Abstract
Ribonucleosides inhibit human immunodeficiency virus type 1 (HIV-1) replication by mechanisms that have not been fully elucidated. Here, we report the antiviral mechanism for the ribonucleoside analog 5-azacytidine (5-AZC). We hypothesized that the anti-HIV-1 activity of 5-AZC was due to an increase in the HIV-1 mutation rate following its incorporation into viral RNA during transcription. However, we demonstrate that 5-AZC's primary antiviral activity can be attributed to its effect on the early phase of HIV-1 replication. Furthermore, the antiviral activity was associated with an increase in the frequency of viral mutants, suggesting that 5-AZC's primary target is reverse transcription. Sequencing analysis showed an enrichment in G-to-C transversion mutations and further supports the idea that reverse transcription is an antiviral target of 5-AZC. These results indicate that 5-AZC is incorporated into viral DNA following reduction to 5-aza-2′-deoxycytidine. Incorporation into the viral DNA leads to an increase in mutant frequency that is consistent with lethal mutagenesis during reverse transcription as the primary antiviral mechanism of 5-AZC. Antiviral activity and increased mutation frequency were also associated with the late phase of HIV-1 replication; however, 5-AZC's effect on the late phase was less robust. These results reveal that the primary antiviral mechanism of 5-AZC can be attributed to its ability to increase the HIV-1 mutation frequency through viral-DNA incorporation during reverse transcription. Our observations indicate that 5-AZC can affect two steps in HIV-1 replication (i.e., transcription and reverse transcription) but that its primary antiviral activity is due to incorporation during reverse transcription.
Significant progress has been made in the clinical management of human immunodeficiency virus type 1 (HIV-1) infection using antiretroviral drugs (11). Nonetheless, the emergence of drug resistance limits the efficacy of current HIV-1 drugs and drives the need for novel therapeutics (44). Although current therapy relies on combination drug therapy, low levels of viral replication coupled with the high HIV-1 mutation rate makes drug resistance difficult to prevent (41, 43). The development of novel anti-HIV-1 drugs that have a high barrier to the emergence of drug resistance would offer new treatment options as the current drugs become ineffective due to the emergence and transmission of drug resistance.
The HIV-1 mutation rate represents a novel drug target that may offer a higher genetic barrier to the emergence of drug resistance than current anti-HIV-1 drugs. The high mutation rate of HIV-1, as well as other RNA viruses, enables virus survival in the face of a rapidly changing host environment (e.g., the host immune response to viral infection) (15). However, because most mutations are detrimental, the high mutation rate leads to a large proportion of noninfectious virions (7). Thus, HIV-1 may have evolved a mutation rate that carefully balances the need for viral adaptation with the need to replicate with enough fidelity to remain viable. Based upon this high mutation rate, a novel therapeutic strategy is to tip this balance in favor of a higher mutation rate so that the virus is unable to replicate with enough fidelity to remain infectious. This strategy, termed lethal mutagenesis, is based on the concept that only a modest increase in the viral mutation rate is needed to render the virus nonviable (14, 27, 38).
The validity of lethal mutagenesis as an antiviral strategy is supported by experimental evidence showing an inverse correlation between the mutation rate and the infectivity of several RNA viruses (e.g., polio virus, foot and mouth disease virus, and Hantaan virus), as well as retroviruses (e.g., spleen necrosis virus and HIV-1) (1, 5, 22, 24, 47). Nucleoside analogs have been shown to effectively increase viral mutation rates (9, 24, 26, 38, 48). However, there has yet to be a nucleoside analog with sufficiently acceptable efficacy and safety to be clinically relevant for the purpose of lethal mutagenesis. One of the concerns regarding the use of mutagenic nucleoside analogs is the potential for toxicity and carcinogenicity, since these compounds can be directly incorporated into the host genome.
Ribonucleoside analogs with mutagenic potential have also been explored for their antiviral activity against riboviruses (10, 23, 50). Ribonucleoside analogs have been used to block retroviral replication, but the mechanism of action is not clear. There are three models that could explain the mechanism by which ribonucleoside analogs inhibit retrovirus replication. First, ribonucleoside analogs could be incorporated into HIV-1 RNA during transcription of the genomic-length RNA (21, 38). Alternatively, the ribonucleoside analog could be incorporated into viral DNA by reverse transcriptase (RT) following its reduction to the 2′-deoxynucleotide form. Finally, ribonucleoside analogs may exert antiviral activity through incorporation into both DNA and RNA. To date, a detailed understanding of how ribonucleosides, including those with mutagenic potential, manifest an antiretroviral effect is not well established.
5-Azacytidine (5-AZC) is a ribonucleoside analog that is used clinically to treat myelodysplastic syndromes (29, 54). 5-AZC has been shown to inhibit HIV-1 infectivity (3). Based on its structure and its effect on the poliovirus mutation rate, 5-AZC was proposed to exert its antiviral activity by increasing the HIV-1 mutation rate, although no data have been published to support lethal mutagenesis as its mechanism of action. As a ribonucleoside, 5-AZC was proposed to increase the HIV-1 mutation rate through direct incorporation into viral RNA. However, 5-AZC could also be incorporated into viral DNA after reduction by cellular ribonucleotide reductase. In this study, we examined the mechanism by which 5-AZC inhibits HIV-1. Our data reveal that 5-AZC exerts its antiviral activity through incorporation into both viral RNA and DNA. Interestingly, the most potent antiviral activity was attributed to incorporation of 5-AZC into viral DNA following its reduction to the 2′-deoxy form. Further investigation into the mechanism of 5-AZC revealed that incorporation of 5-AZC into viral DNA during reverse transcription leads to lethal mutagenesis that is characterized by a significant increase in transition mutations within the provirus.
MATERIALS AND METHODS
Plasmids, cell lines, and reagents.
