Abstract
We previously identified a novel insect picorna-like virus, termed Kakugo virus (KV), obtained from the brains of aggressive honeybee worker bees that had counterattacked giant hornets. Here we examined the tissue distribution of KV and alterations of gene expression profiles in the brains of KV-infected worker bees to analyze possible effects of KV infection on honeybee neural and physiological states. By use of in situ hybridization, KV was broadly detected in the brains of the naturally KV-infected worker bees. When inoculated experimentally into bees, KV was detected in restricted parts of the brain at the early infectious stage and was later detected in various brain regions, including the mushroom bodies, optic lobes, and ocellar nerve. KV was detected not only in the brain but also in the hypopharyngeal glands and fat bodies, indicating systemic KV infection. Next, we compared the gene expression profiles in the brains of KV-inoculated and noninoculated bees. The expression of 11 genes examined was not significantly affected in KV-infected worker bees. cDNA microarray analysis, however, identified a novel gene whose expression was induced in the periphery of the brains of KV-infected bees, which was commonly observed in naturally infected and experimentally inoculated bees. The gene encoded a novel hypothetical protein with a leucine zipper motif. A gene encoding a similar protein was found in the parasitic wasp Nasonia genome but not in other insect genomes. These findings suggest that KV infection may affect brain functions and/or physiological states in honeybees.
Virus infection is sometimes pathogenic and even lethal to the host, but it can also be inapparent, depending on the virus-host interactions. Pathogenic infection, especially lethal infection, generally attracts more attention because of its apparent clinical and/or agricultural importance, but nonpathogenic infection represents another aspect of virus-host interactions, such as symbiotic host-microbe interactions (34). We have studied virus-host interactions by using a nonpathogenic virus, the Kakugo virus (KV), identified from the honeybee.
The honeybee is an insect that is agriculturally and industrially beneficial as well as an important model insect in the field of behavioral genetics and neurophysiology. In an effort to protect the honeybee from pathogenic microbial infections, microbial pathogens of the honeybee have long been studied (2, 3, 44). To date, 20 distinct viruses are known to infect honeybees (3). KV is a novel insect picorna-like virus (Iflavirus) that we originally identified in the brains of aggressive but apparently healthy worker honeybees (Apis mellifera L.) (15, 16). In the honeybee colony, adult females differentiate into a reproductive queen and thousands of sterile worker bees (45). Some worker bees gather at the entrance of their hives and exhibit attacking behavior against their natural enemies, such as the giant hornet, to protect their colony (attacker bees). KV was identified from such worker bees that had attacked a giant hornet (Vespa mandarinia japonica) presented as a decoy, suggesting a possible relationship between KV infection and worker bee aggressive behavior. A phylogenetic analysis of KV (14) and deformed wing virus (DWV) (5) revealed that KV is closely related to but distinct from DWV, which is associated with wing deformities of adult bees (3). An epidemiological analysis suggested that in colonies where KV infection is not severe, KV infection is attacker specific, whereas in colonies where KV infection is epidemic, KV is detected in various worker populations (14). Therefore, the relationship between KV infection and worker aggressive behavior remains to be solved at present. The relationship between DWV and aggressiveness is currently under debate (35, 40).
Some microbe-honeybee interactions are suggested to influence honeybee behaviors. For example, infection by sacbrood virus or Nosema apis induces precocious foraging in worker bees (4, 42). Israeli acute paralysis virus infection may be linked to colony collapse disorder, in which most of the worker bees suddenly disappear from their colonies (9). These findings together indicate that microbe infection may influence honeybee behaviors in nonlethal infectious phases, but the possible influence of microbe infection on honeybee brain function is not well studied.
Identifying the host cells/tissues that are infected by a virus is an effective strategy for understanding the influence of the virus on its host. The results of our previous study using reverse transcription-PCR (RT-PCR) indicated that KV is detected in the head, thorax, and abdomen of worker bees (14), but the details of KV infection in honeybee cells and tissues are not yet known. Knowledge of the KV distribution in the brain and other organs may elucidate the influence of KV infection on honeybee brain functions and physiological states. In the present study, we examined KV-infected cells and tissues histologically and analyzed the changes in gene expression profile in the brains of KV-infected worker bees, using naturally and experimentally KV-infected bees. Our present study analyzed, for the first time, the distribution of a honeybee virus and its molecular effects in infected bee brains, comparing natural infection and artificial infection, providing the first clues to understanding the molecular and neural bases of the effects of virus-honeybee interactions.
