Abstract
Adeno-associated virus type 2 (AAV 2) is the only eukaryotic virus capable of site-specific integration; the target site is at chromosome 19q13.4, a site termed AAVS1. The biology of AAV latency has been extensively studied in cell culture, yet the precise mechanism and the required cellular factors are not known. In this study, we assessed the relative frequencies of stable site-specific integration by characterization of cell clones containing integrated AAV vectors. By this assay, two proteins involved in nonhomologous end joining (NHEJ), DNAPKcs and ligase IV, exhibit differential effects on AAV site-specific integration. DNAPKcs is not required; its presence increases the frequency of junction formation indicative of site-specific integration, but seems to reduce the ratio of site-specific integration to random integration (i.e., the latter is even more enhanced). In contrast, site-specific integration is significantly reduced relative to random integration in cells deficient in ligase IV expression. Furthermore, we show that single-stranded AAV vectors are better substrates for site-specific integration than are self-complementary AAV vectors; the absence of DNAPKcs did not affect the targeted integration of these double-stranded AAV vectors. Together, these data suggest that NHEJ proteins participate in site-specific integration, and indicate a role for the single-stranded form of AAV DNA in targeted integration.
Adeno-associated virus (AAV) is a ubiquitous human virus (∼80% of adults are seropositive [4]); like other nuclear DNA viruses, it causes persistent infections. Productive infection by AAV requires coinfection with either an adenovirus or herpesvirus (1, 21). In cell culture, an AAV latent infection is readily established by infection with a high multiplicity of infection (MOI) (>250 infectious particles/cell) in the absence of a helper virus coinfection, and such latent infections have been reported to persist for over 100 passages (3). Because of the stability of latent infections, it has been possible to clone latently infected cells and to determine the molecular characteristics of the persistent viral genome. More than 65 to 90% of such clones have been determined to have the AAV genome integrated at a specific site on chromosome 19q13.4 (16, 26). The degree of specificity of the integration site is unique among human viruses. Integration is not specific at the nucleotide level, but a specific target sequence has been determined, which includes a binding site for the AAV Rep protein (RBS) and a so-called terminal resolution site (TRS) which is cleaved in one strand by the Rep protein during AAV DNA replication (19). In addition to the target site, which has been termed AAVS1, site-specific integration has been demonstrated to require the AAV rep protein (either Rep 68 or 78) and a sequence in the inverted terminal repeat homologous to AAVS1 (31, 33). A third Rep binding site (RBS) is found in the promoter at map position 5, which has been reported to greatly enhance site-specific integration (24).
Although there have been numerous studies of the mechanisms involved in site-specific integration, many aspects remain to be elucidated. In particular, when during the cell cycle site-specific integration occurs is unknown, and the cellular proteins which are involved have not been identified. Such integration might occur as the consequence of nonhomologous end joining (NHEJ) or homologous recombination (HR). Although the former seems to be a more likely pathway, because integration is into a region of very limited homology, the fact that there is some homology between the minimal essential target sequence and the AAV inverted terminal repeat (ITR) suggests that homologous recombination cannot be arbitrarily dismissed as a possibility. Another unknown feature of site-specific integration is the molecular state of the AAV substrate, i.e., whether the substrate is single or double stranded. Transfection of AAV-containing plasmids does lead to site-specific integration (24); thus, a circular form of duplex AAV DNA can serve as the initial substrate, although it has been more challenging to detect significant levels of such integration after transfection with linear duplex AAV DNA (J. Dyall and K. I. Berns, unpublished data).
There have been several studies of the fate of AAV vectors used in gene therapy. The studies have been performed primarily in mice and on occasion in vitro. AAV vectors lack the Rep gene and thus do not preferentially integrate into AAVS1 (25). However, random integration at a low frequency, less than 10−7 (5), does take place and is of concern because of the potential for insertional mutagenesis and the attendant possibility of oncogenesis (20, 22). Such studies have indicated that NHEJ is involved and that different tissues seem to have variable capacities to support integration (14). Other studies have studied recombination between the ITRs at the ends of AAV DNA. Of particular note have been such studies of AAV vectors containing self-complementary genomes (8). These genomes contain complementary sequences separated by an ITR sequence, which itself is a hairpin sequence. Consequently, these genomes can hairpin on themselves to form a duplex structure which contains three hairpinned ITRs, the one in the middle and the two on the ends. The hairpinned ends are substrates for recombination mediated by NHEJ.
If NHEJ were involved in AAV integration, either random or site specific, it would seem likely that an animal deficient in a major component of NHEJ would show less evidence of vector integration. The DNA-dependent protein kinase catalytic subunit is an integral component of NHEJ and is the locus of the underlying genetic defect in immunodeficient SCID mice. Song et al. (30) compared AAV integration both in vitro using cell extracts from wild-type C57BL/6 and SCID mice (derived from C57BL/6) and in vivo. The in vitro assay had been developed as an assay for site-specific integration. Interestingly, the presence of a DNA-dependent protein kinase catalytic subunit (DNAPKcs) in the wild-type extract appeared to inhibit AAV integration. The in vivo results were comparable. Hepatectomies were performed on animals 2 weeks after administration of AAV vectors carrying green fluorescent protein (GFP) as the transgene. Approximately 75% of the liver was removed, and regeneration was allowed to take place. In the SCID mice, a significant percentage (greater than 40%) of hepatocytes still expressed GFP, while GFP fluorescence was lower (less than 10%) in the regenerated wild-type livers. These results were interpreted to mean that either integration was more frequent in SCID mice and/or more stable; thus, DNAPKcs was inhibitory to persistent vector integration (which allowed transgene expression).
In this paper, we report experiments designed to assess the effects of mutants which lead to defects in NHEJ on site-specific integration by AAV. We have also compared frequency and stability of site-specific integration relative to those of integration at other sites for both AAV containing single-stranded genomes and those with self-complementary genomes.
MATERIALS AND METHODS
Cell lines.
M059J (CRL-2366) and M059K (CRL-2365) cell lines were purchased from American Type Culture Collection (Manassas, VA). Cells were maintained in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F-12 medium supplemented with 0.05 mM nonessential amino acids, 10% fetal bovine serum, and 100 U/ml of penicillin and streptomycin. They were maintained in a 37°C humidified incubator with 5% CO2.