The HIV-1-based vector HIG was constructed by cloning the internal ribosome entry site (IRES)-enhanced green fluorescent protein (GFP) sequence from pIRES2-eGFP (Clontech, Mountain View, CA) in the XhoI site of pNL4-3.HSA.R+.E− (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, Germantown, MD) (8, 25). The resulting vector, pHIG, has a vector cassette with the murine heat-stable antigen (HSA) gene, IRES, and GFP gene. The Env expression vector pIIINL4env was a kind gift from Eric Freed (NCI Drug Resistance Program, NCI, Frederick, MD). The U373-MAGICXCR4 and CEM-GFP cell lines were also obtained from the AIDS Reagent Program (from M. Emerman and J. Corbeil, respectively) (19, 53). The phycoerythrin (PE)-conjugated antibody to mouse HSA was obtained from BD Pharmingen (San Diego, CA). The CellTiter-Glo and CellTiter 96 cell proliferation kits were obtained from Promega (Madison, WI). 5-AZC was purchased from Sigma-Aldrich (St. Louis, MO), dissolved in dimethyl sulfoxide (DMSO) at 1 M stock concentration, and stored at −20°C. The High Pure PCR Template Preparation Kit was from Roche Applied Science (Indianapolis, IN), and the pCR2.1 TOPO cloning vector was from Invitrogen (Carlsbad, CA).
Transfections, infections, drug treatments, and mutant frequency.
To produce the HIG (HSA, IRES, GFP) vector as a virus, a calcium phosphate transfection protocol was used. Briefly, 1.5 × 106 HEK 293T cells were transfected with 10 μg pNL4-3.HIG.E- and 1 μg pIIINL4env. The cells were washed the next day with phosphate-buffered saline (PBS) and resuspended in culture medium (Dulbecco's modified Eagle's medium plus 10% HyClone FetalClone III [FC3; ThermoScientific]). The cell culture supernatants were harvested 48 h posttransfection and passed through a 0.2-μm filter. For drug treatment during virus particle production, 5-AZC or 5-aza-2′-deoxycytidine (5-aza-dC) was added for 12 h prior to the harvesting of cell culture supernatants. The amount of drug that could potentially be carried over while in the producer cell supernatant was calculated to be negligible after the ensuing dilutions. U373-MAGICXCR4 cells were used as permissive target cells for infections and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FC3 and in the presence of 0.2 mg/ml neomycin, 0.1 mg/ml hygromycin B, and 1.0 μg/ml puromycin. The cells were infected at a multiplicity of infection (MOI) of 0.3 in the absence of selectable medium. For the treatment of target cells, 5-AZC was added 2 h prior to infection and maintained for 24 h postinfection. The mutant frequency was determined from the percentage of target cells expressing one of the reporter genes relative to that of the entire infected population (i.e., [HSA+/GFP−] plus [HSA−/GFP+] relative to the total percentage of infected cells). Mutant frequencies were normalized to virus replication in the absence of drug treatment.
Virus particle capsid protein (p24) assay.
Enzyme-linked immunosorbent assay plates (96 well) were incubated overnight with 1:1,000 rabbit p24 antiserum (AIDS Reagent Program, catalog no. 4250). The plates were then washed with PBS-0.5% Tween 20 (PBST) and blocked for 1 h with 3% milk in PBST. Samples and standards were prepared by adding a 1:1 volume of PBS-0.1% Empigen (Sigma-Aldrich, St. Louis, MO) and incubating them at 56°C for 30 min. The samples were then added to the plate and incubated at 37°C for 1 h, and the wells were washed with PBST and then incubated with mouse anti-p24 in PBST-1% milk at 22°C for 1 h. Incubation was then done with a horseradish peroxidase-conjugated anti-mouse secondary antibody in PBST-1% milk for 30 min, and the wells were washed with PBST and PBS before the addition of the tetramethylbenzidine substrate. The reactions were stopped by the addition of 1 M sulfuric acid, and the absorbance was determined at 450 nm.
Cellular toxicity analysis.
Both 293T and U373-MAGICXCR4 cells were diluted out on 96-well plates to determine the linear range for cell numbers in each assay. Five thousand 293T cells and 6,000 U373-MAGICXCR4 cells were plated, and the CellTiter-Glo and Cell Titer96 assays were performed according to the manufacturer's protocol. Briefly, the CellTiter-Glo assay generates a luminescent signal that is proportional to the ATP levels, while the CellTiter-96 assay measures the amount of a colorimetric product produced after a substrate is reduced by mitochondrial reductase.
Flow cytometry analysis.
Infected target cells were prepared for flow cytometry by harvesting cells with trypsin-EDTA. The cells were then centrifuged at 200 × g for 5 min, and the cell pellets were resuspended at a final concentration of 5 × 106 cells/ml in PBS-2% FC3. The cells (2.5 × 105) were incubated for 20 min on ice with 1:250 anti-HSA-PE (BD Pharmingen). The cells were then washed with PBS-2% FC3, centrifuged at 200 × g for 5 min, and resuspended in PBS containing 1% FC3 and 1% paraformaldehyde.
The cells were analyzed using a FACScan (BD Biosciences) with CellQuest software. The cells were gated for morphology, and a minimum of 10,000 cells were counted. Excitation was done at 488 nm; fluorescence channel 1 (FL1) detected GFP emission at 507 nm, and FL2 detected PE emission at 578 nm. Compensation was set at FL2-99% FL1 to remove detection of GFP signal from FL2.
Cell sorting and proviral-DNA sequence analysis.
Target cells were collected, and single cells were sorted using a FACSAria (BD Biosciences) 48 h postinfection. Approximately 10,000 cells from the HSA+/GFP− quadrant were collected. Total genomic DNA was purified from these cells (Roche High Pure PCR Template Preparation Kit) and used as a template for nested PCR to amplify the GFP gene (outer primer pair, 5′-CTGAAGGATGCCCAGAAGG-3′ and 5′-TGCTTCTAGCCAGGCACAAGC-3′; inner primer pair, 5′-TTACATGTGTTTAGTCGAGG-3′ and 5′-GCTACTTGTGATTGCTCCATA-3′). The resulting PCR products were purified, ligated to pCR 2.1 (Invitrogen), and transformed into the DH5α strain of Escherichia coli. Plasmid DNA was purified from the cells (Invitrogen Quick Plasmid Miniprep Kit) and used for DNA sequencing analysis. Sequence alignment was performed using the SeqMan program of the Lasergene 7 software package (DNAStar, Madison, WI).
Analysis of 5-AZC with replication-competent HIV-1.
CEM-GFP cells (1.5 × 106) were infected with the HIV-1 NL4-3 molecular clone at an MOI of 0.05. 5-AZC or DMSO alone was added to each culture at a 1:1,000 dilution. The cultures were monitored every 2 days by flow cytometry to determine the percentage of infected cells.
Statistical analysis.
All statistical analyses, graphical representation, and curve fitting were done using GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA).
