Abstract
The quest to create a human immunodeficiency virus type 1 (HIV-1) vaccine capable of eliciting broadly neutralizing antibodies against Env has been challenging. Among other problems, one difficulty in creating a potent immunogen resides in the substantial overall sequence variability of the HIV envelope protein. The membrane-proximal region (MPER) of gp41 is a particularly conserved tryptophan-rich region spanning residues 659 to 683, which is recognized by three broadly neutralizing monoclonal antibodies (bnMAbs), 2F5, Z13, and 4E10. In this study, we first describe the variability of residues in the gp41 MPER and report on the invariant nature of 15 out of 25 amino acids comprising this region. Subsequently, we evaluate the ability of the bnMAb 2F5 to recognize 31 varying sequences of the gp41 MPER at a molecular level. In 19 cases, resulting crystal structures show the various MPER peptides bound to the 2F5 Fab′. A variety of amino acid substitutions outside the 664DKW666 core epitope are tolerated. However, changes at the 664DKW666 motif itself are restricted to those residues that preserve the aspartate's negative charge, the hydrophobic alkyl-π stacking arrangement between the β-turn lysine and tryptophan, and the positive charge of the former. We also characterize a possible molecular mechanism of 2F5 escape by sequence variability at position 667, which is often observed in HIV-1 clade C isolates. Based on our results, we propose a somewhat more flexible molecular model of epitope recognition by bnMAb 2F5, which could guide future attempts at designing small-molecule MPER-like vaccines capable of eliciting 2F5-like antibodies.
Eliciting broadly neutralizing antibodies (bnAbs) against primary isolates of human immunodeficiency virus type I (HIV-1) has been identified as a major milestone to attain in the quest for a vaccine in the fight against AIDS (12, 28). These antibodies would need to interact with HIV-1 envelope glycoproteins gp41 and/or gp120 (Env), target conserved regions and functional conformations of gp41/gp120 trimeric complexes, and prevent new HIV-1 fusion events with target cells (21, 57, 70, 71). Although a humoral response generating neutralizing antibodies against HIV-1 can be detected in HIV-1-positive individuals, the titers are often very low, and virus control is seldom achieved by these neutralizing antibodies (22, 51, 52, 66, 67). The difficulty in eliciting a broad and potent neutralizing antibody response against HIV-1 is thought to reside in the high degree of genetic diversity of the virus, in the heterogeneity of Env on the surface of HIV-1, and in the masking of functional regions by conformational covering, by an extensive glycan shield, or by the ability of some conserved domains to partition to the viral membrane (24, 25, 29, 30, 38, 39, 56, 68, 69). So far, vaccine trials using as immunogens mimics of Env in different conformations have primarily elicited antibodies with only limited neutralization potency across different HIV-1 clades although recent work has demonstrated more encouraging results (4, 12, 61).
The use of conserved regions on gp41 and gp120 Env as targets for vaccine design has been mostly characterized by the very few anti-HIV-1 broadly neutralizing monoclonal antibodies (bnMAbs) that recognize them: the CD4 binding-site on gp120 (bnMAb b12), a CD4-induced gp120 coreceptor binding site (bnMAbs 17b and X5), a mannose cluster on the outer face of gp120 (bnMAb 2G12), and the membrane proximal external region (MPER) of gp41 (bnMAbs 2F5, Z13 and 4E10) (13, 29, 44, 58, 73). The gp41 MPER region is a particularly conserved part of Env that spans residues 659 to 683 (HXB2 numbering) (37, 75). Substitution and deletion studies have linked this unusually tryptophan-rich region to the fusion process of HIV-1, possibly involving a series of conformational changes (5, 37, 41, 49, 54, 74). Additionally, the gp41 MPER has been implicated in gp41 oligomerization, membrane leakage ability facilitating pore formation, and binding to the galactosyl ceramide receptor on epithelial cells for initial mucosal infection mediated by transcytosis (2, 3, 40, 53, 63, 64, 72). This wide array of roles for the gp41 MPER will put considerable pressure on sequence conservation, and any change will certainly lead to a high cost in viral fitness.
Monoclonal antibody 2F5 is a broadly neutralizing monoclonal anti-HIV-1 antibody isolated from a panel of sera from naturally infected asymptomatic individuals. It reacts with a core gp41 MPER epitope spanning residues 662 to 668 with the linear sequence ELDKWAS (6, 11, 42, 62, 75). 2F5 immunoglobulin G binding studies and screening of phage display libraries demonstrated that the DKW core is essential for 2F5 recognition and binding (15, 36, 50). Crystal structures of 2F5 with peptides representing its core gp41 epitope reveal a β-turn conformation involving the central DKW residues, flanked by an extended conformation and a canonical α-helical turn for residues located at the N terminus and C terminus of the core, respectively (9, 27, 45, 47). In addition to binding to its primary epitope, evidence is accumulating that 2F5 also undergoes secondary interactions: multiple reports have demonstrated affinity of 2F5 for membrane components, possibly through its partly hydrophobic flexible elongated complementarity-determining region (CDR) H3 loop, and it has also been suggested that 2F5 might interact in a secondary manner with other regions of gp41 (1, 10, 23, 32, 33, 55). Altogether, even though the characteristics of 2F5 interaction with its linear MPER consensus epitope have been described extensively, a number of questions persist about the exact mechanism of 2F5 neutralization at a molecular level.
One such ambiguous area of the neutralization mechanism of 2F5 is investigated in this study. Indeed, compared to bnMAb 4E10, 2F5 is the more potent neutralizing antibody although its breadth across different HIV-1 isolates is more limited (6, 35). In an attempt to shed light on the exact molecular requirements for 2F5 recognition of its primary gp41 MPER epitope, we performed structural studies of 2F5 Fab′ with a variety of peptides. The remarkable breadth of possible 2F5 interactions reveals a somewhat surprising promiscuity of the 2F5 binding site. Furthermore, we link our structural observations with the natural variation observed within the gp41 MPER and discuss possible routes of 2F5 escape from a molecular standpoint. Finally, our discovery of 2F5's ability to tolerate a rather broad spectrum of amino acids in its binding, a spectrum that even includes nonnatural amino acids, opens the door to new ways to design small-molecule immunogens potentially capable of eliciting 2F5-like neutralizing antibodies.
MATERIALS AND METHODS
gp41 MPER sequence alignment analysis.
The Epilign sequence alignment and manipulation tool from the Los Alamos National Laboratory HIV database (http://www.hiv.lanl.gov) was used to probe for gp41 MPER sequence variability. A search was performed for protein residues 659 to 683 of HIV-1 Env. It resulted in 1,766 aligned sequences. A Clustal W format file of all aligned sequences was then used with the program WebLogo (16, 60) to qualitatively represent variability at each amino acid position of the gp41 MPER. Subsequently, aligned sequences were manually divided by their clade information (clade A, 94 sequences; clade B, 529 sequences; clade C, 469 sequences; clade D, 61 sequences; clade F, 19 sequences; clade G, 60 sequences; clade J, 2 sequences; clade AC, 41 sequences; clade AE, 71 sequences; clade AG, 75 sequences; clade BF, 68 sequences; clade BG, 4 sequences) and represented using WebLogo to show gp41 MPER sequence variability by clade.
2F5 Fab′ production and crystal complex formation.
The bnMAb 2F5 amino acid sequence and the preparation and purification of its 2F5 Fab′ fragment have been described elsewhere (9). 2F5 immunoglobulin G was a gift from Sanofi-Pasteur. Peptides were gifts from either A. Pedyczak and P. Chong (Sanofi-Pasteur) or were obtained commercially. Peptides dissolved in 20 mM Tris, pH 8.0, buffer at a concentration of 10 mM were added to a 8 to 10 mg/ml 2F5 Fab′ protein solution in the same buffer in a molar ratio ranging from 3:1 to 50:1. Crystal screening was performed using the commercially available Hampton Research Crystal Screen and ammonium sulfate grid screens. Initial crystal hits were identified from Crystal Screen I condition 41 (0.1 M HEPES, pH 7.5, 10% vol/vol 2-propanol, 20% [wt/vol] polyethylene glycol 4000) and from the ammonium sulfate grid screen condition 8 (0.1 M citric acid, pH 5.0, 1.6 M ammonium sulfate). Crystal complexes were refined using the hanging drop method at room temperature, and most 2F5 Fab′-peptide crystals used for X-ray diffraction experiments were obtained from a solution of 0.1 M citric acid, pH 5.6, and 1.6 M ammonium sulfate. As shown in Table 1, some 2F5 Fab′-peptide mixtures did not form any crystals at all or only those of the free Fab′ under the conditions of the crystal screening procedure outlined above.
