Abstract
The EBNA1 protein of Epstein-Barr virus (EBV) plays several important roles in EBV latent infection, including activating DNA replication from the latent origin of replication (oriP) and activating the transcription of other latency genes within the EBV chromatin. These functions require EBNA1 binding to the DS and FR elements within oriP, respectively, although how these interactions activate these processes is not clear. We previously identified interactions of EBNA1 with the related nucleosome assembly proteins NAP1 and TAF-I, known to affect the replication and transcription of other chromatinized templates. We have further investigated these interactions, showing that EBNA1 binds directly to NAP1 and to the β isoform of TAF-I (also called SET) and that these interactions greatly increase the solubility of EBNA1 in vitro. These interactions were confirmed in EBV-infected cells, and chromatin immunoprecipitation with these cells showed that NAP1 and TAF-I both localized with EBNA1 to the FR element, while only TAF-I was detected with EBNA1 at the DS element. In keeping with these observations, alteration of the NAP1 or TAF-Iβ level by RNA interference and overexpression inhibited transcriptional activation by EBNA1 in FR reporter assays. In addition, EBNA1-mediated DNA replication was stimulated when TAF-I (but not NAP1) was downregulated and was inhibited by TAF-Iβ overexpression. The results indicate that the interaction of EBNA1 with NAP1 and TAF-I is important for transcriptional activation and that EBNA1 recruits TAF-I to the DS element, where it negatively regulates DNA replication.
Epstein-Barr virus (EBV) is a ubiquitous human gammaherpesvirus which establishes latent infections in B lymphocytes and, to a lesser degree, in epithelial cells (66, 78). Since latent infection can involve immortalization of the infected cell, EBV latent infection is causatively associated with several types of B-cell lymphomas and carcinomas. During latent infection, the circularized viral chromosomes are maintained as multicopy episomes that are assembled into nucleosomes with spacing typical of cellular chromatin (15, 59). Two viral components, the latent origin of DNA replication (oriP) and the Epstein-Barr nuclear antigen 1 protein (EBNA1), are required for stable maintenance of the viral genome (75), which replicates once per cell cycle and segregates equally to the daughter cells in mitosis (2, 16).
oriP is comprised of two functional elements separated by approximately 1 kb, namely, the dyad symmetry (DS) element and the family of repeats (FR) (52). The DS contains four EBNA1 recognition sites and functions as an initiation site for DNA replication within oriP when bound by EBNA1 (21, 24, 76). The FR is a cluster of 20 EBNA1 binding sites and governs the mitotic segregation of the EBV genomes or oriP-containing plasmids (39, 52). In addition, FR acts as a transcriptional enhancer when bound by EBNA1, activating the expression of other EBV latency genes and of reporter genes placed under FR control (22, 52, 74).
The mechanisms by which EBNA1 binding to the DS and FR elements activates DNA replication and transcription, respectively, are currently unclear, although considerable information is accumulating on the mechanism of initiation of DNA replication from the DS element. For example, it has been shown that EBNA1 dimers must assemble on at least two of the adjacent binding sites in the DS element to activate replication, that the spacing between these sites must be maintained, and that EBNA1 dimers assemble cooperatively on these sites (24, 64, 76). The assembly of EBNA1 on the DS element does not melt the DNA but causes sharp bending of the DS region and localized distortion within the EBNA1 binding sites due to insertion of an EBNA1 peptide along the minor groove of the DNA recognition site (6, 17, 18, 29). In addition, several studies have shown that the host cell origin recognition complex (ORC) and minichromosome maintenance complex are recruited to oriP, most likely through interactions between EBNA1 and ORC (8, 14, 33, 47, 56). A third cellular factor, the telomere repeat-associated factor TRF2, has also been shown to bind the DS element in an EBNA1-dependent manner, where it appears to facilitate ORC recruitment (4, 12). Alterations to the chromatin structure at the DS element are also likely to be important for origin activation. For example, it has been shown that the DS element is flanked by positioned nucleosomes that are destabilized and undergo transient deacetylation at G1/S (80) and that EBNA1 can destabilize nucleosomes formed at the DS element (5).
The transcriptional activation function of EBNA1 is known to require two distinct EBNA1 regions, an N-terminal sequence from amino acids 61 to 83 and the central Gly-Arg repeat from amino acids 325 to 376 (7, 36, 73). The 61-83 region mediates an interaction with the host bromodomain protein Brd4 (38). This appears to contribute to transcriptional activation, since Brd4 silencing decreases EBNA1-mediated transcription and Brd4 can be detected with EBNA1 at the FR (38). The 325-376 region has been found to interact with P32/TAP, a promiscuous binder of basic proteins, and EBP2, a nucleolar protein whose interaction with EBNA1 in mitosis is important for EBNA1-mediated segregation (27, 61, 71). However, neither of these interactions seems likely to mediate EBNA1-specific transcriptional activation.
To gain a more comprehensive understanding of EBNA1-host protein interactions important for EBNA1 functions in EBV latency, we used two proteomic approaches, EBNA1 affinity column profiling and in vivo tandem affinity purification tagging, to screen for human proteins that are specifically recognized by EBNA1 (28, 62). Both approaches identified ubiquitin-specific protease 7 (USP7), casein kinase 2, and protein arginine methyltransferase 5 (PRMT5) as specific binding partners of EBNA1. In addition, the affinity column approach identified interactions with three related histone chaperone proteins, namely, nucleosome assembly protein 1 (NAP1), template-activating factor TAF-Iα, and TAF-Iβ (also called SET). NAP1 and TAF-Iβ were shown to interact specifically with EBNA1 and not with a charge-matched control protein, and the recovery of NAP1 (but not TAF-Iα and TAF-Iβ) was diminished when affinity column profiling was performed with an EBNA1 mutant lacking the 325-376 region (28).
NAP1, TAF-Iα, and TAF-Iβ belong to the nucleosome assembly family of proteins that have a highly conserved central domain (the NAP domain) and acidic C-terminal sequences (50). All of these proteins bind histones and have roles in nucleosome assembly and disassembly and chromatin remodeling. TAF-Iα and TAF-Iβ are two alternatively spliced isoforms that differ only at the extreme N terminus. They were first identified as factors that stimulate the transcription and replication of adenovirus core particles in vitro (40, 46) and were shown to form homo- and heterodimers (42, 45). TAF-Iα and TAF-Iβ are also components of the inhibitor of histone acetylation (INHAT) complex and therefore can inhibit acetylation of some proteins (43, 57). Conversely, TAF-Iβ can bind p300 and CBP and stimulate acetylation by CBP (34, 60).
NAP1 is well known for its ability to assemble nucleosomes in vitro but also has been implicated in several other processes (82). NAP1 can function similarly to TAF-I in the activation of replication and transcription of adenovirus core particles (35) and in the recruitment of p300 and CBP acetyltransferases to chromatin and stimulation of their acetylation activity (60). NAP1 is also utilized by a number of viruses, binding some viral proteins to promote viral transcription (51, 58, 60, 69). NAP1 is highly homologous (70%) to NAP2, which is found only in higher eukaryotes. Not surprisingly, these proteins appear to share several functions. Both NAP1 and NAP2 bind histones and can transfer them onto DNA, both are involved in nucleocytoplasmic shuttling, and both can bind and stimulate p300 (44, 48, 54, 60).
The association of nucleosome assembly proteins with histone modifications and nucleosome remodeling, known to influence transcription and DNA replication, suggests that their interactions with EBNA1 may be important for the replication and transcriptional activation functions of EBNA1. In this study, we further defined the nature of the interactions of EBNA1 with NAP1, TAF-Iα, and TAF-Iβ and examined EBNA1 interactions with NAP2. We also investigated the functional significance of these interactions through RNA interference, overexpression, and chromatin immunoprecipitation (ChIP) approaches. The results point to the importance of the nucleosome assembly proteins in transcriptional activation by EBNA1 and to a specific role for TAF-I in regulating EBNA1-mediated DNA replication.
MATERIALS AND METHODS
Cell lines.
CNE2Z is an EBV-negative nasopharyngeal carcinoma cell line (31). Raji is an EBV-positive Burkitt's lymphoma cell line. D98/Raji cells were generated by fusion of D98 and Raji cells and retain the Raji cell EBV (23). AGS-rEBV cells are AGS gastric carcinoma cells that have been infected with recombinant EBV in culture (63) (kindly provided by Lawrence Young).