RESULTS
5-AZC inhibits HIV-1 replication and increases the frequency of viral mutants.
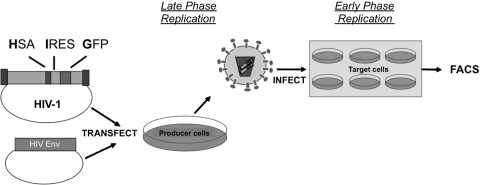
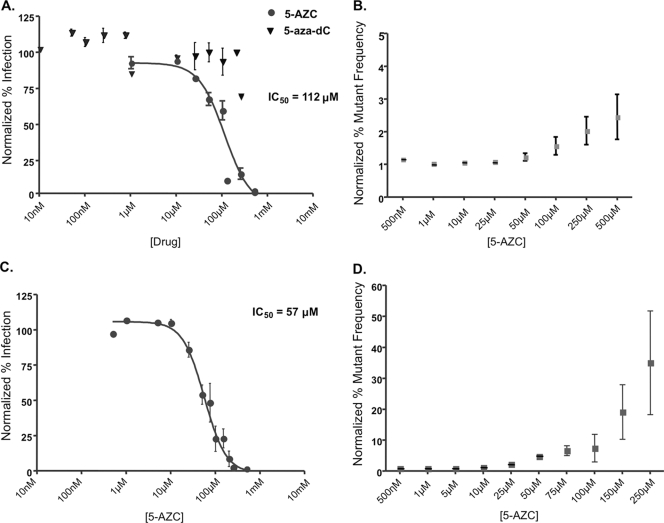
To elucidate the stage of viral replication that 5-AZC inhibits, we used a single-round vector assay that enables differentiation of early and late stages of viral replication (Fig. 1). Specifically, cells cotransfected with envelope-deficient HIV-1 plasmid and a plasmid encoding the HIV-1 envelope support the late stages of viral replication by enabling viral transcription, translation, and particle production. In contrast, virus-infected target cells support the early stages of viral replication by enabling viral entry, reverse transcription, and integration. The stage at which 5-AZC exerts its antiviral activity can be determined by specifically treating either the virus-producing cells, to examine late stages of replication, or the target cells, to examine the early stages of viral replication. From previous studies, it was expected that ribonucleosides exert their antiviral activity in the late phase of the HIV replication cycle (39, 49). In support of this, 5-AZC led to a concentration-dependent decrease in HIV-1 infectivity with a half-maximal inhibitory concentration (IC50) of 112 μM (Fig. 2A). Importantly, there was no concentration-dependent effect of the deoxynucleoside 5-aza-dC during the late phase of viral replication, suggesting that the antiviral activity is dependent on 5-AZC's influence on viral RNA. Additionally, this decrease in infectivity was not due to changes in virus production (see Fig. S1 in the supplemental material). Unexpectedly, 5-AZC led to a more potent inhibition of HIV-1 infectivity in the early phase of the HIV-1 replication cycle with an IC50 of 57 μM (Fig. 2C).
FIG. 1.
HIV-1 vector for monitoring viral infectivity and mutant frequency in the presence of antiretroviral drugs. An envelope- and Nef-deficient HIV-1 vector was constructed with a gene cassette containing the mouse HSA, an IRES element, and the GFP gene. This vector was cotransfected into 293T cells, along with a HIV envelope expression plasmid for producing vector virus. Forty-eight hours posttransfection, the cell culture supernatants were collected, filtered, and used to infect permissive U373-MAGICXCR4 target cells at an MOI of 0.3. Forty-eight hours postinfection, cells were harvested, stained with a PE-conjugated HSA antibody, and analyzed by flow cytometry. Virus-producing cells or permissive target cells were pretreated with drug or DMSO for 12 h after either transfection or 2 h prior to infection, respectively, and treatment continued 24 h postinfection. FACS, fluorescence-activated cell sorting.
FIG. 2.
Concentration-dependent effects of 5-AZC on HIV-1 infectivity and mutant frequency. 293T or U373-MAGICXCR4 cells were treated with the indicated concentrations of compound prior to transfection (late-phase replication) (A and B) or infection (early-phase replication) (C and D). Infected cells (MOI, 0.3) were analyzed by flow cytometry. Only the ribonucleoside 5-AZC had a concentration-dependent effect on viral infectivity in treatment during late-phase replication (A), and this coincided with an increase in the viral mutant frequency (B). (C) A more potent antiviral effect was observed with 5-AZC treatment during early-phase replication. (D) This antiviral activity coincided with a much more dramatic increase in the viral mutant frequency. The data shown are means ± standard deviations of five independent experiments.
Since 5-AZC could cause mutations through its incorporation into viral RNA during transcription, we next examined whether 5-AZC increased HIV-1 mutant frequency. To do this, the single-cycle assay shown in Fig. 1 was used to determine HIV-1 mutant frequency when either virus-producing cells or target cells were treated with 5-AZC. The single-cycle assay enables the determination of the mutant frequency through the use of two reporter genes, the HSA and GFP genes. The HSA gene serves as a reporter for infectivity, while the other reporter gene, the GFP gene, is used as a mutation target. Specifically, cells that express one target gene but not the other are cells that are infected but harbor a mutation that abrogates expression of one of the target genes. Since 5-AZC is a ribonucleoside analog, it was expected that an increase in the mutant frequency would be observed in the late phase of the HIV-1 replication cycle. As expected, 5-AZC increased the HIV-1 mutant frequency 1.5-fold when virus-producing cells were treated at the respective IC50 (Fig. 2B). This indicates that 5-AZC increases the mutant frequency during the late phase of HIV-1 replication. Figure 2D shows that 5-AZC also increased the mutant frequency threefold when target cells were also treated with the respective IC50. These data suggest that 5-AZC may inhibit HIV-1 infectivity by increasing the mutant frequency at two distinct stages of the HIV-1 replication cycle.