MATERIALS AND METHODS
Naturally KV-infected bee samples for in situ hybridization.
European honeybee Apis mellifera L. colonies maintained at the University of Tokyo were used for this study. Worker bees parasitized by Varroa destructor were collected as candidates infected with KV. To select KV-infected worker bees, the worker bees were anesthetized on ice and the brains, thoraces, and abdomens were dissected. The brains were frozen in OCT Tissue-Tek compound (Sakura Fine Technical, Tokyo, Japan) on dry ice and then stored at −80°C for in situ hybridization. Total RNA (0.5 μg) was isolated from parts of thoraces and abdomens of the same samples by use of Trizol reagent (Invitrogen, Carlsbad, CA), treated with RNase-free DNase I (Invitrogen), and then reverse transcribed with or without Superscript III (Invitrogen), using oligo(dT) primers. Real-time PCR was performed using gene-specific primers and fluorescent probes for Kakugo RNA or honeybee cytoplasmic actin in a total of 10 μl as described previously (14). Relative Kakugo RNA content was normalized by actin mRNA contents, and then worker bees with values of more than 5 were used as KV-infected worker bees for in situ hybridization.
Virus preparation.
Homogenates of worker tissues that contained KV were prepared as follows: the whole heads of 70 attacker bees infected with KV were homogenized with 10 volumes of phosphate-buffered saline (PBS; 10 mM phosphate buffer, pH 7.4, containing 137 mM NaCl and 3 mM KCl), using a glass homogenizer. The homogenates were then centrifuged at 1,500 × g for 15 min at 4°C, and 1 μl of the resulting supernatant was injected into the heads of worker bees at the neck on the ventral side or into larvae. The thoraces of the worker bees or whole bodies of larvae were collected 9 days after inoculation and were used to prepare supernatants as described above. The presence of Kakugo RNA in the supernatants was confirmed by RT-PCR analysis, and the supernatants were stocked at −20°C and −80°C for short- and long-term storage, respectively. Aliquots of the supernatants were diluted 10-fold with PBS to be used as inocula for injection into worker bees.
Experimental KV inoculation.
Worker bees were collected randomly from inside two independent hives. The worker bees were injected in the thorax with 1 μl of either the inoculum or PBS as described previously (16, 33). Tissue homogenate prepared from uninfected forager bees or larvae was used as a control inoculum (see Fig. 5B). The inoculated bees were kept in separate cages at 35°C and fed with 50% honey-water. They were collected at different days after inoculation. For in situ hybridization, the dissected brains, hypopharyngeal glands (HGs), and abdomens were frozen in OCT Tissue-Tek compound on dry ice and then stored at −80°C. For quantitative RT-PCR (qRT-PCR) analysis, the tissues were stored at −80°C.
FIG. 5.
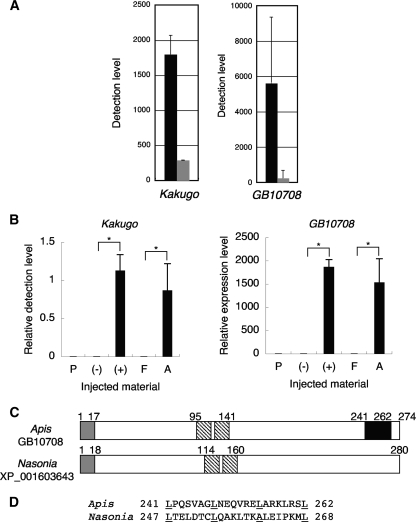
Identification of a novel gene expressed preferentially in KV-infected brains. (A) Detection levels of Kakugo and GB10708 by cDNA microarray analysis in the MBs of worker bees injected with KV (black bars) or PBS (gray bars). Data are indicated as means ± standard errors (Kakugo; n = 44) or means ± standard deviations (GB10708; n = 2). (B) qRT-PCR analysis of Kakugo and GB10708, using total RNA extracted from the MBs of individual bees injected with KV-containing (+) or non-KV-containing (−) homogenate from larvae, KV-containing homogenate from attacker bee heads (A), non-KV-containing homogenate from forager bee heads (F), or PBS (P). The amount of transcript was divided by that of actin. Relative Kakugo content was normalized to the value for one of the KV-infected bees. The GB10708 expression level was normalized to the value for one of the uninfected bees. Data are shown as means ± standard errors (n = 5 for groups P, +, and A and n = 4 for groups − and F). *, P < 0.05 by Student's t test. (C) Structure of Apis GB10708 and Nasonia XP_001603643. Open boxes, open reading frame; gray boxes, signal peptides; shaded boxes, leucine-rich repeat-like motifs; black box, leucine zipper pattern. Numbers indicate amino acid positions corresponding to each domain. (D) Comparison of amino acid sequence in the leucine zipper pattern of GB10708 with the corresponding region of Nasonia XP_001603643. Underlined amino acid residues show the positions of the consensus sequence for the leucine zipper pattern.