Plasmids and siRNAs.
pET15b-NAP1, pET15b-NAP2, and pET14b-TAF-Iα were gifts from J. Pelletier (McGill University), P. Rodriguez (University of Pittsburgh) (54), and K. Nagata (University of Tsukuba) (42), respectively. A TAF-Iβ human clone was purchased from ATCC and inserted between the BamHI and NdeI sites of pET15b (Novagen). cDNAs for NAP1, NAP2, TAF-Iα, and TAF-Iβ were amplified using the appropriate primers and inserted between the BamHI and EcoRI sites of the mammalian vector pCMVmyc, downstream of a myc tag. Small interfering RNA (siRNA) for NAP1 (GCCGAUAUUUUCCAGUUCUUACAACA) was purchased from Integrated DNA Technologies Inc. siRNAs for NAP2 (UCAGGUGAUGCAGAAUCCUCGAGUU), TAF-I (oligo 1, UCUCUCCAAAGAAUUUCAUCUGAAU; and oligo 2, UUUACUGACUUUGGCCGCCUGCGCC), and EBNA1 (GGAGGUUCCAACCCGAAAUTT) were synthesized by Invitrogen. siRNA against green fluorescent protein (GFP) (GAACUUCAGGGUCAGCUUGCCG) was used as a negative control.
Expression and purification of recombinant proteins.
His-tagged NAP1, NAP2, TAF-Iα, and TAF-Iβ were expressed in Escherichia coli BL21(DE3)pLysS from pET15b or pET14b plasmids. Freshly transformed colonies were grown in 2 liters of Luria broth at 37°C to an A600 of ∼0.7, and expression of recombinant proteins was induced overnight at 15°C by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.5 mM. Sonicated cell lysates were subjected to a Qiagen Ni-nitrilotriacetic acid column for purification according to the manufacturer's protocol. The recombinant proteins were dialyzed against buffer A (50 mM HEPES [pH 7.5], 200 mM NaCl, 10% glycerol, 0.5 mM EDTA, 0.1 mM dithiothreitol). EBNA1 (lacking most of the Gly-Ala repeat region) was expressed as a hexahistidine fusion from a baculovirus and purified from insect cell nuclei as previously described (18). The recombinant EBNA1 protein was dialyzed in buffer A with 1 M NaCl.
Western blotting and antibodies.
Rabbit polyclonal antibodies to TAF-Iβ (Abcam) and actin (Santa Cruz Biotechnology) were purchased and used according to the manufacturers' protocols. Anti-EBNA1 monoclonal antibody OT1x (kindly provided by Jaap Middeldorp) was used for Western blotting at a 1:5,000 dilution, while anti-EBNA1 rabbit serum R4 (28) was used in ChIP assays. Polyclonal rabbit anti-hNAP-2 antibody was a gift from Pedro Rogriguez (55) and was used at a 1:1,000 dilution for Western blotting. A monoclonal antibody against NAP1 was kindly provided by Yoshiko Ishimi and used for Western blotting at a 1:500 dilution. Rabbit antibodies were generated against full-length NAP1 and TAF-Iα purified from bacteria and were used for coimmunoprecipitation and ChIP assays. For Western blotting, antibodies were diluted in blocking buffer containing 5% milk in TBS-T buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Tween 20). Blots were washed three times with TBS-T buffer and then incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) for 1 h. Signals were detected by enhanced chemiluminescence (ECL) assay (Perkin Elmer Life and Analytical Sciences).
Coimmunoprecipitation.
Coimmunoprecipitation was performed on EBV-positive Raji Burkitt's lymphoma cells. Raji cells were lysed in hypotonic buffer, and the nuclei were harvested and extracted in RIPA buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors). The cell extract was precleared by incubation with protein A-agarose (Santa Cruz) for 30 min at 4°C, followed by 1 hour of incubation at 4°C with ExactaCruz F IP matrix (Santa Cruz). Antibodies against NAP1, NAP2, and TAF-Iα and -β were conjugated to ExactaCruz F IP matrix as described by the manufacturer and then incubated with 1 mg of nuclear lysate at 4°C overnight. The immunocomplexes were collected by centrifugation, washed in cold phosphate-buffered saline, and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting.
Glycerol gradient sedimentation assays.
Purified NAP1 (110 μg), TAF-Iα (73 μg), or TAF-Iβ (73 μg) was incubated with or without EBNA1 (100 μg) for 60 min at room temperature in buffer A in a final volume of 500 μl. A 12-ml glycerol gradient containing 10 to 30% buffer A was generated using a Gradient master apparatus (Biocomp). Protein samples applied to the gradient were subjected to centrifugation at 34,000 rpm in an SW41 rotor (Beckman) for 20 h at 4°C. Twenty-three fractions of 500 μl each were collected from the top of the gradient and analyzed by SDS-PAGE and silver staining. An identical gradient was also performed with aldolase (158 kDa) and catalase (232 kDa) (Amersham) as molecular mass standards.
EBNA1 solubility assays.
Purified NAP1 (11 μg), TAF-Iα (7.3 μg), TAF-Iβ (7.3 μg), and EBNA1 (10 μg) were incubated separately in 50 μl of buffer A in which the salt concentration was varied from 50 to 250 mM. The same amount of NAP1, TAF-Iα, or TAF-Iβ was also coincubated with 10 μg of EBNA1 in 50 μl of buffer A in which the salt concentration was varied from 50 to 250 mM. After 1 h at room temperature, samples were spun at 13,000 rpm in a microcentrifuge to separate the pellet and supernatant fractions. Pellets were then resuspended in 50 μl of buffer A, and the whole supernatant and pellet fractions were analyzed by SDS-PAGE and Coomassie blue staining.
CAT reporter transcriptional activation assays.
Transcriptional activation assays were performed using the pFRTKCAT plasmid (kindly provided by Bill Sugden) under conditions either silencing or overexpressing nucleosome assembly proteins. For assays involving silencing, 1 × 105 CNE2Z cells were plated in one well of a six-well plate at ∼30% confluence and then transfected with 40 pmol siRNA against NAP1, NAP2, TAF-I, or siGFP (negative control), using Lipofectamine 2000 (Invitrogen). This transfection was repeated 24 h later. Forty-eight hours after the first siRNA treatment, cells were transfected with 1.0 μg of pFRTKCAT reporter plasmid (52), 5 ng of pc3OriPEBNA1 or pc3OriP (61), and 0.5 μg of plasmid CMVPLAP expressing secreted alkaline phosphatase (SEAP) (72), using Fugene HD (Roche). At 48 h posttransfection, cells were harvested to measure chloramphenicol acetyltransferase (CAT) activity as previously described (73). The amount of acetylated product produced at each time point was used to determine the acetylation rate for each lysate. SEAP levels were measured at 405 nm as described previously (72). Changes in CAT activity were standardized to changes in SEAP to correct for any general effects of the treatments on gene expression. Standard two-tailed Student t tests were performed to determine statistically significant differences between treated cells and the siGFP negative control.
For the transactivation assays involving protein overexpression, 4 × 105 CNE2Z cells were plated in a 6-cm dish. The next day, cells were cotransfected with 1.0 μg of pFRTKCAT reporter, 0.5 μg of SEAP plasmid, 5 ng of pc3OriPEBNA1 or pc3OriP, and 3 μg of pMyc-CMV plasmid expressing myc-tagged NAP1, NAP2, TAF-Iα, TAF-Iβ, or nothing (empty vector). At 48 h posttransfection, cells were harvested and CAT and SEAP levels were determined as described above.
Transient DNA replication assays.
For replication experiments involving protein silencing, 4 × 105 CNE2Z cells in one 6-cm dish were transfected twice on subsequent days with 80 pmol siRNA against NAP1, NAP2, TAF-I, or GFP (negative control), using Lipofectamine 2000. Forty-eight hours after the first siRNA treatment, cells were transfected with 4 μg of pc3OriPEBNA1 or pc3OriP, using Fugene HD. Seventy-two hours later, cells were harvested and plasmids were isolated by Hirt's method as described previously (7, 26). The extracted plasmids were linearized with XhoI, and 9/10 of the linearized DNA was further digested with DpnI. The remained 1/10 of the linearized samples was used as an input control for the recovery efficiency of the plasmids. Finally, the plasmid samples were separated in 1% agarose gels, transferred to Hybond-XL membranes (Amersham), and probed with 32P-labeled pc3OriPEBNA1. Bands were visualized by autoradiography and quantified by PhosphorImager analysis using ImageQuant software (Molecular Dynamics).