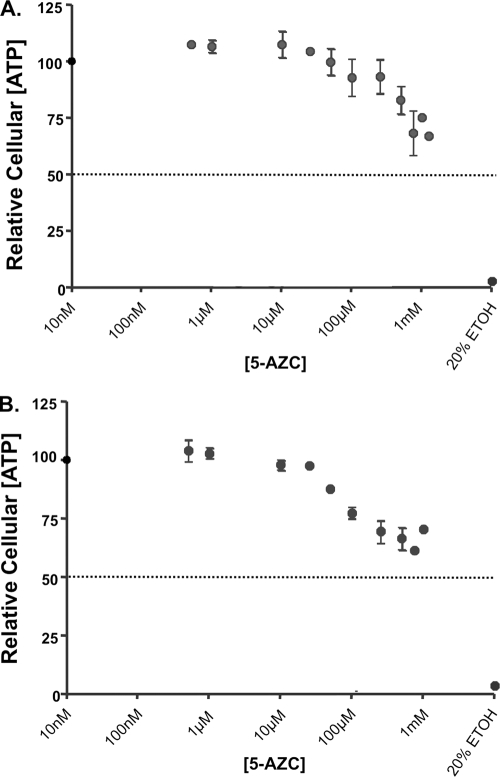
Although the loss of infectivity coincided with an increase in the mutant frequency, it is possible that the loss of infectivity could be attributed to compound toxicity. To investigate this, the toxicity of 5-AZC was analyzed for two different endpoints, including relative cellular ATP levels, as well as mitochondrial reductase activity (data not shown), when either virus-producing cells or permissive target cells were treated with 5-AZC. Figure 3 shows that loss of viral infectivity was not correlated with cellular toxicity. Similar results were observed when mitochondrial reductase activity was used as an endpoint for toxicity (data not shown).
FIG. 3.
Cytotoxicity of 5-AZC. 293T (A) or U373-MAGICXCR4 (B) cells were treated at the indicated concentrations of 5-AZC and analyzed for cell viability by measuring cellular ATP levels. The data represent the means ± standard deviations of four independent experiments.
Analysis of HIV-1 mutation spectra during viral replication in the presence of 5-AZC.
Based on the data shown in Fig. 2, we hypothesized that 5-AZC increased the mutation frequency by two different mechanisms. First, 5-AZCTP could induce mutations through its direct incorporation into viral RNA. Second, 5-AZC could be incorporated into viral DNA after its 2′-OH reduction by ribonucleotide reductase. Since incorporation during either RNA or DNA synthesis would result in mutations in the provirus, we examined the mutation spectra of the proviral GFP gene after treating either virus-producing cells or target cells with 5-AZC. To do this, cells infected by mutant HIV-1 were isolated by cell sorting. In this case, cells expressing HSA but not GFP (HSA+/GFP−) were sorted, and the mutant GFP gene was sequenced.
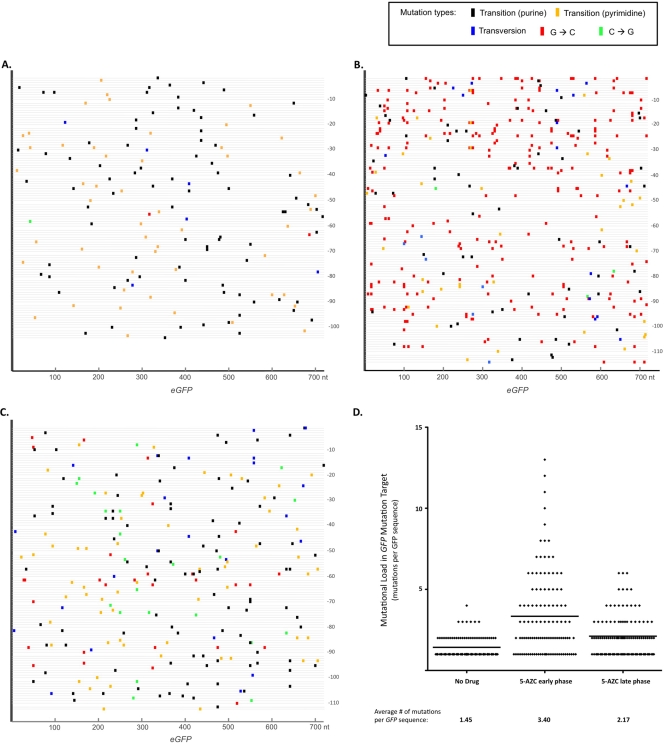
Figure 4 shows the locations and mutation types of over 100 individual GFP gene sequences recovered from target cells. The mutation spectra revealed a dramatic enrichment in G-to-C transversion mutations from virus exposed to 5-AZC in the early phase of replication compared to untreated virus-producing cells (Fig. 4A and B). Besides a dramatic change in the mutation spectra, 5-AZC increased the average number of mutations per nucleotide sequenced by 2.3-fold compared to virus obtained from untreated cells (Table 1). This indicates that 5-AZC not only affects the mutation type but also increases the mutation frequency. The finding that treatment of permissive target cells dramatically altered the HIV-1 mutation spectra suggests that 5-AZC was likely incorporated into viral DNA during reverse transcription after its reduction by cellular ribonucleotide reductase, since a similar mutation spectrum has been reported in cells exposed to 5-aza-dC (30, 47).
FIG. 4.
Mutation spectra of GFP gene sequences from vector proviral DNAs. The HSA+/GFP− cell population from infected U373-MAGI cells was sorted from cells treated with 5-AZC (Fig. 1). The sequence results from at least 100 mutants are shown for no drug (A), 5-AZC treatment of permissive target cells (early phase) (B), and 5-AZC treatment of virus-producing cells (late phase) (C). Each GFP gene sequence analyzed is represented by a thin gray horizontal line. The location of a mutation in the GFP gene sequence is indicated by a colored box perpendicular to the line relative to the 5′ end of the 720-bp open reading frame. Transition mutations are represented by either black (purine) or yellow (pyrimidine) rectangular boxes. G-to-C and C-to-G transversion mutations are indicated by red and green rectangular boxes, respectively. Transversion mutations (other than G-to-C and C-to-G) are indicated by blue rectangular boxes. (D) Mutational loads in GFP genes from proviral DNAs. A summary of the mutational loads (all mutation types) in the GFP gene from either no drug, 5-AZC early phase, or 5-AZC late phase is shown, with each diamond representing a proviral GFP sequence. The calculation of the average number of mutations per GFP target gene sequence is shown.
TABLE 1.
