Abstract
Hepatitis C virus subtype 3a is a highly prevalent and globally distributed strain that is often associated with infection via injection drug use. This subtype exhibits particular phenotypic characteristics. In spite of this, detailed genetic analysis of this subtype has rarely been performed. We performed full-length viral sequence analysis in 18 patients with chronic HCV subtype 3a infection and assessed genomic viral variability in comparison to other HCV subtypes. Two novel regions of intragenotypic hypervariability within the envelope protein E2, of HCV genotype 3a, were identified. We named these regions HVR495 and HVR575. They consisted of flanking conserved hydrophobic amino acids and central variable residues. A 5-amino-acid insertion found only in genotype 3a and a putative glycosylation site is contained within HVR575. Evolutionary analysis of E2 showed that positively selected sites within genotype 3a infection were largely restricted to HVR1, HVR495, and HVR575. Further analysis of clonal viral populations within single hosts showed that viral variation within HVR495 and HVR575 were subject to intrahost positive selecting forces. Longitudinal analysis of four patients with acute HCV subtype 3a infection sampled at multiple time points showed that positively selected mutations within HVR495 and HVR575 arose early during primary infection. HVR495 and HVR575 were not present in HCV subtypes 1a, 1b, 2a, or 6a. Some variability that was not subject to positive selection was present in subtype 4a HVR575. Further defining the functional significance of these regions may have important implications for genotype 3a E2 virus-receptor interactions and for vaccine studies that aim to induce cross-reactive anti-E2 antibodies.
Hepatitis C virus (HCV) infection is a major global health issue leading to persistent viral infection in the majority of those infected and is associated with progressive liver disease, cirrhosis, and hepatocellular carcinoma. Six major genotypes of HCV have been described that have evolved in geographically distinct regions and that share approximately. 80% nucleotide homology with one another. HCV viral genotypes have been further classified into subtypes (25). HCV subtype 3a infection is now the most common subtype in the United Kingdom (11), although it is globally distributed and frequently associated with intravenous drug use.
The classification of HCV viral strains by genotype and subtype has proven informative not only in terms of the epidemic and evolutionary history of the virus but also in terms of clinical outcomes. In particular, the response rates to current gold standard therapy (9) and the prevalence of hepatic steatosis (20) are significantly higher for subtype 3a than for genotype 1 infections. The reasons for this are not understood but must relate to viral genetic and phenotypic differences between strains, or to differences in the ability of hosts to exert an effective immune response against particular viral sequences, or to a combination of both factors.
To date, detailed assessment of the HCV genome has largely focused on HCV genotype 1. Indeed, only a few full-length HCV subtype 3a viral sequences are currently published and available within the major HCV databases (Los Alamos; http://hcv.lanl.gov/components/hcv-db/combined_search/searchi.html and euHCVdb; http://euhcvdb.ibcp.fr/euHCVdb/) (16).
To characterize HCV subtype 3a in detail, we performed whole-genome analysis of a cohort of patients with persistent HCV subtype 3a infection. We subsequently focus on the highly variable regions observed in the envelope protein E2 in both acute and chronic infection, since it was apparent that these regions were not restricted to the well-documented hypervariable region 1 (HVR1) that is found at the 5′ end of E2 in all HCV genotypes.
Viral genomic variability can be assessed at a number of different levels; first, intergenotypic variability may arise in genomic regions that are conserved within the same subtype but are distinct between subtypes. Second, there is intragenotypic variability, which may be defined as regions of viral variability within the same genotype or subtype. Finally, intrahost variability is where viral genomic variability occurs within the same viral subtype and also the same host when individual clonal sequences are assessed. Although intergenotypic variability may simply be a feature of the existence of geographically distinct HCV subtypes, intragenotypic and intrahost variability may reflect viral regions subject to specific selection pressures, with important functional implications.
We observed two distinct regions of intrahost and intragenotypic hypervariability within genotype 3a envelope 2 (E2)—in addition to the previously described HVR1—that we have named HVR495 and HVR575. We show that these regions are subject to positive selection pressure, sometimes very early in acute infection. Although HVR575 has been previously recognized as a site of intergenotypic variation (18), the identification of this region as a hypervariable site within genotype 3a and as a site under early selection pressure leading to variability within the same host has not been previously described.
MATERIALS AND METHODS
Patients.
Plasma samples were obtained and immediately stored at −80°C from 40 treatment naive patients with chronic HCV infection; 18 with subtype 3a, 13 with subtype 1a, and 9 with subtype 1b (John Radcliffe Hospital, Oxford, United Kingdom) and from 4 patients with acute subtype 3a infection sampled at multiple time points longitudinally (San Bortolo Hospital, Vicenza, Italy). Acute HCV was defined as an alanine transaminase (ALT) of >1,000 IU/ml with detectable HCV RNA in the presence of specific risk factors for acute HCV infection (three patients reported a history of recent intravenous drug use, and one had recently undergone orthopedic surgery) and the absence of any other cause of an acute hepatitis. In two of four patients the development of HCV antibody seroconversion was also demonstrated. Local ethical approval was obtained, and all patients gave written informed consent for study participation.
Viral RNA extraction and sequencing.
