Abstract
Viruses often evolve resistance to antiviral agents. While resistant strains are able to replicate in the presence of the agent, they generally exhibit lower fitness than the wild-type strain in the absence of the inhibitor. In some cases, resistant strains become dependent on the antiviral agent. However, the agent rarely, if ever, elevates dependent strain fitness above the uninhibited wild-type level. This would require an adaptive mechanism to convert the antiviral agent into a beneficial growth factor. Using an inhibitory scaffolding protein that specifically blocks φX174 capsid assembly, we demonstrate that such mechanisms are possible. To obtain the quintuple-mutant resistant strain, the wild-type virus was propagated for approximately 150 viral life cycles in the presence of increasing concentrations of the inhibitory protein. The expression of the inhibitory protein elevated the strain's fitness significantly above the uninhibited wild-type level. Thus, selecting for resistance coselected for dependency, which was characterized and found to operate on the level of capsid nucleation. To the best of our knowledge, this is the first report of a virus evolving a mechanism to productively utilize an antiviral agent to stimulate its fitness above the uninhibited wild-type level. The results of this study may be predictive of the types of resistant phenotypes that could be selected by antiviral agents that specifically target capsid assembly.
While viruses often acquire resistance to antiviral agents, resistance mutants generally exhibit lower fitness than the wild-type strain in the absence of the inhibitor (6, 16, 17, 25) and can develop a dependency on the antiviral agent (1, 19). However, the molecular mechanism of dependency rarely, if ever, involves the productive use of the antiviral agent to elevate fitness above the uninhibited wild-type level. Many studies are conducted with animal viruses, often in clinical settings, which can impose restraints on the experimental durations. Thus, prolonged exposure to antiviral agents may be required for the emergence of a multiply mutant strain that has evolved mechanisms to productively utilize inhibitors.
Due to its rapid replication, bacteriophage φX174 has become an attractive model system for evolutionary studies (2, 3, 23, 24). Selective pressures can be applied for hundreds of infection cycles in a relatively short period of time. Using the atomic structure of assembly intermediates as a guide (8, 9, 15), viral scaffolding proteins that inhibit virion assembly have been designed (4). The molecular mechanism of inhibition was characterized, and resistance mutants were isolated via one-step genetic selections (4). In this study we report the isolation of a more robust resistant mutant. The quintuple mutant was generated by propagating φX174 for approximately 150 life cycles in the presence of increasing concentrations of the inhibitory protein, which was derived from the external scaffolding D protein. This protein forms asymmetric dimers that direct procapsid assembly. A conformational switch at glycine residue 61 (G61) in α-helix 3 is critical for productive dimer formation. In one subunit, α-helix 3 is bent 30°, whereas it assumes a straight conformation in the other subunit. All amino acid substitutions for G61 inhibit the ability to undergo the requisite conformational switch. The severity of the conferred dominant lethal phenotypes directly correlates with the side chain sizes of the substituted amino acids (4, 5).
The inhibitory proteins most likely remove assembly intermediates by lowering the thermodynamic barriers that normally prevent off-pathway reactions (4, 5). Both off-pathway reactions and proper assembly involve D-D protein interactions across what will become the twofold axes of symmetry in the virion (8, 9). In the procapsid crystal structure, α-helix 3 of the D2, D3, and D4 subunits mediates these interactions. Mutants resistant to the dominant lethal proteins were isolated in one-step genetic selections, and mutations mapped to either the coat or internal scaffolding proteins. These mutations may indirectly reinstate the avidity of the D protein electrostatic bonding partners required for productive morphogenesis (4, 5). However, the resistance phenotype is weak. To isolate a more robust phenotype, wild-type φX174 was continually cultured through exponential phase cells expressing an inhibitory D protein. Results from this analysis indicate that the selection for resistance coselected for a level of dependence. The inhibitory protein stimulates resistant strain fitness significantly above the uninhibited wild-type level and appears to be required for efficient capsid nucleation. These results suggest that the virus evolved a mechanism to convert this potent antiviral agent into a beneficial factor and may be predictive of the types of resistant phenotypes that could be selected by antiviral agents that specifically target capsid assembly.
MATERIALS AND METHODS
Phage plating, media, buffers, and stock preparation.
The reagents, media, buffers, and protocols have been previously described (11).
Bacterial strains, phage strains, and plasmids.
The Escherichia coli C strains C122 (sup0) and BAF30 (recA) have been previously described (11, 12). The host cell slyD mutation confers resistance to E protein-mediated lysis (18). The construction of the cloned wild-type and inhibitory D genes, pND and pG61D, respectively, has been previously reported (4).
