yeast extracts. The FG repeat domains of Nup159 and Nup42, FxFG repeat domain of Nsp1, the GLFG repeat domains of Nup100, and Nup116, and Nup59, a Nup lacking dense FG repeats, were expressed in E. coli as N-terminal GST fusions. GST-Nup fusions were purified and bound to glutathione beads for capture assays. Cell extracts containing Ty3 VLPs were prepared from yeast strain yTM443 transformed with Ty3 plasmid pDLC201 and induced to express Ty3 by growth in galactose as a carbon source. Glutathione-coated Sepharose beads were incubated with purified GST-Nup fusions followed by incubation with yeast cell extracts containing the Ty3 VLPs. Bound proteins were eluted with glutathione. Immunoblot analysis of elution fractions using anti-GST revealed that similar amounts of GST-Nup fusions were bound to the beads (Fig. 2, top panel). Immunoblot analysis with anti-CA, which reacts with Gag3 polyprotein, intermediate processing forms, and CA, showed no detectable retention of Gag3 by GST alone or GST-Nup59, GST-FGNup159, GST-FGNup42, or GST-FxFGNsp1. We observed a small amount of CA binding to GST-Nup59 and GST-FGNup42. Binding of Gag3, processing intermediate p27, and CA to both GST-GLFGNup100 and GST-GLFGNup116 repeats was readily detected (Fig. 2, middle panel). These results suggested that Gag3 binds specifically to Nups that contain GLFG repeats, and not to Nups with other classes of FG repeats. Gag3 could associate with NPCs prior to assembly, or later, as part of the VLP. In the latter case, in addition to CA other mature VLP proteins such as IN should be detectable. Immunoblot analysis using anti-IN showed that IN was present in the same elutions as Gag3, consistent with association of VLPs with the GST-GLFG repeats (Fig. 2, bottom panel).
FIG. 2.
Ty3 particles associate with GLFG domains of Nups in vitro. GST fusions to Nups or domains of functional FG repeats were expressed in E. coli and purified using glutathione-Sepharose beads, and similar amounts were used as bait in the affinity capture assay with extracts of yTM443 cells induced to express Ty3 for 24 h (see Materials and Methods). Elution fractions were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblot analysis was performed using anti-GST (top panel), anti-CA (middle panel), and anti-IN (bottom panel) IgG.
Gag3 directly interacts with the GLFG Nups.
The foregoing experiments demonstrated interaction of Ty3 Gag3 with the Nup GLFG repeats, but because these experiments involved whole-cell extracts, they did not address whether the interaction was direct or indirect. In order to investigate whether Ty3 Gag3 or IN associates directly with GLFG Nups, purified recombinant His6-tagged Ty3 Gag3 and IN were tested for GST-GLFGNup116 interaction in vitro (Fig. 3A). Similar amounts of His6-tagged Gag3 and IN proteins were used in the affinity capture experiment. GST alone failed to interact with His6-tagged recombinant proteins. The measurement of His6-tagged IN binding to GST-GLFGNup116 was not above background. However, His6-tagged Gag3 bound GST-GLFGNup116 efficiently. This result suggested that particle Gag3 or its subdomains interact with GLFG motifs in vivo via Gag3 and that the IN interaction was more likely to be indirect (Fig. 3A).
FIG. 3.
Interactions of Ty3 VLPs with GLFG repeats are mediated by Gag3. (A) Interactions of IN and Gag3 and subdomains with recombinant GST-GLFG. Extracts of E. coli containing similar amounts of His6-IN, His6-Gag3, and His6-tagged Gag3 subdomains were used in the affinity capture assay. Total cell extracts (lanes 1) and elutions from GST alone (lanes 2) or GST-GLFGNup116 (lanes 3) were probed using an antibody against polyhistidine. The ratio of total extract to total elution loaded was 1:50 except for the His6-IN samples, for which the ratio was 1:42. (B) Interaction of wt and Ty3 ΔSP particles with GST-GLFG. wt Ty3 or Ty3 ΔSP was expressed in yTM443. Cell extracts were used in affinity binding assays and probed as described in the legend for Fig. 2.
In order to determine which domain of Gag3 binds GST-GLFGNup116, His6-tagged Gag3 subdomains (CA, CA NTD, CA CTD, SP-NC, and NC [see Table S2 in the supplemental material]) were constructed and expressed as described above for full-length Gag3. Similar amounts of each recombinant subdomain were used in the affinity capture assay. The His6-Gag3 and His6-SP-NC bound GST-GLFGNup116. However, no binding was observed for His6-NC alone or His6-CA and its subdomains (Fig. 3A). This result suggested that Gag3 binding to Nup116 may be unique to structures available in the intact Gag3 protein or that it may be mediated by the SP domain. If SP were the sole determinant of the Gag3 Nup116 GLFG domain binding, then an SP deletion mutant (ΔSP) of Gag3 would lose the ability to bind GLFGs. To test this possibility, wt Ty3 and its ΔSP derivative were expressed in yTM443 and extracts from these cells were used to perform the affinity capture assay. Gag3 ΔSP bound to GLFGNup116 repeats (Fig. 3B), indicating that Gag3 interaction with Nup116 GLFG is not mediated solely by the SP subdomain.