Plasma (500 μl) was concentrated by high-speed centrifugation (23,600 × g for 1 h) at 4°C. Viral RNA was extracted by using a QIAmp viral RNA minikit (Qiagen). Reverse transcription (RT) and first-round PCR were performed in a single reaction (Superscript III OneStep RT-PCR system with Platinum Taq enzyme; Invitrogen). Subtype 3a, 1a, and 1b specific primers included both previously described and newly designed in-house primers (Table 1) . For subtype 3a, primers 277-For and F4-Rev amplified a 4-kb product that coded Core, E1, and E2 structural proteins. Primers 2412F and 9192R amplified a 7-kb product that coded nonstructural proteins (NS2 to NS5). The RT-PCR cycling conditions were as follows: 55°C for 30 min and 94°C for 2 min, followed by 39 cycles of 94°C for 15 s, 60°C and 58°C for 30 s (7- and 4-kb reactions, respectively), 68°C for 1 min/kb, and a final extension of 68°C for 10 min. Second-round PCR used High Fidelity Taq DNA polymerase (Roche), in nested PCRs (Core, 277-For and 977-Re [600 bp]; E1-745-For and 1585-Rev [840 bp]; E2-1435-For and 2982-Rev [1,547 bp]; F4-F10 primer pairs [∼1 kb each]). PCR conditions were set according to the manufacturer's instructions. For genotype 1 samples, primers UTR-246.for and NS4a-5315.rev amplified a 5,063-bp fragment (17). A second-round PCR was performed to produce a 1,260-bp fragment of E2 using the inner primers outerCE1.for and BlyR1.4 (31). Second-round PCR fragments were gel purified (Qiagen) and sequenced bidirectionally with second-round PCR inner primers and additional inner primers using Prism BigDye (Applied Biosystems) on an ABI 3100 DNA automated sequencer. Each sequence was edited by using X11 software and aligned by using Se-Al (http://tree.bio.ed.ac.uk) to obtain full-length genomic sequence for each subtype 3a patient and E2 sequences of genotype 1 patients.
TABLE 1.
Primers used to obtain viral sequence of HCV subtype 3a
| Primer | Sequence (5′-3′) | Binding site (nt)a | Source or reference |
|---|---|---|---|
| F4 For | TGGGATGGGCGCTGAAATGGGA | 2470 | In-house |
| F4 Rev | CTGGGTAGCCGTAGAAAGCACCT | 3520 | In-house |
| F5 For | ACAGCATACGCCCAGCAAACTAGG | 3429 | In-house |
| F5 Rev | TAGAATGTGGCACAGTGATGCTGC | 4399 | In-house |
| F6 For | GATGAATGTCATGCCCAAGACGCTAC | 4287 | In-house |
| F6 Rev | GCCATGATGTATTTTGTGATGGGGTGTG | 5254 | In-house |
| F7 For | TGTCTCGTGCGGCTTAAGCCAA | 5169 | In-house |
| F7 Rev | GTGACAGTTAGAGAACTCAGCAATG | 6178 | In-house |
| F8 For | GGAGGGAGCGGTACAGTGGATGA | 6068 | In-house |
| F8 Rev | CACAACCTTTGTTTCAGACTCCACCCG | 7068 | In-house |
| F9 For | TGAGCTAGTGGACGCCAACTTGTTATG | 7013 | In-house |
| F9 Rev | GTTCTTCGCCATGATGGTGGTTGGAAT | 8001 | In-house |
| F10 For | CGAAGTTCGGGTATAGTGCGAAGGA | 7897 | In-house |
| F10 Rev | TGCCCGATGTCTCCAAGCTCGTA | 9091 | In-house |
| HCV_2412 F2 | CACCTCCACCARAACATYGT | 2412 | In-house |
| HCV_9192R | GGAGTGAGTTTGAGCTTGGT | 9195 | In-house |
| UTR_277-For | CCTTGTGGTACTGCCTGATAG | 279 | 2 |
| 977-Rev | GTCHTCRGCCTCATACACAAT | 975 | 2 |
| 745-For | TACATCCCGCTCGTCGGC | 747 | In-house |
| E2-1585-Rev | ATGTGCCACGAGCCATTGGT | 1587 | 2 |
| 1435-For | GGCAACTGGGCCAAGGTCGC | 1437 | In-house |
| 2982-Rev | ATAAAGCAGGCTTGTTAG | 2967 | In-house |
| 2237-For | TCAAGGTGAGGATGTTTGTG | 2221 | In-house |
| 2340-Rev | GAATGCAGCAGCGGATGTTGC | 2324 | In-house |
| Utr-246.for | GACTGCTAGCCGAGTAGTGTTG | 248 | 17 |
| NS4a-5315.rev | CGACCTCYARGTCNGCYCACATRC | 5310 | 17 |
| outerCE1.for | ATGATGATGAACTGGTCNCCYAC | 1308 | In-house |
| BlyR1.4 | CTAYCAGCARCATCATCC | 2251 | 31 |
nt, nucleotide.
E2 cloning.
The entire E2 PCR product (1,105 nucleotides) was cloned (TOPO TA cloning kit [Invitrogen]) for patients with acute genotype 3a HCV infection (It13, It14, It16, and It17) at multiple (n = 4 to 6) time points (It13 [total 75 clones, mean number of clones per time point = 19]; It14 [total 91 clones, mean number clones per time point = 20.5; It16 [total 85 clones, mean number clones per time point = 22.3]; It17 [total 109 clones, mean number clones per time point 18.1]). The entire E2 protein was also cloned for patients with chronic HCV infection; 9 with subtype 3a, 9 with subtype 1a, and 7 with subtype 1b (6 to 26 clones per patient; mean = 18.7 clones). Colonies were grown overnight and plasmid DNA purified by using a Montage plasmid miniprep kit (Millipore). EcoRI digestion identified positive clones, which were sequenced bidirectionally using M13For, M13Rev, and additional internal primers. Sequences were aligned and edited by using Sequencher software (GeneCodes Corp.).