HeLa cells were maintained in Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum and 100 U/ml of penicillin and streptomycin in a 37°C humidified incubator with 5% CO2.
Ligase IV (GM16089) cells were purchased from the Coriell Institute (Camden, NJ) and maintained in minimum essential medium (Gibco, Carlsbad, CA) supplemented with 2 mM l-glutamine, 100 U/ml penicillin and streptomycin, and 10% fetal bovine serum.
Immunoblot analysis.
Whole-cell proteins were isolated using cell lytic reagent (Sigma, St. Louis, MO), according to the manufacturer's instructions. The proteins were loaded onto a 7.5% Tris-HCl precast sodium dodecyl sulfate (SDS)-polyacrylamide gel (Bio-Rad, Hercules, CA). The samples were transferred onto an Immun-Blot polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA) and blocked for 1 h in 5% nonfat milk (Carnation; Nestle, Vevey, Switzerland) in 1× Tris-buffered saline-Tween 20 (TTBS; 20 mM Tris, 500 mM NaCl, 0.05% Tween 20). Thereafter, it was incubated with primary antibody (DNAPKcs antibody Ab-1; NeoMarkers, Fremont, CA) diluted 1:5,000 in 1× TTBS, washed three times with 1× TTBS for 10 min each, and then incubated with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Bio-Rad, Hercules, CA) diluted 1:20,000 in 1× TTBS for 1 h. Subsequently, the membrane was washed three times in TTBS for 10 min and incubated in 12 ml of Immun-Star horseradish peroxidase chemiluminescent reagent (Bio-Rad, Hercules, CA) for 5 min. The membrane was wrapped in cling film and exposed to film for the appropriate time for good signal development.
Construction of recombinant AAV (rAAV) vectors.
P5UF11 vector, previously described by Zhang et al. (34), was packaged into AAV type 2 (AAV 2) capsid in HEK 293 cells via cotransfection with helper/packaging plasmids pAAVRC and pADhelper.
P5PGKHygroGFP was constructed as follows: pMSCVHyg (Clontech, Mountain View, CA) was digested with XhoI and HindIII to isolate a 1,872-bp fragment containing the PGK promoter and hygromycin-coding sequence. This fragment was cloned into the pdsP5AAV-CB-EGFP vector backbone isolated by digesting with XhoI and HindIII. This new construct, called dsP5PGKhygo, was digested with XhoI and SacI to isolate the PGK and hygromycin sequences and cloned into P5UF11 digested with XhoI and SacI. The vector was packaged into AAV 2 capsids, using HEK 293 cells and helper vector pDG (12).
pSVAV2 was constructed from pAV2 and pSVRep (a gift from Falck-Pedersen). The pSVRep was digested with EcoRV and HindIII to isolate a fragment containing the SV40 promoter and Rep78 sequences. This fragment was inserted into pAV2 digested with BmgBI and HindIII. Packaging of this construct was described previously by Zhang et al. (34).
dsP5AAVNeoR, a self-complementary AAV vector, was created by cloning the P5 and neomycin sequences into pdsAAV-CB-EGFP (from Arun Srivastava) (32). The P5 sequence was amplified using P5UF11 and inserted into a KpnI site of pdsAAV-CB-EGFP to generate a pdsP5AAV-CB-EGFP construct. The neomycin cassette was isolated from P5UF11 by using XhoI and SacII and inserted into the backbone of pdsP5AAV-CB-EGFP digested with XhoI and SacII. The construct pdsP5AAVNeoR was verified by restriction digestion. The vector was packaged into AAV 2 capsids by using helper vector pACG2 (17).
dsAAVSVRep78, a self-complementary AAV vector, was created by isolating a 1,982-bp fragment containing the SV40 and Rep78 sequences from pSVRep and inserting it into the backbone of pdsAAV-CB-EGFP digested with KpnI and HincII. The constructed vector was verified by restriction digestions and packaged into AAV 2 capsids by using helper vector pACG2.
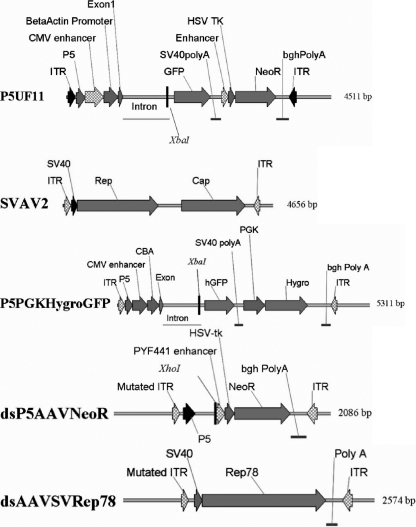
See Fig. 1 for vector structures. All vectors were packaged into AAV 2 capsids and purified using a single-step gravity column method described by Auricchio et al. (2).
FIG. 1.
Schematic representation of vector constructs. The single-stranded AAV vectors include P5UF11, SVAV2, and P5PGKHygroGFP. The self-complementary AAV vectors include dsP5AAVNeoR and dsAAVSVRep78. The important restriction sites (XbaI, XhoI) used for Southern analysis are marked. There are no EcoRV restriction sites in these vectors.
Junction assay: detecting AAVS1 integration by PCR-Southern blotting.
A combined PCR and Southern blotting approach was used to determine if the vectors used in the experiments were capable of integrating into AAVS1. In this approach, PCR was used to amplify AAV-AAVS1 junction sequences from genomic DNA isolated from cells infected with the various AAV vectors. Primers were used to allow isolation of junction sequences by PCR, as follows. The forward primer was specific for the right AAV ITR (ITR1, 5′-AGGAACCCCTAGTGATGGAG-3′), and the reverse primer falls on AAVS1 near a BamHI restriction site on the host sequence (AAVS1dRBS, 5′-CACCACGTGATGTCCTCTGA-3′). The PCR conditions were set up using HotStarTaq master mix (Qiagen, Valencia, CA) as follows: initial heating at 94°C for 15 min (hot start), followed by 30 cycles at 94°C for 1 min, 57°C for 1 min, 72°C for 2 min, and a final elongation at 72°C for 10 min. The PCR products were separated on a 1% agarose gel and then transferred onto a positively charged nylon filter membrane (Hybond N+; Amersham). The filter was hybridized with a 441-bp digoxigenin (DIG)-labeled AAVS1 probe generated using the primer combination of AAVS1p1 (5′-ACCCTATGCTGACACCCCGT-3′) and AAVS1p2 (5′-CGCAGAAGCCAGTAGAGCT-3′), using noninfected HeLa DNA as the template.
