Abstract
Glycoprotein L (gL) is one of four glycoproteins required for the entry of herpes simplex virus (HSV) into cells and for virus-induced cell fusion. This glycoprotein oligomerizes with gH to form a membrane-bound heterodimer but can be secreted when expressed without gH. Twelve unique gL linker-insertion mutants were generated to identify regions critical for gH binding and gH/gL processing and regions essential for cell fusion and viral entry. All gL mutants were detected on the cell surface in the absence of gH, suggesting incomplete cleavage of the signal peptide or the presence of a cell surface receptor for secreted gL. Coexpression with gH enhanced the levels of cell surface gL detected by antibodies for all gL mutants except those that were defective in their interactions with gH. Two insertions into a conserved region of gL abrogated the binding of gL to gH and prevented gH expression on the cell surface. Three other insertions reduced the cell surface expression of gH and/or altered the properties of gH/gL heterodimers. Altered or absent interaction of gL with gH was correlated with reduced or absent cell fusion activity and impaired complementation of virion infectivity. These results identify a conserved domain of gL that is critical for its binding to gH and two noncontiguous regions of gL, one of which contains the conserved domain, that are critical for the gH/gL complex to perform its role in membrane fusion.
Glycoprotein L (gL) is one of the four glycoproteins required for the entry of herpes simplex virus (HSV) into cells and for virus-induced cell fusion (26, 33). The others are gB, gD, and gH (30). The functional unit containing gL is a heterodimer formed with gH (gH/gL) (15). Because mature gL has no membrane-spanning domain, other than a cleavable signal peptide, it is secreted unless it is coexpressed with gH, a type 1 glycoprotein that anchors gL to the cell membrane (2). Also, gH is not properly processed or transported out of the endoplasmic reticulum unless it is coexpressed with gL (15).
Most, if not all, herpesviruses express orthologs of gB, gH, and gL, which are believed to form the core membrane-fusing machinery necessary for viral entry and cell fusion. For some herpesviruses, such as Epstein-Barr virus and human cytomegalovirus, the gH/gL oligomer may contain additional viral subunits that can influence binding of the complex to cell receptors and determine cell tropism (14, 34, 35). For HSV, however, only gD and gB have been shown to have receptor-binding activities that are required for entry (27, 31). Although HSV gH has an RGD motif and the gH/gL heterodimer can bind to certain integrins, this binding seems not to be necessary for viral entry (3, 22).
The initial interaction of HSV with cells can be the reversible attachment of virus to cell surface heparan sulfate, mediated by viral glycoprotein gB and/or gC (29). Then, gD can bind to one of its receptors, including herpesvirus entry mediator (HVEM), a member of the tumor necrosis factor receptor family; nectin-1 or nectin-2, cell adhesion molecules belonging to the immunoglobulin superfamily; or specific sites in heparan sulfate generated by 3-O-sulfotransferases (31). In addition to binding to heparan sulfate, gB can also bind to other cell surface receptors, including paired immunoglobulin-like receptor alpha (PILRα) (27). Binding of both gD and gB to one of their respective receptors appears to be required for triggering the membrane-fusing activity of gB and/or gH/gL, which leads to viral entry.
A recent X-ray structure of HSV type 1 (HSV-1) gB suggests that it is a class III viral fusogen similar in domain organization, but not primary sequence, to the G protein of vesicular stomatitis virus (13). It has been proposed that HSV-1 gH has features characteristic of class I viral fusogens, such as putative heptad repeats and fusion peptides (6, 9-11). Also, peptides matching the sequence of gH can interact with lipids and/or induce the fusion of lipid vesicles (4, 5, 8). Hemifusion between cells and between virus and cell can be induced by gH/gL and gD in the absence of gB (32). Many questions remain about the respective roles of gH/gL and gB in inducing membrane fusion.
The four conserved cysteines in gL were found to be essential for gL-gH association and function (1). Mutational analyses of gL by C-terminal deletions showed that the first 147 amino acids of gL are sufficient for association with gH but that the first 161 amino acids are necessary for cotransport of gH and gL to the cell surface (17, 23) and for gL activity in cell fusion and viral entry (17). Lastly, certain anti-gL monoclonal antibodies (MAbs) can inhibit cell fusion but not viral entry, despite demonstrable binding of the MAbs to virus, suggesting that gL may play a different role in each process (21). These MAbs were mapped to the C-terminal region of gL (21, 23). The diagram at the bottom of Fig. 1 shows the locations of the gL features mentioned above and of the signal peptide.
FIG. 1.
Effects of insertional mutations on HSV-1 gL and gH cell surface expression. CHO cells were transfected with plasmids expressing gH and WT gL or a gL mutant. Cell surface expression of gL and gH was quantified by CELISA using polyclonal R88 antiserum (filled circles) and MAb 52S (open triangles), respectively. A linear representation of the gL polypeptide is shown below the graph, with coded bars identifying features of gL. The bars represent the signal peptide (uncolored hatched), the N-terminal 161-amino-acid fragment necessary for the formation of functional gH/gL complexes (dark gray), highly conserved residues within this fragment (cross-hatched dark- gray bar), and epitopes recognized by a panel of anti-gL MAbs (light-gray and uncolored vertically striped bars). The values presented for cell surface expression of each mutant gL and of cotransfected WT gH are means from three independent experiments expressed as percentages of WT gL (or of gH cotransfected with WT gL) values, after subtraction of background values obtained in the absence of gL expression and as a function of the position of the insertion. Standard deviations are presented in Fig. 2 and 3 for similar experiments.