TABLE 1.
List of peptide sequences (with associated MPER residue variation) that were used for cocrystallization experiments with 2F5 Fab′
| Peptide | Amino acid sequence | Variation at position: | X-ray diffraction dataa |
|||||
|---|---|---|---|---|---|---|---|---|
| Resolution range (Å) | Rsym (%) | Complete (%) | Rwork (%) | Rfree (%) | PDB ID | |||
| 1 | ELDKWAS | HXB2b | 12.0-2.00 | 3.5 (31.3) | 90.0 (93.3) | 23.2 | 25.8 | 2F5B |
| 2 | ALDKWAS | 662 | 12.0-2.10 | 3.3 (38.6) | 97.4 (96.9) | 22.1 | 23.6 | 1U8H |
| 3 | ELAKWAS | 664 | ||||||
| 4 | ELEKWAS | 664 | 50.0-2.24 | 6.0 (36.8) | 85.6 (83.2) | 22.8 | 23.8 | 1U8Q |
| 5 | ELNKWAS | 664 | ||||||
| 6 | ELQKWAS | 664 | ||||||
| 7 | ELDAWAS | 665 | ||||||
| 8 | DLDRWAS | 662, 665 | 17.0-2.60 | 8.8 (35.6) | 88.3 (88.0) | 22.3 | 24.9 | 1U8L |
| 9 | ELDHWAS | 665 | 80.0-2.24 | 5.6 (24.6) | 96.9 (100) | 21.0 | 23.9 | 1U95 |
| 10 | ELD(Orn)WAS | 665 | 80.0-2.24 | 4.6 (11.8) | 91.3 (97.4) | 22.9 | 22.7 | 3IDJ |
| 11 | ELD(Nrg)WAS | 665 | 80.0-2.24 | 5.7 (31.3) | 93.2 (99.9) | 22.9 | 23.6 | 3IDM |
| 12 | ELD(Paf)WAS | 665 | 50.0-2.25 | 6.3 (23.6) | 97.4 (99.1) | 22.0 | 23.8 | 3IDN |
| 13 | ELDEWAS | 665 | ||||||
| 14 | ELDKAAS | 666 | ||||||
| 15 | ELDKYAS | 666 | 50.0-2.40 | 5.4 (36.4) | 99.2 (96.3) | 21.8 | 22.6 | 1U8M |
| 16 | ELDKFAS | 666 | 50.0-2.56 | 9.6 (37.1) | 92.2 (93.9) | 20.5 | 23.1 | 1U8N |
| 17 | ELDKHAS | 666 | 20.0-3.02 | 9.5 (34.7) | 94.8 (93.7) | 21.1 | 21.4 | 1U8O |
| 18 | ELDKIAS | 666 | ||||||
| 19 | ELDKLAS | 666 | ||||||
| 20 | ELDK(Nle)AS | 666 | ||||||
| 21 | ALDKWQN | 662, 667, 668 | 30.0-2.10 | 8.3 (39.2) | 99.9 (99.8) | 21.2 | 23.0 | 3IDI |
| 22 | ALDKWD | 662, 667 | 20.0-1.86 | 4.2 (29.9) | 98.5 (99.5) | 22.4 | 22.7 | 3IDG |
| 23 | ELDKWNSL | 667 | 50.0-2.60 | 13.5 (35.2) | 95.8 (90.3) | 21.3 | 24.0 | 2PW1 |
| 24 | ELDKWKSL | 667 | 50.0-2.55 | 22.0 (39.0) | 99.0 (98.5) | 21.8 | 23.7 | 2PW2 |
| 25 | ELDKWAN | 668 | 17.0-2.00 | 5.9 (39.9) | 97.1 (89.0) | 23.4 | 24.7 | 1U8I |
| 26 | ELDKWAG | 668 | 80.0-2.24 | 13.6 (27.3) | 91.3 (96.5) | 22.9 | 23.8 | 1U8J |
| 27 | ECDKWCS | 663, 667 | 20.0-3.23 | 12.6 (30.2) | 97.2 (97.8) | 21.2 | 22.2 | 1U8P |
| 28 | E(Dap)DKWES | 663, 667 | 80.0-2.24 | 10.9 (24.8) | 94.0 (87.9) | 22.5 | 24.9 | 1U92 |
| 29 | EDDKW(Dap)S | 663, 667 | 80.0-2.24 | 9.4 (20.0) | 91.4 (88.1) | 22.7 | 23.1 | 1U91 |
| 30 | EEDKW(Dap)S | 663, 667 | 80.0-2.37 | 9.6 (34.5) | 92.0 (95.3) | 24.0 | 23.6 | 1U93 |
| 31 | E(Dap)DKWDS | 663, 667 | ||||||
| 32 | KLDNWAS | 662, 665 | ||||||
| 33 | ALDNWNN | 662, 665, 667, 668 | ||||||
Collection and refinement statistics are given for each 2F5 Fab′-peptide complex that formed crystals along with the PDB identifier (ID). The absence of data indicates that no crystal complex was obtained. Values in parentheses are those for the highest-resolution bin.
Reference sequence.
X-ray diffraction analysis.
All diffraction data were collected on crystals cooled to 100 K after being soaked in a cryoprotectant of 25% glycerol mixed with mother liquor on a home source Rigaku FR-C rotating copper anode with a Mar345 detector and/or at National Synchrotron Light Source synchrotron station X8C. Data were processed with the program packages DENZO and SCALEPACK (46). Most 2F5 Fab′-peptide complexes adopted space group P212121 (a of ∼59.0 Å, b of ∼65.0 Å, and c of ∼175.6 Å) and grew as rectangular prisms. Due to the long c-axis, high and low resolution data sets were collected and subsequently merged. The structure of the 2F5 Fab′-peptide 1 crystal complex was determined by molecular replacement using the program AMoRe with the Protein Data Bank (PDB) entry 1CLZ as the search model (43). The constant and variable regions were used as independent models. The correct solution showed one Fab′ molecule per asymmetric unit and a correlation coefficient of 35.3 (R = 47.3%) using data to 3.3 Å. The CNS package was used for refinement (8). Real-space refinement was done using the programs O and Coot (19, 26). Density for the peptides was clear and could be fitted unambiguously. After numerous cycles of real-space, positional, and B-factor refinements were performed, waters were included where peaks of >3.5 sigma were found in a difference map at an appropriate distance from a donor or acceptor atom. Most peptide complexes gave crystals isomorphous and isostructural to the 2F5 Fab′-peptide 1 complex crystals. Figure images were generated using the program PyMol (17).
RESULTS
MPER variation.
Table 2 qualitatively depicts the sequence variation of the HIV-1 gp41 MPER. Fifteen amino acid positions out of the 25 contained in this region are conserved and show little to no variation: L660, L661, L663, D664, W666, L669, W670, W672, F673, I675, W678, L679, W680, Y681, and I682. Except for aspartic acid at position 664, all conserved positions are occupied by nonpolar or hydrophobic residues. Interestingly, one-third of the conserved positions are tryptophan residues. Looking more closely, it is clear that sequence variation in the gp41 MPER is highly influenced by HIV-1 clade. Notably, position 662 is particularly variant in clades B and D, whereas position 665 varies significantly in clades C and D. Moreover, position 667 accommodates different residues in clade C, and position 677 varies considerably in clade D.
TABLE 2.
Natural sequence variation in HIV MPER and 2F5 neutralization across different cladesa
Footnotes for Table 2 on facing page.