For replication assays involving overexpression, 1 × 106 CNE2Z cells in one 10-cm dish were cotransfected with 4 μg of pc3OriPEBNA1 or pc3OriP plasmid and 4 μg of pMycCMV expressing myc-tagged NAP1, NAP2, TAF-Iα, or TAF-Iβ. At 72 h posttransfection, plasmid DNA was isolated by Hirt's extraction and processed as described above.
ChIP assays.
Raji cells were subjected to 1% paraformaldehyde cross-linking for 15 min and then incubated with hypotonic buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) on ice for 30 min. After Dounce homogenization, nuclei were collected by centrifugation and lysed in RIPA buffer. Chromatin was sheared by sonication to an average DNA length of 500 to 1,000 bp, using a Branson 450 sonifier, and precleared by incubation with 50% (vol/vol) salmon sperm DNA-protein A-agarose (Upstate Biochemicals). Fifty micrograms of sheared chromatin was then incubated with 2.0 μg of rabbit immunoglobulin G (IgG) (Santa Cruz), anti-EBNA1 R4 rabbit antibody, or a rabbit antibody against either NAP1 or TAF-Iα overnight at 4°C with rotation. Immune complexes were recovered by incubation with 50 μl of salmon sperm DNA-protein A-agarose with rotation for 1 h at 4°C. After reversal of the cross-links, phenol-chloroform extraction, and ethanol precipitation, immunoprecipitated DNA was resuspended in 50 μl of 10 mM Tris-Cl (pH 8.0). Quantitative real-time PCR was performed using 1/50 of the ChIP DNA and Platinum SYBR green qPCR SuperMix-UDG (Invitrogen) in a Rotorgene qPCR system (Corbett Research). Real-time PCR was also performed on samples directly after the shearing step (input samples), using 1/2,500 of each sample, and values obtained for ChIP samples were normalized to those for input samples with the same primer sets. The primers for amplification of the DS element and the BZLF1 promoter region are as described by Deng et al. (13), while primers for the FR region correspond to oligonucleotides SC3F and SC3B of Schepers et al. (56). In experiments involving silencing of EBNA1, D98/Raji and AGS-rEBV cells (in 6-cm dishes) were subjected to three rounds of transfection with 100 pmol of siRNA against EBNA1, and then ChIP assays were performed as described above.
RESULTS
EBNA1 interacts with NAP1, NAP2, and TAF-I in EBV-infected cells.
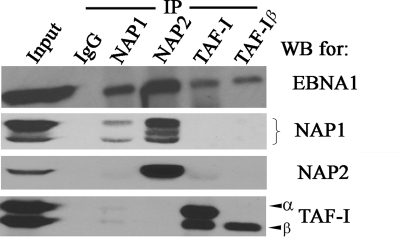
We have previously shown that related nucleosome assembly proteins, NAP1 and TAF-I (both α and β subunits), can interact with EBNA1, as they were isolated from HeLa cell lysates on EBNA1 affinity columns. To determine if these interactions occur in EBV-infected cells, coimmunoprecipitation experiments were performed on endogenous proteins in EBV-positive Raji Burkitt's lymphoma cells. Immunoprecipitation was performed with equal amounts of Raji nuclear lysates with antibodies against NAP1, TAF-I (both α and β subunits are recognized), or TAF-Iβ or with a nonspecific rabbit IgG antibody. Western blots for EBNA1 showed that it coimmunoprecipitated with all three specific antibodies but not with the nonspecific antibody, indicating that EBNA1 was able to interact specifically with these nucleosome assembly proteins under physiological conditions (Fig. 1). TAF-Iα antibody recovered both TAF-Iα and TAF-Iβ in addition to EBNA1. However, despite the fact that TAF-Iα and -β are known to heterodimerize, TAF-Iβ immunoprecipitation recovered EBNA1 and TAF-Iβ but not TAF-Iα. Failure to recover TAF-Iα is likely a property of the TAF-Iβ antibody, which reacts with the N-terminal dimerization region. The results indicate that EBNA1 can interact with the β subunit of TAF-I in the absence of the α subunit.
FIG. 1.
Coimmunoprecipitation of EBNA1 with nucleosome assembly proteins. Nucleosome assembly proteins were immunoprecipitated from Raji nuclear lysates by use of antibodies against NAP1, NAP2, TAF-I (α and β forms), or TAF-Iβ, and recovered proteins were Western blotted (WB) for EBNA1, NAP1, NAP2, or TAF-I. Immunoprecipitation was also performed with a nonspecific antibody (IgG) as a negative control. A 1/20 sample of the nuclear lysate used for immunoprecipitation is also shown (input).
NAP2 is 70% homologous to NAP1 and is ubiquitously expressed (30), prompting us to ask whether EBNA1 also interacts with NAP2. Note that the major peptides identified as NAP1 in matrix-assisted laser desorption ionization-time-of-flight analyses from the EBNA1 affinity column experiments (28) are also present in NAP2, and additional peptides were recovered that match only NAP1 or NAP2 (but not both), suggesting that both NAP1 and NAP2 were present in the same band (data not shown). Immunoprecipitation with NAP2-specific antibody recovered EBNA1, although it is possible that this interaction was mediated by NAP1, since NAP1 was also recovered (Fig. 1).
EBNA1 directly interacts with NAP1 and TAF-Iβ.
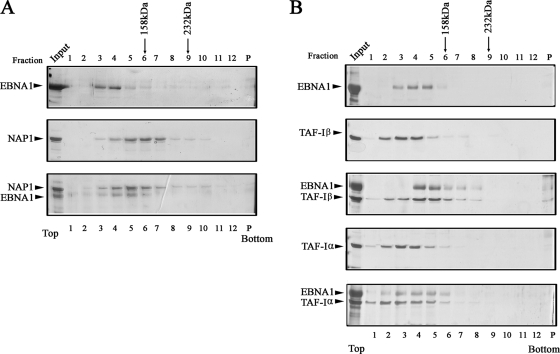
Next, we wanted to determine if the interactions that we observed between EBNA1 and NAP1, NAP2, TAF-Iα, and TAF-Iβ were direct. To this end, we purified each of the proteins and incubated EBNA1 with each nucleosome assembly protein at a 1:1 molar ratio. The complexes were then analyzed by glycerol gradient sedimentation followed by SDS-PAGE of gradient fractions. The positions of the proteins from these samples were compared to identical analyses performed on each protein alone, to identify any shifts in the gradient positions of the proteins that resulted in cosedimentation, as would occur if the proteins directly interacted. Comparison of the sedimentation of NAP1 and EBNA1, alone and together (Fig. 2A), showed little change in the position of NAP1 (the larger of the two proteins), which migrated as a 150-kDa species. Since a NAP1 monomer is 58 kDa, its migration suggests that NAP1 interacts with itself, and this is consistent with previous reports of NAP-1 dimerization (41). However, in the presence of NAP1, EBNA1 shifted from a peak at gradient fractions 3 and 4 (a position consistent with its dimeric nature) (19) to a peak at fraction 5. Importantly, this EBNA1 shift resulted in cosedimentation with NAP1, indicating a direct interaction between the two proteins.
FIG. 2.
Glycerol gradient sedimentation analysis of EBNA1-nucleosome assembly protein complexes. Equal molar ratios of EBNA1 and NAP1 (A) or EBNA1 and TAF-Iβ or TAF-Iα (B) were preincubated, analyzed on a glycerol gradient, and compared to the same proteins analyzed individually. Equal-volume fractions were collected from the top of each gradient and analyzed by SDS-PAGE and silver staining, and the pellet at the bottom of the tube (P) is also shown. Sedimentation positions are shown (at the top) for the molecular mass markers aldolase (158 kDa) and catalase (232 kDa), which were analyzed on an identical gradient. A sample of the protein(s) loaded on the gradients is also shown (input).
A similar analysis of EBNA1 with TAF-Iβ (Fig. 2B) showed a shift in the sedimentation position of TAF-Iβ to that of the larger EBNA1 protein upon incubation with EBNA1. Thus, like NAP1, TAF-Iβ was able to bind directly to EBNA1. In contrast, the sedimentation profiles of TAF-Iα and EBNA1 did not change when they were coincubated, suggesting that TAF-Iα does not stably bind to EBNA1 and that any interaction of EBNA1 with TAF-I heterodimers occurs through TAF-Iβ.