The interactions between gL and gH required for proper intracellular transport, processing, and cell surface expression make it difficult to investigate the functional role of one of these glycoproteins in cell fusion and viral entry independently of the other. We generated a panel of gL linker-insertion mutants to identify regions critical for gH binding and transport and regions essential for cell fusion and viral entry. One aim was to determine whether these roles of gL could be dissociated or were linked. Characterization of 12 unique gL linker-insertion mutants showed that (i) a conserved domain of gL is critical for the physical interaction of gL with gH and for the normal processing of gH, (ii) two noncontiguous regions of gL, one of which contains the highly conserved domain, are critical for the normal conformation and function of gH/gL heterodimers, and (iii) wild-type (WT) and mutant gLs can be detected on the cell surface in the absence of gH, suggesting the possibility of an independent role for uncomplexed gL. These results support and extend previous studies suggesting that gL has a larger role in membrane fusion than serving as a chaperone for gH and that specific mutations in gL can influence the function of the gH/gL heterodimer.
MATERIALS AND METHODS
Cells and viruses.
The cell lines used included Chinese hamster ovary (CHO-K1; ATCC) cells, CHO cells stably expressing human HVEM (20) or nectin-1 (7), Vero cells, and Vero-gL cells carrying the HSV-1 gL gene and used for the propagation and titration of the gL-negative mutant HSV-1(KOS)gL86 (20). The CHO cell line and derivatives were grown in Ham's F-12 medium supplemented with 10% fetal bovine serum, and the Vero cell line and derivatives were grown in Dulbecco minimal essential medium containing 10% fetal bovine serum.
Random linker-insertion mutagenesis of HSV-1 gL.
The GPS-LS linker scanning system (New England Biolabs) was used as recommended by the manufacturer and was applied to the HSV-1 gL insert excised from pPEP101 (24) to generate random linker-insertion mutations of gL. After religation of the library of inserts into pCAGGS and transformation of bacteria, 12 unique gL linker-insertion mutants were isolated and sequenced using PrimerN (30-mer) and PrimerS (30-mer), provided by the manufacturer. After removal of the transposon, each mutant plasmid was resequenced to verify the position of each insertion.
CELISA.
To quantify the cell surface expression of gH and gL, several previously described anti-gH (28) and anti-gL (21) antibodies were used. One of the anti-gH MAbs (52S) binds to gH in the presence or absence of gL or other viral proteins (12, 15, 17), and the other (53S) binds to the gH/gL heterodimer but not to gH or gL expressed alone (17). Both MAbs were used at 1:10,000 dilution. The anti-gL rabbit serum was used at 1:5,000 dilution. The anti-gL MAbs were used at 1:5,000 (VIII-62-15), 1:100,000 (VIII-82-24), 1:20,000 (VIII-87-1), 1:2,000 (VIII-200-1), and 1:100,000 (VIII-820-8) dilutions. CHO cells seeded in 96-well plates were transfected with 20 ng of plasmid expressing WT gH or empty vector, a plasmid expressing WT gL or a gL mutant, and 0.13 μl of Lipofectamine 2000 diluted in Opti-MEM (Gibco). The cells were washed once with phosphate-buffered saline (PBS) 24 h after transfection, and a cell-based enzyme-linked immunosorbent assay (CELISA) was performed as described previously (19). Briefly, after incubation of the transfected cells with the antibodies, the cells were washed, fixed, and incubated with biotinylated goat anti-rabbit immunoglobulin G or biotinylated rabbit anti-mouse immunoglobulin G (Sigma), followed by streptavidin-horseradish peroxidase (GE Healthcare) and horseradish peroxidase substrate (BioFX).
Coimmunoprecipitation and Western blotting.
CHO cells seeded in 24-well plates were transfected with 300 ng of a plasmid expressing WT HSV-1 gH (pPEP100), 300 ng of empty vector (pCAGGS) or a plasmid expressing WT gL (pPEP101) or a gL mutant, and 1.8 μl of Lipofectamine 2000 (Invitrogen) diluted in Opti-MEM (Gibco). The cells were washed with PBS 24 h after transfection and lysed with 300 μl of lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40) containing a protease inhibitor mixture (Roche Diagnostics). Fifty microliters of lysate was saved for analysis by Western blotting. The remaining 250-μl samples were mixed with 5 μl of the anti-gH MAb 52S (28) and then with protein A/G beads (Thermo Scientific) for immunoprecipitation. The lysate and recovered immunoprecipitates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 20% gels under reducing conditions, and Western blot analysis was done using rabbit serum R137 at 1:10,000 dilution. This antiserum recognizes both gH and gL (23).