The left column displays the overall sequence variability of HIV-1 Env residues 659 to 683 (gp41MPER) as well as sequence variability across different HIV-1 clades as a WebLogo representation (16, 60). Briefly, the height of each logo represents the relative frequency of a given residue at a particular position (see Materials and Methods for details). Single-letter representations of amino acids are color coded based on their chemical properties: basic, blue; acidic, red; polar residues G, S, T, Y, and C, green; polar residues N and Q, purple; and hydrophobic, black.
b The right column displays the ability of 2FS to neutralize HIV-1 viruses of different clades as previously reported by Binley et al. (6). To stay consistent with the original report, the data have been color coded according to the percentage of viruses that were neutralized: no viruses neutralized, white; 31 to 60% of viruses neutralized, yellow; 61 to 90% of viruses neutralized, orange; and >90% of viruses neutralized, red.
The primary epitopes of bnMAbs 2F5, Z13, and 4E10 have been mapped to the gp41 MPER. A buried surface area analysis of gp41 residues from the crystal structures of 2F5 (PDB code 3D0L), 4E10 (PDB code 2FX7), and Z13 (PDB code 3FN0) reveal that the five residues with the largest buried surfaces are L661, D664, K665, W666, and W670 for 2F5; W672, F673, T676, L679, and W680 for 4E10; and W670, N671, D674, I675, and T676 for Z13 (13, 27, 48). Clearly, the core residues recognized by 2F5 and 4E10 are more conserved than those recognized by Z13, which helps to explain the increased neutralization breadth of the former pair of bnMAbs (44, 75). In the case of 2F5, core residues are generally conserved although significant variation is observed at position K665 in the case of HIV-1 clades B, C, and D.
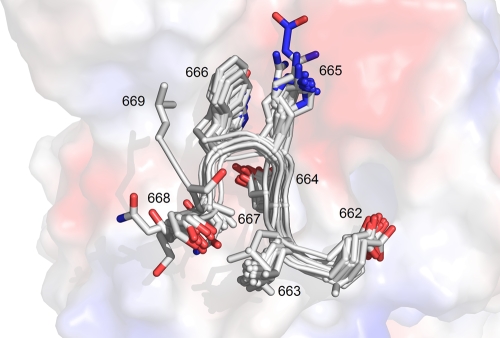
Table 1 contains a list of 7-mer peptides that were tested for crystal complex formation with 2F5 Fab′ and statistics of data collection, processing, and structure refinement for these peptides that formed complex crystals. Peptide 1 (ELDKWAS) represents the consensus HXB2 reference of the primary 2F5 epitope amino acid sequence from HIV protein gp41 (42). The structure of the complex of this peptide with 2F5 Fab′ was previously reported and described in detail (27, 45, 47). Briefly, the antibody was shown to recognize the gp41 MPER in the conformation of a type I β-turn centered on the DKW core residues. The present study has as its objective to investigate how tight the spatial restrictions are that govern the interaction between the gp41 core epitope and the corresponding 2F5 paratope. First, alanine substitutions were made at all positions of the ELDKWAS peptide. Subsequently, if such a substitution was tolerated and a crystal complex could be obtained, substitutions with amino acids found with a high frequency in the gp41 MPER were attempted. When alanine substitutions were not tolerated, only conservative substitutions were investigated. Figure 1 is an overlay of all peptides reported in this study and depicts the central β-turn conformation adopted by residues at position 664 through 666.
FIG. 1.
Overlay of all MPER representing peptides (except for constrained peptides) characterized in complex with 2F5 Fab′ reported in the present structural study. All peptides are seen to adopt a type 1 β-turn conformation. Peptides are shown in gray as a cartoon representation with side chain positions shown as stick models. An electrostatic potential of the 2F5 Fab′ paratope is shown in the background, with red, blue, and white regions representing negatively charged, positively charged, and noncharged areas, respectively.
Asp664 substitutions.
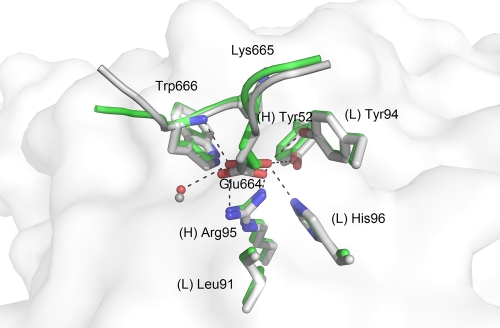
To study the extent of variability allowed in the DKW core itself, we started our analysis by investigating peptides carrying mutations in this core. Converting the Asp664 side chain to a methyl group (peptide 3, ELAKWAS) prevented complex formation with 2F5, consistent with the lack of observable binding found in alanine scanning mutagenesis studies on gp41 peptides (36). Similarly, substitutions D664N (peptide 5, ELNKWAS) and D664Q (peptide 6, ELQKWAS) did not form complexes with 2F5 Fab′. A crystal complex did form for peptide 4, ELEKWAS, which has the conservative D664E substitution (Fig. 2). To achieve Fab′-peptide complex formation, the concentration of this peptide had to be 10-fold higher than the standard used, e.g., with the consensus ELDKWAS peptide. The structure of the ELEKWAS complex reveals that the side chain carboxylate makes the same polar contacts as aspartate with heavy chain (H) residue Arg(H)-95 and light chain (L) residue His(L)-96 of the antibody, the amide bond of the peptide's Trp666, and an adjacent water molecule. In addition, a slight shift in the glutamate side chain position compared to the aspartate side chain allows it to come into closer proximity with residues Tyr(H)-52 and Tyr(L)-94 of the antibody. Furthermore, the backbone of the peptide does not exactly overlie that of reference peptide 1, and the B-factor of the α-carbon atom is 70.1 Å2 as opposed to 23.7 Å2 when aspartate is present. The entropic penalty paid to stabilize the longer glutamic acid side chain and, especially, limits on space leading to molecular crowding may be responsible for the reduced affinity of this peptide. These results indicate that the correct positioning of the negative charge at position 664 is essential for epitope recognition.
FIG. 2.
Crystal structures of 2F5 Fab′ in complex with peptides probing substitutions at position 664 of the MPER. The reference peptide ELDKWAS is shown in light gray, whereas peptide ELEKWAS is shown in green. A white rendering of the 2F5 Fab′ paratope surface is shown in the background.
Lys665 substitutions.
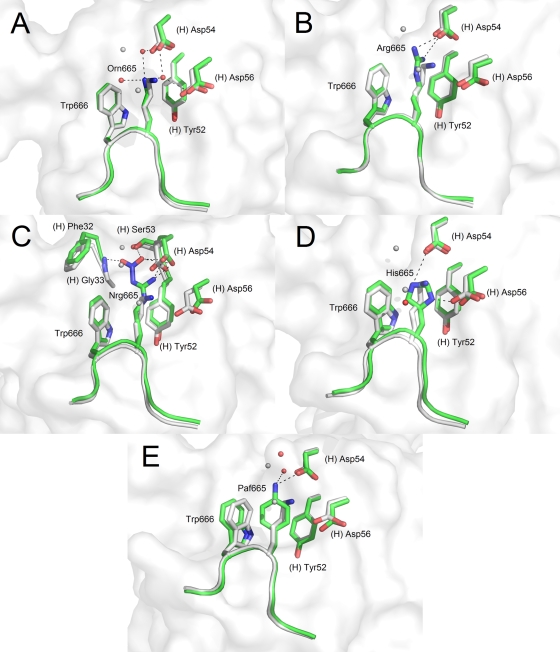
In a similar fashion to the Asp664 substitutions, we synthesized a peptide changing Lys665 to an alanine residue (peptide 7, ELDAWAS). Again, we were unable to generate a complex of this peptide with 2F5 Fab′. Replacing position 665 with a negatively charged glutamic acid (peptide 13, ELDEWAS) also failed to produce complex crystals.
The conservative substitution of Lys665 for arginine in peptide 8 (DLDRWAS) led to complex crystal formation with 2F5 Fab′ (Fig. 3B). As expected, the extended alkyl chain of arginine occupies the same space as that of lysine from the reference peptide, forming hydrophobic alkyl-π stacking interactions with the peptide tryptophan and heavy chain Tyr(H)-52. The positive charge at the end of the arginine side chain interacts in the same way with the Asp(H)-54 carboxylate as the corresponding positive charge of lysine but is no longer able to interact with Asp(H)-56. It should also be noted that two water molecules making hydrogen bonding interactions near Lys665 observed in the reference peptide structure are absent in the Fab′-DLDRWAS structure.
FIG. 3.