In situ hybridization.
In situ hybridization was performed using frozen sections (10 μm thick) as described previously (39). Digoxigenin-labeled antisense or sense RNA probes corresponding to nonstructural (positions +8389 to +8765) and structural (positions +3141 to +3588) regions of Kakugo RNA and to positions +106 to +437 of GB10708 cDNA were prepared by in vitro transcription from plasmid clones, using a DIG RNA labeling kit (Roche, Switzerland). Sections of the brains and the HGs were viewed with a BX50 microscope (Olympus, Japan). Sections of the abdomens were viewed and photographed with an Axio Imager Z1 microscope and MosaiX (Carl Zeiss, Germany).
qRT-PCR for various genes in the brain.
Worker bees were injected with homogenate containing KV or PBS. Six days after the inoculation, total RNA was isolated from the mushroom bodies (MBs) of the worker bees by use of Trizol reagent (Invitrogen). Total RNA (0.5 to 1.0 μg) was treated with RNase-free DNase I and then reverse transcribed with or without Superscript III (Invitrogen), using an oligo(dT) primer. PCR was then performed as previously described (14, 16), with Light Cycler and LightCycler-DNA master hybridization probes (Roche) and gene-specific primers and fluorescent probes (Table 1) in a total volume of 10 μl. PCR conditions, other than for the IP3R gene, were up to 40 cycles of 94°C for 1 s, 55°C for 10 s, and 72°C for 10 s. For the IP3R gene, PCR conditions were up to 40 cycles of 94°C for 1 s, 60°C for 10 s, and 72°C for 10 s. A cDNA clone of known concentration was used for each standard. For normalization, the amounts of transcripts were divided by that of actin.
TABLE 1.
Primers and probes used for qRT-PCR
| Gene | Sequence (5′-3′)a |
GenBank accession no. | |
|---|---|---|---|
| Primer | Hybridization probe | ||
| Kakugo | F, GACTGAACCAAATCCGATGTC | F-CCTTTGTCTTCATTAAAGCCACCTGG | AB070959 |
| R, TCTCAAGTTCGGGACGCATTC | L-CATCAGGTAAGCGATGGTTGTTTGA | ||
| actin | F, GAAATGGCAACTGCTGCATC | F-CCATGAAAATTAAGATCATCGCGCCAC | AB023025 |
| R, TCCACATCTGTTGGAAGGTG | L-CGAGAAGAAATATTCCGTATGGATTGGTG | ||
| pka | F, CCGATTTGAAAGACTTACTACG | F-TACTGGCGAACCACTTGTGACCCTTGATATCGTTT | AJ271674 |
| R, GCCTCGATTTTCTTTTGGA | L-CACCAGCCTTCAAATTCCCATACCTTTTGGTAAGAT | ||
| mblk-1 | F, GGCCCCTACATACAAAAGATGATCGC | F-CGTTCAAAGGCATGGAGACGCAAGATTATC | NM_001011629 |
| R, TGGGGATGCGTCGACTGGTG | L-CATTCCAGAGGTGATGAGAAGGCTGATGA | ||
| CaMKII | F, GCGAGAATTCTACAGCGAAGC | F-GCATTGAAGCACCCATGGATCTG | AB013287 |