DIG-labeling of AAVS1 probe.
PCR DIG probe synthesis kit (Roche, Nutley, NJ) was used to generate and label a 441-bp AAVS1 fragment according to manufacturer's instructions.
Hybridization of PCR products.
Hybridization of the PCR products was performed using DIG Easy Hyb (Roche, Nutley, NJ) overnight at 42°C. Posthybridization, the nylon filter was washed twice in 2× SSC (0.3 M sodium chloride, 0.03 M sodium citrate [pH 7.0]; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature for 10 min, with each wash followed by washing in 0.5× SSC and 0.1% SDS at 68°C for 30 min. After hybridization and stringency washes, the blot was processed according to manufacturer's instructions.
Single cell cloning of infected cells.
Cells infected with the different viruses were selected for 2 weeks in the presence of Geneticin (Gibco, Carlsbad, CA) (HeLa, 600 μg/ml; MO59J and M059K, 100 μg/ml) or hygromycin B (Gibco, Carlsbad, CA) (ligase IV, 75 μg/ml). Resistant colonies were pooled by trypsinization, mixed, counted, and seeded at 1 cell per well into several 96-well plates. The individual colonies from the 96-well plates were expanded, in the presence of G418 or hygromycin, until they were confluent in T25 flasks or 100-mm dishes, at which time they were harvested for genomic DNA isolation and Southern hybridization analysis.
Southern blot analysis.
DNA from individual clones resistant to G418 or hygromycin B was isolated using the DNAeasy tissue kit (Qiagen, Germantown, MD). Ten to twenty micrograms of DNA was digested with EcoRV and XbaI (for single-stranded AAV vectors) or XhoI (for self-complementary AAV vectors) and EcoRV for 12 to 16 h and loaded onto a 0.8% agarose gel. The DNA was transferred from the gel onto a nylon filter membrane, and hybridization was carried out at 65°C overnight with 32P-labeled probes for either the AAVS1 target or the transgene (AAVS1, neomycin-GFP, or neomycin). The DNA probes were radiolabeled using RadPrime DNA labeling kit (Invitrogen, Carlsbad, CA) following manufacturer's instructions. The AAVS1 probe consisted of a 3.5-kb fragment generated by digesting an AAVS1 plasmid (pRVK) with EcoRI and KpnI. The neomycin-GFP probe (2 kb) was generated by digesting P5UF11 with XbaI and BamHI. The 1-kb neomycin probe (for self-complementary AAV vectors) was generated by digesting P5UF11 with XhoI and BamHI. The 1.9-kb Hygro probe was generated by digesting P5PGKHygro with XhoI and SacI. Hybridization was performed in Church buffer (0.5 M sodium phosphate, 7% SDS, 1 mM EDTA, 1% bovine serum albumin) at 65°C overnight. The following day, the blots were washed in 2× SSC and 0.1% SDS (65°C for 15 min), 1× SSC and 0.1% SDS (65°C for 15 min), 0.5× SSC and 0.1% SDS (65°C for 30 min), and a final wash in 0.1× SSC and 0.1% SDS (65°C for 15 min). The nylon filter was rinsed in 2× SSC, wrapped in Saran wrap, and exposed to film for appropriate times for signal development, using autoradiography.
Statistical analysis.
Statistical analysis was performed with Minitab, release 14 (Minitab Inc., State College, PA) on clonal data to compare groups for the two characteristics (random and specific integration) using a two-by-two chi-square contingency table analysis. Differences were considered to be statistically significant when the P value was <0.05.
RESULTS
Single-stranded AAV vectors are better substrates for site-specific integration.
Wild-type AAV has a single-stranded DNA genome, as do most rAAV vectors. However, it is feasible to create AAV vectors with self-complementary genomes which are thought to fold on themselves after uncoating to form duplex DNAs which can be transcribed without the necessity for second-strand DNA synthesis. Self-complementary AAV (scAAV) vectors are more efficient in transduction and known to be substrates for DNAPKcs. Thus, it was of interest to assess whether such vectors can integrate in a site-specific manner and, if they can, whether they do so more efficiently than single-stranded vectors.
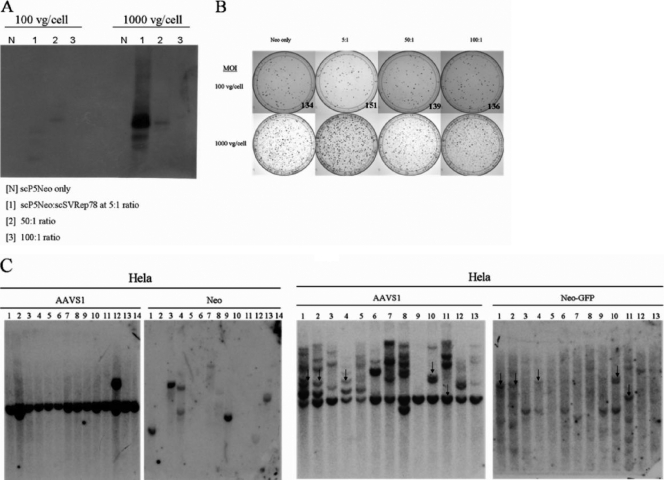
It has not been shown if scAAV vectors are capable of site-specific integration. To this end, HeLa cells were coinfected with two self-complementary vectors (dsP5AAVNeoR and dsAAVSVRep78) (Fig. 1) at different ratios and infection doses (number of viral particles/cell). The scAAV vector, dsP5AAVNeoR, was used for selection, and the helper vector, dsAAVSVRep78, was used to provide the Rep protein for targeting the dsP5AAVNeoR vector to AAVS1. Different ratios of the two vectors were used to identify which ratio provides optimal integration detectable by the junction assay analyzed 1 week postinfection (Fig. 2A). In addition, a colony-forming assay was used to see if Rep coinfection expression increases the number of G418-resistant colonies, suggestive of targeted integration. In agreement, a ratio of 5:1 resulted in both an apparent modest increase in colony formation (Fig. 2B) and a strong detectable junction product (Fig, 2A). It should be noted that many G418-resistant colonies were present in the absence of dsAAVSVRep78 and at various vector and MOI ratios that did not show much, if any, junction formation by PCR (Fig. 2A). This in itself suggests that random vector integration was likely to predominate, in accord with data from clonal analysis described below. Moreover, increasing the dose of scAAV infection augmented both colony formation and junction formation. Overall, the results indicated that scAAV vectors are capable of integrating in HeLa cells. In order to analyze to what extent integration was site specific requires cloning of G418-resistant colonies. To assess this, the ratio of site-specific integration to random integration in HeLa cells was determined. HeLa cells were infected either with single-stranded AAV vectors P5UF11:SVAV2 (50:1) at an MOI of 105 (number of viral particles/cell) or with dsP5AAVNeoR:dsAAVSVRep78 (50:1) at an MOI of 105 (number of viral particles/cell). After 2 weeks of G418 selection, resistant cells were cloned and assessed by Southern blot analysis. In this experiment, we observed that 45% of the clones from cells infected with single-strand vector contained a detectable vector sequence linked to AAVS1 sequences and, hence, presumably integrated at that site (Fig. 2C, right). In contrast, none of the G418-resistant clones arising after self-complementary vector infection had evidence for site-specific integration (Fig. 2C, left). These results suggest the importance of single-stranded DNA in the mechanism of AAV site-specific integration.