Crystal structures of 2F5 Fab′ in complex with peptides probing substitutions at position 665 of the MPER. The reference peptide ELDKWAS is shown in light gray, whereas peptides ELD(Orn)WAS (A), ELDRWAS (B), ELD(Nrg)WAS (C), ELDHWAS (D), and ELD(Paf)WAS (E) are shown in green. A white rendering of the 2F5 Fab′ paratope surface is shown in the background.
Another allowed substitution is a histidine residue in place of Lys665 (peptide 9, ELDHWAS) (Fig. 3D). Crystallization of this peptide with 2F5 Fab′ was done using sodium citrate, pH 5.6, and at this pH, the histidine is protonated and positively charged, making contact with the Asp(H)-56 side chain carboxylate. Alternate rotamer conformations of the imidazole ring could also lead to the positive charge making contacts with Asp(H)-54, which is also an interaction partner for Lys665 in the reference peptide. Another interesting feature in this structure is the tight semiperpendicular stacking between the imidazole and indole rings of the peptide (the distance between the imidazole δ carbon and the indole carbons of the six-membered ring varies between 3.3Å and 3.8Å) and partially parallel stacking arrangement with Tyr(H)-52 of the Fab′ (the distance between the histidine β, γ, and ɛ carbons and the phenol ring carbons varies between 3.7 Å and 4.1 Å).
Ornithine is an analogue of lysine, but its alkyl chain is one methylene group shorter than that of lysine. When lysine was replaced with ornithine at position 665 (peptide 10, ELD[Orn] WAS) a new water network formed around position 665 (Fig. 3A). In the ELDKWAS structure, the positively charged amino group hydrogen bonds directly with the negatively charged side chains of 2F5 Fab′ residues Asp(H)-54 and Asp(H)-56. When ornithine is substituted for lysine, a water molecule now forms a bridge between the amino group at the tip of the shorter side chain of ornithine and Asp(H)-54 as well as Asp(H)-56. Two additional water molecules also create secondary bridging between the retracted ornithine amino group and side chains of the Fab′, including Ser(H)-53.
Nitroarginine corresponds to an extended arginine that carries an additional nitro group on its guanidinium group. In peptide 11 (ELD[Nrg]WAS), Lys665 was replaced with nitroarginine (Fig. 3C). Mirroring the results of the closely related K665R substitution, the positively charged guanidinium group again forms a hydrogen bond with the negatively charged Asp(H)-54 side chain and stacking interactions with the surrounding ring systems. As in the case of the K665R substitution, however, no direct contact is observed to the side chain of Asp(H)-56. Furthermore, the additional negatively charged nitro group forms a hydrogen bond with Ser(H)-53 as well as the main chain peptide nitrogen of Gly(H)-33. The nitro group is now stationed in the same region previously occupied by two water molecules in the peptide with the K665-ornithine substitution.
p-Aminophenylalanine is a nonnatural amino acid that was put in place of Lys665 in peptide 12 (ELDPafWAS), for which a crystal complex was obtained (Fig. 3E). The structure revealed that the plane of the cyclic side chain is exactly superimposable with the imidazole ring of histidine in peptide 9 (ELDHWAS; K665H substitution). The amino group of this side chain now interacts with Asp(H)-54 and is bridged to Ser(H)-33 via an additional water molecule, in the same way as was seen in peptide 10 (ELD[Orn]WAS). Taken together, these results suggest that the 2F5 paratope can accommodate alternative interactions in the 665 binding region as long as key hydrophobic stacking interactions and hydrogen bonding networks are conserved. However, no substitutes at position 665 reproduce exactly the interactions observed between 2F5 and Lys665.
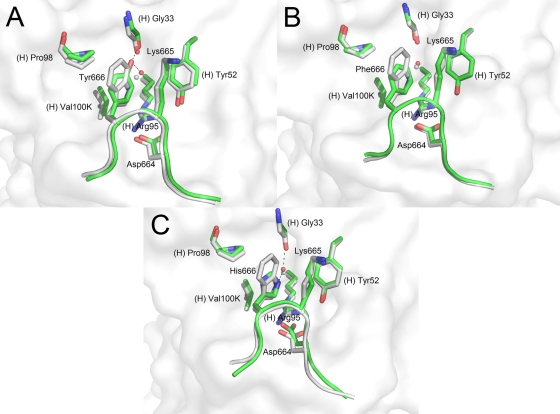
Trp666 substitutions.
Consistent with alanine scanning mutagenesis studies, peptide 14 (ELDKAAS) did not form a crystal complex (36). We synthesized a series of peptides with conservative amino acid substitutions at position 666: peptide 15 (ELDKYAS), peptide 16 (ELDKFAS), peptide 17 (ELDKHAS), peptide 18 (ELDKIAS), peptide 19 (ELDKLAS), and peptide 20, with a nonnatural amino acid substitution with norleucine (ELDK[Nle]AS). Of these, only peptides carrying substitutions with amino acids having aromatic side chains formed complexes with 2F5 Fab′ (Fig. 4) while substitutions with amino acids with hydrophobic alkyl side chains, i.e., isoleucine, leucine, and norleucine, did not. In the three structures for which a crystal complex was obtained, the different rings all occupy the same space as the indole ring plane of Trp666. Hydrophobic alkyl-π stacking interactions were observed between the ring at position 666 and the Lys665 alkyl side chain, as well as van der Waals contacts with Pro(H)-98 and Val(H)-100K of 2F5. Furthermore, the position of a key water molecule located below the indole ring in the reference structure is conserved in all three structures. In the reference peptide structure, this water molecule bridges the ɛ nitrogen of the Trp666 indole ring with the backbone carbonyl Gly(H)-33.
FIG. 4.
Crystal structures of 2F5 Fab′ in complex with peptides probing substitutions at position 666 of the MPER. The reference peptide ELDKWAS is shown in light gray, whereas peptides ELDKYAS (A), ELDKFAS (B), and ELDKHAS (C) are shown in green. A white rendering of the 2F5 Fab′ paratope surface is shown in the background.
In the case of the ELDKYAS peptide, the terminal tyrosine hydroxyl interacts directly with the backbone carbonyl of Gly(H)-33 as well as with the conserved water molecule below the ring. Interestingly, for the W666F substitution, the conserved water molecule is observed even though phenylalanine lacks a polar functional group to coordinate via hydrogen bonding. Finally, replacement of the Trp666 position with histidine required a buffer with a pH of >7.0 to form a crystal complex with 2F5 Fab′. Obviously, a histidine residue is tolerated in position 666 only when uncharged. In this structure, the uncharged imidazole ring occupies a similar position as the indole, phenyl, and phenol rings and makes a similar water-bridged contact to Gly(H)-33. Also of notice in this structure, the Asp664 side chain is slightly shifted toward the imidazole ring, making tighter contacts with the peptide backbone amide. Depending on the rotamer conformation of histidine, the Asp664 side chain could also directly interact via hydrogen bonding with the side chain of His666.
These results indicate that hydrophobic alkyl-π stacking interactions between a planar aromatic residue at position 666 and the extended alkyl chain of the previous residue at position 665 are an important structural feature of the DKW core β-turn and key to the stability of the epitope-2F5 Fab′ interaction. Linear alkyl side chains (norleucine) and branched alkyl side chains (isoleucine and leucine) are unable to form tight stable van der Waals interactions with the lysine side chain and residues of the 2F5 paratope, hence, disrupting β-turn stability.
Substitutions outside of the DKW core.