Evolution of a more robust resistance phenotype.
BAF30 pG61D and BAF30 pND cells were grown to a concentration of 1.0 × 108 cells/ml at 37°C in TK medium supplemented with 20 μg/ml of ampicillin. Cells were concentrated 100-fold, resuspended in 30% glycerol, and frozen at −80°C. Prior to infection, 10 ml of TK medium with 20 μg/ml ampicillin was inoculated with 25 μl of frozen cells and incubated with aeration for 1 h in a 125-ml flask, at which time the cells were in exponential phase. MgCl2 and CaCl2 were added to respective concentrations of 10 mM and 0.5 mM. Cells were infected at a multiplicity of infection (MOI) of less than 0.01 and incubated at 37°C for 40 min. An aliquot of the infection culture was then used to inoculate a fresh flask of exponential cells. After each passage, titers of the cultures were determined to calculate approximate fitness, and plaque morphologies were noted on cells expressing the inhibitory protein. A portion of each infection culture was frozen at −80°C. If titers indicated that infections were conducted at an MOI above 0.01, all further passages were discarded. The concentration of the inducer, IPTG (isopropyl-β-d-thiogalactopyranoside), varied during passages. When it appeared that a selective sweep had occurred in the population, the induction level of the cloned gene was increased.
Fitness assays.
The protocol for the fitness assays and the calculation of fitness have been previously described (3). Fitness is expressed as doublings per hour.
Determination of D protein expression levels.
To determine levels of mutant G61D external scaffolding protein expression, a 20-ml culture of TK medium, supplemented with 20 μg/ml ampicillin and IPTG at concentrations varying from 0 to 1.3 mM, was inoculated at 1/100 with a fresh overnight culture of lysis-resistant cells and grown to a concentration of 1.0 × 108 cells/ml. Cells were infected with either wild-type or nullD φX174 at an MOI of 3.0 and incubated for 1 h; cells were then concentrated, resuspended in 0.8 ml of 20 mM Tris-HCl (pH 8.0), boiled, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Band intensity was analyzed with ImageJ software (NIH). The relative level of G61D protein expression was estimated by comparison with wild-type- and nullD-infected cells, with and without exogenous D protein expression.
Attachment and eclipse assays.
Attachment assays were performed as previously described (13). The protocol for the eclipse assay was modified from a previously published protocol (14). Exponential cells were concentrated and resuspended in HFB buffer with 10 mM MgCl2 and 0.5 mM CaCl2. After infection, samples were incubated at 10°C for 30 min. Unattached virions were removed by centrifugation. The cell pellets with attached phage were resuspended in 1.0 ml of ice-cold HFB with 10 mM MgCl2 and 0.5 mM CaCl2 and placed into a 37°C water bath to initiate the eclipse reaction. At selected time points, samples were diluted 1/10 in borate-EDTA buffer, which releases uneclipsed phages from cell membranes, and titers were determined for surviving particles.
In vivo characterization of the morphogenetic pathway and the kinetics of virus production.
The protocols for generating extracts of infected lysis-resistant cells, ultracentrifugation parameters, particle (virions, procapsid, and degraded procapsid) detection, and in vivo kinetics have been previously described (20, 21).
RESULTS
Generation of the multiply mutant resistant strain.
To evolve the multiply mutant resistant strain, wild-type φX174 was continuously cultured in exponential phase E. coli BAF30 (recA) cells expressing the strongly inhibitory G61D external scaffolding D protein for 59 40-min passages. As a control, φX174 was cultured through cells expressing the wild-type D protein. To minimize other possible selective pressures, all passages were conducted at a low MOI. During the first eight passages, the cloned gene, under lac induction, was expressed at a low level, one that reduced fitness to approximately one-half that observed in cells not expressing the inhibitory protein.
Throughout the course of the experiment, fitness was measured, and plaque morphologies were observed on cells expressing the inhibitory protein. When it appeared that fitness had increased, indicative of a selective sweep of at least one beneficial mutation, the induction level of the cloned inhibitory gene was increased. In parallel, the induction of the cloned wild-type gene was increased in the control cultures; however, the fitness of that control population remained constant throughout the experiment. After 24 passages, the expression of the cloned gene reached its maximal level, resulting in approximately 1:1 mutant-to-wild-type protein ratios (20). Once the passages were completed, the entire genome sequence of an aliquot from each population was determined to reveal all mutations that had swept to high frequency. The population cultured through cells expressing the wild-type D protein acquired a single mutation in amino acid 100 of the coat protein. However, five different mutations were identified in the population cultured in cells expressing the inhibitory protein. These mutations mapped to the coat, DNA pilot, and external scaffolding proteins (Table 1).