We also performed glycerol gradient assays with NAP2. NAP2 on its own was found to migrate in the glycerol gradient at the same position as EBNA1 alone, and the migration of these two proteins did not change when they were combined. Since the two proteins comigrate even when not combined, we could not make conclusions about whether or not they were interacting by using this method (data not shown).
NAP-1 and TAF-Iβ affect EBNA1 solubility.
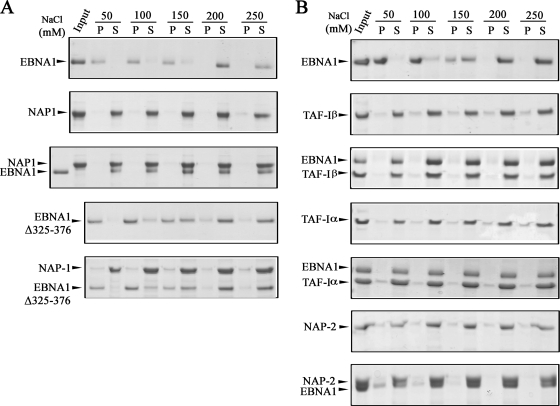
EBNA1 is a highly basic protein that is known to require relatively high salt concentrations to remain soluble in vitro (19). During the course of our in vitro studies, we noticed an effect of the nucleosome assembly proteins on the solubility of EBNA1, which was then examined in more detail. To this end, purified EBNA1 was incubated on its own or at a 1:1 molar ratio with purified NAP1, NAP2, TAF-Iα, or TAF-Iβ, with various salt concentrations. Soluble and insoluble proteins were then separated by centrifugation, and the entire soluble and pellet fractions were analyzed by SDS-PAGE and Coomassie staining (Fig. 3). EBNA1 alone was insoluble at NaCl concentrations of 150 mM or less and became soluble at 200 mM NaCl, resulting in a shift from the pellet to the supernatant fraction (Fig. 3A and B, top panels). This is in contrast to the solubility profiles of NAP1, TAF-Iα, and TAF-Iβ, each of which was largely soluble at all salt concentrations tested (50 to 250 mM). The solubility profile of EBNA1 changed significantly when it was incubated with NAP1 (Fig. 3A, third panel), TAF-Iβ, or TAF-Iα (Fig. 3B, third and fifth panels), in that EBNA1 became soluble at salt concentrations as low as 50 mM and fractionated with NAP1, TAF-Iα, and TAF-Iβ in the supernatant. Therefore, these results provide additional evidence that EBNA1 interacts directly with these nucleosome assembly proteins and show that these interactions can alter the properties of EBNA1.
FIG. 3.
Effects of nucleosome assembly proteins on EBNA1 solubility. (A) EBNA1 and NAP1 were incubated alone or together (at an equal molar ratio) in buffer containing 50 to 250 mM NaCl. Soluble (S) and precipitated (P) proteins were then separated by centrifugation and analyzed by SDS-PAGE and Coomassie blue staining. A sample of the protein(s) prior to incubation is shown as “input.” The same experiment was also performed with an EBNA1 mutant lacking amino acids 325 to 376 (EBNA1Δ325-376). (B) Same experiment as in panel A, except that TAF-Iβ, TAF-Iα, or NAP2 was used in place of NAP1.
In comparison to the other nucleosome assembly proteins, NAP2 had only a partial effect on EBNA1 solubility (Fig. 3B, bottom panel). NAP2 itself was soluble at all the salt concentrations tested and caused ∼50% of the EBNA1 to shift to the soluble fraction in 50 mM salt and ∼75% of EBNA1 to be soluble in 100 mM salt. This suggests a weaker interaction of EBNA1 with NAP2 than with NAP1.
We previously observed that less NAP1 was recovered from an EBNA1 affinity column containing the Δ325-376 EBNA1 mutant, which lacks the large Gly-Arg-rich region, than from a wild-type EBNA1 column, suggesting that this region is important for NAP1 binding (28). We further examined the importance of the 325-376 region for the NAP1 interaction by repeating the solubility assays with EBNA1Δ325-376. EBNA1Δ325-376 alone had a solubility profile similar to that of wild-type EBNA1 (Fig. 3A, fourth panel). However, unlike the case for wild-type EBNA1, the solubility profile of EBNA1Δ325-376 did not change when it was incubated with NAP1, with the protein remaining insoluble at 50 and 100 mM NaCl (Fig. 3A, bottom panel). These results confirm that the 325-376 region is important for EBNA1 binding to NAP1.
Nucleosome assembly proteins contribute to EBNA1-mediated transcriptional activation.
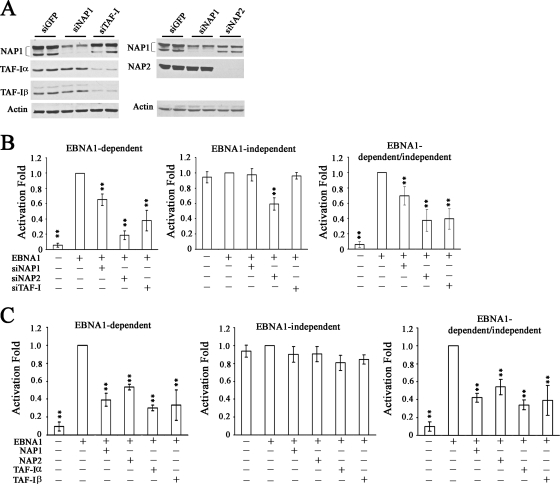
Interactions of EBNA1 with the EBV oriP DS and FR sequences are known to activate DNA replication and to enhance transcription of other EBV latency genes, respectively (52, 77). Since both of these processes are likely to be regulated by nucleosome positioning and histone modifications, we examined whether NAP1, NAP2, and TAF-I affected these EBNA1 functions. We first tested the effects of silencing these proteins on transcriptional activation by EBNA1, using a standard reporter assay in which expression of a CAT gene is under the control of the oriP FR transcriptional element (52). Assays were performed in an EBV-negative nasopharyngeal carcinoma cell line, CNE2Z, which is a physiologically relevant background for EBNA1, since development of this carcinoma is closely tied to EBV latent infection. siRNAs specific to NAP1, NAP2, and a region of TAF-I conserved in both the α and β subunits were used to downregulate these proteins individually prior to cotransfection of the cells with the FR-CAT reporter plasmid, a SEAP reporter plasmid that is not regulated by EBNA1, and an EBNA1 expression plasmid. As shown in Fig. 4A, introduction of the siRNAs decreased the endogenous expression of the target protein(s) compared to that with the control siRNA against GFP. As expected, the TAF-I siRNA decreased the expression of both TAF-Iα and TAF-Iβ without affecting NAP1, and NAP1 siRNA decreased NAP1 without affecting TAF-I or NAP2. However, NAP2 siRNA partially decreased NAP1 in addition to dramatically decreasing NAP2 levels.
FIG. 4.
Effects of nucleosome assembly proteins on transcriptional activation by EBNA1. (A) Western blots of extracts of CNE2Z cells after transfection with siRNA against GFP (negative control), NAP1, NAP2, or TAF-I. Duplicate samples are shown. (B) After transfection with the indicated siRNA (or with siGFP in the second columns), cells were transfected with an EBNA1 expression plasmid or an empty plasmid (first column in each histogram), an FR-CAT reporter plasmid that is EBNA1 dependent, and a SEAP reporter plasmid that is independent of EBNA1. Effects on CAT expression (left) and SEAP expression (middle) were determined separately. CAT levels were also normalized to SEAP levels to account for any nonspecific transcriptional effects (right). **, P < 0.001 relative to the EBNA1 positive control. (C) CNE2Z cells were transfected with a plasmid overexpressing NAP1, NAP2, TAF-Iα, or TAF-Iβ or with empty plasmid (second columns) prior to transfection with EBNA1 expression, CAT reporter, and SEAP reporter plasmids and calculation of transcriptional activities as in panel B.