The Gag3 NTD is important for interaction with GLFG repeats.
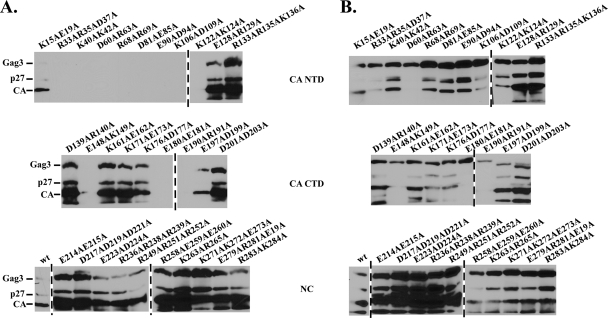
In spite of the fact that recombinant Gag3 bound to GST-GLFGNup116 repeats, Gag3 subdomains, including His6-CA, -CA NTD, -CA CTD, and -NC expressed in E. coli failed to show significant interactions with GST-GLFGNup116 repeats. It is known that Gag3, but not CA, forms structured particles when expressed in E. coli (31). This raised the possibility that individual subdomains of CA and NC expressed in E. coli may fold improperly and lose GLFG binding capability. Therefore, an alternative approach to identifying potential GLFG-binding domains was adopted. Roles of Gag3 subdomains were previously characterized using alanine-scanning mutagenesis (30, 32). A collection of such mutants provided an opportunity to probe Gag3 for residues required for interaction with GST-GLFGNup116 in complexes approximating native VLPs. As described above for wt Gag3, Ty3 elements with mutations in Gag3 were expressed in strain yTM443 by growth in galactose-containing medium for 24 h. Cell extracts containing similar amounts of Ty3 proteins (Fig. 4B) were assayed for binding to GST-GLFGNup116 as described above for wt Ty3 Gag3. Mutants D60A/R63A and E148A/K149A did not assemble (32) and did not interact with GST-GLFGNup116. Mutant E180A/E181A formed long filaments and also showed no interaction. This assay revealed a cluster of mutations in the CA NTD that reduced binding (Fig. 4A, top panel). Although these mutants formed particles (32), they showed reduced processing. This suggested that perhaps only mature particles interact with NPC. In order to specifically test whether failure to process Gag3 resulted in lack of mutant interaction with GLFG repeats, a Ty3 PR catalytic mutant was tested. These unprocessed VLPs interacted with GST-GLFG (data not shown) indicating that loss of GLFG interaction of VLPs with decreased Gag3 processing was not attributable to lack of processing per se. Thus, residues in the Gag3 NTD appear to be essential for wt interactions between VLPs and GLFG repeats. None of the alanine scanning mutations in NC resulted in loss of GST-GLFGNup116 binding (Fig. 4A, bottom panel). These results, together with the previous results, suggest that multiple subdomains in Gag3 have the potential for independent interactions with GLFG repeats, and that residues in Gag3 NTD, SP and possibly NC contribute to these interactions. The implication of the Gag3 NTD in Nup interaction is consistent with the predicted location of the NTD on the outside of the VLP (32).
FIG. 4.
Mutations in the Gag3 NTD result in loss of the GST-GLFGNup116 interaction. Ty3 elements containing alanine-scanning mutations in GAG3 (30, 32) were expressed in yTM443, and cell extracts were used in affinity capture assays as described in the legend for Fig. 2. Immunoblot analysis with anti-CA IgG was performed to analyze the elution fractions (A) and total cells extracts (B). Ty3 Gag3 contained mutations in the CA NTD (top panel), CA CTD (middle panel), or NC (bottom panel).
NC displays NLS activity dependent upon GLFG and Kap123.