TABLE 1.
The mutations identified within the multiply mutant resistant strain
| Nucleotide substitution |
Conferred amino acid substitution |
|||
|---|---|---|---|---|
| Position | Change | Protein type | Position | Change |
| 493 | A→G | External scaffolding | 34 | Asp→Gly |
| 1133 | G→C | Coat | 44 | Asp→His |
| 1616 | G→A | Coat | 205 | Asp→Asn |
| 1682 | T→C | Coat | 227 | Ser→Pro |
| 3340 | A→G | DNA pilot | 136 | Asp→Gly |
The fitness and phenotype of the resistant strain.
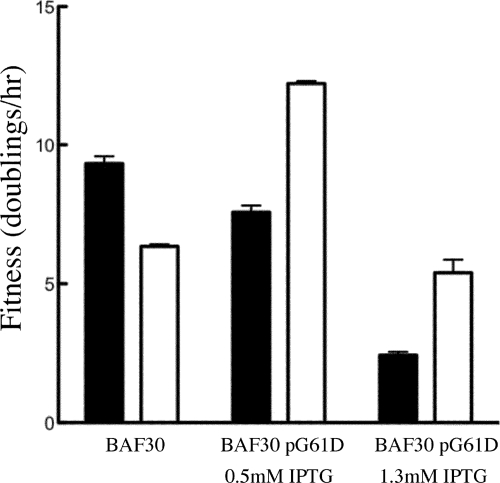
To eliminate complications associated with assaying the fitness of a mixed population, single plaques were isolated and sequenced to identify a strain containing all five mutations listed in Table 1. Fitness assays for the ancestor and evolved strains were conducted in BAF30 cells without the cloned inhibitory gene and in cells expressing various levels of the inhibitory G61D external scaffolding protein (Fig. 1). All assays were conducted in replicates of five and analyzed to determine statistical significance (GraphPad Prism software). BAF30 is a recA derivative of E. coli C122, in which the ancestral strain has been propagated for over 40 years. The recA cell line was used for continuous culturing to minimize possible recombination events between the phage and plasmid. The fitness of the ancestral strain was lower in BAF30 (9.4 ± 0.3 doublings/h) than in E. coli C122 (19.9 ± 0.6 doublings/h).
FIG. 1.
Fitness values for the ancestral (black box) and multiply mutant resistant (white box) strains. Fitness is measured as doublings per hour. Cells containing plasmids were induced to express the cloned D protein with 0.5 and 1.3 mM IPTG concentrations. All infections were conducted at low MOIs and in replicates of five.
The evolved strain was significantly more fit than the ancestor in the presence of the inhibitory protein but had lower fitness in its absence (Fig. 1). Ancestral strain fitness decreased with increased inhibitory protein expression. However, the evolved strain exhibited optimal fitness when the inhibitory protein was expressed at approximately 25% of the native species (see below). Thus, selecting for resistance coselected for a level of dependence.
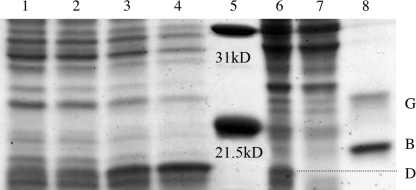
To determine the relative level of mutant D protein required to promote optimal fitness, protein levels were examined in wild-type lysis-resistant infected cells. Infections were conducted with and without the plasmid encoding the inhibitory protein. IPTG concentrations varying from 0 to 1.3 mM were used to control cloned gene expression. Whole-cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 2), and the gels were analyzed by ImageJ software (NIH). The major spike protein G served as an internal control for each infection, allowing the relative D/G protein ratios to be determined. To estimate the levels of D protein expressed from the plasmid, D/G protein ratios from infections in cells with the induced cloned gene were compared to the D/G protein ratio obtained from infected cells without the plasmid. No significant differences in D protein levels were observed between the infection without the plasmid and the infection with an uninduced cloned gene. IPTG concentrations of 0.5 and 1.3 mM resulted in 25% and 100% increases, respectively, in total D protein levels. The results of experiments conducted with the nullD mutant yielded similar results (data not shown).
FIG. 2.