Mutation spectra in the enhanaced GFP reporter gene from background and 5-AZC treatments
| Mutation from: | Mutation to (%): |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No druga |
5-AZC early phaseb |
5-AZC late phasec |
||||||||||
| A | C | G | T | A | C | G | T | A | C | G | T | |
| A to | 1 | 38 | 0 | 0 | 13 | 0 | 2 | 30 | 4 | |||
| C to | 1 | 1 | 11 | 0 | 1 | 7 | 1 | 10 | 13 | |||
| G to | 18 | 1 | 1 | 3 | 66 | 5 | 10 | 13 | 1 | |||
| T to | 1 | 23 | 0 | 0 | 5 | 0 | 1 | 13 | 1 | |||
| Indel | 4 | 0 | 2 | |||||||||
Total no. of sequences, 104; total no. of mutations, 152; no. of nucleotides sequenced, 74,880; mutation frequency, 0.2030%.
Total no. of sequences, 114; total no. of mutations, 386; no. of nucleotides sequenced, 82,080; mutation frequency, 0.4703%; Δ in mutation frequency, 2.3167 (P < 0.001).
Total no. of sequences, 112; total no. of mutations, 243; no. of nucleotides sequenced, 80,640; mutation frequency, 0.3013%; Δ in mutation frequency, 1.4845 (P < 0.001).
Similar G-to-C transversion mutations were observed when virus-producing cells were treated with 5-AZC (Fig. 4C). However, in addition to an increase in G-to-C mutations, there was also an increase in C-to-G mutations, consistent with previous observations that 5-AZC could alter the mutation spectrum of spleen necrosis virus (47). Additionally, 5-AZC treatment increased the average number of mutations per nucleotide sequenced by 1.5-fold compared to untreated virus-producing cells (Table 1).
Analysis of the average number of mutations per GFP gene sequenced revealed a general trend toward an increased mutational load when 5-AZC early and late phase treatments were compared with the no drug treatment (Fig. 4D). The GFP proviral sequences analyzed had mutational loads ranging from 0 to over 10 mutations per GFP target gene. The high mutation load is unlikely to produce infectious virus and suggests that 5-AZC's ability to increase the HIV-1 mutation rate leads to a significant decrease in viral replication capacity.
Susceptibility of replication-competent HIV-1 to 5-AZC.
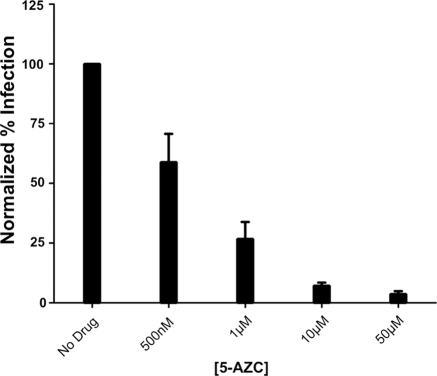
Although the results so far suggested that 5-AZC inhibits HIV-1 infectivity by increasing the mutation rate, we wanted to confirm that these results would extend to replication-competent HIV-1. Additionally, we hypothesized that 5-AZC would inhibit replication-competent virus at concentrations lower than that used in the single-cycle assay. This hypothesis was based on two factors. First, unlike the experiments described thus far, both phases of the replication cycle are exposed to 5AZC during treatment of cells infected with replication-competent HIV-1. Second, if the antiviral activity of 5-AZC is due to an increase in the mutation frequency, then multiple rounds of replication would allow the accumulation of mutations, resulting in a greater proportion of lethally mutagenized viruses. The results in Fig. 5 show that concentrations significantly lower than the concentrations required to inhibit HIV-1 in the single-cycle assay were required to inhibit replication-competent HIV-1.
FIG. 5.
5-AZC inhibits replication-competent HIV-1. The HIV-1 NL4-3 molecular clone was transfected into 293T cells to produce an infectious virus stock that was used to infect CEM-GFP cells. The cells were treated with 5-AZC at the indicated concentrations. The cells were split every 2 days, and fresh medium and 5-AZC were added. Flow cytometry was used to determine the percentage of infected cells every 2 days. The data represent the means ± standard errors of parallel experiments done in triplicate from 8 days posttreatment and are representative of three independent experiments.
DISCUSSION
The emergence and transmission of drug resistance, as well as the halting of the Merck vaccine trial, emphasize the continual need for the development of novel HIV-1 drugs, as well as a return to fundamental aspects of vaccine development (20, 34). As an antiviral strategy, lethal mutagenesis offers a novel drug target (i.e., the viral mutation rate) and is likely to have a high barrier to drug resistance. However, little progress has been made in identifying compounds with enough therapeutic potential to be used clinically to promote HIV-1 lethal mutagenesis. The ribonucleoside analog ribavirin is the only clinically approved ribonucleoside identified so far that may act as a lethal mutagen to inhibit viral replication, specifically, that of hepatitis C virus (10). While ribavirin is not effective against HIV-1, other ribonucleoside analogs, such as 5-AZC, have been shown to have anti-HIV activity in cell culture (3). However, little is known about its mechanism of action.
In this study, we examined the antiviral activity, mechanism of action, and toxicity of 5-AZC. Since 5-AZC is a ribonucleoside analog, it was hypothesized that its antiviral activity would primarily be attributed to its incorporation into viral RNA and the subsequent increase in the HIV-1 mutation frequency. In support of this, several previous studies have shown that 5-AZC can be incorporated into RNA (6, 16, 36, 46). One study demonstrated that 5-AZC was a weak competitive inhibitor with a 20-fold-lower affinity than CTP for RNA polymerase II (35). However, our results show that the most potent antiviral activity of 5-AZC can actually be attributed to its effect on the early phase of HIV-1 replication, which includes reverse transcription. In fact, while 5-AZC increased the HIV-1 mutation frequency in both the late and early phases of HIV-1 replication, it had a greater effect on the early phase of replication. These data suggest that 5-AZC exerts its antiviral activity at both phases of replication through an increase in the mutation frequency. Although 5-AZC led to a modest increase in the mutant frequency, similar increases in mutation rates have been shown to be sufficient to lethally mutagenize other RNA viruses (24, 26, 33, 38, 48). In fact, the theory of lethal mutagenesis suggests that small increases in viral mutation rates should lead to a disproportionately larger decrease in viral infectivity (13, 17).
Both phosphorylation and 2′-OH reduction are prerequisites for 5-AZC's clinical use as a DNA hypomethylating agent (reviewed in references 29 and 54). Similarly, our results suggest that 5-AZC is likely to be phosphorylated and reduced prior to its incorporation into viral DNA by RT. Phosphorylation of 5-AZC is likely performed by uridine-cytidine kinase, as this enzyme is responsible for the phosphorylation of CMP, UMP, and dCMP, as well as many pyrimidine analogs used for cancer and antiviral therapy (28). Additionally, 5-AZC was shown to be a suitable substrate for uridine-cytidine kinase (37).