| R, CTTCAAGCAATCCACAGTCTC | L-CAACGTGAACGTGTCGCGTCCG | ||
| IP3R | F, AGAATATCCTCTGGTGATGGA | F-AGGGTATATTTGGTAGCAGCGAGGAATG | AB006152 |
| R, CCATGTTCTTCTGATGCTTTAG | L-GTGGCGTTAGATTTAGATGGACAAGGTG | ||
| hr38 | F, TTTCCAGTCGGCCAGTCTGG | F-CTTCGCGATCACGTCACCTATAACGCCGAGG | AB253737 |
| R, TGCACCGACAGACTCCTCAG | L-GCAGAGGAAAGCGCACTACCTGTCCCGTTTG | ||
| jhdk | F, TTCTTCGATTTCAATCAGAGTGGCG | F-GAACCCATCCGAATGGCAAGAAGCAT | XM_393500 |
| R, CGGCTCGTCCGAGGAGAAATATTG | L-CATGTCTGTGACGTTCCAGTTGTTCGACG | ||
| nos | F, ATATCACTACACCACCAACGC | F-GCATCCATTGCTACAAATCCAAAAGAACAAGC | AB204558 |
| R, TGAATTTCCATCCGTAAGACC | L-CAACTCAATCTTTTAGCTTCTGATCCAGCAGTTTATGAGG | ||
| GB10708 | F, AGGAATGATTGGTACCGGTGCG | F-GAATGTGAGCAGGATTCGGAGCGGC | XM_397526 |
| R, GGATCTATTTCCGTGATCCGATTACG | L-CGTTCGCCAAGCTTTCCGAGTCGTT | ||
| ef1α | F, ATCGTGGTGATCGGCCACGT | F-GTATTCGGAGGCCCGTTTCGAGG | X52884 |
| R, GCCGTGCCATCCGGAGATCG | L-GATCAAGAAAGAGGTGTCATCGTACA | ||
| rp49 | F, GGCTATTCGTCCAGTATACAG | F-GTAAAAAACGTAAATCCATTGTTGAACG | AF441189 |
| R, GAGAACGTAACCTTGCACTGG | L-GCTCAACAACTTTCAATAAGAGTAACT | ||
| rps8 | F, AGGGATGGGTATTTCTCGCG | F-ACCATTGGGTCGTAAACGAGGTG | AF080430 |
| F, CAAATTGTTCCTCAAGAGCAG | L-GAAACTGACTGAAGCTGAAGAAGAA | ||
Forward (F) and reverse (R) primers and probes labeled with fluorescein isothiocyanate (F) and LC-Red640 (L) were used to examine the respective genes.
cDNA microarray analysis.
We previously prepared a honeybee cDNA microarray, on which 5,209 cDNA fragments for genes that are expressed in honeybee brains were printed, for several screening experiments (38, 46). Twenty-two spots corresponding to partial Kakugo cDNA and one spot corresponding to partial GB10708 cDNA were included in the cDNA microarray. For hybridization, total RNA (750 ng) extracted from the MBs of 10 worker bees was amplified using an amino-allyl MessageAmp aRNA amplification kit (Ambion, Austin, TX) and was labeled with the fluorescent dye Cy5 (Amersham Bioscience, Piscataway, NJ). To prepare control RNA for normalization, total RNA extracted from the whole brains of 36 worker bees was also amplified and labeled with Cy3. Hybridization was performed twice for KV-infected samples and twice for uninfected samples, using different lots of RNA samples. Normalization and data analysis were performed using Genespring software (Agilent Technologies, Santa Clara, CA). Intensity-dependent bias was corrected using the Lowess function (31).
RESULTS
Distribution of KV in naturally infected worker bee brains.