FIG. 2.
scAAV vectors integrate site-specifically in HeLa cells. (A) Junction PCR assay of HeLa cells infected with dsP5AAVNeoR only or with dsAAV-SVRep78 at different ratios and MOIs (viral genomes per cell [vg/cell]) analyzed 1 week postinfection. (B) Colony-forming assay. Ten thousand infected HeLa cells were seeded into 100 mM dishes and selected with G418 for 2 weeks. Postselection, the resistant colonies were stained with crystal violet and counted. The counts are shown only for the lower dose (100 vg/cell). Visually, more colonies are seen at the 5:1 ratio at both doses. (C) Comparison of ssAAV and scAAV specific integration. Left panel, HeLa scAAV clones (50:1; dsP5AAVNeoR:dsAAVSVRep78; MOI, 105). Right panel, HeLa ssAAV clones (50:1; P5UF11:SVAV2; MOI, 105). The arrows indicate the clones that have both AAVS1 and AAV signals colinked.
AAV-AAVS1 junction formation in the absence of DNAPKcs.
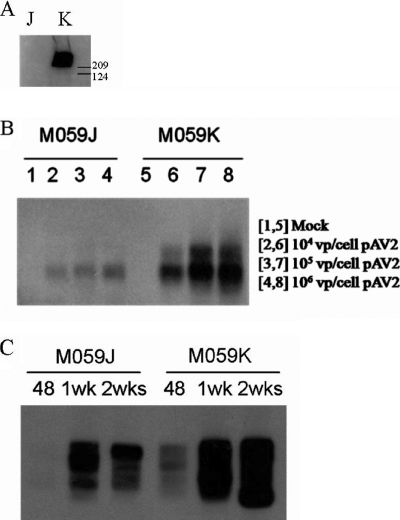
In work by Song et al. (30), it was demonstrated using an in vitro assay that the presence of DNAPKcs decreased the formation of AAV-AAVS1 junctions. To determine if a similar effect takes place in the context of a cell culture infection, M059J and M059K cells were infected with wild-type AAV 2 at different MOIs (104, 105, and 106 viral particles per cell). M059K cells express functional DNAPKcs, whereas M059J cells do not express DNAPKcs, verified by Western analysis (Fig. 3A). AAV-AAVS1 junction formation was assessed by PCR in these cells at 48 h after wild-type AAV 2 infection. Junction formation was greatly reduced in M059J (DNAPKcs-defective) cells at all MOIs tested relative to that in the M059K (DNAPKcs-proficient) cells, as judged by the intensity of the AAVS1 signal after PCR amplification (Fig. 3B).
FIG. 3.
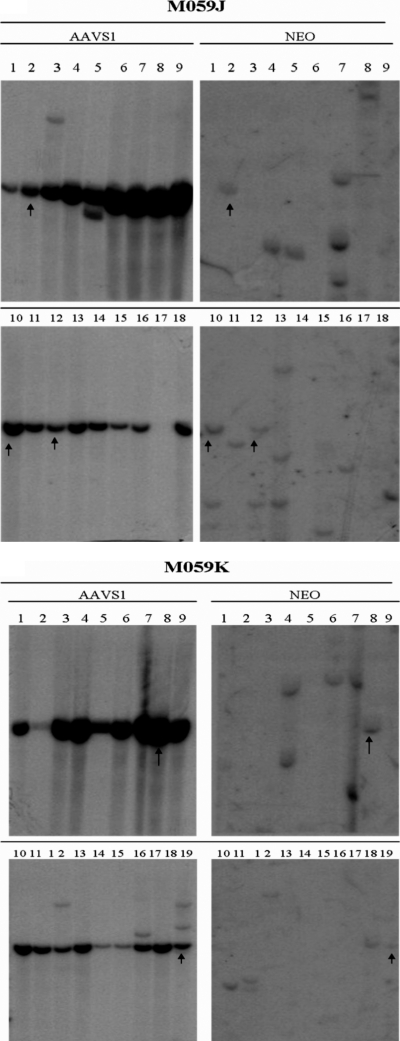
AAV-AAVS1 junction product formation in M059J and M059K cells. (A) Western blot for DNAPKcs from whole-cell extracts from M059J (J) and M059K (K) cells. Sizes in kilodaltons are marked on the side. DNAPKcs is approximately 460 kDa in size. (B) Junction formation at different doses of wild-type AAV 2 infection. The cell lines were infected with different doses (number of viral particles [vp] per cell) of wild-type AAV 2. Genomic DNA from infected cells was isolated 48 h postinfection for junction assay. (C) Time course of junction formation. Both cells were infected with 106 viral genomes per cell (50:1 ratio of P5UF11 to SVAV2). Genomic DNA was isolated at 48 h, 1 week, or 2 weeks postinfection for junction assay.