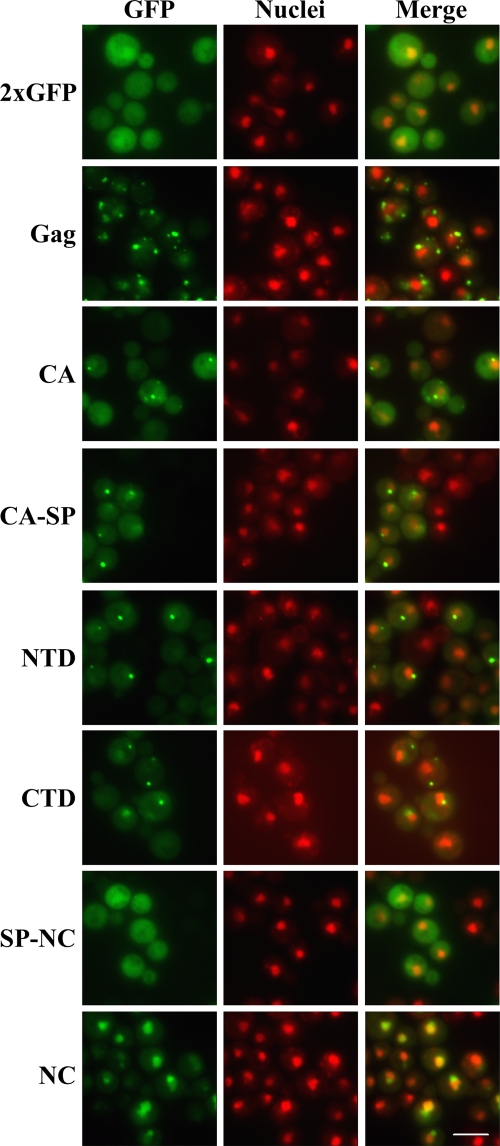
Although extracts of cells containing mutant VLPs suggested a role for the Gag3 NTD in GLFG interactions, an interaction of SP-NC, as suggested by recombinant protein binding studies, would presumably require some degree of VLP dissociation, possibly in the NPC. Evidence for interaction of NC and GLFG Nups was only indirect. SP-NC, not NC alone, interacted with GLFG Nups, but SP was not essential for Gag3-GLFG Nup interaction. It seemed possible that some of these interactions were sensitive to conformational effects and possibly particle maturation as well. Therefore we tested Gag3 and its subdomains CA, CA-NTD, CA-CTD, CA-SP, SP-NC, and NC tagged with a tandem copy of GFP (2×GFP) for the ability to mediate nuclear entry in vivo. After 2 h of expression, the Gag3 fusion formed multiple clusters in the cytoplasm of cells, similar to the early postinduction pattern of the full-length Ty3-GFP element. CA-, CA NTD-, CA CTD-, and CA-SP-2×GFP fusions formed cytoplasmic clusters as well, although fewer in number than Gag3-2×GFP. Some diffuse cytoplasmic protein was also present in these cells. SP-NC-2×GFP fusion protein was diffuse in the cytoplasm but also localized to the nucleus. Nuclear accumulation of the NC-2×GFP reporter resembled the previously described nucleolar localization of IN (35) (Fig. 5). Together these results suggest that Gag3 makes initial NPC contact, but that in the intact molecule, signals for nuclear entry are either occluded or trapped in complexes which physically impede nuclear pore translocation.
FIG. 5.
NC localizes to the nucleus of wt yeast cells. Yeast strain BY4741 was transformed with Gag3-2×GFP or Gag3 subdomain-2×GFP fusion expression plasmids and induced for expression of 2×GFP fusions by growth in galactose-containing medium for 2 h. Direct fluorescence microscopy was performed. Gag3 and its subdomains are green, and nuclei were stained with Hoechst and are pseudocolored red. Bar, 5 μm.
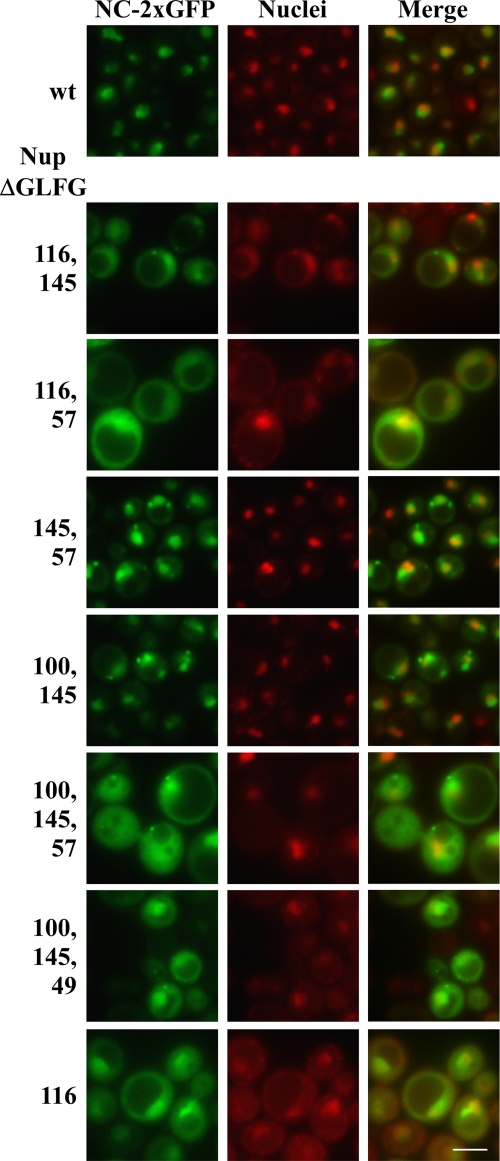
In order to determine whether the GLFG Nups required for transposition were also required for NC-2xGFP nuclear localization and accumulation, NC-2×GFP was tested for localization in Nup mutants previously tested for transposition. Expression of NC-2×GFP was induced in the wt and nup mutant strains for 2 h and localization of NC-2×GFP was examined. Eight of the nine single mutants (nup100ΔGLFG, nup145ΔGLFG, nup49ΔGLFG, nup57ΔGLFG, nup1ΔFxFG, nup2ΔFxFG, nup60ΔFxF, and nsp1ΔFGΔFxFG) showed NC-2×GFP localization that was indistinguishable from wt (Fig. 6 and data not shown). The nup116ΔGLFG mutant did not abolish nuclear localization of NC-2×GFP but showed greater cytoplasmic signal than wt cells or other single ΔFG nup mutants. Because of the apparent redundancy of GLFG domain function, double GLFG deletion mutants were also tested (nup57ΔGLFG nup49ΔGLFG, nup116ΔGLFG nup145ΔGLFG, nup116ΔGLFG nup57ΔGLFG, nup116Δ GLFG nup49ΔGLFG, nup100ΔGLFG nup145ΔGLFG, nup100ΔGLFG nup57ΔGLFG, nup100ΔGLFG nup49ΔGLFG, nup145ΔGLFG nup57ΔGLFG, and nup145ΔGLFG nup49ΔGLFG). These divided into phenotypic groups, one of which showed diffuse cytoplasmic and nuclear NC-2×GFP fluorescence and one of which showed a more concentrated nuclear signal. The nup116ΔGLFG allele combined with any other GLFG deletion resulted in a diffuse cytoplasmic pattern of NC-2×GFP that was similar to what was observed for nup116ΔGLFG alone. Some of these mutant cells also contained NC-2×GFP clusters in the cytoplasm. Mutants that contained combinations of any two other GLFG deletions, but not nup116ΔGLFG, also