After phosphorylation, it is inferred that 5-aza-CDP is reduced by cellular ribonucleotide reductase before its incorporation into viral DNA. It has been shown that 10 to 20% of 5-AZC is incorporated into cellular DNA, suggesting a metabolic pathway through ribonucleotide reductase (36). To further support a role for ribonucleotide reductase in 5-AZC metabolism, the ribonucleotide reductase inhibitor hydroxyurea was found to block the epigenetic hypomethylation activity of 5-AZC in vivo (12). Furthermore, our mutational data support the conversion of 5-AZC and its incorporation into DNA. Specifically, our data show a significant increase in G-to-C transversion mutations in proviral DNA after target cells were treated with 5-AZC (Fig. 4B and Table 1), and a similar increase in G-to-C mutations was reported for cellular DNA exposed to 5-aza-dC (30). To our knowledge, there have been no biochemical studies that have looked at the interaction between 5-AZCTP (or 5-aza-dCTP) and purified RT; however, it is well established that the DNA polymerase activity of RT is specific for deoxynucleoside triphosphates (dNTPs) by preferentially excluding ribonucleoside triphosphates (rNTPs) from entering the polymerase active site (18). Moreover, previous reports have shown that DNA polymerase α has similar affinities and rates of incorporation for 5-aza-dCTP and dCTP (2).
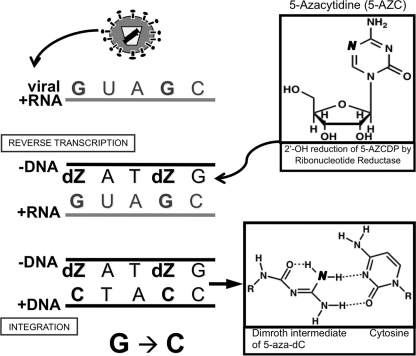
Based on the data presented here, we propose a model that accounts for 5-AZC's antiviral effect on the early phase of the HIV-1 replication cycle (Fig. 6). In this model, 5-AZC is first reduced to 5-aza-dCDP by ribonucleotide reductase. Next, 5-aza-dCTP is incorporated into viral DNA during reverse transcription. Once incorporated into DNA, the 5-aza-cytosine triazine ring (i.e., the base) can undergo a ring-opening step that would enable it to base pair with cytosine (30). Thus, as shown in Fig. 6, cytosine would be incorporated into the plus-strand DNA opposite 5-aza-dC. Finally, our model shows that integration is a key step in repairing the 5-AZC-induced mutations. The model proposes that 5-aza-dC is excised by host DNA repair machinery during integration. 5-Aza-dC would be replaced with guanosine, since it can base pair with the cytosine located in the plus-strand DNA opposite the abasic site. When transcribed, the minus-strand DNA then results in viral progeny carrying G-to-C mutations. In contrast, 5-aza-dC could be incorporated into the positive-strand viral DNA across from guanosines present in the minus strand. However, during integration, the DNA repair machinery would likely excise the 5-aza-dC and replace it with a cytosine, which would not lead to a mutation.
FIG. 6.
Model of 5-AZC mutagenesis during minus-strand DNA synthesis in HIV-1 reverse transcription. Ribonucleotide reductase converts 5-AZCDP to 5-aza-dCDP. After the incorporation of 5-aza-dC (dZ) triphosphate into minus-strand viral DNA, a spontaneous cytosine ring opening to a Dimroth intermediate occurs, which allows the base to pair with dC. During integration, 5-aza-dC is excised by DNA repair machinery and replaced with a guanosine, since it base pairs with the cytosine present opposite the abasic site in the plus-strand DNA. When transcribed, the guanosine in the minus strand codes for a cytosine, thereby leading to an overall G-to-C mutation.
The use of ribonucleoside analogs as lethal mutagens offers the benefit of being able to predict the type of mutations that give rise to virus lethality (39, 49). The increase in G-to-C mutations caused by 5-AZC could be predicted based on the base-pairing properties of 5-AZC and 5-aza-dC. Thus, it is possible that ribonucleoside analogs could be designed to specifically target certain nucleotides for mutation. Ribonucleoside analogs may be superior to current HIV-1 drugs in their ability to delay the emergence of drug resistance. For high-level resistance to emerge against ribonucleoside analogs that function like 5-AZC, it is likely that mutations would have to be acquired in both RT and RNA polymerase II. Although RT is likely to accumulate mutations, there is little pressure on RT to select for mutations that would exclude mutagenic nucleoside analogs, since these drugs do not appear to prevent RT-mediated polymerization. This is in contrast to nucleoside analogs, such as azidothymidine, which prevent replication by chain termination and therefore efficiently select for any mutations in RT that restore viral-DNA synthesis.
A limitation to the development of ribonucleosides as potential antiretroviral agents is the relatively high concentrations needed to observe an antiviral effect (Fig. 2A and C). However, the high concentrations of 5-AZC shown here may be attributed in part to the cell lines used in this study. A previous meta-analysis study documented that cell lines have up to fivefold-higher concentrations of rNTPs than primary cells and that cellular rNTPs are 10- to 100-fold more abundant that dNTPs (51). This suggests that during late-phase replication, 5-AZCTP must compete with an intracellular concentration of CTP of 109 to 455 μM. Similarly, dividing cells were found to have intracellular dCTP concentrations ranging from 27 to 50 μM (51). Because the current study investigated treatment with the ribonucleoside 5-AZC, which has the potential to participate in a number of nucleos(t)ide metabolic pathways, it is difficult to predict the intracellular concentrations of the rNTP and dNTP forms of 5-AZC. Nonetheless, these particular cell lines were chosen because of technical reasons during the virological assays, as well as the sheer number of cells needed for the sequencing experiments.
Differences in dNTP pools in the cell types used here may also account for the discrepancy in IC50 values when the single-cycle assay (Fig. 2) is compared to the multiple-round assay (Fig. 5), although differences in transport pathways and metabolisms could also play a role. Future studies will be needed to precisely measure intracellular 5-AZCTP and 5-aza-dCTP levels when cells are exposed to ribonucleosides. It may also be of interest to measure endogenous dNTP pool levels when treating cells with nucleoside analogs, because this has been shown to cause alterations in natural dNTP pool levels (52) and thus may contribute to an increase in the retroviral mutation rate (4, 31-33, 40, 42). However, the presence of the specific G-to-C transversion in the presence of 5-AZC seems to argue against this notion (Fig. 4B and Table 1).