Transcriptional assays were performed on the cells treated with each of the specific siRNAs and the results compared to those for cells treated with siGFP. EBNA1-mediated transcription was measured by assaying for CAT, while transcriptional effects that are independent of EBNA1 were determined by SEAP assays. We consistently found that EBNA1-mediated transcriptional activity was significantly reduced when NAP1 expression was decreased (P < 0.01) and more dramatically reduced upon downregulation of NAP-2 or TAF-I expression (Fig. 4B, left panel) (P < 0.01), perhaps due to the better silencing of these proteins than that of NAP1. NAP1 and TAF-I downregulation did not significantly affect the expression of SEAP, showing that the effects were specific for EBNA1-mediated transcriptional activation (Fig. 4B, middle panel). In contrast, NAP2 silencing did significantly decrease EBNA1-independent SEAP expression, although not as dramatically as EBNA1-dependent transcription. The EBNA1-dependent results were then normalized to the EBNA1-independent results to account for any general effects of silencing the nucleosome assembly proteins (Fig. 4B, right panel). We concluded that NAP1, TAF-I, and possibly NAP2 can positively contribute to transcriptional activation by EBNA1.
We also examined the effect of overexpressing NAP1, NAP2, TAF-Iα, or TAF-Iβ on EBNA1-mediated transcriptional activation by including a plasmid expressing each myc-tagged nucleosome assembly protein (or myc tag alone) along with the reporter and EBNA1 expression plasmids. The results from three independent experiments with duplicate samples showed that EBNA1-dependent transcriptional activation was two- to threefold lower when cells overexpressed NAP1, NAP2, TAF-Iα, or TAF-I β than when they expressed the myc tag alone (Fig. 4C, left panel) (P < 0.01). However, overexpression of these proteins did not affect expression of SEAP from the EBNA1-independent SEAP reporter (Fig. 4C, middle panel), showing that the effect has specificity for EBNA1. These results further support a role for the nucleosome assembly proteins in contributing to EBNA1-mediated transcriptional activation and indicate that the level of these proteins is important for regulation of this EBNA1 function.
TAF-I contributes to EBNA1-mediated replication.
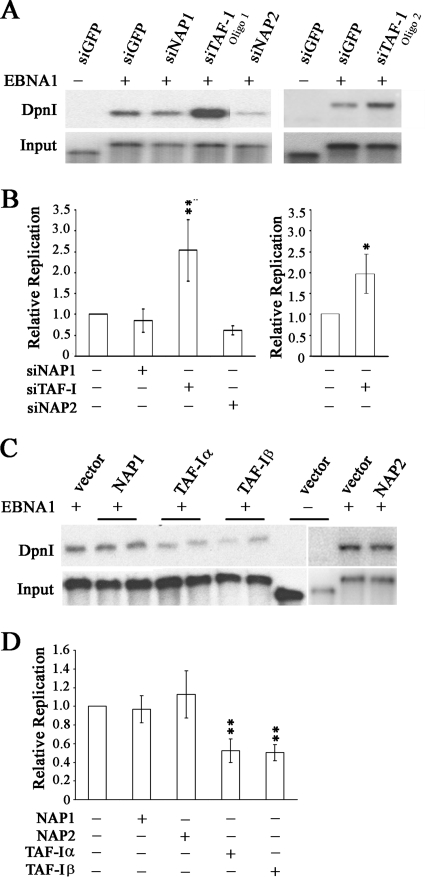
To investigate the roles of NAP-1, NAP-2, TAF-Iα, and TAF-Iβ in EBNA1 replication activity, we performed transient replication assays in CNE2Z cells after downregulation of the nucleosome assembly proteins with siRNA treatment or with the control GFP siRNA. The replication of an oriP plasmid expressing EBNA1 (or without EBNA1 as a negative control) was then assayed at 3 days posttransfection by DpnI resistance assays, in which the level of the DpnI-resistant plasmid (reflecting replication in the mammalian cells) was compared to the total amount of plasmid recovered from the cells. As shown in Fig. 5A, silencing of TAF-I caused a pronounced increase in EBNA1-mediated plasmid replication, while silencing of NAP1 or NAP2 had less obvious effects. Composite results from multiple experiments confirmed that TAF-I silencing significantly stimulated EBNA1 replication activity (P < 0.01) but that NAP1 and NAP2 silencing had no significant effect (Fig. 5B). To verify that the stimulation of DNA replication seen after TAF-I silencing was not due to off-target effects of the siRNA, we repeated the experiments using a second siRNA sequence against TAF-Iβ (oligo 2). As shown in the right panels of Fig. 5A and B, this siRNA had similar effects on the replication of oriP plasmids to those with the initial TAF-I siRNA, confirming that the effects were due to the loss of TAF-I.
FIG. 5.
Effects of nucleosome assembly proteins on DNA replication activity of EBNA1. (A) CNE2Z cells were treated with siRNA against NAP1, NAP2, TAF-I, or GFP (negative control) and then transfected with an oriP plasmid expressing EBNA1 or lacking EBNA1 (first lane only). For TAF-I, two different siRNA sequences were used (oligo 1 and oligo 2). Three days later, oriP plasmids were harvested from the cells and linearized. One-tenth of the sample was saved as a recovery control (input), and then the remaining sample was digested with DpnI to digest any transfected plasmid that had not replicated in the human cells. Samples were analyzed by Southern blotting and probing for the oriP plasmid. The DpnI-resistant plasmid band is shown in the top panel. (B) Quantification of multiple experiments performed as in panel A. For TAF-I, the left panel is for oligo 1 and the right panel is for oligo 2. Values are shown relative to that for the sample with EBNA1 and siGFP within each experiment, which was set to 1.0. P values of <0.01 (**) and <0.05 (*) are indicated. (C) CNE2Z cells were transfected with plasmids overexpressing NAP1, NAP2, TAF-Iα, or TAF-Iβ or with empty plasmid (vector) along with the oriP plasmid expressing EBNA1 or lacking EBNA1. Replication of the oriP plasmids was then determined as in panel A. (D) Quantification of multiple experiments performed as in panel C. Values are shown relative to that for EBNA1 with an empty overexpression plasmid within each experiment, which was set to 1.0, and P values of <0.01 are indicated.
We further examined the contributions of NAP1, NAP2, TAF-Iα, and TAF-Iβ to EBNA1-mediated replication by conducting transient replication assays in the presence of plasmids that overexpress the myc-tagged nucleosome assembly proteins or the myc tag alone (Fig. 5C and D). Overexpression of either TAF-Iα or TAF-Iβ consistently caused a twofold repression of EBNA1-mediated replication (P < 0.01), while overexpression of NAP1 or NAP2 did not significantly change EBNA1 replication activity. These results support the effects of RNA interference, indicating that TAF-I negatively regulates EBNA1-mediated replication but that NAP1 and NAP2 do not contribute to this process.
Association of nucleosome assembly proteins with oriP functional elements.
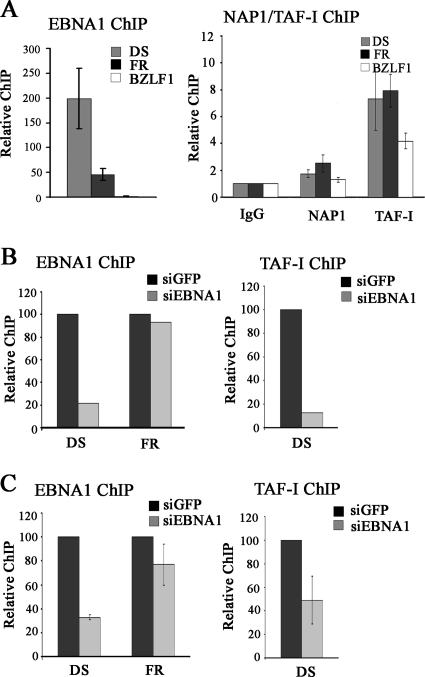
Transcriptional activation by EBNA1 requires EBNA1 binding to the FR element of oriP, while DNA replication from oriP requires EBNA1 binding to the DS element, and both of these elements are constitutively bound by EBNA1 in EBV-infected cells (10, 53). Therefore, proteins that are functioning directly with EBNA1 to modulate transcription and replication should localize with EBNA1 on the FR and DS sequences, respectively. To better understand the mechanisms by which NAP1 contributes to EBNA1-mediated transcriptional activation and TAF-I contributes to both the transcription and replication functions of EBNA1, we performed ChIP assays with Raji cells to determine whether NAP1 or TAF-I localized with EBNA1 to the FR and DS elements within the EBV episome. NAP1 and TAF-I are both expected to have general chromatin interactions in addition to being targeted to specific sites. Therefore, ChIP assays were also performed to examine association with the BZLF1 promoter region, which is not regulated by EBNA1. In all cases, the levels of the DNA fragments recovered from sheared Raji cell DNA were determined by quantitative real-time PCR.