had cytoplasmic NC foci, frequently several per cell, but differed in that they did not show a diffuse cytoplasmic NC-2×GFP fluorescence. Triple GLFG deletions and triple deletions including nsp1ΔFGΔFxFG, which act similarly to a GLFG deletion in this context (51) (nup100ΔGLFG nup145ΔGLFG nup57ΔGLFG, nup100ΔGLFG nup145ΔGLFG nup49ΔGLFG, nup100ΔGLFG nup57ΔGLFG nsp1ΔFGΔFxFG, and nup100ΔGLFG nup145ΔGLFG nsp1ΔFGΔFxFG) resulted in diffuse cytoplasmic fluorescence and clusters. Similar phenotypes were also seen in double GLFG deletion mutants that included nup116ΔGLFG (Fig. 6 and data not shown). Notably, all of these symmetric ΔGLFG mutants still had some NC in the nucleus. These results suggest a collective role of GLFGs in NC nuclear entry, and indicate a specific role for Nup116. Because the defect in NC-2×GFP nuclear accumulation correlated with a reduction in transposition, these results suggest that NC may enter the nucleus as part of the preinitiation complex (PIC). Supporting this model, similar effects of these mutations on IN-2×GFP (data not shown) were also observed.
FIG. 6.
GLFG repeats facilitate nuclear accumulation of NC. wt and nup deletion strains transformed with the NC-2×GFP plasmid were induced for NC-2×GFP expression for 2 h, and direct fluorescence microscopy was performed. NC-2×GFP is green, and nuclei were stained with Hoechst and pseudocolored red. Bar, 5 μm.
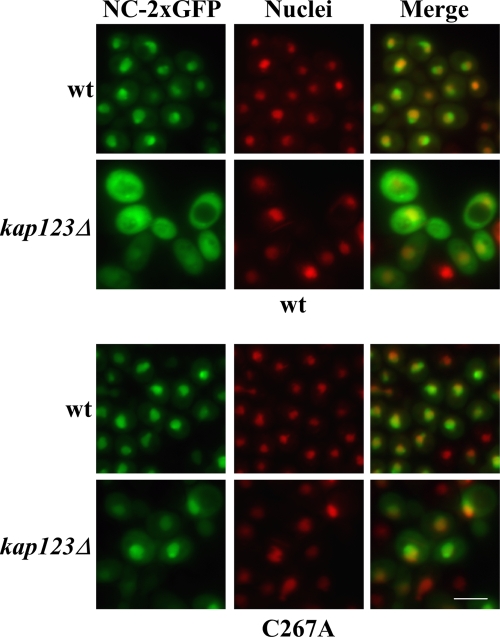
Fluorescence experiments showed that NC has NLS activity and that nuclear entry is dependent upon GLFG domains. However, the in vitro interaction assay results suggested that NC does not interact directly with GLFG Nups. We therefore further investigated whether NC NLS activity was dependent upon a karyopherin. NC possesses a number of positively charged amino acid clusters, especially in the NTD, but no conventional NLS sequence has been identified. There are 14 Kapβs in S. cerevisiae (20, 43), which act to escort substrates through the NPC via interactions with FG Nups. Seven derivatives of the wt S. cerevisiae BY4741 strain, each with one of the seven nonessential karyopherin genes deleted (kap108Δ, kap114Δ, kap120Δ, kap122Δ, kap123Δ, kap142Δ/msn5Δ, and los1Δ) were tested for NC-2×GFP nuclear localization and for Ty3 transposition. Only the strain harboring the kap123Δ allele was significantly reduced for NC-2×GFP concentration in the nucleus (Fig. 7, top panel, and data not shown). Thus, Kap123 could contribute to nuclear entry of NC subdomain-containing species. However, Ty3 transposition in the kap123Δ strain was not reduced compared to the wt (data not shown). Therefore, if Kap123 activity is involved in Ty3 nuclear entry, interaction of the NC subdomain with Kap123 is redundant with other activities, so that Kap123 affects localization, but is not limiting for transposition. Gag3 containing mutations in conserved residues of the NC zinc-binding motif concentrates in the nucleus and fails to make normal particles. It seemed possible that NC expressed as an independent domain in the NC-2×GFP construct might also adopt a nonnative conformation allowing nonphysiologic entry mediated by Kap123. NC(C267A)-2×GFP dependence upon Kap123 was therefore also tested. The zinc-binding motif mutant localized to the nucleus independent of Kap123 (Fig. 7, lower panel). This result is consistent with the requirement of Kap123 by a native form of the Ty3 NC subdomain of Gag3. Together these observations suggest that if uncoating occurs in the NPC, exposed subdomains of Gag3 intermediates have redundant contacts with nuclear import components.