Expression levels of the inhibitory G61D external scaffolding protein in wild-type-infected cells. Lane 1, infected cells and no plasmid; lane 2, infected cells with pG61D and no IPTG; lane 3, infected cells with pG61D and 0.5 mM IPTG; lane 4, infected cells with pG61D and 1.3 mM IPTG; lane 5, molecular mass marker; lane 6, BAF30 cells expressing wild-type D protein; lane 7, BAF30 cells without a plasmid; lane 8, purified viral proteins B and G.
Molecular basis of the resistance phenotype.
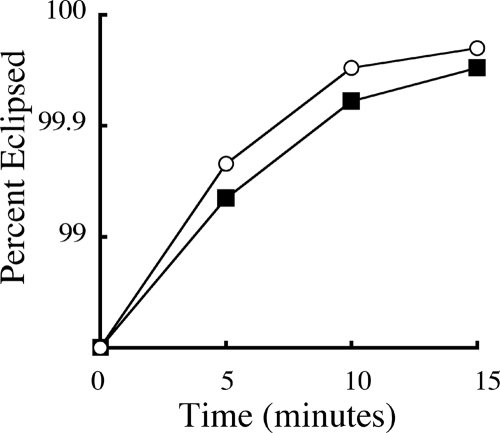
Although the primary selective pressure was most likely the expression of the inhibitory scaffolding protein, which would affect assembly, the contribution of other selective pressures cannot be eliminated a priori. Therefore, the ancestral and evolved strains were characterized for attachment and eclipse efficiency. No significant differences in attachment kinetics were observed (data not shown). In eclipse assays, conducted in triplicate, the evolved strain exhibited modestly faster kinetics (Fig. 3). This benefit is likely conferred by the substitution in the DNA pilot protein.
FIG. 3.
Eclipse kinetics of ancestral (filled squares) and evolved (empty circles) strains.
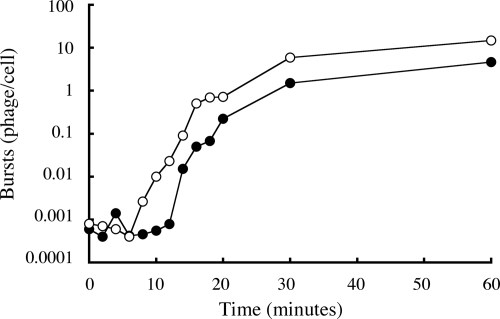
Mutant D proteins are known to affect the timing of procapsid nucleation and the rate of virion assembly in vivo (5, 20, 22). To determine if the evolved strain had altered assembly kinetics, the time course of phage production was followed in lysis-resistant cells with and without inhibitory gene induction. The cloned gene was induced at the level that promoted optimal fitness (Fig. 1). In these experiments phage were preattached to lysis-resistant cells, and unattached particles were removed by centrifugation. Infections commenced at time zero, which represents the beginning of the irreversible eclipse reaction. At each time point, aliquots of the infection were removed and chemically lysed, and titers were determined for progeny production. The low level of phage detected during the lag phase most likely represents uneclipsed virions. As can be seen in Fig. 4, induction of the inhibitory D gene resulted in the faster appearance and increased production of progeny. In the absence of inhibitory gene induction, the longer lag phase and decreased yield could reflect less efficient procapsid nucleation or DNA packaging. Defects in DNA packaging result in the accumulation of procapsids and empty particles (10, 21). To determine if these particles were accumulating, extracts prepared from infected cells with and without inhibitory gene induction were analyzed by rate zonal sedimentation. Neither procapsids nor empty particles were observed (data not shown). These data indicate that the evolved strain is dependent on the inhibitory protein for efficient procapsid nucleation.
FIG. 4.
The kinetics of evolved-strain virion production with (empty circles) and without (filled circles) inhibitory gene induction.
DISCUSSION
As observed in other studies, acquiring resistance to an antiviral agent lowered strain fitness vis-à-vis the uninhibited wild-type levels (6, 16, 25). The acquisition of compensatory mutations occasionally restores replication capacity (7), and coselection for dependence on the antiviral chemical compounds has been documented (1, 19). However, to the best of our knowledge this represents the first documentation of a virus's evolving a mechanism to productively utilize an antiviral agent to stimulate its fitness at a level substantially higher than the uninhibited wild-type level. Furthermore, the results of this study may be predictive of the types of resistance phenotypes that could be selected in the presence of antiviral agents that specifically target capsid assembly.