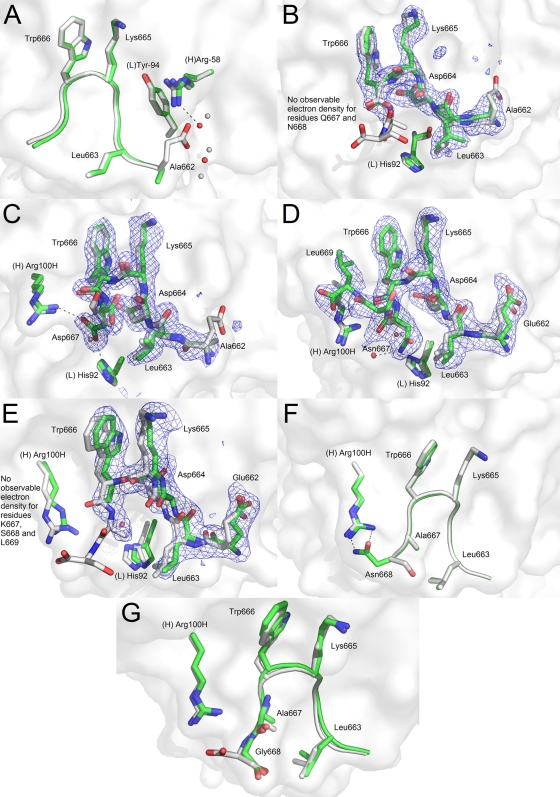
In the ELDKWAS reference structure, Glu662 interacts via hydrogen bonding with Arg(H)-58 and two water molecules. In the case of the D662A substitution (peptide 2, ALDKWAS), the alanine residue lacks any interaction with Arg(H)-58, as was expected (Fig. 5A). The loss of the glutamate side chain leads to a rearrangement in the water network around position 662. In particular, a water molecule appears in place of the glutamate functional group to interact with Arg(H)-58.
FIG. 5.
Crystal structures of 2F5 Fab′ in complex with peptides probing substitutions outside the DKW core, namely, positions 662, 667, and 668 of the MPER. The reference peptide ELDKWAS is shown in light gray, whereas peptides ALDKWAS (A), ALDKWQN (B), ALDKWD (C), ELDKWNSL (D), ELDKWKSL (E), ELDKWAN (F), and ELDKWAG (G) are shown in green. A white rendering of the 2F5 Fab′ paratope surface is shown in the background. For substitutions at position 667, an Fo − Fc electron density composite omit map at a 2.5 sigma level are represented in a blue mesh.
Leu663 is invariant across HIV-1 isolates, and 2F5 interacts with conserved Leu663 through hydrophobic van der Waals contacts with residues His(L)-92 and Phe(L)-93 (45). Therefore, this position was changed to only residues that allowed the cyclization of the peptide in our efforts to favor β-turn formation, as described below.
Position 667 is particularly interesting to study at the molecular level because it shows significant variability across viruses of different clades. For peptide 21 (ALDKWQN), clear electron density is observed for residues Ala662 through Trp666, but it quickly fades after that with no interpretable electron density for residues Gln667 and Asn668, which suggests that these amino acids do not adopt a unique stable conformation (Fig. 5B). This could indicate an inability of these residues to make stabilizing interactions with the antibody or with other components of the linear sequence, such as Leu663. To further investigate the role of various amino acids at position 667, peptides 22 (ALDKWD), 23 (ELDKWNSL), and 24 (ELDKWKSL) were synthesized. In the case of ALDKWD, the side chain of Asp667 interacts with His(L)-92 and Arg(H)-100H (Fig. 5C). It replaces a water molecule interacting with His(L)-92 in the reference ELDKWAS structure. As previously documented, in the present P212121 crystal system, the position of Arg(H)-100H is determined by crystal lattice interactions. Therefore, the interaction between Asp667 and Arg(H)-100H might not be representative of the interaction observed in a biological context (see reference 27 for a complete discussion). Nevertheless, this crystal structure documents that 2F5 is able to accommodate an aspartic acid at position 667. When 2F5 Fab′ is crystallized in complex with the peptide ELDKWNSL, electron density for all residues is observed (Fig. 5D). The asparagine side chain, however, does not engage in the same contacts with residues of the Fab′ that aspartate had formed. Instead, it packs tightly against Leu663, at a distance of ∼3.5 Å, and makes a hydrogen bond interaction with a conserved water, which in turn coordinates with Arg(H)-100H. This is a significant decrease in distance between the 663 and 667 positions, which is ∼4.0 Å in the reference ELDKWAS peptide. It is possible to see from the electron density that Leu663 moves back slightly compared to the reference structure in order to accommodate a bulkier side chain at position 667. Also, this structure shows a significant difference in the direction of the C terminal residues, which now fold back toward Trp666. This can be attributed to the addition of Leu669 to the peptide, which has been previously described to be part of a canonical α-helical turn when the peptide epitope is extended (27). Finally, when a lysine residue is introduced at position 667, no clear electron density is observed for residues Lys667, Ser668, and Leu669 (Fig. 5E). This is similar to the structure of the Fab′ with peptide 21 (ALDKWQN), with the bulky glutamine amino acid at position 667. These data suggest that a residue with a bulky side chain at position 667 might prevent residues located at the C terminus of the DKW core from adopting the proper conformation recognized by 2F5 because of clashing interactions, particularly with Leu663.
In the reference sequence, position 668 is occupied by serine. Previous reports demonstrated that Ser668 does not interact with 2F5 (45). Substitutions at this position are therefore not expected to have major effects on the overall peptide structure as recognized by 2F5. The most commonly observed substitution at this position is S668N. Crystal structures of the 2F5 Fab′ in complex with peptides 25 (ELDKWAN) and 26 (ELDKWAG) revealed no difference in core binding (Fig. 5F and G). Worth mentioning, the asparagine residue of peptide 25 forms hydrogen bonds to Arg(H)-100H, providing this peptide with additional interactions with the Fab′ compared to the reference peptide. However, as was previously mentioned, this interaction with Arg(H)-100H is probably compromised by crystal lattice interactions.
Cyclic peptides.
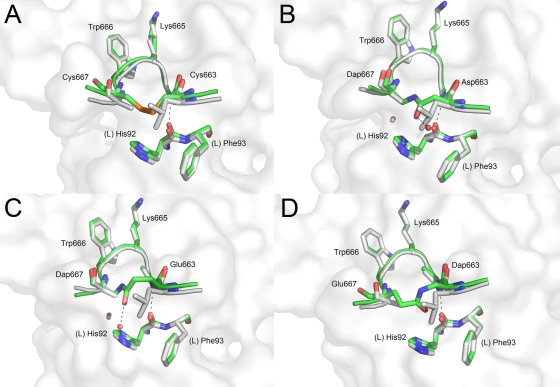
In an attempt to constrain the β-turn motif, peptides 27 (ECDKWCS), 28 [E(Dap)DKWES, where Dap is l-α-diaminopropionic acid], 29 [EDDKW(Dap)S], 30 [EEDKW(Dap)S], and 31 [E(Dap)DKWDS] were synthesized and mixed with 2F5 Fab′. All but the E(Dap)DKWDS peptide formed crystal complexes. Peptide ECDKWCS has Leu663 and Ala667 replaced with cysteine residues (i and i + 4, respectively). Under oxidative conditions, a disulfide bond forms to constrain the β-turn. Figure 6A shows how the essential β-turn conformation and contacts seen in reference peptide 1 are conserved.
FIG. 6.
Crystal structures of 2F5 Fab′ in complex with peptides harboring β-turn stabilizing linkages between positions 663 and 667 of the MPER. The reference peptide ELDKWAS is shown in light gray, whereas peptides ECDKWCS (A), ENDKW(Dap)S (B), EQDKW(Dap)S (C), and E(Dap)DKWQS (D) are shown in green. A white rendering of the 2F5 Fab′ paratope surface is shown in the background.
An alternative method to constrain the β-turn was to create a lactam bridge between residues at the same position. This was done by incorporating an aspartate (peptide 29) and glutamate (peptide 30) residue in the i position and Dap in the i + 4 position, reacting to generate a lactam bridge. Figure 6B and C show the resulting complexes with 2F5 Fab′ using the aspartate and glutamate residue in the i position, respectively. The DKW core residues of both peptides superimpose in the same positions as reference peptide 1 without structural variation, demonstrating that the lactam bridge is sufficient in constraining the β-turn conformation. The extra methylene group (γ-carbon) of the glutamate side chain does not adversely affect the conformation as it bulges out away from the Fab′ while the Dap β-carbon is in the same position as the native alanine β-carbon. Furthermore, in these structures an additional water molecule is observed interacting with the carbonyl of the lactam bond.
When the residues are reversed such that the Dap group is in the i position and a glutamate is at the i + 4 position, a crystal complex forms with 2F5 Fab′ (peptide 28) (Fig. 6D). In this structure the α-carbon of the glutamate is shifted away from the optimal positions of the native alanine structure, and unlike peptides 29 and 30, the methylene groups of the side chain point toward the Fab′ body. The use of an aspartate residue at the i + 4 position [E(Dap)DKWDS] did not allow for complex formation with 2F5 Fab′. The shorter side chain coupled with the backbone shift probably generated a highly constrained peptide unable to maintain a stable β-turn and bind properly to 2F5 Fab′.