Cell fusion assay.
CHO (effector) cells in 96-well plates were transfected with 20 ng each of plasmids expressing the T7 RNA polymerase, gD, or gH; 30 ng of plasmid expressing gB; and 20 ng of empty vector or plasmid expressing either WT (pPEP101) or mutant gL and 0.4 μl of Lipofectamine 2000. CHO-nectin-1 or CHO-HVEM (target) cells in six-well plates were transfected with 400 ng of a plasmid carrying the firefly luciferase gene under the control of the T7 promoter, 1.8 μg of empty vector (pCAGGS), and 7 μl of Lipofectamine 2000. Two hours posttransfection, the effector cells were washed once with Opti-MEM while each target cell population was washed, detached with EDTA (0.2 g/liter in PBS), and suspended in Opti-MEM. The target cell population was overlaid (3 × 104 cells/well) on the effector population. After 10 h, the cells were washed once with PBS and lysed with 50 μl/well of 1× passive lysis buffer (Promega). The expression of luciferase was quantified by adding 50 μl/well of luciferase substrate (Promega) and measuring the light output with a Wallac 1420 plate reader (Perkin-Elmer).
Complementation assay and analysis of virions for gH incorporation.
The complementation assay was performed essentially as described elsewhere (16). Vero cells in six-well plates were transfected with 1.0 μg of a plasmid expressing vector alone, WT gL, or one of the mutant forms of gL. At 24 h after transfection, the Vero cells were infected with HSV-1(KOS)gL86, which has the gL gene deleted but was complemented by propagation in a gL-expressing cell line. Inoculations were carried out at a multiplicity of infection of 8 in Dulbecco minimal essential medium supplemented with 5% heat-inactivated serum for 2 h at 37°C. The medium was then removed, and extracellular virus was inactivated with citrate buffer (pH 3.0) for 1 min, followed by three washes with medium. The cells were then overlaid with 3 ml of fresh medium and incubated for 24 h at 37°C. Complemented virus was harvested 24 h after infection. The culture medium was collected to sample for the titration of released infectious virus and also for the purification of released virions. The cells were lysed for the titration of cell-associated infectious virus. The culture medium containing extracellular virus was subjected to low-speed centrifugation to pellet cells and debris. After removal of a sample for titration, the supernatant was centrifuged at 48,000 × g for 1 h at 4°C over a 10% sucrose cushion. The pellet was dissolved in sample buffer and separated by SDS-PAGE on a 4 to 20% gel under reducing conditions. Western blot analysis was performed using rabbit serum R137 at 1:10,000 dilution and VP5 MAb (EastCoast Bio) at 1:5,000 dilution. The cell lysate and supernatant were titrated on the gL-complementing Vero cell line, Vero-gL, to determine the number of PFU harvested for each gL mutant.
RESULTS
Effects of the gL insertional mutations on cell surface expression of gL and gH.
The random-mutagenesis scheme utilized here introduced a 5-amino-acid insertion or a stop codon. Since truncation mutants of gL had previously been characterized, we focused on the insertion mutants only. Twelve unique insertion mutants were identified. The positions of the insertions are indicated by the one-letter name and position number (counting from the Met that starts translation) of the gL amino acid immediately preceding the inserted 5 amino acids (Table 1). To determine the effects of the insertions on cell surface expression of gL and gH, plasmids encoding the gL mutants were transfected into CHO cells, along with a plasmid expressing WT gH. The live cells were incubated with an anti-gL rabbit antiserum, R88, for detection of gL, and replicate cultures were incubated with the MAb 52S for detection of gH by a CELISA.
TABLE 1.
Binding of monoclonal antibodies to gL mutants
| Position of insertion | Amino acids inserted | Reactivity of MAba: |
||||
|---|---|---|---|---|---|---|
| VIII-62-15 | VIII-82-24 | VIII-87-1 | VIII-200-1 | VIII-820-8 | ||
| G005 | CLNMG | +++ | +++ | +++ | +++ | +++ |
| P048 | CLNIP | +++ | +++ | +++ | +++ | +++ |
| R055 | LFKQR | ++ | ++ | ++ | +++ | ++ |
| F071 | LFKQF | + | ++ | ++ | +++ | ++ |
| E105 | VFKQE | + | ++ | ++ | ++ | + |
| L107 | MFKHL | +++ | +++ | +++ | +++ | +++ |
| F112 | LFKQF | +++ | +++ | +++ | +++ | +++ |
| S135 | CLNNS | +++ | +++ | +++ | +++ | +++ |
| A140 | MFKHA | +++ | +++ | +++ | +++ | +++ |
| T157 | LFKHT | +++ | +++ | +++ | +++ | +++ |
| A185 | CLNKA | − | − | − | − | − |
| P195 | LFKQP | +++ | +++ | +++ | +++ | +++ |
CELISA data were normalized to the reactivity of each MAb with WT gL. The normalized values for each MAb were then compared to values obtained using rabbit antiserum R88 for each gL mutant to determine the relative differences in reactivity. The symbols represent >80% (+++), 50 to 80% (++), 20 to 50% (+), and <20% (−) reactivity of each MAb compared to the R88 value for each gL mutant.