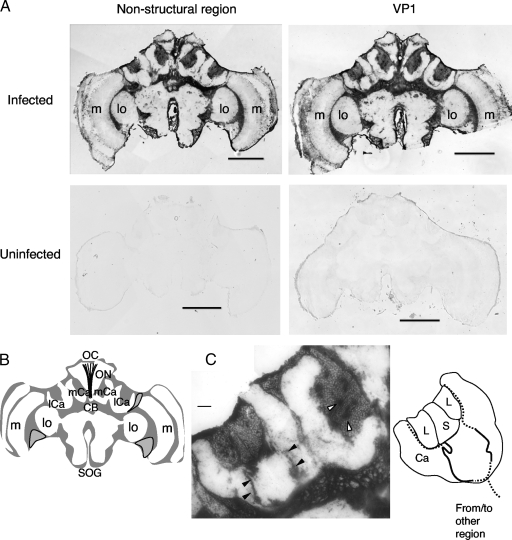
Insect brains consist of some distinct regions, such as the optic lobes (OLs), which process visual information; the antennal lobes, which process olfactory information; and the MBs and central complex, which are higher-order centers that are important for memory, learning, and sensory integration. To analyze the distribution of KV in the brains of virus-infected bees, brain sections of worker bees naturally infected with KV were analyzed by in situ hybridization using a digoxigenin-labeled RNA probe corresponding to the nonstructural regions of Kakugo RNA. Although the extent of KV infection varies between colonies, it tends to be higher in colonies parasitized by Varroa destructor, which transmits KV (14). Therefore, we randomly collected worker bees from colonies parasitized by Varroa mites and selected the KV-infected worker bees by qRT-PCR, using RNAs extracted from the thorax and abdomen. Signals were broadly detected in various brain regions, including the somata in the MBs, OLs, and subesophageal ganglion (SOG) and the ocellar nerves (n = 3) (Fig. 1A; see Fig. S1 in the supplemental material). In contrast, no signal was detected in brain sections of uninfected bees (Fig. 1A) or in those hybridized with a sense probe (data not shown). The same results were obtained using an RNA probe corresponding to the VP1 region of Kakugo RNA, indicating that the signals were due to Kakugo RNA (Fig. 1A). These results suggest that KV infects broad regions of the brain. On the other hand, KV was not detected in the somata of several parts of the brain, such as the ventral area between the medulla and OL lobula of some bees, indicating that KV does not uniformly infect all cells in the brain. In some cases, signals were also detected in some of the neuropil (a part of peduncles) in the MBs and in the central complex (Fig. 1C; see Fig. S1C to F in the supplemental material), suggesting that KV is present not only in neuronal cell bodies but also in axons.
FIG. 1.
Detection of KV in worker honeybee brain naturally infected with KV. (A) Frontal sections of KV-infected and uninfected bee brains hybridized with digoxigenin-labeled RNA antisense probes for the nonstructural or VP1 region of Kakugo. Representative data from multiple sections from three individuals are shown. (B) Schematic representation of the worker brain. Gray and white regions indicate neuronal somata and the neuropil, respectively. Outlined regions contain somata without a Kakugo RNA signal, as shown in panel A. (C) High-magnification micrograph of the left MB hybridized with the VP1 antisense probe in panel A (left) and schematic representation of the lateral calyx (right). Open and closed arrowheads indicate Kakugo signals detected as plaque-like and neuronal axon-like structures, respectively. Solid and dotted lines indicate the locations of the signals and putative axonal positions. OC, ocellus; ON, ocellar nerve; mCa, median calyx; lCa, lateral calyx; cb, central body; SOG, subesophageal ganglion; lo, lobula; m, medulla; L, large-type Kenyon cells; S, small-type Kenyon cells; Ca, calyx. Bars, 500 μm (A) and 100 μm (C).
It was not certain whether the Kakugo RNA detected by in situ hybridization represents the virion RNA and/or the replicated RNA in the cytoplasm. Dot-like signals larger than a cell body, which possibly represent plaque-like structures, were sometimes detected inside the MB calyces, where the cell bodies of the Kenyon cells are located (Fig. 1C), and in the OLs (see Fig. S1B in the supplemental material), suggesting that KV infection progresses in these areas.
Time-dependent changes in KV distribution in experimentally inoculated worker bee brains.