ChIP assays conducted with EBNA1 antibody confirmed that EBNA1 was bound to the DS and FR elements but not to the BZLF1 promoter (Fig. 6A, left panel), with better recovery of the DS element, as previously reported (13, 38, 56). In contrast, NAP1 antibody preferentially recovered the FR DNA fragment over either the DS (P = 0.018) or BZLF1 (P = 0.001) fragment. TAF-I antibody (which binds both TAF-Iα and TAF-Iβ) recovered all three DNA fragments to some degree but resulted in significantly more immunoprecipitation of both the DS and FR fragments than of the BZLF1 fragment (P = 0.009 for the DS element and P = 00005 for the FR element). Therefore, TAF-I is preferentially localized with EBNA1 on both the DS and FR elements, while NAP1 is preferentially detected at the FR element. These results support the functional assays indicating that TAF-I modulates EBNA1 functions at both the FR and DS elements and that NAP1 contributes to EBNA1 function only at the FR element.
FIG. 6.
Localization of NAP1 and TAF-I to oriP elements by ChIP. (A) ChIP experiments were performed with Raji cells, using antibodies against EBNA1 (left), NAP1, TAF-I, or nonspecific rabbit IgG (right). Recovered DNA fragments were quantified by real-time PCR, using primer sets for the oriP DS and FR elements and the BZLF1 promoter region. The amplification signals were normalized to those from the same cell lysates prior to IP, using the same primer pairs. Signals from NAP1 and TAF-I antibody samples were expressed relative to that for the control IgG samples, which was set to 1. The results shown are from three independent experiments, with PCR quantification performed in triplicate for each experiment. (B and C) D98/Raji (B) and AGS-rEBV (C) cells were treated with siRNA against EBNA1 or GFP, and ChIP assays were performed for EBNA1 (left) and TAF-I (right) as in panel A.
We also investigated the requirement of EBNA1 for the recruitment of NAP1 and TAF-I to oriP elements by downregulating EBNA1 in EBV-positive cells, using siRNA treatment. These experiments could not be performed with Raji cells due to their low transfection efficiency and instead were performed with D98/Raji fusion cells and AGS-rEBV gastric carcinoma cells, both of which contain EBV. In both cases, downregulation of the cellular levels of EBNA1 resulted in decreased EBNA1 binding to the DS element, as determined by ChIP (Fig. 6B and C, left panels). However, EBNA1 silencing did not significantly decrease the level of EBNA1 bound to the FR element (Fig. 6B and C, left panels), which is known to contain higher-affinity EBNA1 binding sites that are more stably bound than those in the DS element (3, 19, 32). Therefore, our analysis of the EBNA1 requirement for recruitment of nucleosome assembly proteins to oriP elements was limited to TAF-I recruitment to the DS element. ChIPs performed using antibody against TAF-I showed that EBNA1 silencing resulted in a decreased association of TAF-I with the DS element in both D98/Raji and AGS-rEBV cells, consistent with EBNA1-mediated recruitment of TAF-I (Fig. 6B and C, right panels).
DISCUSSION
We have previously shown that three nucleosome assembly proteins, NAP1, TAF-Iα, and TAF-Iβ, are recovered from cell extracts on EBNA1 affinity columns (28). We have now confirmed the interactions of EBNA1 with NAP1 and TAF-Iβ by glycerol gradient sedimentation and, in the process, have shown that these interactions are direct. NAP1 and TAF-Iβ also altered the biochemical properties of EBNA1 in vitro, such that its solubility under low-salt conditions was dramatically increased, a property that might reflect the ability of these proteins to act as chaperones for EBNA1 in vivo. An interaction between EBNA1 and TAF-Iα was not detected by glycerol gradient sedimentation, but like NAP1 and TAF-Iβ, TAF-Iα did increase the solubility of EBNA1 in vitro, indicating an interaction with EBNA1. Since gradient sedimentation analyses would require a more stable interaction than the solubility assays, the results indicate that EBNA1 can interact to some degree with either TAF-Iα or TAF-Iβ but interacts more stably with TAF-Iβ. This is consistent with our previous affinity column experiments that identified TAF-Iβ as interacting more specifically with EBNA1 than TAF-Iα does. Since it is known that TAF-Iα and TAF-Iβ can heterodimerize (40, 42), the retention of TAF-Iα on EBNA1 affinity columns could have been indirect, mediated by TAF-Iβ. In support of this model, coimmunoprecipitation assays showed that EBNA1 could be isolated in a complex with TAF-Iβ that did not contain TAF-Iα, further pointing to TAF-Iβ as the more important TAF-I interaction.
Since NAP2 is a close homologue of NAP1 that is found in the same cells, we examined possible interactions of EBNA1 with NAP2. NAP2 antibody immunoprecipitated EBNA1 from Raji cells, but NAP1 was also recovered. Since NAP1 and NAP2 are known to interact (60), it is not clear whether the recovered EBNA1 was bound to NAP2 or to NAP1. On the other hand, NAP1 antibody recovered EBNA1 in the absence of NAP2. The results of glycerol gradient sedimentation with EBNA1 and NAP2 were ambiguous, since both proteins comigrated even when analyzed individually. However, NAP2 did increase the solubility of EBNA1 in vitro, although to a lesser degree than that with NAP1. Taken together, the results suggest that the more prominent interaction of EBNA1 with the NAPs is through NAP1.
The fact that EBNA1 interacts with both NAP1 and TAF-I suggests that the interaction occurs through a region conserved in the two proteins. These proteins share a NAP domain and C-terminal acidic sequences, either of which might mediate the EBNA1 interaction. NAP1 also contains a long N-terminal sequence, but this does not appear to be important for the EBNA1 interaction, since an N-terminally truncated form of NAP1 that is present in human cell extracts is retained on EBNA1 affinity columns along with full-length NAP1 (28). In addition, TAF-Iβ lacks the N-terminal region present in NAP1 and TAF-Iα contains only a very short N-terminal sequence. The observation that TAF-Iα interacts less stably with EBNA1 than TAF-Iβ does suggests that this N-terminal sequence is inhibitory to binding, perhaps due to steric hindrance of the adjacent NAP domain.
Downregulation of NAP1, NAP2, and TAF-I (both α and β subunits) resulted in decreased transcriptional activation by EBNA1. Silencing of NAP1 and TAF-I occurred specifically without affecting the levels of the other nucleosome assembly proteins and did not affect the transcription of a reporter gene that was not under EBNA1 control. Therefore, NAP1 and TAF-I appear to contribute directly to EBNA1-mediated transcriptional activation, and in keeping with these results, both proteins were preferentially detected at the EBNA1-controlled FR transcriptional element. Due to technical limitations, we have not been able to determine whether the recruitment of NAP1 and TAF-Iβ requires EBNA1, but this is the most likely explanation. The results for NAP2 are more complicated to interpret because (i) NAP2 silencing also decreased NAP1 levels and (ii) NAP2 silencing also inhibited transcription of the EBNA1-independent reporter, although not as dramatically as that of the EBNA1-dependent reporter. Therefore, although NAP2 could contribute directly to the transcriptional activity of EBNA1, it could also act indirectly through NAP1 and through general effects on chromatin structure. Note that while the degree to which downregulation of NAP1, NAP2, and TAF-I inhibited EBNA1-mediated transcription was only 1.5- to 3-fold, this is what is typically seen for effects of NAP1 and TAF-I on specific reporter genes under the control of their binding partners (65, 69).
Overexpression of any of the nucleosome assembly proteins was found to specifically inhibit EBNA1-dependent transcription. The fact that both silencing and overexpression of these proteins inhibited EBNA1-dependent transcription suggests that a squelching mechanism is at play, in which the formation of a particular complex containing the nucleosome assembly proteins is disrupted by either treatment. Similar observations were made with Xenopus NAP1, as overexpressing and silencing this protein both resulted in similar embryonic defects (1, 20). NAP1 is known to multimerize, forming complexes ranging from dimers to hexadecamers, in a concentration-dependent manner, where the largest complexes are impaired for histone binding (67). Therefore, altering the level of NAP1 in the cell may affect the nature of the NAP1 complexes formed, and hence their ability to interact with histones. In addition, nucleosome assembly proteins are known to mediate interactions between the p300 coactivator and specific transcription factors, which are important for their stimulatory effects on these transcription factors (51, 60, 69). Both overexpression and silencing of the nucleosome assembly proteins would be expected to inhibit such ternary interactions and therefore could account for the effects we observed on EBNA1-mediated transcriptional activation.