FIG. 7.
wt NC nuclear localization is Kap123 dependent, but zinc-binding domain mutant NC C267A nuclear localization is not. BY4741 and its kap123Δ derivative were transformed with wt NC-2×GFP or NC(C267A)-2×GFP. Cells were induced for NC-2×GFP expression for 2 h, and direct fluorescence microcopy was performed. Nuclei were stained with Hoechst and pseudocolored red. Bar, 5 μm.
DISCUSSION
The current study was undertaken to determine whether perinuclear Ty3-P body clusters observed by fluorescence and EM studies are physically associated with nuclear pore complexes and whether the FG Nups which dominate the NPC channel impede or facilitate access of Ty3 components to the nucleus. Using mutants that have NPCs clustered in the NE, we found that VLP clusters are physically associated with NPCs, arguing that VLPs dock on the NPCs during the normal Ty3 replication cycle. Effects of deleting the FG domains of Nups individually and by class were complex. Recombinant Gag3 interacted with GLFG Nups, and mutations in the Gag3 NTD, which is predicted to lie on the outer surface of the particle, disrupted interaction of VLPs and GLFG Nups. Evidence that multiple subdomains of Gag interact with GLFG domains is consistent with previous suggestions that large cargoes require multiple contacts with FG domains for docking or movement through the NPC (44). For example, 60S ribosomal subunit export involves multiple transport receptors (23, 61). The absence of GLFG Nups decreased transposition, consistent with a significant role for these in transposition. SP-NC, but not NC, interacted with GLFG Nups in vitro, but because SP is likely not exposed in the mature VLP and deletion of SP did not affect VLP association with GLFG Nups, a physiological interaction between SP-NC and GLFG Nups, if it occurs, might be subsequent to VLP docking. Although nuclear concentration of Gag3 zinc-binding domain mutant C267A-2×GFP has been observed (30), nuclear entry of wt Gag3 has not, presumably because multimerization impedes translocation. Investigation of subdomains of Gag3 that might be exposed in the wt remodeled particle showed that ectopically expressed SP-NC was found throughout the cytoplasm and nucleus but NC localized to nuclei. NC-2×GFP, similar to Ty3 IN-2×GFP, concentrated in a subcompartment of the nucleus. Localization of NC-2×GFP was dependent upon Kap123, but transposition was not. Thus, an NC NLS could function redundantly with the IN NLS in vivo. Mutations affecting the IN NLS block transposition in vivo, but also integration in vitro (35). Although localization of both NC-2×GFP and IN-2×GFP is very efficient, no Ty3 protein has been observed in the nucleus in the course of physiologic transposition in a wt strain. This is consistent with extreme restriction of nuclear entry by Gag3 multimers, but could also reflect localization of PICs to the inner surface of the nuclear envelope.
Two negative findings were striking in our study. First, despite the apparent accumulation of VLPs at NPCs, deletion of the asymmetric FG Nup subclass on the cytoplasmic NPC face did not significantly diminish Ty3 transposition. Thus, Ty3 does not require docking on cytoplasmic FG Nups to transit the nuclear pore. Second, simple depletion of more than half of the total wt mass of FG domains (5.27 MDa of repeats) from the pore channel, such as occurs in the ΔNΔCΔnsp1ΔFGΔFxFG mutant to leave 2.47 MDa in repeat mass (51), did not increase Ty3 transposition, indicating that nuclear pore transit is an active process rather than one which is simply impeded by the filamentous network of FG Nups.