Wild-type φX174 was evolved in cells expressing an inhibitory external scaffolding D protein for approximately 150 phage generations. The evolved strain acquired five mutations, mapping to the coat, external scaffolding, and DNA pilot proteins. In contrast, only one unrelated coat protein mutation was identified when the ancestral strain was cultured in cells expressing the wild-type D protein. This coat protein mutation, which was not present in the evolved strain, was previously identified in another evolutionary study in which the only selective pressure was the ability to compete within the population (23). Evidence that the adaptation is primarily to the inhibitory protein and not to the recA cells includes the absence of fitness increase in the control line and the reduced fitness of the evolved virus in the absence of the inhibitor. Thus, the expression of the inhibitory protein imposed the primary selective pressure in these experiments.
The fitness of the ancestral and evolved strains was determined. While ancestral strain fitness decreased as a function of increased inhibitory gene induction, the fitness of the evolved strain dramatically increased. Optimal fitness was observed when the inhibitory protein was present at a concentration equal to approximately 25% of the indigenous protein. The inhibitory protein appears to stimulate the faster appearance and increased production of progeny. As there is no temporal gene expression in φX174, the reduced lag phase before progeny production most likely represents more efficient procapsid nucleation (5, 20, 22).
The location of the identified coat protein mutations is reminiscent of mutations observed in previous studies (4). The S227P mutation was isolated in one-step selections, and the D44H site is directly adjacent to a previously identified resistance mutation in the atomic structure. Thus far, all mutations involved in resistance reside under the D3 subunit in the procapsid lattice. This clustering may indicate that the evolved strain has tailored a coat protein region to accommodate mutant external scaffolding proteins. However, the resistance phenotype is not allele specific. The multiply mutant strain is resistant to all previously identified inhibitory proteins (4), including one with a G→P substitution (data not shown). While other substitutions would restrict the bending of α-helix 3, a proline substitution would lock it in a kinked position. Thus, it is difficult to conceive of a model in which a region of the coat protein is becoming structurally specialized.
As the inhibitory proteins are known to promiscuously promote off-pathway reactions (4, 5), the coat protein resistance mutations may produce early assembly intermediates that are less prone to associate with scaffolding subunits, both wild type and inhibitory. The dependency phenomenon appears to operate on the level of capsid nucleation. Therefore, efficient nucleation may now depend on a small concentration of the more reactive inhibitory proteins. The acquired mutation in the genome-encoded external scaffolding protein may alleviate defects in the capsid elongation reaction. The mutation confers a D → G substitution in α-helix 2. As this particular helix mediates no contacts with other structural or scaffolding proteins, the substitution may provide an alternative locus for conformational flexibility, which compensates for the steric hindrance introduced into α-helix 3. Regardless of the exact mechanism, the virus has evolved a means to productively incorporate an antiviral agent into its life cycle.
Acknowledgments
This research was supported by National Science Foundation grant MCB 054297 to B.A.F. and an NSF Graduate Research Fellowships to J.E.C.
Members of the University of Arizona Virology Laboratory Course 2007 were Jessica A. Birkholz, Hannah A. Dineen, Dylan C. Grippi, Tyler L. Kempton, Judy Kwan, Nikita N. Patel, Pablo Sanchez-Soria, and Brandon M. Toussaint.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Baldwin, C., and B. Berkhout. 2007. HIV-1 drug-resistance and drug-dependence. Retrovirology 4:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull, J. J., M. R. Badgett, and H. A. Wichman. 2000. Big-benefit mutations in a bacteriophage inhibited with heat. Mol. Biol. Evol. 17:942-950. [DOI] [PubMed] [Google Scholar]