Entropy and diversity measurement.
A mathematical measure of entropy was used to evaluate the sequence diversity of the full-length HCV sequences from 18 chronically infected genotype 3a patients and the E2 sequences from 32 patients with subtype 1a, 27 patients with subtype 1b, 20 patients with subtype 2a, 15 patients with subtype 4a, and 15 patients with subtype 6a sequences (derived from in-house bulk sequencing or consensus sequence from multiple clones, with additional sequences from the Los Alamos and euHCVdb HCV databases). Entropy values for each amino acid position were calculated by using the Shannon Heterogeneity In Alignments Tool v1.0 (http://evolve.zoo.ox.ac.uk/software). This program computes the Shannon (23) information entropy score, E, for each codon as follows:
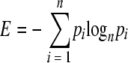 |
where pi is the proportion of sequences that contain residue i at the codon in question. In this analysis there are n = 21 types of residue (20 amino acids plus the stop codons).
Selection analysis.
The program CODEML was used to identify amino-acid sites that had undergone positive selection (Yang et al. [30]; http://abacus.gene.ucl.ac.uk), both in the patients with chronic infection through analysis of E2 bulk sequences and in each acutely infected individual, through analysis of all E2 clonal sequences obtained from each individual patient. The maximum-likelihood method implemented in the CODEML program fits various models of codon evolution to sequence data connected by a phylogenetic tree and considers selection pressures at individual codon sites. The models of codon evolution differ in their distribution of dN/dS values among codons. CODEML detects selection by calculating ω, the ratio of nonsynonymous to synonymous nucleotide substitution. Positive selection at a codon is signified by a ω value that is greater than 1. A likelihood ratio test between the M7 and M8 models was performed in order to test for the presence of selected sites (χ2 distribution with 2 degrees of freedom and P ≤ 0.05) (30). An empirical Bayesian approach, also available in CODEML, was then used to identify individual codons subject to positive selection, with a posterior probability of >90% taken to indicate positive selection.
Nomenclature.
The amino acid nomenclature throughout the manuscript conforms to the system proposed by the Los Alamos database group (15). Amino acids are numbered relative to genotype 1a H77, and genotype 3a insertions are designated with a lowercase letter.
Accession numbers.
Full-length HCV subtype 3a sequences (GQ356200 to GQ356217), E2 subtype 3a clones from chronically infected patients (GQ356218 to GQ356422), subtype 3a clones from acutely infected patients (GQ356423 to GQ356779), E2 subtype 1a and 1b chronically infected patients (bulk and clonal sequencing) (GQ370065 to GQ370362) were evaluated.
RESULTS
Cross-sectional analysis of full-length subtype 3a viral genomes.
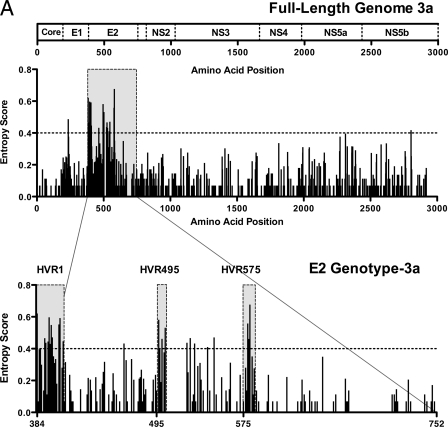
Full-length viral genomic sequences were obtained from 18 treatment-naive patients with chronic HCV subtype 3a infection. A mathematical measure of entropy (Shannon information entropy score) was used to evaluate the sequence diversity of the full-length sequences from these 18 patients. Analysis of aligned sequences showed, as expected, that the regions of highest variability (which we define here as entropy scores of >0.4) were predominantly located in the genomic region coding for the envelope proteins, particularly E2 (Fig. 1A, upper panel). Further analysis of the E2 region of subtype 3a revealed three distinct regions of genomic variability including not only the HVR1 at the N-terminal end of E2 (which is known to be present in all HCV genotypes) but also two further regions. The first of these we named HVR495; this region spans amino acids 495 to 501 and is 7 amino acids long. The second we named HVR575, which represents amino acids 575 to 578e and is 9 amino acids long (Fig. 1A, lower panel).
FIG. 1.
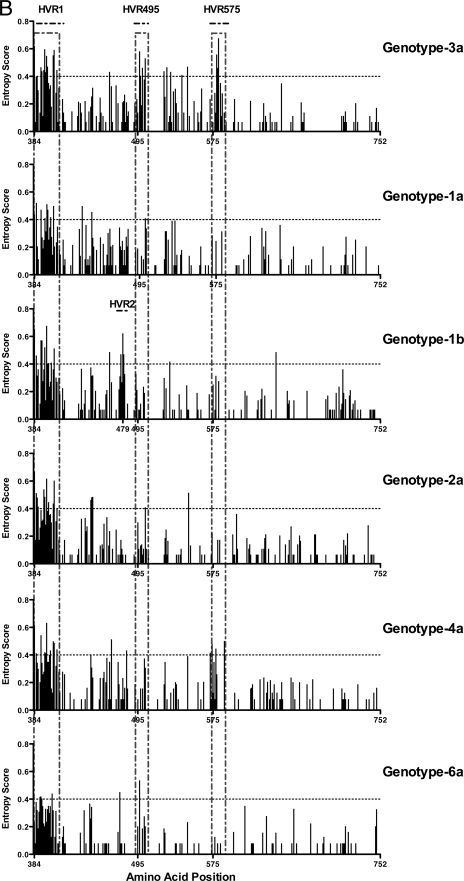
Sequence variability across the HCV subtype 3a genome identifies two novel HVRs within E2. (A) The entropy score (a mathematical measure of variability) at each amino acid site following full-length viral genome sequence analysis of 18 patients with chronic HCV genotype 3a infection is shown. Each bar represents variability at a single amino acid site. The corresponding HCV subtype 3a genome map is given above. Analysis of E2 subtype 3a shows the HVR1 at the N-terminal domain of E2, in addition to two novel hypervariable regions (HVR495 and HVR575). (B) For comparison, we show the E2 entropy scores from 31 patients with HCV subtype 1a, 27 patients with subtype 1b, 20 patients with subtype 2a, 15 patients with subtype 4a, and 15 patients with subtype 6a infection (sequences were determined in house with additional sequences derived from the Los Alamos and euHCVdb HCV databases).