Since our data support a model in which 5-AZC is converted to the corresponding deoxyribonucleoside triphosphate, it is possible that it could be incorporated into the host genome. This raises concerns about the possible genotoxicity of potential mutagenic ribonucleosides. However, toxicity was not significant at the concentrations required to inhibit viral replication. Additionally, a nucleoside analog currently in development, KP1212/KP1461, induces mutations in viral DNA but does not appear to do so in the host cell genome (24, 45). It is likely that the host DNA repair machinery is sufficiently effective to eliminate any of these analogs that are incorporated into genomic DNA. However, the novelty of these drugs warrants further investigation into their potential long-term effects.
Previous studies have demonstrated the anti-HIV activity of 5-AZC (3). Based on the structure of 5-AZC, it was speculated that its antiviral activity was due to its ability to be incorporated into viral RNA during transcription. Here, our data show that while 5-AZC does demonstrate antiviral activity by this mechanism, its more potent anti-HIV activity can be attributed to its reduction to 5-aza-dCTP followed by incorporation into viral DNA during reverse transcription. Thus, 5-AZC inhibits HIV-1 infectivity through its incorporation into both viral RNA and DNA. This incorporation significantly increases the HIV-1 mutation frequency to a point consistent with lethal mutagenesis. Compounds with a similar mechanism of action could represent an important new class of anti-HIV compounds to explore for clinical viability.
Supplementary Material
Acknowledgments
We thank R. Harris for stimulating discussions.
This research was supported by NIH grant GM56615 (to L.M.M.), an Academic Health Center Faculty Development Grant (to S.P. and L.M.M.), and a Funding Agreement from the Center for Drug Design. M.J.D. received support from NIH grant T32DA007097; C.L.C. was supported by T32 CA009138.
Footnotes
Published ahead of print on 2 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Agudo, R., A. Arias, N. Pariente, C. Perales, C. Escarmis, A. Jorge, A. Marina, and E. Domingo. 2008. Molecular characterization of a dual inhibitory and mutagenic activity of 5-fluorouridine triphosphate on viral RNA synthesis. Implications for lethal mutagenesis. J. Mol. Biol. 382:652-666. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard, J., and R. L. Momparler. 1983. Incorporation of 5-aza-2′-deoxycytidine-5′-triphosphate into DNA. Interactions with mammalian DNA polymerase alpha and DNA methylase. Mol. Pharmacol. 24:109-114. [PubMed] [Google Scholar]
- 3.Bouchard, J., M. C. Walker, J. M. Leclerc, N. Lapointe, R. Beaulieu, and L. Thibodeau. 1990. 5-Azacytidine and 5-azadeoxycytidine inhibit human immunodeficiency virus type 1 replication in vitro. Antimicrob. Agents Chemother. 34:206-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, R., M. Yokoyama, H. Sato, C. Reilly, and L. M. Mansky. 2005. Human immunodeficiency virus mutagenesis during antiviral therapy: impact of drug-resistant reverse transcriptase and nucleoside and nonnucleoside reverse transcriptase inhibitors on human immunodeficiency virus type 1 mutation frequencies. J. Virol. 79:12045-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, D. H., Y. Sun, W. B. Parker, J. B. Arterburn, A. Bartolucci, and C. B. Jonsson. 2007. Ribavirin reveals a lethal threshold of allowable mutation frequency for Hantaan virus. J. Virol. 81:11722-11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cihak, A., J. Vesely, and F. Sorm. 1965. Incorporation of 5-azacytidine into liver ribonucleic acids of leukemic mice sensitive and resistant to 5-azacytidine. Biochim. Biophys. Acta 108:516-518. [DOI] [PubMed] [Google Scholar]
- 7.Coffin, J. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 8.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 9.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuevas, J. M., F. Gonzalez-Candelas, A. Moya, and R. Sanjuan. 2009. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J. Virol. 83:5760-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Clercq, E. 2009. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents 33:307-320. [DOI] [PubMed] [Google Scholar]
- 12.DeSimone, J., P. Heller, L. Hall, and D. Zwiers. 1982. 5-Azacytidine stimulates fetal hemoglobin synthesis in anemic baboons. Proc. Natl. Acad. Sci. USA 79:4428-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingo, E. 2003. Quasispecies and the development of new antiviral strategies. Prog. Drug Res. 60:133-158. [DOI] [PubMed] [Google Scholar]
- 14.Domingo, E., C. Escarmís, E. Lázaro, and S. C. Manrubia. 2005. Quasispecies dynamics and RNA virus extinction. Virus Res. 107:129-139. [DOI] [PubMed] [Google Scholar]
- 15.Domingo, E., E. Martinez-Salas, F. Sobrino, J. C. de la Torre, A. Portela, J. Ortin, C. Lopez-Galindez, P. Perez-Brena, N. Villanueva, R. Najera, S. VandePol, D. Steinhauer, N. DePolo, and J. Holland. 1985. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene: 40:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Doskocil, J., V. Paces, and F. Sorm. 1967. Inhibition of protein synthesis by 5-azacytidine in Escherichia coli. Biochim. Biophys. Acta 145:771-779. [PubMed] [Google Scholar]
- 17.Eigen, M. 2002. Error catastrophe and antiviral strategy. Proc. Natl. Acad. Sci. USA 99:13374-13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, G., M. Orlova, M. M. Georgiadis, W. A. Hendrickson, and S. P. Goff. 1997. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl. Acad. Sci. USA 94:407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gervaix, A., D. West, L. M. Leoni, D. D. Richmond, F. Wong-Staal, and J. Corbeil. 1997. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. USA 94:4653-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard, M. P., and G. P. Bansal. 2008. HIV/AIDS vaccines: a need for new concepts? Int. Rev. Immunol. 27:447-471. [DOI] [PubMed] [Google Scholar]
- 21.Graci, J. D., and C. E. Cameron. 2004. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antivir. Chem. Chemother. 15:1-13. [DOI] [PubMed] [Google Scholar]