- 3.Bull, J. J., M. R. Badgett, H. A. Wichman, J. P. Huelsenbeck, D. M. Hillis, A. Gulati, C. Ho, and I. J. Molineux. 1997. Exceptional convergent evolution in a virus. Genetics 147:1497-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherwa, J. E., Jr., A. Uchiyama, and B. A. Fane. 2008. Scaffolding proteins altered in the ability to perform a conformational switch confer dominant lethal assembly defects. J. Virol. 82:5774-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherwa, J. E., Jr., and B. A. Fane. 2009. Complete virion assembly with scaffolding proteins altered in the ability to perform a critical conformational switch. J. Virol. 83:7391-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, J. A., M. G. Thompson, E. Paintsil, M. Ricketts, J. Gedzior, and L. Alexander. 2004. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J. Virol. 78:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dam, E., R. Quercia, B. Glass, D. Descamps, O. Launay, X. Duval, H. G. Krausslich, A. J. Hance, and F. Clavel. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 5:e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dokland, T., R. A. Bernal, A. Burch, S. Pletnev, B. A. Fane, and M. G. Rossmann. 1999. The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus φX174. J. Mol. Biol. 288:595-608. [DOI] [PubMed] [Google Scholar]
- 9.Dokland, T., R. McKenna, L. L. Ilag, B. R. Bowman, N. L. Incardona, B. A. Fane, and M. G. Rossmann. 1997. Structure of a viral procapsid with molecular scaffolding. Nature 389:308-313. [DOI] [PubMed] [Google Scholar]
- 10.Ekechukwu, M. C., D. J. Oberste, and B. A. Fane. 1995. Host and phi X 174 mutations affecting the morphogenesis or stabilization of the 50S complex, a single-stranded DNA synthesizing intermediate. Genetics 140:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fane, B. A., and M. Hayashi. 1991. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage phi X174. Genetics 128:663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fane, B. A., S. Head, and M. Hayashi. 1992. Functional relationship between the J. proteins of bacteriophages phi X174 and G4 during phage morphogenesis. J. Bacteriol. 174:2717-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hafenstein, S. L., M. Chen, and B. A. Fane. 2004. Genetic and functional analyses of the φX174 DNA binding protein: the effects of substitutions for amino acid residues that spatially organize the two DNA binding domains. Virology 318:204-213. [DOI] [PubMed] [Google Scholar]
- 14.Incardona, N. L. 1974. Mechanism of adsorption and eclipse of bacteriophage φX174. III. Comparison of the activation parameters for the in vitro and in vivo eclipse reactions with mutant and wild-type virus. J. Virol. 14:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morais, M. C., M. Fisher, S. Kanamaru, L. Przybyla, J. Burgner, B. A. Fane, and M. G. Rossmann. 2004. Conformational switching by the scaffolding protein D directs the assembly of bacteriophage φX174. Mol. Cell 15:991-997. [DOI] [PubMed] [Google Scholar]
- 16.Paredes, R., M. Sagar, V. C. Marconi, R. Hoh, J. N. Martin, N. T. Parkin, C. J. Petropoulos, S. G. Deeks, and D. R. Kuritzkes. 2009. In vivo fitness cost of the M184V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J. Virol. 83:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pesola, J. M., and D. M. Coen. 2007. In vivo fitness and virulence of a drug-resistant herpes simplex virus 1 mutant. J. Gen. Virol. 88:1410-1414. [DOI] [PubMed] [Google Scholar]
- 18.Roof, W. D., H. Q. Fang, K. D. Young, J. Sun, and R. Young. 1997. Mutational analysis of slyD, an Escherichia coli gene encoding a protein of the FKBP immunophilin family. Mol. Microbiol. 25:1031-1046. [DOI] [PubMed] [Google Scholar]
- 19.Salvati, A. L., A. De Dominicis, S. Tait, A. Canitano, A. Lahm, and L. Fiore. 2004. Mechanism of action at the molecular level of the antiviral drug 3(2H)-isoflavene against type 2 poliovirus. Antimicrob. Agents Chemother. 48:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchiyama, A., M. Chen, and B. A. Fane. 2007. Characterization and function of putative substrate specificity domain in microvirus external scaffolding proteins. J. Virol. 81:8587-8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchiyama, A., and B. A. Fane. 2005. Identification of an interacting coat-external scaffolding protein domain required for both the initiation of φX174 procapsid morphogenesis and the completion of DNA packaging. J. Virol. 79:6751-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchiyama, A., P. Heiman, and B. A. Fane. 2009. N-terminal deletions of the φX174 external scaffolding protein affect the timing and fidelity of assembly. Virology 386:303-309. [DOI] [PubMed] [Google Scholar]
- 23.Wichman, H. A., J. Millstein, and J. J. Bull. 2005. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics 170:19-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wichman, H. A., L. A. Scott, C. D. Yarber, and J. J. Bull. 2000. Experimental evolution recapitulates natural evolution. Philos. Trans. R. Soc. Lond. B 355:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, Y., D. J. Bartels, B. L. Hanzelka, U. Muh, Y. Wei, H. M. Chu, A. M. Tigges, D. L. Brennan, B. G. Rao, L. Swenson, A. D. Kwong, and C. Lin. 2008. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 52:110-120. [DOI] [PMC free article] [PubMed] [Google Scholar]