Comparative analysis of E2 HCV subtype 3a with other viral subtypes.
Next, we assessed whether HVR495 and HVR575 were found in other HCV genotypes. In addition to the HCV subtype 3a patients, E2 sequence was determined in-house in 22 patients with genotype 1 infection (13 subtype 1a and 9 subtype 1b). In addition, 85 patients with full-length genomic sequences—including 18 patients with each of the HCV subtypes 1a and 1b, 9 patients with subtype 2a, 15 patients with subtype 4a, and 15 patients with subtype 6a—were randomly selected from the Los Alamos and euHCVdb databases after exclusion of related and synthetic sequences (Fig. 1B). These were aligned within each subtype. Summing the entropy scores for each amino acid position within each subtype showed that the total entropy score for E2 subtype 3a (30.15) was higher than that for subtypes 1a (26.51), 2a (25.23), 4a (26.86), and 6a (17.33) and similar to subtype 1b (that contains the additional HVR2). HVR495 and HVR575 were not observed in the analysis of the 1a, 1b, 2a, or 6a subtypes. There was a single polymorphic amino acid at position 495 in subtype 6a, and variability (less marked than that observed in subtype 3a infection) was observed within HVR575 in subtype 4a infection with an entropy score of >0.4 at position 575b (Fig. 1B).
Evolutionary analysis of positive selection within HCV subtype 3a E2.
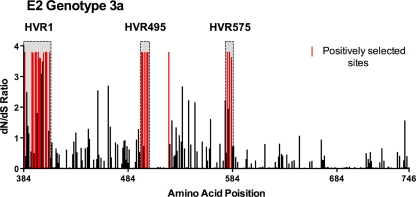
In theory, regions of high variability may arise because some viral genomic regions are simply functionally unconstrained or because variation is induced by selective forces. We therefore performed a selection analysis using the program CODEML to ascertain whether HVR495 and HVR575 were also subject to positive selection. Evolutionary analysis of E2 by CODEML revealed 21 positively selected sites where ω > 1 and with a posterior probability of >90% (Fig. 2, highlighted in red; Table 2), which were concentrated predominantly within HVR1, HVR495, and HVR575. Positively selected sites included amino acids 495, 496, 498, 500, and 501 within HVR495, and 577, 578a, and 578c within HVR575. Only 1 of 21 selected sites detected were located outside these three regions. Neither the polymorphic site at position 495 in subtype 6a nor HVR575 in subtype 4a was subject to positive selection by CODEML analysis.
FIG. 2.
Assessment of positive selection within HCV subtype 3a E2. Assessment of positive selection was performed by using CODEML analysis in 18 patients with chronic subtype 3a infection. dN/dS ratios within E2 are shown; positively selected sites, with a posterior probability of >90%, are highlighted in red.
TABLE 2.
Position and residue of selected amino acids
| HVRa | Amino acid position | Amino acid residue |
|---|---|---|
| HVR1 | 384 | E |
| 392 | S | |
| 392 | A | |
| 394 | H | |
| 395 | S | |
| 397 | S | |
| 398 | G | |
| 399 | I | |
| 402 | L | |
| 404 | S | |
| 405 | P | |
| 408 | R | |
| HVR495 | 495 | D |
| 496 | T | |
| 498 | P | |
| 500 | L | |
| 501 | N | |
| 521 | T | |
| HVR575 | 577 | D |
| 578a | N | |
| 578c | G |
HVR1 spans amino acid positions 384 to 408, HVR495 spans amino acid positions 495 to 501, and HVR575 spans amino acid positions 577 to 578c.
HVR575 contains a genomic insertion unique to HCV subtype 3a.
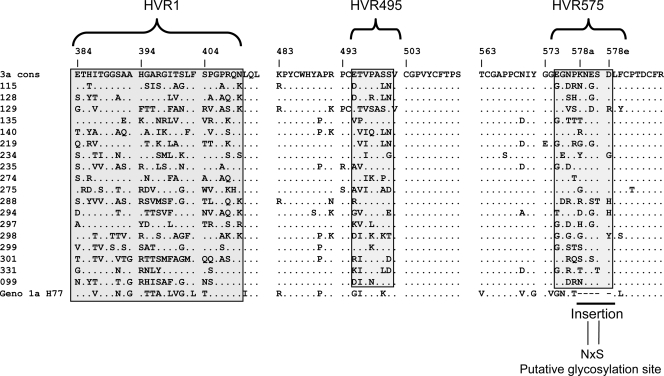
HVR575 contains a 5-amino-acid insertion that is found only in genotype 3a infection. The viral genomes that constitute HVR1, HVR495, and HVR575 in the 18 patients with chronic infection are shown (Fig. 3). Within the 5-amino-acid insertion lies at a putative N-linked glycosylation site (N-X-S/T-X; where X represents any amino acid except proline, N represents asparagine, and S/T represents serine/threonine). Detailed analysis of this insertion shows that, in all 18 patients, the only conserved sites are those absolutely required for glycosylation (i.e., amino acids 578b and 578d), which are always asparagine and serine/threonine, respectively (Fig. 3).