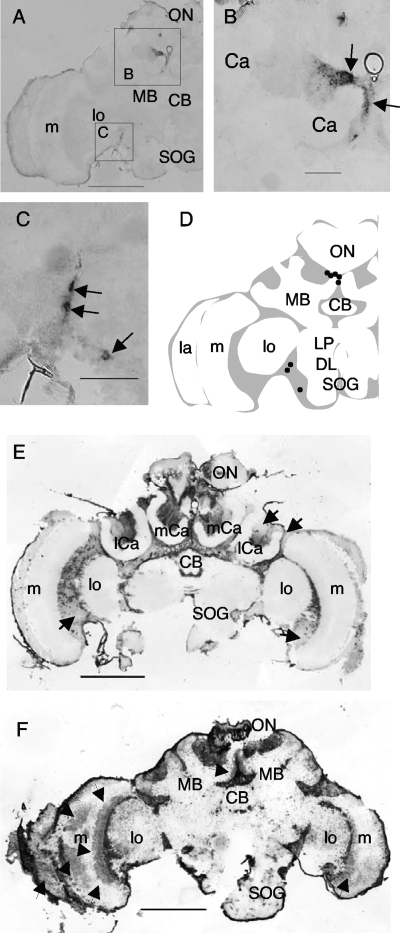
To examine how KV distribution in the brain changes during the infection process, we analyzed the KV distribution over time after infection. Worker bees were inoculated experimentally with KV by injecting a tissue homogenate of KV-infected worker bees into their thoraces (16, 33), and the brains were dissected at different days after injection for in situ hybridization. Artificial KV inoculation did not affect the survival rate for at least 1 week (see Fig. S2A in the supplemental material). Kakugo RNA was not detected by in situ hybridization and qRT-PCR in RNA samples extracted from the brains up to day 3 but was detected on day 4 after inoculation (see Table S1 and Fig. S2B in the supplemental material). Since KV was detected in the brain after being injected into the thorax, KV may circulate in the hemolymph. In contrast, in the worker bees injected with PBS, KV was seldom detected, suggesting that the signals detected at day 4 were due to experimental KV inoculation and amplification. Since the worker bees were collected from colonies parasitized by Varroa and natural infection occurred, KV-positive worker bees that were injected with PBS may have been infected naturally with KV.
The brain areas where the KV signals were detected varied both with time postinoculation and between individual bees. In some of the day 4 samples, the signals were detected in a small number of cell clusters in the brain, which may represent an earlier stage of infection (n = 2) (Fig. 2A; see Fig. S3A in the supplemental material). In one case, Kakugo RNA was detected in the upper part of the median MB and in some somata between the OL and the dorsal lobe (Fig. 2A to C). In another case, Kakugo RNA was detected near the ocellar nerve, suggesting that KV infection starts in various brain regions (see Fig. S2A in the supplemental material). In some day 5 and 6 samples, Kakugo RNA was detected in neuronal somata that were widely distributed in the brain, including the MBs, OLs, and the central region of the brain containing the neurosecretory cells (Fig. 2E). The signal intensity and pattern varied by individual, suggesting that the progression of KV infection varied depending on the individual (Fig. 2E; see Fig. S3B and C in the supplemental material). In addition, signal intensity varied even in the same brain regions of the same samples, and in some regions no significant signals were detected (Fig. 2E). In some day 11 samples, Kakugo RNA was detected even in the neuropil of the OLs and MBs (Fig. 2F; see Fig. S3D in the supplemental material), which might represent a later stage of infection.
FIG. 2.
Time-related changes in KV distribution in worker brains experimentally infected with KV. In situ hybridization was performed with a Kakugo antisense probe and the frontal section of the brains of worker bees injected with KV-containing homogenate. The worker bees were collected on day 4 (A), day 6 (E), or day 11 (F) after injection. Representative data from multiple sections of bees are shown (also see Table S1 in the supplemental material). Arrows in panels E and F indicate Kakugo-negative somata and Kakugo-positive areas in the neuropil, respectively. (B and C) Boxed areas in panel A are magnified. Arrows indicate Kakugo-positive cells. (D) Schematic representation of the positions of Kakugo-positive cells (dots) in the worker brain hemisphere. Gray and white regions indicate neuronal somata and the neuropil, respectively. ON, ocellar nerve; mCa, median calyx; lCa, lateral calyx; CB, central body; LP, lateral protocerebral lobe; DL, dorsal lobe; SOG, subesophageal ganglion; lo, lobula; m, medulla; MB, mushroom body; Ca, calyx. Bars, 500 μm (A, E, and F) and 100 μm (B and C).
Tissue distribution of KV.