In order to generate clones to measure relative integration frequency in these cell lines after infection, we used the bipartite vector infection strategy previously described (34). The M059J and M059K cells were infected at an MOI of 106 viral particles/cell (>90% of cells infected) (data not shown), using a 50:1 ratio of P5UF11 to SVAV2 virus. We have previously demonstrated site-specific integration in HeLa cells using this bipartite vector combination and ratio (34). Junction analysis of M059J and M059K cells infected with the bipartite vectors P5UF11 and pSVAV2 showed a response similar to that of wild-type AAV infection; junction formation was reduced in J cells compared to in K cells at 48 h postinfection (Fig. 3C). Furthermore, by 2 weeks postinfection, more junctions were formed in both cell lines, but J cells still displayed fewer junctions than did K cells. This suggested to us that maybe specific integration was reduced in the absence of DNAPKcs, contrary to the in vitro observation by Song et al. (30). To test this hypothesis, infected M059J and M059K cells were selected with G418 for 2 weeks, and single-cell clones were generated for Southern hybridization analysis.
A greater fraction of clones had vector integrated in AAVS1 in the absence of DNAPKcs.
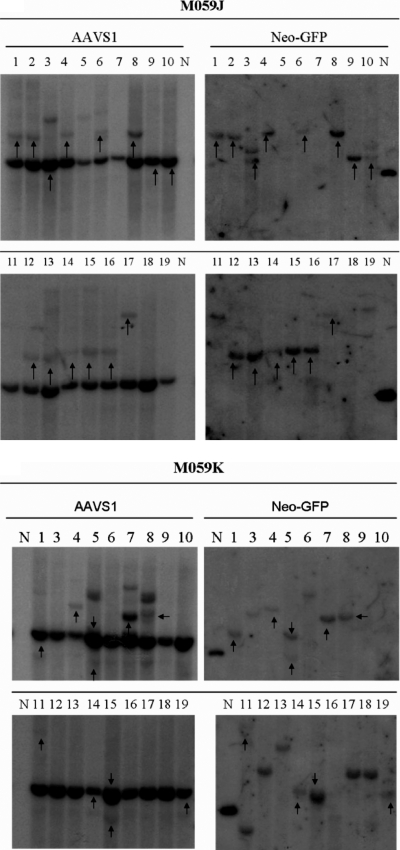
Southern hybridization analysis of single-cell G418-resistant clones from infected J and K cells revealed a surprising result. In fact, more site-specific integration was observed in J-cell clones (72%) than in K-cell clones (52%) after analysis of 18 or 19 individual clones (with detectable vector sequences) for each set, respectively (Fig. 4A and B). This difference, though not statistically significant (P = 0.145), is in accord with the in vitro results of Song et al. (30), who detected less site-specific integration in the absence of DNAPKcs (i.e., in SCID− extracts). Our conclusion is that overall integration, site specific and random, is enhanced by DNAPKcs, but that the ratio of site-specific integration to random integration was increased in the absence of DNAPKcs.
FIG. 4.
Southern hybridization of M059J and M059K clones infected with single-stranded AAV vectors P5UF11 and SVAV2 (50:1 ratio, 106 viral particles [vp]/cell). Representative blots are shown. The left blots show signals for AAVS1 sequences, and the right blots show signals for neomycin. The arrows indicate the clones that had both AAVS1 and neomycin signals colinked. The endogenous AAVS1 band is seen in all clones. The presence of additional AAVS1 bands is indicative of rearrangements of the site, which occurs when it is targeted. In some cases, the endogenous AAVS1 is colinked with AAV, indicative of deletions occurring before integration. If integration occurs with minimal deletions, a higher AAVS1 band colinks with AAV. The samples are numbered. N represents a positive control plasmid which was used to determine specificity of the neomycin probe.
The relative frequency of site-specific integration by scAAV is not affected by DNAPKcs.
Since it has been demonstrated that the DNAPKcs can process both single-stranded (11) and self-complementary AAV vectors (8), we were interested to see if a difference in integration in J and K cells would also apply to scAAV vectors. As described earlier, the scAAV vectors integrated into HeLa cells, but analysis of clones did not indicate much, if any, site-specific integration. Our next objective was to quantitate the efficiency of scAAV integration in the M059J and M059K cell lines.
Although site-specific integration by scAAV was not detected in HeLa cell clones, we were interested in seeing whether the absence of DNAPKcs affected site-specific integration by self-complementary vectors. Using the optimal 5:1 ratio for scAAV vectors, M059J and M059K cells were infected with dsP5AAVNeoR and dsAAVSVRep78 (5:1) at a total dose of 104 viral particles/cell. Infected cells were selected for 2 weeks with G418. Twenty-seven M059J and 28 M059K resistant colonies were expanded in the presence of continuous selection and used for genomic DNA isolation and Southern hybridizations. In accord with the data for HeLa cells, integration was predominantly random in M059K cells. Essentially the same results were obtained with the M059J cell clones (Fig. 5A and B). Roughly 25% of clones, from both J and K cells, had scAAV integrated into AAVS1, with the other 75% randomly integrated. Thus, DNAPKcs had no apparent effect on the relative frequencies of site-specific integration by the self-complementary vectors, although a PCR assay for vector-cell junction formation indicated enhanced junction in M059K cells (data not shown). Thus, once again, there was an enhancement in both site-specific and random integration, this time for self-complementary vectors in cells containing DNAPKcs.
FIG. 5.
Southern hybridization of M059J and M059K clones infected with self-complementary vectors dsP5AAVNeoR and dsAAVSVRep78 (5:1 ratio, 104 viral genomes/cell). Representative blots are shown. The left blots show signals for AAVS1 sequences, and the right blots show signals for neomycin. The arrows indicate the clones that had both AAVS1 and neomycin signals colinked.
These observations indicate that single-stranded DNA AAV vectors are a better substrate for site-specific integration and suggest the importance of single-stranded DNA in the mechanism of AAV site-specific integration. More interestingly, it implies that DNAPKcs may differentially act on the two forms of AAV substrates.
AAV site-specific integration is greatly reduced in a cell line deficient in DNA ligase IV.
An important aspect of integration is the stable joining of the exogenous DNA to the cellular DNA. These linkages are performed by cellular repair proteins called DNA ligases. One such DNA ligase has been investigated for an effect on AAV-AAVS1 junction formation. Ligase IV is involved in repair by single-stranded break repair and NHEJ (17).