Figure 1 shows the level of cell surface gL detected for each mutant, as a percentage of WT gL, in relation to the amino acid position of the insertion. In addition, the level of cell surface gH detected, as a percentage of the gH levels on cells cotransfected with WT gL, is also shown. A linear representation of gL is shown below the x axis and coded to identify specific features of gL mentioned in the introduction. All gL mutants were detected on the cell surface. The two mutants with insertions after F071 and E105 exhibited the lowest levels of gL expression. Moreover, the gH coexpressed with these mutants was undetectable on cell surfaces, suggesting that they failed to associate with gH or, alternatively, associated in such a way that the 52S epitope was destroyed. Two other mutants (with insertions after R055 and T157) exhibited a reduced gH/gL detection ratio (Fig. 1), suggesting some aberration in the association of gL with gH. A fifth mutant (with an insertion after A185) exhibited a reduced level of gL expression, but not of gH expression. The other seven mutants were indistinguishable from WT gL in their levels of expression on the cell surface and in codetection of gH.
A previous study demonstrated that, in the absence of gH, gL was secreted by transfected cells into the medium and could not be detected on cell surfaces by immunofluorescence (2), although others (15) reported low levels of cell surface gL under these conditions. CELISAs were performed using polyclonal antibodies to detect gL on CHO cells expressing WT gL or each gL mutant in the presence or absence of gH coexpression. Figure 2 shows the results, expressed as percentages of the level of WT gL detected when it was cotransfected with gH. The amount of WT gL detected in the absence of gH was approximately 68% of levels in the presence of gH. For most of the gL mutants, the presence of gH increased the levels of gL detected on the cell surface, as for WT gL, consistent with the formation of stable gH/gL heterodimers that were transported to the cell surface. However, the mutants with insertions after F071 and E105 showed no increase in gL detectable on the cell surface when they were coexpressed with gH, consistent with the results in Fig. 1, indicating that gH was not present on the cell surface. Interestingly, the mutant with an insertion after A185 exhibited significantly reduced levels on the cell surface in the absence of gH, but cell surface expression was to a large extent restored in the presence of gH, indicating that the presence of gH could compensate for its impaired association with the cell surface or for its impaired recognition by the antibodies used. Thus, our results indicate a significant level of gL detectable on CHO cell surfaces in the absence of gH (although cell surface expression was enhanced by gH coexpression for all but two mutants).
FIG. 2.
Cell surface expression of WT gL and gL mutants in the absence and presence of gH. CHO cells were transfected with plasmids expressing WT gL or a gL mutant or empty vector and either a plasmid expressing gH or an empty vector. Cell surface expression of gL was quantified by CELISA using polyclonal R88. The values presented for the cell surface expression of each mutant gL are means from three independent experiments expressed as percentages of WT gL values (obtained in the presence of gH after subtraction of background values obtained in the absence of gL expression). Standard deviations are presented as error bars.
Effects of the gL insertional mutations on the conformation of gH/gL complexes and the conformation of gL.
To assess the effects of the insertions on the conformation of the gH/gL complexes, CELISAs were performed on CHO cells coexpressing WT gL or each of the gL mutants and WT gH, using a MAb that recognizes the gH/gL heterodimer but not gH or gL alone (53S). The results are presented in Fig. 3, along with parallel results obtained using MAb 52S, which recognizes gH alone. Expression of most of the gL mutants with gH resulted in approximately WT levels of gH and gH/gL on cell surfaces, as detected by 52S and 53S, respectively. Two mutants (with insertions after F071and E105) failed to promote the cell surface expression of gH, as indicated by the absence of binding of either MAb, consistent with the results shown in Fig. 1 and 2. Another mutant (with an insertion after T157) permitted the cell surface expression of gH but with an overall decrease in detection by both the 52S and 53S MAbs, to the same relative degree. Of interest, coexpression of two mutants (with insertions after P048 and R055) with gH resulted in reduced binding by 53S compared to 52S, indicating that the gH/gL complexes formed with these gL mutants were altered in conformation compared to WT complexes.
FIG. 3.
Binding of 52S and 53S to cell surface gH coexpressed with gL mutants. CHO cells were transfected with plasmids expressing gH and WT gL or a gL mutant or empty vector. The cell surface expression of gH and gH/gL complexes was quantified by CELISA using 52S and 53S, respectively. The values presented for binding of 52S and 53S to gH are means from three independent experiments expressed as percentages of values obtained when gH was coexpressed with WT gL (after subtraction of background values obtained in the absence of gL expression). Standard deviations are presented as error bars.