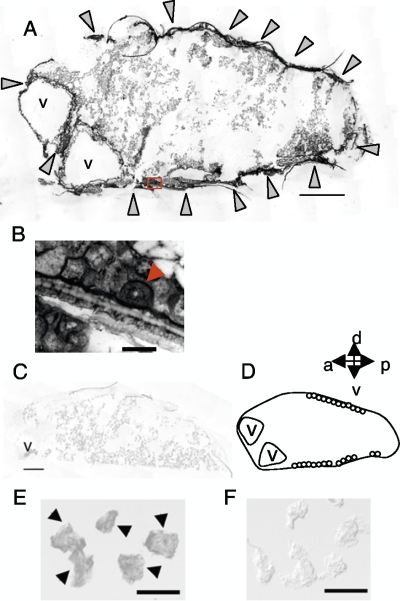
Our previous study, using qRT-PCR, suggested that KV infects various body parts, including the head, thorax, and abdomen (14). To examine KV distribution in tissues other than the brain, in situ hybridization for Kakugo RNA was performed using sections of worker bees from day 11 after KV inoculation. In the abdomen, Kakugo RNA was detected in the fat body cells underneath the cuticle and in the cells between vacuoles (Fig. 3A). The fat body consists of two different types of cells, oenocytes and trophocytes (37), and plays an important role in both metabolism and immunity (17, 22). Kakugo RNA was detected in both oenocytes and trophocytes, with stronger signals observed in the oenocytes (Fig. 3B).
FIG. 3.
Tissue distribution of KV. In situ hybridization was performed with a Kakugo antisense probe and sections of the abdomen (A to C) and the acini of the HG cells (E and F) of worker bees injected with KV-containing homogenate. Representative data from multiple sections of the bees are shown (n = 5 for the abdomen and n = 2 for HGs). Arrowheads indicate the positions of the acini with Kakugo signals. (B) High-magnification micrograph of the boxed area in panel A. The arrowhead indicates an oenocyte. (D) Schematic representation of the abdomen. v, vacuole. Bars, 500 μm (A and C) and 100 μm (B, E, and F).
Kakugo RNA was also detected in the HGs (Fig. 3E). The HGs are head exocrine glands that comprise acini containing 12 or 13 secretory cells and that synthesize and secrete royal jelly proteins in nurse bees and glucose oxidase, which converts nectar into honey, in the foragers (21). The signals were detected in all acini, suggesting that all of the secretory cells of the HGs were infected with KV (Fig. 3E).
Picorna-like viruses replicate their genomic RNA (a positive-strand RNA) in the host cytoplasm by using negative-strand RNAs as a template (32), and thus negative-strand RNAs are generated during replication. Although negative-strand RNAs can be detected in honeybee tissues infected with some bee viruses by use of strand-specific RT-PCR or in situ hybridization (6, 28, 29, 47), negative-strand KV RNAs were not detected by our in situ hybridization assay using the sense probe for Kakugo RNA in either the naturally KV-infected or experimentally inoculated worker bees (data not shown). Thus, we could not identify the cells where KV replicates. It might be that the level of KV negative-strand RNAs was below the detection limit of in situ hybridization.
Alteration in gene expression profile in brains of KV-infected worker bees.
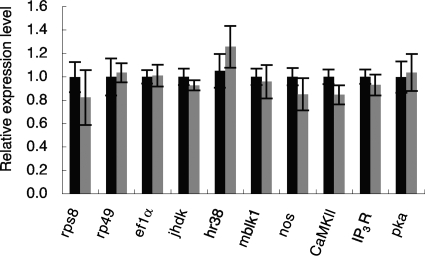
To examine whether KV infection can affect brain function, we analyzed the changes in mRNA expression level of genes that were previously shown to be expressed in worker bee brains. In particular, we focused on the genes expressed preferentially in the MBs because the MBs are a higher-order center of the insect brain, and thus the influence on the expression of MB-preferential genes by KV infection might be crucial for brain function. Among the MB-preferential genes, we selected the following five for analysis: calcium/calmodulin-dependent protein kinase II gene (CaMKII), inositol 1,4,5-triphosphate receptor gene (IP3R), mblk-1, juvenile hormone diol kinase gene (jhdk), and hr38 (18, 19, 39, 41, 46). In addition, we analyzed the cAMP-dependent protein kinase gene, which is involved in neural plasticity (10, 11, 25), and the nitric oxide synthase gene, which is involved in neural plasticity and/or immunity (13, 24, 43). Using qRT-PCR, we compared the expression levels of the genes in the MBs between the worker bees experimentally inoculated with KV and those injected with PBS. These bee brains were infected with KV but not with other viruses, such as DWV, sacbrood virus, and acute bee paralysis virus (see Fig. S4A in the supplemental material). The expression levels of the 11 genes did not differ significantly between the two groups (Fig. 4). In situ hybridization of PKC, CaMKII, IP3R, jhdk, IP3 phosphatase (38), and broad-complex (30) revealed that the MB-preferential expression pattern was not altered by KV infection (data not shown). These results suggest that KV infection does not lead to drastic changes in the mRNA expression levels of at least these 11 genes in the brain.