- 22.Graci, J. D., D. A. Harki, V. S. Korneeva, J. P. Edathil, K. Too, D. Franco, E. D. Smidansky, A. V. Paul, B. R. Peterson, D. M. Brown, D. Loakes, and C. E. Cameron. 2007. Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue. J. Virol. 81:11256-11266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graci, J. D., K. Too, E. D. Smidansky, J. P. Edathil, E. W. Barr, D. A. Harki, J. E. Galarraga, J. M. Bollinger, Jr., B. R. Peterson, D. Loakes, D. M. Brown, and C. E. Cameron. 2008. Lethal mutagenesis of picornaviruses with N-6-modified purine nucleoside analogues. Antimicrob. Agents Chemother. 52:971-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris, K. S., W. Brabant, S. Styrchak, A. Gall, and R. Daifuku. 2005. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antivir. Res. 67:1-9. [DOI] [PubMed] [Google Scholar]
- 25.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodefiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland, J. J., E. Domingo, J. C. De La Torre, and D. A. Steinhauer. 1990. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 64:3960-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes, E. C. 2003. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 11:543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu, C. H., J. Y. Liou, G. E. Dutschman, and Y. C. Cheng. 2005. Phosphorylation of cytidine, deoxycytidine, and their analog monophosphates by human UMP/CMP kinase is differentially regulated by ATP and magnesium. Mol. Pharmacol. 67:806-814. [DOI] [PubMed] [Google Scholar]
- 29.Issa, J. P., and H. Kantarjian. 2005. Azacitidine. Nat. Rev. Drug Discov. (Suppl.) :S6-S7. [DOI] [PubMed]
- 30.Jackson-Grusby, L., P. W. Laird, S. N. Magge, B. J. Moeller, and R. Jaenisch. 1997. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc. Natl. Acad. Sci. USA 94:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jewell, N. A., R. Chen, R. Raices, and L. M. Mansky. 2003. Nucleoside reverse transcriptase inhibitors and HIV mutagenesis. J. Antimicrob. Chemother. 52:547-550. [DOI] [PubMed] [Google Scholar]
- 32.Julias, J. G., T. Kim, G. Arnold, and V. K. Pathak. 1997. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J. Virol. 71:4254-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julias, J. G., and V. K. Pathak. 1998. Deoxyribonucleoside triphosphate pool imbalances in vivo are associated with an increased retroviral mutation rate. J. Virol. 72:7941-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozal, M. J. 2009. Drug-resistant human immunodefiency virus. Clin. Microbiol. Infect. 15(Suppl. 1):69-73. [DOI] [PubMed] [Google Scholar]
- 35.Lee, T. T., and R. L. Momparler. 1977. Kinetic studies with 5-azacytidine-5′-triphosphate and DNA-dependent RNA polymerase. Biochem. Pharmacol. 26:403-406. [DOI] [PubMed] [Google Scholar]
- 36.Li, L. H., E. J. Olin, H. H. Buskirk, and L. M. Reineke. 1970. Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer Res. 30:2760-2769. [PubMed] [Google Scholar]
- 37.Liacouras, A. S., and E. P. Anderson. 1975. Uridine-cytidine kinase. Purification from a murine neoplasm and characterization of the enzyme. Arch. Biochem. Biophys. 168:66-73. [DOI] [PubMed] [Google Scholar]
- 38.Loeb, L. A., J. M. Essigmann, F. Kazazi, J. Zhang, K. D. Rose, and J. I. Mullins. 1999. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. USA 96:1492-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loeb, L. A., and J. I. Mullins. 2000. Lethal mutagenesis of HIV by mutagenic ribonucleoside analogs. AIDS Res. Hum. Retrovir. 16:1-3. [DOI] [PubMed] [Google Scholar]
- 40.Mansky, L. M. 2003. Mutagenic outcome of combined antiviral drug treatment during human immunodeficiency virus type 1 replication. Virology 307:116-121. [DOI] [PubMed] [Google Scholar]
- 41.Mansky, L. M. 1998. Retrovirus mutation rates and their role in genetic variation. J. Gen. Virol. 79:1337-1345. [DOI] [PubMed] [Google Scholar]
- 42.Mansky, L. M., and L. C. Bernard. 2000. 3′-azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J. Virol. 74:9532-9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mansky, L. M., and K. S. Cunningham. 2000. Virus mutators and antimutators: roles in evolution, pathogenesis and emergence. Trends Genet. 16:512-517. [DOI] [PubMed] [Google Scholar]
- 44.Marsden, M. D., and J. A. Zack. 2009. Eradication of HIV: current challenges and new directions. J. Antimicrob. Chemother. 63:7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami, E., A. Basavapathruni, W. D. Bradley, and K. S. Anderson. 2005. Mechanism of action of a novel viral mutagenic covert nucleotide: molecular interactions with HIV-1 reverse transcriptase and host cell DNA polymerases. Antivir. Res. 67:10-17. [DOI] [PubMed] [Google Scholar]
- 46.Paces, V., J. Doskocil, and F. Sorm. 1968. Incorporation of 5-azacytidine into nucleic acids of Escherichia coli. Biochim. Biophys. Acta 161:352-360. [PubMed] [Google Scholar]
- 47.Pathak, V. K., and H. M. Temin. 1992. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J. Virol. 66:3093-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sierra, S., M. Davila, P. R. Lowenstein, and E. Domingo. 2000. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J. Virol. 74:8316-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, R. A., L. A. Loeb, and B. D. Preston. 2005. Lethal mutagenesis of HIV. Virus Res. 107:215-228. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki, T., T. Okada, C. Otsuka, K. Moriyama, D. Loakes, and K. Negishi. 2005. Mutagenic properties of ribonucleotide analogues in reverse transcription with HIV and AMV reverse transcriptases. Nucleic Acids Symp. Ser. 49:97-98. [DOI] [PubMed] [Google Scholar]
- 51.Traut, T. W. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]
- 52.Vela, J. E., M. D. Miller, G. R. Rhodes, and A. S. Ray. 2008. Effect of nucleoside and nucleotide reverse transcriptase inhibitors of HIV on endogenous nucleotide pools. Antivir. Ther. 13:789-797. [PubMed] [Google Scholar]
- 53.Vodicka, M. A., W. C. Goh, L. I. Wu, M. E. Rogel, S. R. Bartz, V. L. Schweickart, C. J. Raport, and M. Emerman. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233:193-198. [DOI] [PubMed] [Google Scholar]
- 54.Yoo, C. B., and P. A. Jones. 2006. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 5:37-50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.