An active role for FG Nups in Ty3 nuclear entry is consistent with previous reports from retrotransposon and retrovirus systems. Nuclear entry of S. pombe wt Tf1 Gag and Tf1 transposition is dependent upon SpNup124, which is related to the FxFG ScNup1 in S. cerevisiae (7, 11). Mutations which relieved this dependence also disrupted particle structure, suggesting that particle structure conferred specific requirements for nuclear translocation (27). In spite of similarities in requirements for FG Nups for nuclear pore translocation, there are likely to be differences between the two retrotransposon systems as well. Ty3 nuclear Gag3 is thus far only observed in the case of mutations that either eliminate particle formation or disrupt nuclear pores (30), whereas Tf1 nuclear Gag is observed in transposing populations. In addition, there is a difference in the FG Nup class that appears to be used by these two elements. Neither Ty3 VLPs nor Gag3 interacted in vitro with GST-FxFG Nups. Although deletion of nuclear pore basket FxFG Nups decreased Ty3 transposition, this effect was actually suppressed by further deletion of the Nsp1 FxFG repeat, indicating that FxFG repeats are not essential for wt Ty3 transposition.
We found that SP-NC, which interacted with GLFG repeats, was cytoplasmic and nuclear, but NC-2×GFP, which as an isolated domain did not interact significantly with GLFG repeats in vitro, localized exclusively to the nucleus. This observation suggested that in addition to the Gag3 NTD interaction with Nups, there might be additional localization signals in SP-NC. Testing of cells expressing NC-2×GFP showed that NLS activity was dependent upon Kap123, but transposition was not. Kap123 is implicated in ribosomal protein nuclear import. Alignment of the substrates for Kap123 nuclear localization identified a common basic motif (47) that is similar to the basic domain of Ty3 NC. Kap121 is apparently redundant with Kap123 (39, 47). Although NC-2×GFP did not localize to the nucleus in the kap123Δ mutant, it is possible that the function of Kap121 was sufficient for the level of NC nuclear localization required in transposition. As noted above, in addition to NC, Ty3 IN-2×GFP localizes to a subcompartment of the nucleus. In the case of IN-2×GFP, the subcompartment was shown to be the nucleolus (35). Ty3 is position specific, and its tDNA and 5S gene targets localize to the nucleolus (55). An attractive aspect of a role for Kap123 in Ty3 nuclear entry is that it might further serve to concentrate incoming PICs to the nucleolus. In addition to Ty3, Ty1 has been shown to have NLS activity in the IN CTD (26, 38).
It has long been thought that retroviruses that infect nondividing cells uncoat upon cytoplasmic contact, essentially to a replicating form competent for nuclear entry. HIV-1 cores contain multiple NLS activities, including ones in MA, VPR, IN, and the DNA flap (reviewed in references 13 and 53). However, the ability of HIV-1 to infect nondividing cells was recently mapped to HIV-1 CA determinants in HIV-1-MoMLV chimeras (59) rather than a characterized NLS. Some retroviruses, in addition to HIV-1, infect nondividing cells, although less efficiently (21, 25). In these viruses NLSs have been reported in MA and NC subdomains of Gag (10) as well as IN (28). Indeed, among both DNA and RNA viruses, there are multiple precedents for nuclear entry-associated remodeling. For example, adeno-, herpes- and hepadnaviruses undergo uncoating or some type of particle rearrangement during nuclear entry (58). It was recently proposed that HIV-1 cores dock at the nuclear pore and undergo flap-dependent uncoating prior to nuclear entry (4). This would be consistent with our findings regarding Ty3 VLPs and nuclear pore association. There may be interesting parallels among FG Nup requirements as well. Mammalian Nup153 and Nup98 with homology to yeast FxFG and GLFG repeat nucleoporins have been identified in multiple HIV-1 siRNA host factor screens (9a). Among the five GLFG Nups in yeast, Nup116 appears to be unique, since deletion of its GLFG repeat domain alone reduced nuclear entry of Ty3 NC, whereas deletions of other GLFG domains had cumulative effects. Interestingly, Nup116 is a yeast ortholog of Nup98, which plays a role in the nuclear entry of HIV-1 cDNA (15).
Based on the interaction of FG Nups with major structural proteins of Ty3, we propose a model in which Ty3 and other retrotransposon particles are restructured during nuclear entry: (i) VLPs are destabilized as they undergo maturation and reverse transcription; (ii) VLPs dock at nuclear pores via direct interactions with specific subclasses of FG Nups, further destabilizing the particle; and (iii) karyopherins bind to newly exposed, internal components of the VLP, promoting loss of outer structural proteins and committing the integration-competent species to nuclear entry. In the case of retrotransposons, which assemble in the cytoplasm and mature into stable particles, it is particularly appealing to consider that nuclear entry and particle rearrangement are coordinated processes.
Supplementary Material
Acknowledgments
This research was supported in part by funds from National Institutes of Health grants R01 GM33281 to S.B.S. and R01 GM051219 to S.R.W. The research was also funded in part by National Cancer Institute contract NO1-CO-12400 to K.N. N.B.-B. and L.J.T. were supported by National Institutes of Health Training Grants 5 T32 AI 07319 and 5 T32 CA 009385, respectively.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
We thank M. Oakes (UCI) for assistance with fluorescence microscopy and M. Rexach (UCSC) for sharing GST fusion clones.