To determine whether the mutant gLs expressed on CHO cell surfaces were grossly altered in conformation, CHO cells were transfected to express both gH and either WT gL or one of the gL mutants, and CELISAs were repeated with a panel of anti-gL MAbs (Table 1). These MAbs were from the VIII series of anti-gL MAbs that were found to map to a C-terminal region (amino acids 168 to 208) of gL (21, 23). One of these studies (23) concluded that major epitopes for these MAbs are located between amino acids 168 and 178 but did not exclude the possibility that additional sequences, downstream of 178, contributed to these epitopes (see Fig. 1, bottom, for the locations of the epitopes). For most mutants, the ratio of MAb binding to polyclonal antibody (R88) binding was essentially equivalent to that observed for WT gL, indicating retention of at least some aspects of WT conformation (Table 1). Mutant A185, located within the C-terminal 168-to-208 domain, showed a complete loss of binding by all of the anti-gL MAbs, indicating disruption of their epitopes. This disruption probably explains the reduced binding of the rabbit anti-gL serum R88, as shown in Fig. 1. The epitopes recognized by the MAbs may also be recognized by a significant fraction of the polyclonal antibodies in serum R88.
Three mutants (with insertions after R055, F071, and E105) showed somewhat reduced binding of several of the anti-gL MAbs relative to R88 binding. Insertions in these mutants appeared to affect the conformation of the MAb epitopes, which are clearly located downstream of the insertions. Since these insertions also impaired the association of gL with gH (Fig. 1 to 3) or altered an epitope dependent on gH/gL heterodimerization (Fig. 3), it seems likely that the conformations of the epitopes recognized by some of the anti-gL MAbs is in part dependent on proper gH/gL heterodimerization.
Effects of the gL insertional mutations on heterodimerization with gH.
Coimmunoprecipitation and Western blot analyses were performed. CHO cells expressing WT gH and either WT gL or one of the gL mutants or no gL were lysed, and the cell lysates were mixed with the anti-gH MAb 52S, which recognizes gH whether or not it is complexed with gL. Figure 4 shows the results of the Western blot analysis for both the coimmunoprecipitates and cell lysates, using polyclonal antibodies capable of recognizing both gH and gL. For most of the mutants, polypeptides corresponding in size to gL and recognized by the gL-specific antibodies were coprecipitated with gH by the anti-gH MAb, with results similar to those observed for WT gL. Four mutants (with insertions after R055, F071, E105, and T157) exhibited reduced levels of coprecipitated gL, particularly in the case of the mutants with insertions after F071 and E105. For these four mutants, there were also reduced levels of both gH and gL in the lysates, as well as in the immunoprecipitates. In cases where the mutant gL did not associate with gH (F071 and E105), gH would likely be targeted for degradation and most of the gL could be secreted. Two of the insertions that permitted formation of gH/gL heterodimers (R055 and T157) also appeared to cause reduced stability of both gH and gL, consistent with reduced levels of gH/gL on cell surfaces (Fig. 3). Interestingly, the mutant with its insertion after P048 was not distinguishable from WT gL according to the coimmunoprecipitation results and Western blot analysis shown in Fig. 4, despite effects of this insertion on gH/gL conformation as assessed by 53S binding (Fig. 3).
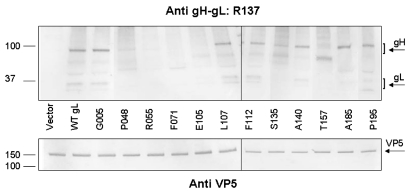
FIG. 4.
Coimmunoprecipitation of gL mutants with WT gH. CHO cells were cotransfected with a plasmid expressing gH and a plasmid expressing WT gL or a gL mutant or an empty-vector control. Proteins in cell lysates were immunoprecipitated with anti-gH MAb 52S and separated by electrophoresis on 4 to 20% polyacrylamide gels under nonreducing conditions. Western blots were performed using the rabbit anti-gH/gL serum R137. The upper blot shows immunoprecipitates, and the lower blot shows the lysates. The vertical lines indicate borders between two separate sets of blots (L107/F112) or lanes that were deleted or rearranged. Comparisons should be made between lanes in the upper and lower blots. The numbers to the left are molecular mass markers with values expressed as thousands of kDa. The positions of the immature and mature forms of gH and gL are indicated by brackets and labels on the right.
Effects of the gL insertional mutations on cell fusion activity.
To assess cell fusion activity by a quantitative luciferase-based assay, WT gL or the 12 gL mutants were coexpressed with HSV-1 gB, gD, and gH in CHO cells (effectors), which were mixed with nectin-1-expressing or HVEM-expressing CHO cells (targets). CELISAs were performed in parallel to quantify cell surface expression of gL in the effector cell populations (the results were not significantly different from those shown in Fig. 1 for CHO cells cotransfected with gH and gL only). Figure 5 presents cell surface expression for all the gL mutants in comparison with cell fusion results obtained with both the HVEM-expressing and nectin-1-expressing target cell populations. Cell fusion activity was virtually absent for three mutants (with insertions after R055, F071, and E105), was drastically impaired for two others (with insertions after P048 and T157), and was slightly impaired for two additional mutants (with insertions after L107 and F112) regardless of the fusion receptor expressed by the target cells. The mutant with an insertion after A185 did not differ significantly from WT gL in cell fusion activity, indicating that disruption of the MAb epitopes (Table 1) was without effect on this activity. The remaining four mutants were also nearly indistinguishable from WT gL in cell fusion activity.