DISCUSSION
Conformational flexibility of the gp41 MPER (α-helix, kinked α-helix, 310-helix, or β-turn) has been suggested to be a characteristic feature of gp41 and to play an important role in HIV membrane fusion (7, 27, 45, 47, 48, 59, 65). This dynamic aspect of the MPER, in addition to its amphipathic properties, and its ability to oligomerize and to interact with other proteins will put a significant constraint on its amino acid composition (2, 3, 40, 53, 63, 64, 72). Indeed, sequence variation within the MPER (especially for residues L660, L661, L663, D664, W666, L669, W670, W672, F673, I675, W678, L679, W680, Y681. and I682), is minimal, especially compared to other regions of the Env protein. The relatively conserved nature of the MPER sequence helps to explain in part its recognition by three neutralizing antibodies of broad activity with epitopes on the gp41 MPER (2F5, Z13, and 4E10). Furthermore, the neutralizing breadth of these three bnMAbs (4E10 > 2F5 > Z13) seems to correlate directly with the conserved nature of the linear stretch of amino acids they recognize.
As shown in this study as well as in previous reports, 2F5 recognizes a core 664DKW666 β-turn motif (27, 45, 47). gp41 sequence analysis revealed that the aspartic acid and tryptophan residues of the 664DKW666 core are highly invariant and completely conserved, respectively, among all HIV isolates (Table 2). The crystal structures presented in this study expose a strict requirement for a negative charge at position 664. Indeed, no crystal complex was isolated when the peptide epitope had D664A, D664N, or D664Q substitutions. Moreover, even though the D664E substitution yielded a complex crystal, the longer side chain of glutamate seems to destabilize the peptide conformation (higher overall B factors for the peptide), with a loss in affinity as evidenced by the much higher concentration of peptide required to obtain complex crystal growth. Overall, it seems clear that 2F5 has gained an ability to recognize an invariant aspartic acid at position 664. Position 666 is able to accommodate residues with planar side chains (such as tyrosine, phenylalanine, and histidine), but no crystal complex forms with residues possessing branched nonpolar side chains at this position. Clearly, this indicates the requirement for specific hydrophobic alkyl-π stacking interactions between the planar side chain at position 666 with an extended alkyl chain at position 665, as well as van der Waals contacts with Pro(H)-98 and Val(H)-100K of 2F5. Although the conserved nature of positions 664 and 666 do not require 2F5 to be promiscuous in its recognition, the insights gained from the structures of 2F5 Fab′ with peptides having substitutions at these positions are useful for understanding key binding interactions and should help in immunogen design efforts.
On the other hand lysine 665, another residue participating in the β-turn motif recognized by 2F5, varies considerably in the known HIV-1 sequences. Overall, lysine 665 is seen to be most commonly replaced by serine, glutamine, or threonine residues, with serine being the prevalent residue at position 665 in HIV-1 clade C isolates. The present study does not report on the structural determination of peptides with these substitutions at position 665. However, it is clear from the interaction between the positively charged side chain amine of lysine 665 and the two negatively charged residues Asp(H)-54 and Asp(H)-56 at the apex of the CDR H2 that the recognition of 2F5 for its epitope relies heavily on these ionic interactions. In addition to the lack of a positive charge, the presence of a serine, glutamine, or threonine residue at position 665 could lead to the disruption of proper hydrophobic stacking interactions with Trp666 and hence a destabilization of the β-turn. Therefore, we suggest that one of the major limitations of 2F5 neutralization breadth resides in its dependency for an extended positively charged residue at variable position 665.
All peptides with 665 substitutions that were able to form crystal complexes possessed both an extended chain capable of hydrophobic stacking interactions as well as a nitrogen-containing functional group, positively charged in most structures, capable of interacting with either Asp(H)-54 or Asp(H)-56 of 2F5. It may be promising that compounds other than naturally occurring amino acids can be used to mimic the extended lysine and tryptophan conformations. Such peptide mimicry perhaps can be a foundation for the synthesis of a synthetic small-molecule vaccine. However, none of the substitutes used at position 665 in this study were able to recreate the exact interaction of a positive charge with both Asp(H)-54 and Asp(H)-56, and these peptides might therefore show limitations in their ability to elicit 2F5-like antibodies.
Position 662 is quite variable in gp41 MPER and is predominantly occupied in HIV-1 isolates by alanine and glutamic acid residues. The ability of 2F5 to make hydrogen bonding interactions with the carboxylate of Glu662 via Arg(H)-58 and Tyr(L)-94 suggests that this antibody has matured to interact with a negative charge at this position. However, the present crystal structure of 2F5 Fab′ in complex with a peptide harboring the E662A substitution reveals that the antibody is able to adapt to the loss of the functional group at position 662. This analysis corresponds to neutralization assays which show that 2F5 is able to neutralize effectively isolates with mutations at position 662 although the efficacy at which it does so might be reduced (6). Finally, position 667 is most commonly occupied by a small alanine residue although it is particularly variable in clade C and F isolates, where residues like lysine, asparagine, glutamine, and aspartic acid are often observed. The crystal structures presented here reveal the ability of 2F5 to adapt its binding to accommodate A667D and A667N substitutions, but substitutions with residues possessing elongated side chains such as lysine and glutamine seem to disrupt proper binding at the C terminus of the DKW core. As reported in Binley et al. (6), most viruses able to evade neutralization by 2F5 have mutations in the linear DKW core. However, it has also been previously observed that sequences with a conserved DKW motif but possessing 667 substitutions such as isolate SG364 (662ALDKWNQ668) are able to escape 2F5 neutralization (6). Combined with our structural studies, these observations lead us to propose that a secondary 2F5 neutralization escape mechanism involves substitution from an alanine at position 667 to a residue with a bulky side chain, as is often observed in HIV-1 clade C isolates.
Since 2F5 is such a potent HIV-1 neutralizing antibody for isolates possessing the LDKWA core, many efforts have been put forward to create immunogens possessing a constrained β-turn motif of this sequence in order to elicit 2F5-like antibodies (4, 14, 18, 20, 31). One of the approaches to constrain a β-turn conformation involves creating a connection between the i and i + 4 β-turn residues in a peptide, namely, Leu663 and Ala667, either via disulfide linkage or lactam covalent bonds. The present crystal structures of such constrained peptides show the ability of 2F5 to interact with such peptides in a way that reproduces the interactions observed in the native peptide. In previous studies, these β-turn-constrained gp41 peptides were tested to determine whether they were sufficient to elicit antibodies capable of neutralizing HIV-1; however, even though these peptides generated very-high-titer antisera, HIV-1 neutralization was not observed (34). Nuclear magnetic resonance analysis revealed that these constrained peptides formed a stable β-turn conformation in solution (34). However, the specific conformations of the DKW side chains that are adopted in solution are probably quite different from those found in the crystal structure. Indeed, a direct cation-π interaction between the terminal amino group of the Lys665 and Trp666 in solution differs from the stacking interaction observed in the crystal structure whereby the methylene groups of the extended Lys665 are stacked in an alkyl-π fashion against the Trp666 indole ring. This suggests that both proper DKW β-turn conformation and DKW side chain positions are crucial for designing immunogens capable of eliciting 2F5-like neutralizing antibodies.
Our structural analysis reveals multiple water molecules undergoing key interactions with the 2F5 paratope and the gp41 peptide epitope. Of particular interest are two conserved water molecules that mediate the binding of the gp41 epitope to the 2F5 paratope in all structures; one of them is located below the indole nitrogen of Trp666 and makes hydrogen bonding interactions with the backbone carbonyls of 2F5 Gly(H)-33 and Arg(H)-96, and the second one is placed next to Asp664 and hydrogen bonds to the 2F5 backbone carbonyl of Leu(L)-91 and the side chain NH group of Asn(H)-100L. These findings suggest that a synthetic immunogen looking to elicit 2F5-like neutralizing antibodies could take advantage of these water-binding sites by incorporating water displacement in its design, thereby decreasing the entropy of binding while maintaining the interaction enthalpy.