Footnotes
Published ahead of print on 16 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alber, F., S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2007. The molecular architecture of the nuclear pore complex. Nature 450:695-701. [DOI] [PubMed] [Google Scholar]
- 2.Allen, N. P. C., S. S. Patel, L. Huang, R. J. Chalkley, A. Burlingame, M. Lutzmann, E. C. Hurt, and M. Rexach. 2002. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1:930-946. [DOI] [PubMed] [Google Scholar]
- 3.Amberg, D. C., D. J. Burke, and J. N. Strathern. 2005. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 4.Arhel, N. J., S. Souquere-Besse, S. Munier, P. Souque, S. Guadagnini, M. Prévost, T. D. Allen, and P. Charneau. 2007. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO 26:3025-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2007. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 6.Aye, M., B. Irwin, N. Beliakova-Bethell, E. Chen, J. Garrus, and S. Sandmeyer. 2004. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics 168:1159-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasundaram, D., M. J. Benedik, M. Morphew, V. D. Dang, and H. L. Levin. 1999. Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol. Cell. Biol. 19:5768-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beliakova-Bethell, N. S., C. Beckham, T. H. Giddings, Jr., M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9a.Bushman, F. D., N. Malani, J. Fernandes, I. D'Orso, G. Cagney, T. L. Diamond, H. Zhou, D. J. Hazuda, A. S. Espeseth, R. Koenig, S. Bandyopadhyay, T. Ideker, S. P. Goff, N. J. Krogan, A. D. Frankel, J. A. T. Young, S. K. Chanda. 2009. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butterfield-Gerson, K. L., L. Z. Scheifele, E. P. Ryan, A. K. Hopper, and L. J. Parent. 2006. Importin-β family members mediate alpharetrovirus Gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 80:1798-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, V. D., and H. L. Levin. 2000. Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol. Cell. Biol. 20:7798-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning, D. P., S. S. Patel, V. Uversky, A. L. Fink, and M. Rexach. 2003. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA 100:2450-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Rijck, J., L. Vandekerckhove, F. Christ, and Z. Debyser. 2007. Lentiviral nuclear import: a complex interplay between virus and host. Bioassays 29:441-451. [DOI] [PubMed] [Google Scholar]
- 14.De Souza, C. P. C., and S. A. Osmani. 2007. Mitosis, not just open or closed. Eukaryot. Cell 6:1521-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebina, H., J. Aoki, S. Hatta, T. Yoshida, and Y. Koyanagi. 2004. Role of Nup98 in nuclear entry of human immunodeficiency virus type 1 cDNA. Microbes Infect. 6:715-724. [DOI] [PubMed] [Google Scholar]
- 16.Frey, S., and D. Görlich. 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130:512-523. [DOI] [PubMed] [Google Scholar]
- 17.Fried, H., and U. Kutay. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen, L. J., D. L. Chalker, K. J. Orlinsky, and S. B. Sandmeyer. 1992. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J. Virol. 66:1414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, L. J., D. L. Chalker, and S. B. Sandmeyer. 1988. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol. Cell. Biol. 8:5245-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harel, A., and D. J. Forbes. 2004. Importin β: conducting a much larger cellular symphony. Mol. Cell 16:319-330. [DOI] [PubMed] [Google Scholar]
- 21.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 75:9526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath, C. V., C. S. Copeland, D. C. Amberg, V. Del Priore, M. Snyder, and C. N. Cole. 1995. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell Biol. 131:1677-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung, N. J., K. Y. Lo, S. S. Patel, K. Helmke, and A. W. Johnson. 2008. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell 19:735-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irwin, B., M. Aye, P. Baldi, N. Beliakova-Bethell, H. Cheng, Y. Dou, W. Liou, and S. Sandmeyer. 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15:641-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenna, M. A., C. B. Brachmann, S. E. Devine, and J. D. Boeke. 1998. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol. Cell. Biol. 18:1115-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, M. K., K. C. Claiborn, and H. L. Levin. 2005. The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in Gag that regulate nuclear localization and particle formation. J. Virol. 79:9540-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kukolj, G., R. Katz, and A. Skalka. 1998. Characterization of the nuclear localization signal in the avian sarcoma virus integrase. Gene 223:157-163. [DOI] [PubMed] [Google Scholar]