FIG. 3.
The amino acid sequence of HVR1, HVR495, and HVR575 within HCV subtype 3a E2. Amino acids within the E2 HVR1, HVR495, and HVR575 are shown. The 5-amino-acid insertion is shown with a bar and is contained within HVR575. The E2 H77 1a sequence is shown for comparison.
HVR495 and HVR575 show clonal variability within a single host that occurs independently of variability observed in HVR1.
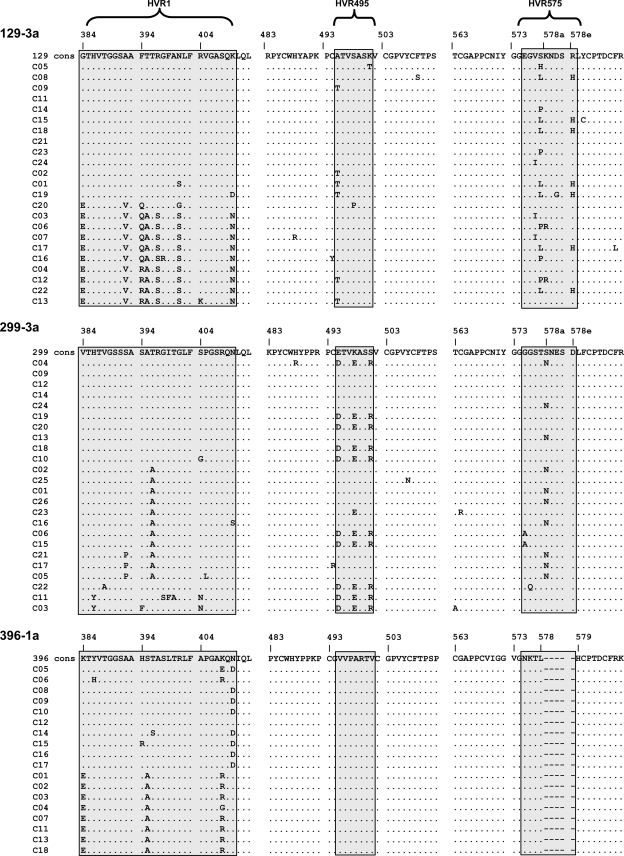
Having shown that HVR495 and HVR575 are highly variable among individuals infected with the same subtype, we next assessed the intrahost variability of these regions. E2 clonal sequence analysis was performed in 22 patients with chronic HCV infection (9 subtype 3a, 9 with subtype 1a, and 7 with subtype 1b). A total of 6 to 26 (mean 18.7) clones were derived from each patient. In subtype 3a patients HVR495 and HVR575 contained multiple variants within a single host; of the nine patients evaluated, all showed clonal variation within HVR575, and seven of nine patients showed clonal variation within HVR495. In contrast, in genotype 1a and 1b these regions were highly conserved except in a single patient (patient 381) that had an aspartic acid to asparagine mutation at position 576 in 5 of the 10 clones sequenced. Clonal analysis of two representative patients of subtype 3a (patients 129 and 299) and one patient with subtype 1a (patient 396) are shown (Fig. 4).
FIG. 4.
Intrahost variability of HVR495 and HVR575. Amino acids within HVR1, HVR495, and HVR575 are derived from 18 to 25 clones after E2 sequencing in three representative patients: patient 129, subtype 3a; patient 299, subtype 3a; and patient 396, subtype 1a. The consensus sequence is given in the top line. A dot indicates that the amino acid within a single clone is identical to the consensus amino acid.
The structure of HCV E2 is currently not known. In theory, then, it is possible that HVR495 and HVR575 lie in close proximity to HVR1 and form a common functional unit. We therefore assessed whether sequence variation within HVR1 was connected to variation seen within HVR495 and HVR575. In patient 129 amino acid variation is seen at multiple sites within HVR575 that is not connected to the variation seen in HVR1 within the same clones. Similarly, patient 299 shows amino acid variation at multiple sites within HVR495 that is not connected to the variability seen in HVR1 within the same clones.
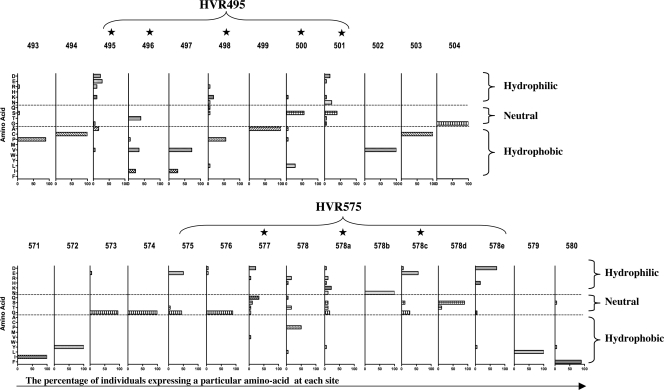
Amino acid compositions of HVR495 and HVR575.
We assessed the amino acid composition of HVR495 and HVR575 in terms of hydropathicity and hydrophilicity. The percentage of individuals expressing a particular amino acid at each site within HVR495 and HVR575 was determined, and amino acids were classified into hydrophobic, neutral, and hydrophilic categories. HVR495 and HVR575 show highly variable central amino acids that are largely hydrophilic or neutral, surrounded by highly conserved hydrophobic amino acids (Fig. 5). The central, highly variable amino acids are those that are identified as being under positive selection by the evolutionary analysis performed by CODEML (indicated by a star in Fig. 5) and include amino acids 495, 498, 500, and 501 within HVR495 and amino acids 577, 578a, and 578c within HVR575.