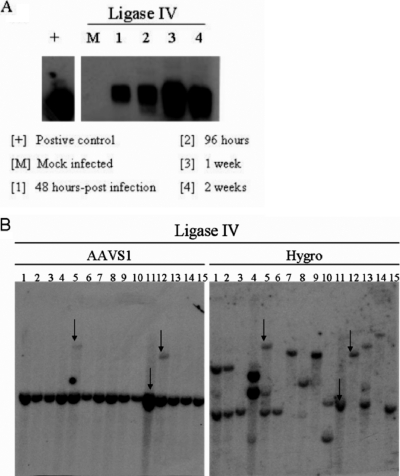
Ligase IV cells were infected with wild-type AAV 2 at 105 viral particles/cell. Junction formation was assessed from genomic DNA isolated from these cells 48 h, 96 h, 1 week, and 2 weeks postinfection. The results demonstrated that, even in cells with reduced expression of ligase IV, junction formation between AAV DNA and the AAVS1 cellular sequence was readily apparent (Fig. 6A).
FIG. 6.
(A) Time course of junction formation in ligase IV hypomorphic cell lines. Cells were infected with wild-type AAV 2 at 105 viral particles/cell. Genomic DNA was isolated at the indicated time points for junction assay. (B) Southern hybridization of ligase IV clones infected with single-stranded AAV vectors P5PGKHygroGFP and SVAV2 (50:1 ratio, 105 vg/cell). Representative blots are shown. The left blots show signals for AAVS1 sequences, and the right blots show signals for hygromycin. The arrows indicate the clones that have both AAVS1 and hygromycin signals colinked.
To see if AAV site-specific-integration frequency was decreased in these cells, we decided to analyze clones from ligase IV mutant cells, to see if AAV specific-integration frequency is decreased in these cells. Southern analysis demonstrating that 20% of the ligase IV clones displayed site-specific integration (Fig. 6B). This observed frequency is lower (P < 0.05) than the well-documented 50 to 70% frequency of AAV specific integration in comparable cell lines. These data strongly suggest a key role for ligase IV in the mechanism of AAV site-specific integration and support the notion that NHEJ is an important pathway in AAV targeted integration.
DISCUSSION
Many aspects of the AAV life cycle are not completely understood. One such aspect is the intracellular processing of AAV genomes after infection and their persistence in the absence of helper adenovirus. It is known that in the absence of adenovirus, wild-type AAV can establish latency in cell culture by integrating its genome into a specific site in the human genome on chromosome 19q13.4, a site termed AAVS1. What remains elusive though is our understanding of both the structural character of the substrate for integration and the host requirements for this targeted specificity of integration. Over the last several years, many groups have reported that cellular proteins involved in DNA repair can affect the transduction and expression of rAAV genomes. Cells lacking ATM, a key cellular DNA damage sensor, are more permissive to AAV transduction (27). Several other groups have reported a role for Mre11/Nbs1/Rad50 as an inhibitor of AAV double-stranded DNA formation and, hence, AAV expression (6, 29).
Because of the very limited homology between AAV and characterized integration sites in AAVS1, NHEJ has been suggested as being involved in AAV targeted integration. We were interested in analyzing AAV site-specific integration in cells lacking components involved in this repair pathway. To this end, the roles of DNAPKcs and ligase IV were evaluated. Our results (summarized in Table 1) clearly demonstrate that DNA ligase IV has a major effect on specific integration of single-stranded AAV, whereas the results with DNAPKcs were less definitive although suggestive.
TABLE 1.
Summary of Southern blot resultsa
| Cell line | Vector | MOI | Vector ratio | No. of clones with AAV sequences | % Integrationb |
|
|---|---|---|---|---|---|---|
| AAVS1 | Random | |||||
| M059J | P5UF11:SVAV2 | 106 | 50:1 | 25 | 72 | 28 |
| dsP5AAVNeoR:dsSVRep78 | 104 | 5:1 | 21 | 23.8 | 76.2 | |
| M059K | P5UF11:SVAV2 | 106 | 50:1 | 25 | 52 | 48 |
| dsP5AAVNeoR:dsSVRep78 | 104 | 5:1 | 17 | 23.5 | 76.5 | |
| Ligase IV | P5PGKHygro:SVAV2 | 105 | 50:1 | 25 | 20 | 80 |
| HeLa | P5UF11:SVAV2 | 105 | 50:1 | 11 | 45 | 54 |
| dsP5AAVNeoR:dsSVRep78 | 105 | 50:1 | 8 | 0 | 100 | |
Genomic DNA from neomycin- or hygromycin-resistant clones was analyzed with Southern blotting by hybridizing to AAVS1 and neomycin/hygromycin-specific probes.
Colinking of AAVS1 and neomycin/hygromycin-specific signals was used to determine the percent AAVS1 and random integration. The percent AAVS1 and random integration was determined using the number of clones with detectable vector sequences.
Role of DNAPKcs in NHEJ repair and AAV processing.
DNAPKcs is involved in both double-stranded break repair by NHEJ and immunoglobulin V(D)J rearrangement. It is recruited by Ku70/80 heterodimers bound at sites of double-stranded break repair forming a complex called DNAPK. Upon association with Ku heterodimers, the catalytic activity of DNAPKcs is activated and presumably serves to phosphorylate other DNA repair proteins. Some of the targets of DNAPKcs include DNAPKcs itself, RPA, Ku70/80, ligase IV, and Artemis. The exact role of DNAPKcs in NHEJ repairs remains poorly understood, despite the important function it serves in maintaining genomic stability.