FIG. 5.
Cell fusion activities of gL mutants in relation to cell surface expression. CHO cells were transfected with plasmids expressing WT gL or each of the gL mutants or empty vector; other HSV-1 glycoproteins (gB, gD, and gH); and T7 polymerase (effector cells). CHO-HVEM and CHO-nectin-1 cells were transfected with a plasmid carrying the firefly luciferase gene under the control of the T7 promoter (target cells). One set of effector cells was used for CELISA, and the other two sets were mixed with target cells expressing either HVEM or nectin-1 for assessment of cell fusion activity by quantification of luciferase activity. The results are expressed as percentages of WT gL activity after subtraction of background values obtained in the absence of gL expression and are means and standard deviations of three independent experiments.
Effects of the gL insertional mutations on cell entry.
To address whether the mutant forms of gL were functional for viral entry, the mutants were tested using a virus complementation assay. Infectious complemented virus was quantified by plaque assay both from the medium of the cells that produced it and from lysates of these cells (Fig. 6). Yields of complemented virus were about 1,000 times higher after complementation with WT gL than in the absence of complementation (with no gL transfected into the cells). Most of the infectious virus produced was cell associated after complementation with WT gL or any of the mutants, as is true for WT HSV-1. The three mutants that had no cell fusion activity (with insertions after R055, F071, and E105) also failed to complement the gL-null virus for cell entry. Of the two mutants that had impaired cell fusion activity, the one with its insertion after T157 had no complementation activity for viral entry, whereas the one with its insertion after P048 had about 10% of the complementation activity of WT gL.
FIG. 6.
Complementation of gL-negative virus entry by gL mutants. Vero cells, transfected with a plasmid expressing vector alone, WT gL, or one of the mutant forms of gL were infected with the gL-negative virus HSV-1(KOS)gL86 at 24 h after transfection. Complemented virus was harvested 24 h after infection and titrated on the gL-complementing Vero cell line Vero-gL to determine the number of PFU per culture harvested for each gL mutant from the medium and from cell lysates, as indicated. Results representative of two independent experiments are shown on a log scale. The horizontal dotted lines indicate the values obtained in the absence of gL expression (background due to revertants in the virus stock), and the dashed lines indicate the values obtained by complementation with transfected WT gL.
To assess the ability of gL-complemented virus to incorporate gH into virions, we purified complemented virus from the medium of infected cells and performed Western blots to detect gH, and also the major capsid protein VP5 as a loading control (Fig. 7). The results showed that gH could be detected in the virions complemented with gL that retained cell fusion activity and viral entry activity. However, little if any gH could be detected in the virions produced in the absence of gL or in the presence of gL mutants that were impaired for, or devoid of, cell fusion activity (Fig. 5) and complementation activity (Fig. 6). Thus, the gL mutations that were most deleterious for cell fusion activity also impaired incorporation of gH/gL into virions. Therefore, it could not be determined whether gH/gL complexes that did form (with gL having insertions after P048, R055, or T157) would have had any activity in viral entry.
FIG. 7.
Incorporation of gH into virions produced in the presence of WT gL or gL mutants or in the absence of gL. Culture supernatants containing complemented virions were harvested, and the virions were collected after centrifugation through 10% sucrose. Half the volume of the sample was processed for SDS-PAGE and an anti-VP5 Western blot, and the other half was separately processed for SDS-PAGE and an anti-gH blot. The VP5 signal indicates the amount of virus recovered, and the gH signal indicates the amount of gH incorporated into the virus. The labels are as in Fig. 4, with the addition of a label for VP5. The vertical lines separate the two different blots that compose each panel. The band detected by the anti-gH/gL antiserum in virions complemented with gL carrying the T157 insertion was seen at lower levels in the other virions. Its identity is not known, but it is too small to be either mature or immature full-length gH.
DISCUSSION
Three classes of gL insertion mutants are described in this study. The first includes mutants with insertions that had little or no effect on gL expression, association with gH, or gH/gL function. It was not surprising that insertions into the C terminus (A185 and P195) were tolerated, inasmuch as they mapped to the region that can be deleted without loss of gL function (17, 23). It was also not surprising that the insertion into the signal peptide (G005) had little effect; it did not have much effect on the predicted efficiency of signal peptidase cleavage at the WT cleavage site. On the other hand, insertions L107, F112, S135, and A140 also had little if any effect on gL expression and function despite their locations between two regions where insertions had severely negative effects on gL activities. Thus, these insertions themselves and the increase in length of the polypeptide chain between positions L107 and A140 were well tolerated, suggesting flexibility of the three-dimensional structure in this region.
The other two classes of insertion mutants include all those that abrogated or reduced gL function. In two cases (the second class), the insertions (F071 and E105) prevented any detectable interaction of gL with gH, thus accounting for the absence of cell fusion activity and the failure to complement viral infectivity. This is consistent with previous studies emphasizing that the functional unit for HSV-induced cell fusion and viral entry is the gH/gL heterodimer, in part because associations between gH and gL are necessary for the proper processing of both and for their incorporation into virions (12, 15, 17, 23, 26). It is of interest that insertions F071 and E105 map within the borders of the only region in gL that is highly conserved among alphaherpesvirus members of the gL family (Fig. 1 and 8). This suggests that this conserved domain is a large part of the gL interface with gH, for HSV at least and perhaps also for other alphaherpesviruses.