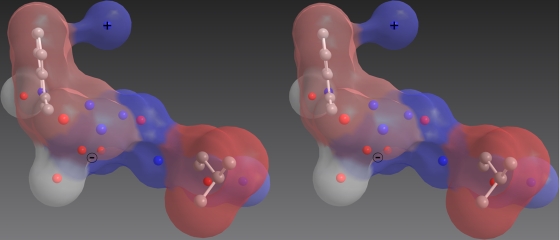
While it remains uncertain why the MPER 664DKW666 β-turn conformation is required for successful HIV fusion to CD4+ T cells, this MPER motif has been the target of much scrutiny for vaccine design. The current study highlights the importance in immunogen design of the correct positioning of the negative charge of Asp664 and the hydrophobic interactions of Trp666, from which 2F5 gets most of its neutralization potency and breadth. Positions 665 and 667 were identified at the molecular level as particular determinants of 2F5 neutralization potency and also of neutralization escape. We suggest that evolving 2F5 in vitro to rely less on variable residues and more on conserved MPER residues (such as Leu661 or Leu663) might be a way to increase both neutralization breath and potency, as was described for bnMAbs Z13 and Z13e1 (44). Finally, we emphasize that other components of the 2F5 interaction with HIV-1 in vivo, such as a canonical α-helical turn at the C terminus of the DKW core, a phosphate ion at the base of the CDR H3 loop, and possible interactions of CDR H3 residues with membrane components, albeit very challenging, nevertheless, have to be included in a successful immunogen. However, the correct presentation of the main determinant of 2F5 binding, the DKW core, should remain the priority in the design of immunogens intended to elicit 2F5-like neutralizing antibodies (Fig. 7).
FIG. 7.
Representation of spatial and chemical requirements for MPER residues for 2F5 core recognition as deduced from the present complex crystal structures. The wall-eyed stereo representation is a translucent Connolly molecular surface colored by group hydrophobicity (with a spectrum from red to blue showing hydrophobic to hydrophilic areas, respectively, and white representing two conserved water molecules.) Also, a ball and stick representation shows the position of crucial nitrogen (blue), oxygen (red), and carbon (gray) atoms. The model design was based on (i) the conserved negative and positive charges present at positions 664 and 665, respectively, (ii) the requirement for an aromatic ring at position 666, (iii) the conserved alkyl functional group at position 663, (iv) important amide and carbonyl atoms taking part in direct interactions with 2F5, and (v) two conserved water molecules mediating the epitope/paratope interaction. This visualization was generated using the program ChemBio 3D by CambridgeSoft.
Acknowledgments
We thank Annie Cunningham for her excellent help with protein purification and crystallization, David Isenman, Brian Barber, and Jason Ho for discussions and valuable advice, and Arthur Pedyczak and Pele Chong for their gift of peptides. We are grateful to the staff at Brookhaven National Laboratory X8C for assistance with data collection.
This research was supported by grants from the National Science and Engineering Research Council of Canada, the Canadian Universities Research Programme (sponsored by Sanofi-Pasteur), and the Protein Engineering Network of Centres of Excellence, Canada (to E.F.P.); J.-P.J. is grateful to the Canadian Merit Scholarship Foundation, les Fonds québécois de la recherche sur la nature et les technologies, and the Canadian Institute of Health Research for scholarships. A joint grant from the Canadian Institutes for Health Research and National Sciences and Engineering Research Council of Canada enabled use of beamline X8C at the National Synchrotron Light Source.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Alam, S. M., M. McAdams, D. Boren, M. Rak, R. M. Scearce, F. Gao, Z. T. Camacho, D. Gewirth, G. Kelsoe, P. Chen, and B. F. Haynes. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfsen, A., and M. Bomsel. 2002. HIV-1 gp41 envelope residues 650-685 exposed on native virus act as a lectin to bind epithelial cell galactosyl ceramide. J. Biol. Chem. 277:25649-25659. [DOI] [PubMed] [Google Scholar]
- 3.Alfsen, A., P. Iniguez, E. Bouguyon, and M. Bomsel. 2001. Secretory IgA specific for a conserved epitope on gp41 envelope glycoprotein inhibits epithelial transcytosis of HIV-1. J. Immunol. 166:6257-6265. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, G. F., P. K. Velasco, A. K. Holmes, T. Wrin, S. C. Geisler, P. Phung, Y. Tian, D. A. Resnick, X. Ma, T. M. Mariano, C. J. Petropoulos, J. W. Taylor, H. Katinger, and E. Arnold. 2009. Broad neutralization of human immunodeficiency virus type 1 (HIV-1) elicited from human rhinoviruses that display the HIV-1 gp41 ELDKWA epitope. J. Virol. 83:5087-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellamy-McIntyre, A. K., C. S. Lay, S. Baar, A. L. Maerz, G. H. Talbo, H. E. Drummer, and P. Poumbourios. 2007. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J. Biol. Chem. 282:23104-23116. [DOI] [PubMed] [Google Scholar]
- 6.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biron, Z., S. Khare, A. O. Samson, Y. Hayek, F. Naider, and J. Anglister. 2002. A monomeric 3(10)-helix is formed in water by a 13-residue peptide representing the neutralizing determinant of HIV-1 on gp41. Biochemistry 41:12687-12696. [DOI] [PubMed] [Google Scholar]
- 8.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 9.Bryson, S., A. Cunningham, J. Ho, R. Hynes, M. Klein, D. Isenman, B. Barber, R. Kunert, H. Katinger, and E. F. Pai. 2001. Cross-neutralizing human monoclonal anti-HIV-1 antibody 2F5: preparation and crystallographic analysis of the free and epitope-complexed forms of its Fab′ fragment. Prot. Pep. Let. 8:413-418. [Google Scholar]
- 10.Bryson, S., J. P. Julien, D. E. Isenman, R. Kunert, H. Katinger, and E. F. Pai. 2008. Crystal structure of the complex between the F(ab)′ fragment of the cross-neutralizing anti-HIV-1 antibody 2F5 and the F(ab) fragment of its anti-idiotypic antibody 3H6. J. Mol. Biol. 382:910-919. [DOI] [PubMed] [Google Scholar]
- 11.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 12.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA 102:14943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardoso, R. M., M. B. Zwick, R. L. Stanfield, R. Kunert, J. M. Binley, H. Katinger, D. R. Burton, and I. A. Wilson. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22:163-173. [DOI] [PubMed] [Google Scholar]
- 14.Coeffier, E., J. M. Clement, V. Cussac, N. Khodaei-Boorane, M. Jehanno, M. Rojas, A. Dridi, M. Latour, R. El Habib, F. Barre-Sinoussi, M. Hofnung, and C. Leclerc. 2000. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine 19:684-693. [DOI] [PubMed] [Google Scholar]
- 15.Conley, A. J., J. A. Kessler II, L. J. Boots, J. S. Tung, B. A. Arnold, P. M. Keller, A. R. Shaw, and E. A. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 18.Eckhart, L., W. Raffelsberger, B. Ferko, A. Klima, M. Purtscher, H. Katinger, and F. Ruker. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 19.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 20.Ferko, B., D. Katinger, A. Grassauer, A. Egorov, J. Romanova, B. Niebler, H. Katinger, and T. Muster. 1998. Chimeric influenza virus replicating predominantly in the murine upper respiratory tract induces local immune responses against human immunodeficiency virus type 1 in the genital tract. J. Infect. Dis. 178:1359-1368. [DOI] [PubMed] [Google Scholar]
- 21.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. USA 102:18514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haynes, B. F., J. Fleming, E. W. St Clair, H. Katinger, G. Stiegler, R. Kunert, J. Robinson, R. M. Scearce, K. Plonk, H. F. Staats, T. L. Ortel, H. X. Liao, and S. M. Alam. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906-1908. [DOI] [PubMed] [Google Scholar]
- 24.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, W. E., and R. C. Desrosiers. 2002. Viral persistance: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499-518. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 27.Julien, J. P., S. Bryson, J. L. Nieva, and E. F. Pai. 2008. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J. Mol. Biol. 384:377-392. [DOI] [PubMed] [Google Scholar]