Many studies have shown evidence for processing of AAV by DNAPKcs (11, 15). The exact functional outcome of AAV processing by DNAPKcs remains unclear. As mentioned above, a recent study has indicated a role for DNAPKcs in signaling AAV DNA damage response under conditions that promote AAV replication (27). Within cells, DNAPKcs is often found associated with Artemis, a cellular endonuclease, and it has been experimentally demonstrated that the DNAPKcs-Artemis complex can cleave hairpin loops, flaps, and gaps. Therefore, it is tempting to think that either during integration and/or after integration, the AAV ITRs are cleaved by the DNAPKcs-Artemis complex, resulting in local chromosomal changes at the site of integration that may lead to our observed differences in integration efficiency. In vivo, AAV hairpin processing is impaired in muscle and kidney, but not liver, in the absence of DNAPKcs and/or Artemis (15). Furthermore, Song et al. (30) noticed a greater percentage of liver cells expressing the transgene 5 weeks posthepatectomy in SCID liver (SCID mice lack DNAPKcs expression) than in C57BL/6 liver. This suggests that processing by DNAPKcs affects either establishment, maintenance, or expression of rAAV integration after induction of cell proliferation. In early studies of AAV integration, it was noted that with continued cell passage, AAV genomes were excised and appeared to be extrachromosomal (as unit length duplex AAV DNA) (7). Restriction enzyme analyses revealed that alteration in integrated genomes had occurred uniquely in the ITR sequences at the junctions with cellular DNA. Since DNAPKcs can act on hairpin structures, it is possible that DNAPK affects both integration and excision. It remains our future challenge to look at the detailed interactions between DNAPKcs and AAV during or after specific integration.
Role of ligase IV in NHEJ repair.
The ligase cell line evaluated here was isolated from a patient suffering from ligase IV syndrome. The patient had a heterozygous mutation (R278H changes in one allele) that impairs the adenylation and ligation functions of ligase IV, yet this particular patient had no overt immunodeficiency, presumably due to the residual activity of ligase IV still present. Moreover, the mutated ligase IV was able to interact with its partner protein, XRCC4. Since ligase IV functions specifically in NHEJ, we examined the site-specific integration process in DNA ligase IV-deficient human cells.
We did not observe a significant decrease in junction formation in the PCR assay. However, upon cloning, it was determined that site-specific integration was reduced to 20%. Once again the junction PCR assay results did not truly reflect the proportion of site-specific integration in cells in which vectors had integrated and continued transgene expression. The PCR assay does measure junctions between vectors and AAVS1; it is not seen in the absence of Rep, but clearly it does not reflect the ratio between site-specific integration and random integration as determined from cloning. Not only was this the case with ligase IV mutant cells, but DNAPKcs-negative cells showed less junction formation by PCR, but, if anything, the relative amounts of site-specific integration were increased. The only conclusion we can draw is that stated above. The observation that junction formation was not diminished in ligase IV-defective cells supports the notion that junction PCR is not an accurate assessment of the ratio of site-specific integration to random integration but, rather, just a qualitative one suggestive of integration into AAVS1. Recently, it was demonstrated that AAV specific integration is accompanied by a partial duplication of the AAVS1 locus (14). This duplication can greatly alter the left and right AAV-AAVS1 junctions; therefore, Southern analysis of clones is the one true way to get quantitative information with regard to frequency and specificity of integration, and the observed 20% specific integration in ligase IV-defective cells may be the result of the residual activity of ligase IV with the R278H mutation (12) and/or the ability of other ligases to participate in NHEJ-mediated repair in the absence of ligase IV (18). It remains unclear if an alternative pathway has a role in AAV specific integration, since the residual 5 to 10% ligase activity (12) may be sufficient to achieve 20% site-specific integration.
Functional significance of ssAAV and scAAV in specific integration.
The observation that self-complementary vectors showed no difference in integration in the presence or absence of DNAPKcs is quite intriguing. It partially addresses the issue of whether the substrate for site-specific integration is a single- or double-stranded molecule. This together with the observations in HeLa cells demonstrates that scAAV vectors are much less efficient than single-stranded AAV vectors at similar MOIs. Does this imply that duplex molecules per se are not good substrates for site-specific integration? In accord with this notion, we have had little success in observing site-specific integration after transfection of cells with linear double-stranded DNA (Dyall and Berns, unpublished). However, plasmids containing AAV sequences integrate at higher frequencies than do self-complementary vectors (24). The latter may reflect structural characteristics of closed circles which contain hairpin sequences (e.g., ITRs) that tend to self pair and pop out of the circle. It remains beyond the scope of this work to elucidate why this difference occurs, although an interesting future experiment would be to include the use of adenovirus E4 and E2 to control the AAV forms in transduced cells and tissues. It has been documented that adenovirus E4 promotes linear concatemers of double-stranded AAV forms, whereas adenovirus E2 increases the abundance of AAV circular forms (10). It would of interest to see if these adenovirus proteins could affect a change in site-specific integration and would answer the question of whether linear or circular double-stranded forms of AAV are important substrates for site-specific integration.
Conclusion.
Cellular recombination proteins clearly must have a role in AAV site-specific integration, because AAV is largely dependent on host factors for all aspects of its life cycle. Analysis of AAV-cellular junction sequences has revealed that the integration process is error prone, strongly suggesting the involvement of NHEJ proteins and/or a short list of other repair proteins in integration.
There seems to be a dynamic interaction between the AAV and cellular proteins, which can greatly affect AAV expression and integration. Interestingly, pull-down assays have identified numerous Rep-interacting DNA repair proteins, including Ku70, Ku80, replication protein A, Rad50, poly(ADP) polymerase I, DNAPK, and proliferating cell nuclear antigen (PCNA) (23). Rep is the only viral trans factor required for specific integration. It now appears likely that at least two proteins involved in NHEJ are involved as well. The extent to which other NHEJ proteins (i.e., Ku70/80, PARP1, Rad50), as well as other repair proteins which do not interact with Rep, have roles in AAV targeted integration remains to be determined.
Results from our studies are encouraging and will contribute to a working understanding of AAV specific integration. Some issues that require further consideration and study include the following: (i) how does the structural conformation of the AAV DNA affect interaction with cellular proteins involved in NHEJ and site-specific integration; and (ii) is targeted integration possible after AAV replication, since replication of AAV produces unique intermediates which may be detected as aberrant DNA due to their hairpin structures? (Integration during productive infection has not been detected.) Determining how the viral Rep protein, the different AAV vectors, and the cellular components interact upon infection or integration will be crucial for developing AAV vectors with the capacity to efficiently target the AAVS1 site and to minimize risks associated with random integration. Our study has indicated that NHEJ is important in AAV site-specific integration and has implications for the potential use of AAV vectors capable of site-specific integration in both ex vivo and in vivo gene therapy modalities.