FIG. 8.
Amino acid sequence alignment of alphaherpesvirus gLs. The alignment begins with the first Met of the gL precursor. The asterisks indicate residues that are 100% conserved and the plus signs indicate residues that are 80% conserved among the viruses shown. VZV, varicella-zoster virus; GHV, gallid herpesvirus 2; EHV, equine herpesvirus 1; BHV-1, bovine herpesvirus 1. The bracket indicates the region of relatively high conservation of amino acid sequence.
In three cases (the third class), the insertions (P048, R055, and T157) reduced or abrogated gL function in cell fusion and complementation of viral infectivity without preventing gL interaction with gH and cell surface expression of gH/gL heterodimers. For all three of these mutants, the insertions permitted near-normal levels of gL cell surface expression with reduced levels of gH cell surface expression for insertions R055 and T157. The gH/gL heterodimers formed appeared to be conformationally aberrant in the cases of the P048 and R055 insertions. Thus, the severe reduction in cell fusion activity of these mutants is not explained simply by the failure of gL to associate with gH, but rather by (i) alterations in other functional domains of gL, (ii) effects of the gL insertions on gH conformation, or (iii) effects of the gL insertions on the overall conformation of the gH/gL heterodimer. The possibility exists, therefore, that gL contributes to the essential function of the gH/gL heterodimer in cell fusion (and in incorporation into virions) rather than just serving as a chaperone for gH processing and transport. This idea has been proposed previously for HSV (16) and for Epstein-Barr virus (25) on the basis of amino acid substitutions that reduced activity to a larger extent than heterodimer formation or transport of gH/gL to cell surfaces. In the case of HSV, these amino acid substitutions were in the vicinity of insertion T157.
Unexpectedly, WT gL and all the gL mutants were detected on the cell surface in the presence or absence of gH, as well as being secreted in the absence of gH. These findings suggest that signal peptidase cleavage may not go to completion in CHO cells, resulting in the signal peptide serving as a membrane anchor for a fraction of the gL produced. For another herpesvirus, Epstein-Barr virus, the signal peptide of gL is largely not removed by cleavage so that the protein can be oriented in membranes as a type 2 glycoprotein or secreted (18). An alternative explanation for cell surface detection of HSV-1 gL in the absence of gH is the potential existence of a cell surface receptor for HSV-1 gL. It has been proposed that herpesvirus gLs may be related to chemokine receptor ligands (36). Further study is required to resolve this issue.
It was somewhat surprising to find that epitopes on gL recognized by MAbs capable of inhibiting cell fusion (21) could be disrupted without any detectable effects on cell fusion activity. An insertion at A185 completely abrogated binding by all the MAbs tested here (Table 1), indicating that their epitopes were disrupted by this insertion and must include at least a short region downstream of amino acids 168 to 178, the minimal region identified in a previous epitope-mapping study (23). The finding that A185 is detected at lower levels on the cell surface by the R88 polyclonal antiserum, relative to WT gL, without a corresponding decrease in the gH level (Fig. 1) also suggests that major epitopes for R88 antibodies lie within the same region. However, the loss of this epitope(s) for the MAbs and the antiserum did not affect cell fusion activity, despite an earlier study showing that these MAbs could block the fusion of cells infected by an HSV-1 syncytial mutant, but not virus entry, even though the MAbs bound to virions (20). Based on these findings, one would predict that a mutant virus carrying the A185 insertion would not have its cell fusion activity inhibited by these MAbs.
The apparently low levels of mutant A185 detected by the rabbit antiserum on cell surfaces in the absence of gH and the restoration of A185 mutant levels in the presence of gH (Fig. 2) suggests several possibilities. Perhaps, in the absence of gH, A185 adopts an altered conformation that reduces the binding of antibodies in the antiserum. The low levels of the A185 mutant detected on the cell surface could also be accounted for by aberrant transport or processing that disrupts entry into the secretory pathway in the absence of gH. A third intriguing possibility is that mutant A185 has greatly reduced ability to bind to the cell surface in the absence of gH. An insertion at position 185 could theoretically disrupt a region on gL responsible for binding of gL to a putative cell surface receptor.
In this study, we identified critical functional regions in HSV-1 gL, including a gH-binding domain and additional contiguous and noncontiguous regions that are important for gH/gL function in cell fusion and viral complementation. Since gL can be either secreted or associated with the cell surface in the absence of gH, the possibility that gL on its own may have another role in virus infection should be explored.
Acknowledgments
We thank N. Susmarski for excellent technical assistance and R. Longnecker for helpful discussions on this project and the manuscript. We also thank R. Eisenberg and G. Cohen (University of Pennsylvania) for the gift of antiserum R137.