FIG. 5.
Hydrophobicity or hydrophilicity of the amino acids that comprise the novel HVRs. The data are derived from E2 amino acid sequence in 18 patients with chronic HCV infection. The HVR495 and HVR575 regions are shown. The numbers given are amino acids relative to the genotype 1a H77 sequence. The percentage of individuals expressing a particular amino acid at each site is given. Amino acids are classified as hydrophobic, neutral, and hydrophilic. The star graphic highlights sites under positive selection as defined by CODEML analysis.
HVR495 and HVR575 are under positive selection during acute infection.
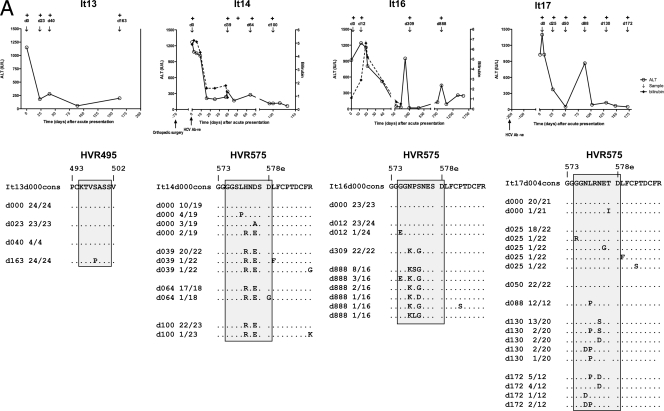
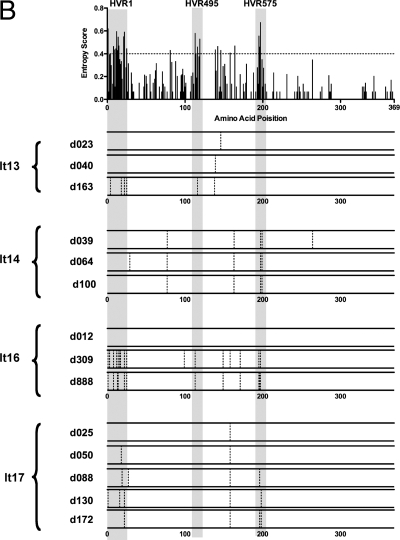
Next, we investigated the evolution of the HVR495 and HVR575 in four individuals with primary HCV genotype3a infection (It13, It14, It16, and It17) presenting with acute hepatitis. Three individuals acquired acute HCV through intravenous drug use, and one acquired acute HCV 70 days after orthopedic surgery. The clinical course of infection is described in Fig. 6A (upper panel graphs). Viral samples were collected at multiple (4-6) time points, and analysis of clonal E2 sequences was performed. Patients It14, It16, and It17 acquired mutations within HVR575 that were detectable in the majority of clones analyzed at 39, 309, and 88 days, respectively, after acute presentation. Dominant mutations were also observed in HVR495 in patients It13 and It16 at 163 and 309 days, respectively, after acute presentation (Fig. 6A, lower panel). Furthermore, CODEML analysis (of all clones derived from all time points) for each acutely infected patient confirmed that mutations within both the HVR495 (amino acid 113 in patient It16) and the HVR575 (amino acids 195, 196, and 198 in patient It17) were under positive selection during acute infection. Although mutations within HVR495 and HVR575 clearly arose early in acute infection, analysis of patient It16 at day 888 after acute infection showed that mutations continued to accumulate within HVR575 after almost a year (day 309) of infection.
FIG. 6.
Evolution of HVR1 and HVR495 and HVR575 during primary HCV infection. (A) The clinical courses of four patients (It13, It14, It16, and It17) with an acute hepatitis secondary to primary HCV infection are shown. HCV RNA was detected by PCR at the time points shown (+). Patients were sampled at multiple time points when HCV RNA positive as indicated by “↓”. E2 quasispecies analysis was performed at each time point. HVR495 or HVR575 are shaded. The amino acid sequence of each variant is shown relative to the consensus sequence at the earliest time point (day 0). A dot indicates that the amino acid of the variant is identical to the consensus amino acid at day 0. The proportion of each variant/total number of variants is given. (B) The upper panel maps the position of the HVRs (shaded regions) derived from 18 patients with chronic HCV subtype 3a infection to the panels below. The data from four patients (It13, It14, It16, and It17) with acute infection are shown in the lower panels. These show mutations (represented by a dotted vertical bar) in the consensus sequence at each time point studied, relative to the consensus sequence at the earliest time point (day 0).
Although sporadic mutations outside HVR1, HVR495, or HVR575 (relative to the earliest time point studied) were observed in a low proportion of sequences during acute infection (Fig. 6A), very few dominant mutations (i.e., found within the majority of clones) within E2 were observed outside HVR1 or HVR495 and HVR575 (Fig. 6B) at any time studied.
DISCUSSION
HCV subtype 3a is the predominant infecting strain in the United Kingdom (11) and is the endemic subtype in parts of Asia. Somewhat surprisingly then, in light of the fact that millions of people are infected with this subtype worldwide, little information is known about the sequence of HCV subtype 3a.