- 28.Klausner, R. D., A. S. Fauci, L. Corey, G. J. Nabel, H. Gayle, S. Berkley, B. F. Haynes, D. Baltimore, C. Collins, R. G. Douglas, J. Esparza, D. P. Francis, N. K. Ganguly, J. L. Gerberding, M. I. Johnston, M. D. Kazatchkine, A. J. McMichael, M. W. Makgoba, G. Pantaleo, P. Piot, Y. Shao, E. Tramont, H. Varmus, and J. N. Wasserheit. 2003. Medicine. The need for a global HIV vaccine enterprise. Science 300:2036-2039. [DOI] [PubMed] [Google Scholar]
- 29.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Y., B. Cleveland, I. Klots, B. Travis, B. A. Richardson, D. Anderson, D. Montefiori, P. Polacino, and S. L. Hu. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 32.Lorizate, M., M. J. Gomara, B. G. de la Torre, D. Andreu, and J. L. Nieva. 2006. Membrane-transferring sequences of the HIV-1 gp41 ectodomain assemble into an immunogenic complex. J. Mol. Biol. 360:45-55. [DOI] [PubMed] [Google Scholar]
- 33.Matyas, G. R., Z. Beck, N. Karasavvas, and C. R. Alving. 2009. Lipid binding properties of 4E10, 2F5, and WR304 monoclonal antibodies that neutralize HIV-1. Biochim. Biophys. Acta 1788:660-665. [DOI] [PubMed] [Google Scholar]
- 34.McGaughey, G. B., M. Citron, R. C. Danzeisen, R. M. Freidinger, V. M. Garsky, W. M. Hurni, J. G. Joyce, X. Liang, M. Miller, J. Shiver, and M. J. Bogusky. 2003. HIV-1 vaccine development: constrained peptide immunogens show improved binding to the anti-HIV-1 gp41 MAb. Biochemistry 42:3214-3223. [DOI] [PubMed] [Google Scholar]
- 35.Mehandru, S., T. Wrin, J. Galovich, G. Stiegler, B. Vcelar, A. Hurley, C. Hogan, S. Vasan, H. Katinger, C. J. Petropoulos, and M. Markowitz. 2004. Neutralization profiles of newly transmitted human immunodeficiency virus type 1 by monoclonal antibodies 2G12, 2F5, and 4E10. J. Virol. 78:14039-14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menendez, A., K. C. Chow, O. C. Pan, and J. K. Scott. 2004. Human immunodeficiency virus type 1-neutralizing monoclonal antibody 2F5 is multispecific for sequences flanking the DKW core epitope. J. Mol. Biol. 338:311-327. [DOI] [PubMed] [Google Scholar]
- 37.Montero, M., N. E. van Houten, X. Wang, and J. K. Scott. 2008. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol. Mol. Biol. Rev. 72:54-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, P. L., E. T. Crooks, L. Porter, P. Zhu, C. S. Cayanan, H. Grise, P. Corcoran, M. B. Zwick, M. Franti, L. Morris, K. H. Roux, D. R. Burton, and J. M. Binley. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore, P. L., E. S. Gray, I. A. Choge, N. Ranchobe, K. Mlisana, S. S. Abdool Karim, C. Williamson, and L. Morris. 2008. The C3-V4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82:1860-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno, M. R., R. Pascual, and J. Villalain. 2004. Identification of membrane-active regions of the HIV-1 envelope glycoprotein gp41 using a 15-mer gp41-peptide scan. Biochim. Biophys. Acta 1661:97-105. [DOI] [PubMed] [Google Scholar]
- 41.Munoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr A. 50:157-163. [Google Scholar]
- 44.Nelson, J. D., F. M. Brunel, R. Jensen, E. T. Crooks, R. M. Cardoso, M. Wang, A. Hessell, I. A. Wilson, J. M. Binley, P. E. Dawson, D. R. Burton, and M. B. Zwick. 2007. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J. Virol. 81:4033-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski, Z., and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 47.Pai, E. F., M. H. Klein, P. Chong, and A. Pedyczak. October 2000. World Intellectual Property Organization patent WO-00/61618.
- 48.Pejchal, R., J. S. Gach, F. M. Brunel, R. M. Cardoso, R. L. Stanfield, P. E. Dawson, D. R. Burton, M. B. Zwick, and I. A. Wilson. 2009. A conformational switch in HIV gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. J. Virol. 83:8451-8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poumbourios, P., W. el Ahmar, D. A. McPhee, and B. E. Kemp. 1995. Determinants of human immunodeficiency virus type 1 envelope glycoprotein oligomeric structure. J. Virol. 69:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purtscher, M., A. Trkola, A. Grassauer, P. M. Schulz, A. Klima, S. Dopper, G. Gruber, A. Buchacher, T. Muster, and H. Katinger. 1996. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS 10:587-593. [DOI] [PubMed] [Google Scholar]
- 51.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robert-Guroff, M., M. Brown, and R. C. Gallo. 1985. HTLV-III-neutralizing antibodies in patients with AIDS and AIDS-related complex. Nature 316:72-74. [DOI] [PubMed] [Google Scholar]
- 53.Saez-Cirion, A., S. Nir, M. Lorizate, A. Agirre, A. Cruz, J. Perez-Gil, and J. L. Nieva. 2002. Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J. Biol. Chem. 277:21776-21785. [DOI] [PubMed] [Google Scholar]
- 54.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez-Martinez, S., M. Lorizate, K. Hermann, R. Kunert, G. Basanez, and J. L. Nieva. 2006. Specific phospholipid recognition by human immunodeficiency virus type-1 neutralizing anti-gp41 2F5 antibody. FEBS Lett. 580:2395-2399. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez-Martinez, S., M. Lorizate, H. Katinger, R. Kunert, and J. L. Nieva. 2006. Membrane association and epitope recognition by HIV-1 neutralizing anti-gp41 2F5 and 4E10 antibodies. AIDS Res. Hum. Retrovir. 22:998-1006. [DOI] [PubMed] [Google Scholar]
- 57.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scanlan, C., D. Calarese, H. K. Lee, O. Blixt, C. H. Wong, I. Wilson, D. Burton, R. Dwek, and P. Rudd. 2005. Antibody recognition of a carbohydrate epitope: a template for HIV vaccine design. Adv. Exp. Med. Biol. 564:7-8. [DOI] [PubMed] [Google Scholar]
- 59.Schibli, D. J., R. C. Montelaro, and H. J. Vogel. 2001. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry 40:9570-9578. [DOI] [PubMed] [Google Scholar]
- 60.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srivastava, I. K., J. B. Ulmer, and S. W. Barnett. 2005. Role of neutralizing antibodies in protective immunity against HIV. Hum. Vaccin. 1:45-60. [DOI] [PubMed] [Google Scholar]
- 62.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 63.Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suarez, T., S. Nir, F. M. Goni, A. Saez-Cirion, and J. L. Nieva. 2000. The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: a novel fusogenic sequence. FEBS Lett. 477:145-149. [DOI] [PubMed] [Google Scholar]
- 65.Sun, Z. Y., K. J. Oh, M. Kim, J. Yu, V. Brusic, L. Song, Z. Qiao, J. H. Wang, G. Wagner, and E. L. Reinherz. 2008. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28:52-63. [DOI] [PubMed] [Google Scholar]
- 66.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 67.Weiss, R. A., P. R. Clapham, R. Cheingsong-Popov, A. G. Dalgleish, C. A. Carne, I. V. Weller, and R. S. Tedder. 1985. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature 316:69-72. [DOI] [PubMed] [Google Scholar]
- 68.Willey, S., and M. M. Aasa-Chapman. 2008. Humoral immunity to HIV-1: neutralisation and antibody effector functions. Trends Microbiol. 16:596-604. [DOI] [PubMed] [Google Scholar]
- 69.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 70.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 71.Yang, X., S. Kurteva, X. Ren, S. Lee, and J. Sodroski. 2005. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 79:12132-12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu, H., A. Alfsen, D. Tudor, and M. Bomsel. 2008. The binding of HIV-1 gp41 membrane proximal domain to its mucosal receptor, galactosyl ceramide, is structure-dependent. Cell Calcium 43:73-82. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zwick, M. B., R. Jensen, S. Church, M. Wang, G. Stiegler, R. Kunert, H. Katinger, and D. R. Burton. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 79:1252-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]