This work was supported by Public Health Service grants CA021776 from the National Cancer Institute and AI036293 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Cairns, T. M., D. J. Landsburg, J. Charles Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2005. Contribution of cysteine residues to the structure and function of herpes simplex virus gH/gL. Virology 332:550-562. [DOI] [PubMed] [Google Scholar]
- 2.Dubin, G., and H. Jiang. 1995. Expression of herpes simplex virus type 1 glycoprotein L (gL) in transfected mammalian cells: Evidence that gL is not independently anchored to cell membranes. J. Virol. 69:4564-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galdiero, M., A. Whiteley, B. Bruun, S. Bell, T. Minson, and H. Browne. 1997. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J. Virol. 71:2163-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galdiero, S., A. Falanga, M. Vitiello, H. Browne, C. Pedone, and M. Galdiero. 2005. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 280:28632-28643. [DOI] [PubMed] [Google Scholar]
- 5.Galdiero, S., A. Falanga, M. Vitiello, M. D'Isanto, M. Cantisani, A. Kampanaraki, E. Benedetti, H. Browne, and M. Galdiero. 2008. Peptides containing membrane-interacting motifs inhibit herpes simplex virus type 1 infectivity. Peptides 29:1461-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galdiero, S., M. Vitiello, M. D'Isanto, A. Falanga, C. Collins, K. Raieta, C. Pedone, H. Browne, and M. Galdiero. 2006. Analysis of synthetic peptides from heptad-repeat domains of herpes simplex virus type 1 glycoproteins H and B. J. Gen. Virol. 87:1085-1097. [DOI] [PubMed] [Google Scholar]
- 7.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 8.Gianni, T., R. Fato, C. Bergamini, G. Lenaz, and G. Campadelli-Fiume. 2006. Hydrophobic α-helices 1 and 2 of herpes simplex virus gH interact with lipids, and their mimetic peptides enhance virus infection and fusion. J. Virol. 80:8190-8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gianni, T., P. L. Martelli, R. Casadio, and G. Campadelli-Fiume. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane α-helix with attributes of an internal fusion peptide, positionally conserved in the Herpesviridae family. J. Virol. 79:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gianni, T., L. Menotti, and G. Campadelli-Fiume. 2005. A heptad repeat in herpes simplex virus 1 gH, located downstream of the α-helix with attributes of a fusion peptide, is critical for virus entry and fusion. J. Virol. 79:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gianni, T., A. Piccoli, C. Bertucci, and G. Campadelli-Fiume. 2006. Heptad repeat 2 in herpes simplex virus 1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J. Virol. 80:2216-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gompels, U. A., and A. C. Minson. 1989. Antigenic properties and cellular localization of herpes simplex virus glycoprotein H synthesized in a mammalian cell expression system. J. Virol. 63:4744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 14.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klyachkin, Y. M., and R. J. Geraghty. 2008. Mutagenic analysis of herpes simplex virus type 1 glycoprotein L reveals the importance of an arginine-rich region for function. Virology 374:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klyachkin, Y. M., K. D. Stoops, and R. J. Geraghty. 2006. Herpes simplex virus type 1 glycoprotein L mutants that fail to promote trafficking of glycoprotein H and fail to function in fusion can induce binding of glycoprotein L-dependent anti-glycoprotein H antibodies. J. Gen. Virol. 87:759-767. [DOI] [PubMed] [Google Scholar]
- 18.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, E., and P. G. Spear. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. USA 104:13140-13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 21.Novotny, M. J., M. L. Parish, and P. G. Spear. 1996. Variability of herpes simplex virus 1 gL and anti-gL antibodies that inhibit cell fusion but not viral infectivity. Virology 221:1-13. [DOI] [PubMed] [Google Scholar]
- 22.Parry, C., S. Bell, T. Minson, and H. Browne. 2005. Herpes simplex virus type 1 glycoprotein H binds to αvβ3 integrins. J. Gen. Virol. 86:7-10. [DOI] [PubMed] [Google Scholar]
- 23.Peng, T., M. Ponce de Leon, M. J. Novotny, H. Jiang, J. D. Lambris, G. Dubin, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Structural and antigenic analysis of a truncated form of herpes simplex virus glycoprotein gH-gL complex. J. Virol. 72:6092-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pertel, P., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 25.Plate, A. E., J. Smajlovic, T. S. Jardetzky, and R. Longnecker. 2009. Functional analysis of glycoprotein L from rhesus lymphocryptovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J. Virol. doi: 10.1128/JVI.00457-09. [DOI] [PMC free article] [PubMed]
- 26.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satoh, T., J. Arii, T. Suenaga, J. Wang, A. Kogure, J. Uehori, N. Arase, I. Shiratori, S. Tanaka, Y. Kawaguchi, P. G. Spear, L. L. Lanier, and H. Arase. 2008. PILRa is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell. Microbiol. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 31.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, D., and T. Shenk. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA 102:18153-18158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyrwicz, L. S., and L. Rychlewski. 2007. Herpes glycoprotein gL is distantly related to chemokine receptor ligands. Antivir. Res. 75:83-86. [DOI] [PubMed] [Google Scholar]