Following full-length sequencing of 18 patients with chronic HCV subtype 3a, we focused our analysis here on the E2 protein. We found that HCV subtype 3a contains not only the common HVR1 at the 5′ end of E2 but also two additional regions of hypervariability, which we have termed HVR495 and HVR575, that are not present in subtypes 1a, 1b, 2a, or 6a. The 15-amino-acid insertion in HCV genotype 3a is contained within the HVR575, and this region, in addition to HVR495, appears to be under selection pressure early in acute genotype 3a HCV infection. Analysis of more datasets may be required to confirm or refute the presence of HVR575 in subtype 4a HCV, where we have demonstrated some variability but were unable to show evidence of positive selection.
The fact that the viral sequence within HVR495 and HVR575 is different in every individual with chronic genotype 3a infection studied makes it highly unlikely that viral variability in this region is driven by T cells, where a T-cell “footprint” will be directed by human leukocyte antigens, and viral variation will be limited to individuals of a particular human leukocyte antigen type. Although speculative, the hydrophobicity profile and associated constant/variable amino acid distribution within these sites suggest the presence of external loops that may act as an antigenic determinant (21, 27) or play a role in viral attachment or entry.
HCV envelope proteins are known to be glycosylated. The E2 proteins of genotypes 1a and 3a contain seven and six potential glycosylation sites, respectively (24). HVR575 contains a putative glycosylation site. N-linked glycosylation has been shown to be required for E2 protein folding (26) and for the formation of E1/E2 complexes (19). Glycosylation also has an important role in shaping immunogenicity against a protein by the shielding of epitopes against targeting antibodies and in maintaining antigenic structure. A number of studies have shown that N-linked glycosylation can limit the antibody response to HCV envelope proteins, and removal of these glycans increases the ability of antibodies to neutralize their target (4, 8, 12). In acute human immunodeficiency virus infection, envelope mutations may lead to the development of new glycosylation sites (that reduce but do not abrogate completely antibody recognition of antigenic determinants in close proximity) and also to the loss of glycosylation sites (29). In HCV infection, glycosylation sites are thought to be highly conserved (18), although, interestingly, we show a dominant mutation developing in the HVR575 glycosylation site (patient It17) 130 days after presentation. Subtypes 1a, 1b, 2b, 4a, and 6a also have a glycosylation site in close proximity to HVR575 (18), suggesting a critical role of the glycosylation site in this region.
Other regions of genotype-specific variability within E2 have been described in chronic HCV infection; this includes a 9-amino-acid region of variability seen in HCV genotype 2 (5) and a 7-amino-acid variable sequence 75 amino acids downstream of HVR1 in HCV genotype 1b infections, termed HVR2 (14). A third region (HVR3) of variability has been described between HVR1 and HVR2 (28), although this is confounded by an analysis that grouped patients of mixed genotypes. The HVR1 region has been intensively studied over recent years and shown to be particularly important as a target for antibody recognition, and there is also some evidence to show that the HVR1 and HVR2 (22) and also the igVR (18) may interplay in a complex fashion to modulate E2 receptor binding. The HVR1 has also been used as a genetic “marker” to identify and quantify circulating viral genetic diversity (quasispecies) within infected hosts and to correlate with various clinical outcomes. Quasispecies dynamics based on HVR1 analysis have been linked to interferon treatment responses (1, 7, 10), spontaneous virus resolution after primary infection (3, 6), and HCV liver pathology (13). Due to the very real practical difficulties in performing full-length viral genome clonal analysis, it is not clear whether differences in HVR1 evolution and clinical outcome relate directly to this viral region or whether this region is serving as a marker for sequence differences elsewhere along the viral genome. However, the finding of HVR495 and HVR575 suggests that these regions should be taken into account in analyses of quasispecies diversity that focus on HCV genotype 3a.
In conclusion, we have identified two regions of hypervariability within E2 in HCV subtype 3a chronically infected individuals. Further analysis shows that these regions are subject to strong intrahost selective pressure that arises early during acute infection. Future studies will need to address the functional significance of these specific regions in subtype 3a infection.
Acknowledgments
This study was supported by the Medical Research Council UK (E.B. and I.H.), the NIHR Biomedical Research Centre Program (E.B. and P.K.), the Wellcome Trust (P.K. and V.F.), the James Martin School for the 21st Century (P.K.), and the Raine Medical Research Foundation of Western Australia (M.L.).
We thank Jane Phillips, Sarah Beer, and Elizabeth Simms (specialist nurses and clinicians at the John Radcliffe Hospital, Oxford, United Kingdom) and the patients who donated blood for the study.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Abbate, I., O. Lo Iacono, R. Di Stefano, G. Cappiello, E. Girardi, R. Longo, D. Ferraro, G. Antonucci, V. Di Marco, M. Solmone, A. Craxi, G. Ippolito, and M. R. Capobianchi. 2004. HVR-1 quasispecies modifications occur early and are correlated to initial but not sustained response in HCV-infected patients treated with pegylated- or standard-interferon and ribavirin. J. Hepatol. 40:831-836. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, E., G. Harcourt, D. Brown, M. Lucas, R. Phillips, G. Dusheiko, and P. Klenerman. 2002. The dynamics of T-lymphocyte responses during combination therapy for chronic hepatitis C virus infection. Hepatology 36:743-754. [DOI] [PubMed] [Google Scholar]
- 3.Chen, S., and Y. M. Wang. 2005. Multigene tracking of quasispecies in viral persistence and clearance of hepatitis C virus. World J. Gastroenterol. 11:2874-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkowska, E., F. Kajumo, E. Garcia, J. Reinus, and T. Dragic. 2007. Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J. Virol. 81:8072-8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan, X., and A. M. Di Bisceglie. 2001. Genetic characterization of hypervariable region 1 in patients chronically infected with hepatitis C virus genotype 2. J. Med. Virol. 64:325-333. [DOI] [PubMed] [Google Scholar]
- 6.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 7.Farci, P., R. Strazzera, H. J. Alter, S. Farci, D. Degioannis, A. Coiana, G. Peddis, F. Usai, G. Serra, L. Chessa, G. Diaz, A. Balestrieri, and R. H. Purcell. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc. Natl. Acad. Sci. USA 99:3081-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fournillier, A., C. Wychowski, D. Boucreux, T. F. Baumert, J. C. Meunier, D. Jacobs, S. Muguet, E. Depla, and G. Inchauspe. 2001. Induction of hepatitis C virus E1 envelope protein-specific immune response can be enhanced by mutation of N glycosylation sites. J. Virol. 75:12088-12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 10.Gaudy, C., A. Moreau, P. Veillon, S. Temoin, F. Lunel, and A. Goudeau. 2003. Significance of pretreatment analysis of hepatitis C virus genotype 1b hypervariable region 1 sequences to predict antiviral outcome. J. Clin. Microbiol. 41:3615-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Health Protection Agency. 2008. Health Protection Agency annual report: hepatitis C in the UK. Health Protection Agency, London, United Kingdom.