While NAP1 and TAF-I had similar effects on the transcriptional activity of EBNA1, only TAF-I had an obvious effect on EBNA1-mediated DNA replication, and in keeping with this observation, only TAF-I was specifically detected at the DS replication element. TAF-I downregulation stimulated DNA replication while overexpression inhibited it, indicating a regulatory role for TAF-I in EBNA1-mediated replication. In addition to its interactions with p300 and related histone acetylases, TAF-I forms part of the INHAT complex and inhibits acetylation of histones and, in some cases, other DNA-binding transcriptional activators (37, 43, 57). Histone acetylation has long been linked to transcriptional activation and is also known to influence the initiation of DNA replication from specific origins (11, 49, 70, 79). In addition, the cell cycle-dependent acetylation of histone H3 at oriP has been reported (80) and has been shown to control the time in S phase at which replication initiates from oriP (81). Therefore, the inhibitory effect of TAF-I on replication from oriP is likely to involve effects on histone acetylation.
An increasing number of examples suggest that viral proteins often usurp cellular nucleosome assembly proteins for their own purposes. For example, TAF-I, which was originally identified as a factor stimulating the replication and transcription of adenovirus core particles in vitro (40, 46), was recently shown to interact with the adenovirus core protein VII and to affect adenovirus early gene transcription in vivo (25). In addition, the herpes simplex virus type 1 tegument protein VP22 was found to bind TAF-I and to inhibit its chromatin assembly activity in vitro (68). NAP1 interacts with the E2 protein of papillomavirus (51) and the Tat protein of human immunodeficiency virus type 1 (HIV-1) (69), in both cases activating transcription from viral elements bound by these proteins. In addition, NAP1 was found to be utilized by human T-cell leukemia virus type 1 in promoting transcription-independent nucleosome disruption at the viral promoter (58). Finally, NAP1 can form a complex with HIV-1 Rev, which stimulates Rev's ability to export HIV RNA (9). We have now shown that the EBNA1 protein of EBV uses NAP1 and TAF-I in the activation of viral gene expression and uses TAF-I to regulate replication from oriP, indicating that the nucleosome assembly proteins also play important roles in EBV latent infection.
Acknowledgments
We gratefully acknowledge Pedro Rodriguez for NAP2 antibodies and the pET15b-NAP2 expression construct, Yukio Ishimi for NAP1 monoclonal antibody, and Lawrence S. Young for AGS-rEBV cells. We also thank Jerry Pelletier for pET15b-NAP1, K. Nagata for pET14b-TAF-Iα, and Alan Cochrane for the SEAP reporter plasmid. Finally, we thank Nirojini Sivachandran for assistance with the revisions.
This work was funded by the Canadian Cancer Society. L.F. is a tier 1 Canada Research Chair in Molecular Virology.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Abu-Daya, A., W. M. Steer, A. F. Trollope, C. E. Friedeberg, R. K. Patient, A. W. Thorne, and M. J. Guille. 2005. Zygotic nucleosome assembly protein-like 1 has a specific, non-cell autonomous role in hematopoiesis. Blood 106:514-520. [DOI] [PubMed] [Google Scholar]
- 2.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambinder, R. F., W. A. Shah, D. R. Rawlins, G. S. Hayward, and S. D. Hayward. 1990. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J. Virol. 64:2369-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atanasiu, C., Z. Deng, A. Wiedmer, J. Norseen, and P. M. Lieberman. 2006. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 7:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avolio-Hunter, T. M., P. N. Lewis, and L. Frappier. 2001. Epstein-Barr nuclear antigen 1 binds and destabilizes nucleosomes at the viral origin of latent DNA replication. Nucleic Acids Res. 29:3520-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochkareva, E., L. Frappier, A. M. Edwards, and A. Bochkarev. 1998. The RPA32 subunit of human replication protein A contains a single-stranded DNA binding domain. J. Biol. Chem. 273:3932-3936. [DOI] [PubMed] [Google Scholar]
- 7.Ceccarelli, D. F., and L. Frappier. 2000. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J. Virol. 74:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochrane, A., L. L. Murley, M. Gao, R. Wong, K. Clayton, N. Brufatto, V. Canadien, D. Mamelak, T. Chen, D. Richards, M. Zeghouf, J. Greenblatt, C. Burks, and L. Frappier. 2009. Stable complex formation between HIV Rev and the nucleosome assembly protein, NAP1, affects Rev function. Virology 388:103-111. [DOI] [PubMed] [Google Scholar]
- 10.Daikoku, T., A. Kudoh, M. Fujita, Y. Sugaya, H. Isomura, and T. Tsurumi. 2004. In vivo dynamics of EBNA1-oriP interaction during latent and lytic replication of Epstein-Barr virus. J. Biol. Chem. 279:54817-54825. [DOI] [PubMed] [Google Scholar]
- 11.Danis, E., K. Brodolin, S. Menut, D. Maiorano, C. Girard-Reydet, and M. Mechali. 2004. Specification of a DNA replication origin by a transcription complex. Nat. Cell Biol. 6:721-730. [DOI] [PubMed] [Google Scholar]
- 12.Deng, Z., C. Atanasiu, J. S. Burg, D. Broccoli, and P. M. Lieberman. 2003. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J. Virol. 77:11992-12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, Z., C. Atanasiu, K. Zhao, R. Marmorstein, J. I. Sbodio, N. W. Chi, and P. M. Lieberman. 2005. Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1. J. Virol. 79:4640-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 15.Dyson, P. J., and P. J. Farrell. 1985. Chromatin structure of Epstein-Barr virus. J. Gen. Virol. 66:1931-1940. [DOI] [PubMed] [Google Scholar]
- 16.Frappier, L. 2004. Viral plasmids in mammalian cells, p. 325-339. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 17.Frappier, L., and M. O'Donnell. 1992. EBNA1 distorts oriP, the Epstein-Barr virus latent replication origin. J. Virol. 66:1786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frappier, L., and M. O'Donnell. 1991. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 88:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frappier, L., and M. O'Donnell. 1991. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J. Biol. Chem. 266:7819-7826. [PubMed] [Google Scholar]
- 20.Friedeberg, C., G. Scarlett, J. McGeehan, A. Abu-Daya, M. Guille, and G. Kneale. 2006. Identification of a structural and functional domain in xNAP1 involved in protein-protein interactions. Nucleic Acids Res. 34:4893-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gahn, T. A., and C. L. Schildkraut. 1989. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell 58:527-535. [DOI] [PubMed] [Google Scholar]
- 22.Gahn, T. A., and B. Sugden. 1995. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J. Virol. 69:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glaser, R., and M. Nonoyama. 1974. Host cell regulation of induction of Epstein-Barr virus. J. Virol. 14:174-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison, S., K. Fisenne, and J. Hearing. 1994. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 68:1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haruki, H., M. Okuwaki, M. Miyagishi, K. Taira, and K. Nagata. 2006. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 80:794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 27.Holowaty, M. N., Y. Sheng, T. Nguyen, C. Arrowsmith, and L. Frappier. 2003. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 278:47753-47761. [DOI] [PubMed] [Google Scholar]
- 28.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987-29994. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh, D.-J., S. M. Camiolo, and J. L. Yates. 1993. Constitutive binding of EBNA1 protein to the Epstein-Barr virus replication origin, oriP, with distortion of DNA structure during latent infection. EMBO J. 12:4933-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu, R. J., M. P. Lee, L. A. Johnson, and A. P. Feinberg. 1996. A novel human homologue of yeast nucleosome assembly protein, 65 kb centromeric to the p57KIP2 gene, is biallelically expressed in fetal and adult tissues. Hum. Mol. Genet. 5:1743-1748. [DOI] [PubMed] [Google Scholar]
- 31.Huang, D. P., J. H. Ho, Y. F. Poon, E. C. Chew, D. Saw, M. Lui, C. L. Li, L. S. Mak, S. H. Lai, and W. H. Lau. 1980. Establishment of a cell line (NPC/HK1) from a differentiated squamous carcinoma of the nasopharynx. Int. J. Cancer 26:127-132. [DOI] [PubMed] [Google Scholar]