- 29.Kuznetsov, Y. G., M. Zhang, T. M. Menees, A. McPherson, and S. Sandmeyer. 2005. Investigation by atomic force microscopy of the structure of Ty3 retrotransposon particles. J. Virol. 79:8032-8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen, L. S. Z., N. Beliakova-Bethell, V. Bilanchone, M. Zhang, A. Lamsa, R. DaSilva, G. W. Hatfield, K. Nagashima, and S. Sandmeyer. 2008. Ty3 nucleocapsid controls localization of particle assembly. J. Virol. 82:2501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen, L. S. Z., Y. Kuznetsov, A. McPherson, G. W. Hatfield, and S. Sandmeyer. 2008. TY3 GAG3 protein forms ordered particles in Escherichia coli. Virology 370:223-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen, L. S. Z., M. Zhang, N. Beliakova-Bethell, V. Bilanchone, A. Lamsa, K. Nagashima, R. Najdi, K. Kosaka, V. Kovacevic, J. Cheng, P. Baldi, G. W. Hatfield, and S. Sandmeyer. 2007. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J. Virol. 81:6957-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, O., C. V. Heath, D. C. Amberg, T. C. Dockendorff, C. S. Copeland, M. Snyder, and C. N. Cole. 1995. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell 6:401-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim, R. Y. H., N. P. Huang, J. Köser, J. Deng, K. H. A. Lau, K. Schwarz-Herion, B. Fahrenkrog, and U. Aebi. 2006. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. 103:9512-9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, S. S., M. H. Nymark-McMahon, L. Yieh, and S. B. Sandmeyer. 2001. Integrase mediates nuclear localization of Ty3. Mol. Cell. Biol. 21:7826-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLane, L. M., K. F. Pulliam, S. E. Devine, and A. H. Corbett. 2008. The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acids Res. 36:4317-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menees, T. M., and S. B. Sandmeyer. 1994. Transposition of the yeast retroviruslike element Ty3 is dependent on the cell cycle. Mol. Cell. Biol. 14:8229-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, S. P., L. A. Rinckel, and D. J. Garfinkel. 1998. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol. Cell. Biol. 18:1105-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosammaparast, N., Y. Guo, J. Shabanowitz, D. F. Hunt, and L. F. Pemberton. 2002. Pathways mediating the nuclear import of histones H3 and H4 in yeast. J. Biol. Chem. 277:862-868. [DOI] [PubMed] [Google Scholar]
- 40.Oakes, M., Y. Nogi, M. W. Clark, and M. Nomura. 1993. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol. Cell. Biol. 13:2441-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pante, N., and M. Kann. 2002. Nuclear pore complex is able to transport macromolecules with diameters of ∼39 nm. Mol. Biol. Cell 13:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel, S. S., B. J. Belmont, J. M. Sante, and M. F. Rexach. 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129:83-96. [DOI] [PubMed] [Google Scholar]
- 43.Pemberton, L., and M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187-198. [DOI] [PubMed] [Google Scholar]
- 44.Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 46.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rout, M. P., G. Blobel, and J. D. Aitchison. 1997. A distinct nuclear import pathway used by ribosomal proteins. Cell 89:715-725. [DOI] [PubMed] [Google Scholar]
- 48.Schliephake, A. W., and A. Rethwilm. 1994. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 68:4946-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siniossoglou, S., C. Wimmer, M. Rieger, V. Doye, H. Tekotte, C. Weise, S. Emig, A. Segref, and E. C. Hurt. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84:265-275. [DOI] [PubMed] [Google Scholar]
- 50.Strawn, L. A., T. Shen, and S. R. Wente. 2001. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J. Biol. Chem. 276:6445-6452. [DOI] [PubMed] [Google Scholar]
- 51.Strawn, L. A., T. Shen, N. Shulga, D. S. Goldfarb, and S. R. Wente. 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6:197-206. [DOI] [PubMed] [Google Scholar]
- 52.Ström, A. C., and K. Weis. 2001. Importin-beta-like nuclear transport receptors. Genome Biol. 2:reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki, Y., and R. Craigie. 2007. The road to chromatin: nuclear entry of retroviruses. Nat. Rev. Microbiol. 5:187-196. [DOI] [PubMed] [Google Scholar]
- 54.Terry, L. J., E. B. Shows, and S. R. Wente. 2007. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318:1412-1416. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, M., R. A. Haeusler, P. D. Good, and D. R. Engelke. 2003. Nucleolar clustering of dispersed tRNA genes. Science 302:1399-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran, E. J., and S. R. Wente. 2006. Dynamic nuclear pore complexes: life on the edge. Cell 125:1041-1053. [DOI] [PubMed] [Google Scholar]
- 57.Weis, K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441-451. [DOI] [PubMed] [Google Scholar]
- 58.Whittaker, G. R. 2003. Virus nuclear import. Adv. Drug Deliv. Rev. 55:733-747. [DOI] [PubMed] [Google Scholar]
- 59.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamashita, M., O. Perez, T. J. Hope, and M. Emerman. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PloS Pathog. 3:1502-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao, W., D. Roser, A. Köhler, B. Bradatsch, J. Bassler, and E. Hurt. 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell 26:51-62. [DOI] [PubMed] [Google Scholar]
- 62.Yu, S. F., K. Edelmann, R. K. Strong, A. Moebes, A. Rethwilm, and M. L. Linial. 1996. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 70:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeitler, B., and K. Weis. 2004. The FG-repeat asymmetry of the nuclear pore complex is dispensable for bulk nucleocytoplasmic transport in vivo. J. Cell Biol. 167:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.