- 12.Helle, F., A. Goffard, V. Morel, G. Duverlie, J. McKeating, Z. Y. Keck, S. Foung, F. Penin, J. Dubuisson, and C. Voisset. 2007. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J. Virol. 81:8101-8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honda, M., S. Kaneko, A. Sakai, M. Unoura, S. Murakami, and K. Kobayashi. 1994. Degree of diversity of hepatitis C virus quasispecies and progression of liver disease. Hepatology 20:1144-1151. [PubMed] [Google Scholar]
- 14.Kato, N., Y. Ootsuyama, S. Ohkoshi, T. Nakazawa, H. Sekiya, M. Hijikata, and K. Shimotohno. 1992. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 189:119-127. [DOI] [PubMed] [Google Scholar]
- 15.Kuiken, C., C. Combet, J. Bukh, I. T. Shin, G. Deleage, M. Mizokami, R. Richardson, E. Sablon, K. Yusim, J. M. Pawlotsky, and P. Simmonds. 2006. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology 44:1355-1361. [DOI] [PubMed] [Google Scholar]
- 16.Kuiken, C., P. Hraber, J. Thurmond, and K. Yusim. 2008. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 36:D512-D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, Z., D. M. Netski, Q. Mao, O. Laeyendecker, J. R. Ticehurst, X. H. Wang, D. L. Thomas, and S. C. Ray. 2004. Accurate representation of the hepatitis C virus quasispecies in 5.2-kilobase amplicons. J. Clin. Microbiol. 42:4223-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCaffrey, K., I. Boo, P. Poumbourios, and H. E. Drummer. 2007. Expression and characterization of a minimal hepatitis C virus glycoprotein E2 core domain that retains CD81 binding. J. Virol. 81:9584-9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meunier, J. C., A. Fournillier, A. Choukhi, A. Cahour, L. Cocquerel, J. Dubuisson, and C. Wychowski. 1999. Analysis of the glycosylation sites of hepatitis C virus (HCV) glycoprotein E1 and the influence of E1 glycans on the formation of the HCV glycoprotein complex. J. Gen. Virol. 80(Pt. 4):887-896. [DOI] [PubMed] [Google Scholar]
- 20.Mihm, S., A. Fayyazi, H. Hartmann, and G. Ramadori. 1997. Analysis of histopathological manifestations of chronic hepatitis C virus infection with respect to virus genotype. Hepatology 25:735-739. [DOI] [PubMed] [Google Scholar]
- 21.Parker, J. M., D. Guo, and R. S. Hodges. 1986. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 25:5425-5432. [DOI] [PubMed] [Google Scholar]
- 22.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon, C. E. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:379-423. [Google Scholar]
- 24.Shaw, M. L., J. McLauchlan, P. R. Mills, A. H. Patel, and E. A. McCruden. 2003. Characterisation of the differences between hepatitis C virus genotype 3 and 1 glycoproteins. J. Med. Virol. 70:361-372. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds, P., A. Alberti, H. J. Alter, F. Bonino, D. W. Bradley, C. Brechot, J. T. Brouwer, S. W. Chan, K. Chayama, D. S. Chen, et al. 1994. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology 19:1321-1324. [PubMed] [Google Scholar]
- 26.Slater-Handshy, T., D. A. Droll, X. Fan, A. M. Di Bisceglie, and T. J. Chambers. 2004. HCV E2 glycoprotein: mutagenesis of N-linked glycosylation sites and its effects on E2 expression and processing. Virology 319:36-48. [DOI] [PubMed] [Google Scholar]
- 27.Strynadka, N. C., M. J. Redmond, J. M. Parker, D. G. Scraba, and R. S. Hodges. 1988. Use of synthetic peptides to map the antigenic determinants of glycoprotein D of herpes simplex virus. J. Virol. 62:3474-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troesch, M., I. Meunier, P. Lapierre, N. Lapointe, F. Alvarez, M. Boucher, and H. Soudeyns. 2006. Study of a novel hypervariable region in hepatitis C virus (HCV) E2 envelope glycoprotein. Virology 352:357-367. [DOI] [PubMed] [Google Scholar]
- 29.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao, E., and J. E. Tavis. 2005. A general method for nested RT-PCR amplification and sequencing the complete HCV genotype 1 open reading frame. Virol. J. 2:88. [DOI] [PMC free article] [PubMed] [Google Scholar]