- 32.Jones, C. H., S. D. Hayward, and D. R. Rawlins. 1989. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J. Virol. 63:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julien, M. D., Z. Polonskaya, and J. Hearing. 2004. Protein and sequence requirements for the recruitment of the human origin recognition complex to the latent cycle origin of DNA replication of Epstein-Barr virus oriP. Virology 326:317-328. [DOI] [PubMed] [Google Scholar]
- 34.Karetsou, Z., G. Martic, G. Sflomos, and T. Papamarcaki. 2005. The histone chaperone SET/TAF-Ibeta interacts functionally with the CREB-binding protein. Biochem. Biophys. Res. Commun. 335:322-327. [DOI] [PubMed] [Google Scholar]
- 35.Kawase, H., M. Okuwaki, M. Miyaji, R. Ohba, H. Handa, Y. Ishimi, T. Fujii-Nakata, A. Kikuchi, and K. Nagata. 1996. NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells 1:1045-1056. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy, G., and B. Sugden. 2003. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 23:6901-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutney, S. N., R. Hong, T. Macfarlan, and D. Chakravarti. 2004. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression. J. Biol. Chem. 279:30850-30855. [DOI] [PubMed] [Google Scholar]
- 38.Lin, A., S. Wang, T. Nguyen, K. Shire, and L. Frappier. 2008. The EBNA1 protein of Epstein-Barr virus functionally interacts with Brd4. J. Virol. 82:12009-12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lupton, S., and A. J. Levine. 1985. Mapping of genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 5:2533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto, K., K. Nagata, M. Ui, and F. Hanaoka. 1993. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 268:10582-10587. [PubMed] [Google Scholar]
- 41.McBryant, S. J., and O. B. Peersen. 2004. Self-association of the yeast nucleosome assembly protein 1. Biochemistry 43:10592-10599. [DOI] [PubMed] [Google Scholar]
- 42.Miyaji-Yamaguchi, M., M. Okuwaki, and K. Nagata. 1999. Coiled-coil structure-mediated dimerization of template activating factor-I is critical for its chromatin remodeling activity. J. Mol. Biol. 290:547-557. [DOI] [PubMed] [Google Scholar]
- 43.Miyamoto, S., T. Suzuki, S. Muto, K. Aizawa, A. Kimura, Y. Mizuno, T. Nagino, Y. Imai, N. Adachi, M. Horikoshi, and R. Nagai. 2003. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol. Cell. Biol. 23:8528-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosammaparast, N., C. S. Ewart, and L. F. Pemberton. 2002. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21:6527-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muto, S., M. Senda, Y. Akai, L. Sato, T. Suzuki, R. Nagai, T. Senda, and M. Horikoshi. 2007. Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc. Natl. Acad. Sci. USA 104:4285-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagata, K., H. Kawase, H. Handa, K. Yano, M. Yamasaki, Y. Ishimi, A. Okuda, A. Kikuchi, and K. Matsumoto. 1995. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. USA 92:4279-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norseen, J., A. Thomae, V. Sridharan, A. Aiyar, A. Schepers, and P. M. Lieberman. 2008. RNA-dependent recruitment of the origin recognition complex. EMBO J. 27:3024-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okuwaki, M., K. Kato, H. Shimahara, S. Tate, and K. Nagata. 2005. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell. Biol. 25:10639-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pappas, D. L., Jr., R. Frisch, and M. Weinreich. 2004. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 18:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, Y. J., and K. Luger. 2006. Structure and function of nucleosome assembly proteins. Biochem. Cell Biol. 84:549-558. [DOI] [PubMed] [Google Scholar]
- 51.Rehtanz, M., H.-M. Schmidt, U. Warthorst, and G. Steger. 2004. Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol. Cell. Biol. 24:2153-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reisman, D., and B. Sugden. 1986. trans-Activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol. Cell. Biol. 6:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971-3984. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez, P., D. Munroe, D. Prawitt, L. L. Chu, E. Bric, J. Kim, L. H. Reid, C. Davies, H. Nakagama, R. Loebbert, A. Winterpacht, M. J. Petruzzi, M. J. Higgins, N. Nowak, G. Evans, T. Shows, B. E. Weissman, B. Zabel, D. E. Housman, and J. Pelletier. 1997. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics 44:253-265. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez, P., J. Pelletier, G. B. Price, and M. Zannis-Hadjopoulos. 2000. NAP-2: histone chaperone function and phosphorylation state through the cell cycle. J. Mol. Biol. 298:225-238. [DOI] [PubMed] [Google Scholar]
- 56.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 58.Sharma, N., and J. K. Nyborg. 2008. The coactivators CBP/p300 and the histone chaperone NAP1 promote transcription-independent nucleosome eviction at the HTLV-1 promoter. Proc. Natl. Acad. Sci. USA 105:7959-7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw, J., L. Levinger, and C. Carter. 1979. Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J. Virol. 29:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shikama, N., H. M. Chan, M. Krstic-Demonacos, L. Smith, C. W. Lee, W. Cairns, and N. B. La Thangue. 2000. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 20:8933-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shire, K., P. Kapoor, K. Jiang, M. N. Hing, N. Sivachandran, T. Nguyen, and L. Frappier. 2006. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J. Virol. 80:5261-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stewart, S., C. W. Dawson, K. Takada, J. Curnow, C. A. Moody, J. W. Sixbey, and L. S. Young. 2004. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway. Proc. Natl. Acad. Sci. USA 101:15730-15735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Summers, H., J. A. Barwell, R. A. Pfuetzner, A. M. Edwards, and L. Frappier. 1996. Cooperative assembly of EBNA1 on the Epstein-Barr virus latent origin of replication. J. Virol. 70:1228-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Telese, F., P. Bruni, A. Donizetti, D. Gianni, C. D'Ambrosio, A. Scaloni, N. Zambrano, M. G. Rosenfeld, and T. Russo. 2005. Transcription regulation by the adaptor protein Fe65 and the nucleosome assembly factor SET. EMBO Rep. 6:77-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thorley-Lawson, D. A., and A. Gross. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350:1328-1337. [DOI] [PubMed] [Google Scholar]
- 67.Toth, K. F., J. Mazurkiewicz, and K. Rippe. 2005. Association states of nucleosome assembly protein 1 and its complexes with histones. J. Biol. Chem. 280:15690-15699. [DOI] [PubMed] [Google Scholar]
- 68.van Leeuwen, H., M. Okuwaki, R. Hong, D. Chakravarti, K. Nagata, and P. O'Hare. 2003. Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J. Gen. Virol. 84:2501-2510. [DOI] [PubMed] [Google Scholar]
- 69.Vardabasso, C., L. Manganaro, M. Lusic, A. Marcello, and M. Giacca. 2008. The histone chaperone protein nucleosome assembly protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology 5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogelauer, M., L. Rubbi, I. Lucas, B. J. Brewer, and M. Grunstein. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10:1223-1233. [DOI] [PubMed] [Google Scholar]
- 71.Wang, Y., J. E. Finan, J. M. Middeldorp, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18-29. [DOI] [PubMed] [Google Scholar]
- 72.Woolaway, K., K. Asai, A. Emili, and A. Cochrane. 2007. hnRNP E1 and E2 have distinct roles in modulating HIV-1 gene expression. Retrovirology 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr virus nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wysokenski, D. A., and J. L. Yates. 1989. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J. Virol. 63:2657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 74:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 78.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]
- 79.Zhou, J., C. Chau, Z. Deng, W. Stedman, and P. M. Lieberman. 2005. Epigenetic control of replication origins. Cell Cycle 4:889-892. [DOI] [PubMed] [Google Scholar]
- 80.Zhou, J., C. M. Chau, Z. Deng, R. Shiekhattar, M. P. Spindler, A. Schepers, and P. M. Lieberman. 2005. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 24:1406-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou, J., A. R. Snyder, and P. M. Lieberman. 2009. Epstein-Barr virus episome stability is coupled to a delay in replication timing. J. Virol. 83:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zlatanova, J., C. Seebart, and M. Tomschik. 2007. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 21:1294-1310. [DOI] [PubMed] [Google Scholar]