Acknowledgments
This work was supported by NIH grant P01 DK58327.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Atchison, R. W., B. C. Casto, and W. M. Hammon. 1965. Adenovirus-associated defective virus particles. Science 149:754-756. [DOI] [PubMed] [Google Scholar]
- 2.Auricchio, A., M. Hildinger, E. O'Connor, G. P. Gao, and J. M. Wilson. 2001. Isolation of highly infectious and pure adeno-associated virus type 2 vectors with a single-step gravity-flow column. Hum. Gene Ther. 12:71-76. [DOI] [PubMed] [Google Scholar]
- 3.Berns, K. I., T. C. Pinkerton, G. F. Thomas, and M. D. Hoggan. 1975. Detection of adeno-associated virus (AAV)-specific nucleotide sequences in DNA isolated from latently infected Detroit 6 cells. Virology 68:556-560. [DOI] [PubMed] [Google Scholar]
- 4.Blacklow, N. R., M. D. Hoggan, and W. P. Rowe. 1968. Serologic evidence for human infection with adenovirus-associated viruses. J. Natl. Cancer Inst. 40:319-327. [PubMed] [Google Scholar]
- 5.Carter, B. J. 2005. Adeno-associated virus vectors in clinical trials. Hum. Gene Ther. 16:541-550. [DOI] [PubMed] [Google Scholar]
- 6.Cervelli, T., J. A. Palacios, L. Zentilin, M. Mano, R. A. Schwartz, M. D. Weitzman, and M. Giacca. 2008. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J. Cell Sci. 121:349-357. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, A. K., M. D. Hoggan, W. W. Hauswirth, and K. I. Berns. 1980. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J. Virol. 33:739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2006. Host cell DNA repair pathways in adeno-associated viral genome processing. J. Virol. 80:10346-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, V. W., R. J. Samulski, and D. M. McCarty. 2005. Effects of adeno-associated virus DNA hairpin structure on recombination. J. Virol. 79:6801-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan, D., P. Sharma, L. Dudus, Y. Zhang, S. Sanlioglu, Z. Yan, Y. Yue, Y. Ye, R. Lester, J. Yang, K. J. Fisher, and J. F. Engelhardt. 1999. Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression. J. Virol. 73:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan, D., Y. Yue, and J. F. Engelhardt. 2003. Consequences of DNA-dependent protein kinase catalytic subunit deficiency on recombinant adeno-associated virus genome circularization and heterodimerization in muscle tissue. J. Virol. 77:4751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girard, P. M., B. Kysela, C. J. Harer, A. J. Doherty, and P. A. Jeggo. 2004. Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: the impact of two linked polymorphisms. Hum. Mol. Genet. 13:2369-2376. [DOI] [PubMed] [Google Scholar]
- 13.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 14.Henckaerts, E., N. Dutheil, N. Zeltner, S. Kattman, E. Kohlbrenner, P. Ward, N. Clement, P. Rebollo, M. Kennedy, G. M. Keller, and R. M. Linden. 2009. Site-specific integration of adeno-associated virus involves partial duplication of the target locus. Proc. Natl. Acad. Sci. USA 106:7571-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inagaki, K., C. Ma, T. A. Storm, M. A. Kay, and H. Nakai. 2007. The role of DNA-PKcs and Artemis in opening viral DNA hairpin termini in various tissues in mice. J. Virol. 81:11304-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 87:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, J., R. J. Samulski, and X. Xiao. 1997. Role for highly regulated rep gene expression in adeno-associated virus vector production. J. Virol. 71:5236-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieber, M. R. 2008. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283:1-5. [DOI] [PubMed] [Google Scholar]
- 19.Linden, R. M., E. Winocour, and K. I. Berns. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA 93:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, D. G., G. D. Trobridge, L. M. Petek, M. A. Jacobs, R. Kaul, and D. W. Russell. 2005. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J. Virol. 79:11434-11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra, L., and J. A. Rose. 1990. Adeno-associated virus DNA replication is induced by genes that are essential for HSV-1 DNA synthesis. Virology 179:632-639. [DOI] [PubMed] [Google Scholar]
- 22.Nakai, H., Y. Iwaki, M. A. Kay, and L. B. Couto. 1999. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J. Virol. 73:5438-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nash, K., W. Chen, M. Salganik, and N. Muzyczka. 2009. Identification of cellular proteins that interact with the adeno-associated virus Rep protein. J. Virol. 83:454-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philpott, N. J., J. Gomos, K. I. Berns, and E. Falck-Pedersen. 2002. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl. Acad. Sci. USA 99:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponnazhagan, S., D. Erikson, W. G. Kearns, S. Z. Zhou, P. Nahreini, X. S. Wang, and A. Srivastava. 1997. Lack of site-specific integration of the recombinant adeno-associated virus 2 genomes in human cells. Hum. Gene Ther. 8:275-284. [DOI] [PubMed] [Google Scholar]
- 26.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanlioglu, S., P. Benson, and J. F. Engelhardt. 2000. Loss of ATM function enhances recombinant adeno-associated virus transduction and integration through pathways similar to UV irradiation. Virology 268:68-78. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz, R. A., C. T. Carson, C. Schuberth, and M. D. Weitzman. 2009. Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J. Virol. 83:6269-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz, R. A., J. A. Palacios, G. D. Cassell, S. Adam, M. Giacca, and M. D. Weitzman. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 81:12936-12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song, S., Y. Lu, Y. K. Choi, Y. Han, Q. Tang, G. Zhao, K. I. Berns, and T. R. Flotte. 2004. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. USA 101:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surosky, R. T., M. Urabe, S. G. Godwin, S. A. McQuiston, G. J. Kurtzman, K. Ozawa, and G. Natsoulis. 1997. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 71:7951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Z., H. I. Ma, J. Li, L. Sun, J. Zhang, and X. Xiao. 2003. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 10:2105-2111. [DOI] [PubMed] [Google Scholar]
- 33.Weitzman, M. D., S. R. Kyostio, R. M. Kotin, and R. A. Owens. 1994. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. USA 91:5808-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, C., N. G. Cortez, and K. I. Berns. 2007. Characterization of a bipartite recombinant adeno-associated viral vector for site-specific integration. Hum. Gene Ther. 18:787-797. [DOI] [PubMed] [Google Scholar]