FIG. 4.
Comparison of mRNA expression levels in MBs of KV-infected and uninfected bees. qRT-PCR analysis was performed using total RNA extracted from the MBs of single worker bees injected with KV-containing homogenate or PBS (n = 5 for each, except for IP3R; for IP3R, n = 10 and 14 for PBS and KV-containing homogenate injection, respectively). The amounts of the transcripts were divided by that of actin and normalized using the values for one of the KV-infected bees. Data are shown as means ± standard errors.
To comprehensively examine the influence of KV infection on the gene expression profiles in the brain, we used a cDNA microarray, which we previously prepared using the partial cDNAs of genes expressed in the brain (38), to compare gene expression profiles in the MBs of KV-infected worker bees with those in the MBs of uninfected worker bees (the KV level was <0.01% that of the KV-infected bees) (see Fig. S4B in the supplemental material). In this experiment, we used naturally KV-infected worker bees that were not infected with other viruses to examine the changes in gene expression profiles under more natural conditions (see Fig. S4B and C in the supplemental material). The expression of one gene (honeybee genome project name, GB10708; GenBank accession number XM_397526) was drastically increased in the KV-infected bees (Fig. 5A; see Fig. S5 in the supplemental material). Expression of GB10708 was also higher in the other brain regions (the OLs and antennal lobes) of the KV-infected worker bees (data not shown). Elevated expression of the gene in the KV-infected worker brains was also observed, by qRT-PCR analysis, in the MBs of the worker bees experimentally inoculated with KV (Fig. 5B). The MBs examined were not infected with other viruses (see Fig. S4D in the supplemental material). The expression level of GB10708 increased gradually until day 6 after the experimental inoculation (see Fig. S6 in the supplemental material), correlating with Kakugo amplification (see Fig. S2B in the supplemental material), further supporting the observation that GB10708 induction is related to KV infection. According to the NCBI database, GB10708 cDNA is 1,166 bp in length and encodes a hypothetical protein of approximately 30 kDa (GenBank accession number XP_397526; locus LOC408807). We used 5′ rapid amplification of cDNA ends (16) to obtain a 1,157-bp cDNA clone which encodes a protein of 274 amino acids. Recently, GB10708 was also identified as the hypothetical protein HP30, which is induced by bacterial infection in the hemolymph of adult honeybees (33). GB10708 has a signal peptide at the N terminus (Fig. 5C), suggesting that it is a secreted protein (33), and has a leucine zipper motif (36) and a leucine-rich repeat-like motif, suggesting that it interacts with other proteins. We searched for proteins that share a motif structure similar to that of GB10708 and found a hypothetical protein from the parasitic wasp Nasonia vitripennis (GenBank accession no. XP_001603643) (Fig. 5C). The molecular mass (31 kDa) and the position of the leucine-rich repeat-like motif are conserved, though the overall amino acid identity is not very high (22%) and the leucine residue in the leucine zipper motif is not conserved in the Nasonia protein (Fig. 5D), suggesting that GB10708 is a Hymenoptera-specific protein and that its interacting molecule may be different between Apis and Nasonia. In situ hybridization analysis of the GB10708 transcript revealed that it was expressed predominantly in the peripheral regions of the brains of both naturally KV-infected and experimentally KV-inoculated worker bees (day 6 after inoculation), whereas the signal was faint in the brains of uninfected worker bees (Fig. 6), suggesting that it is induced locally in the brain by KV infection.
FIG. 6.
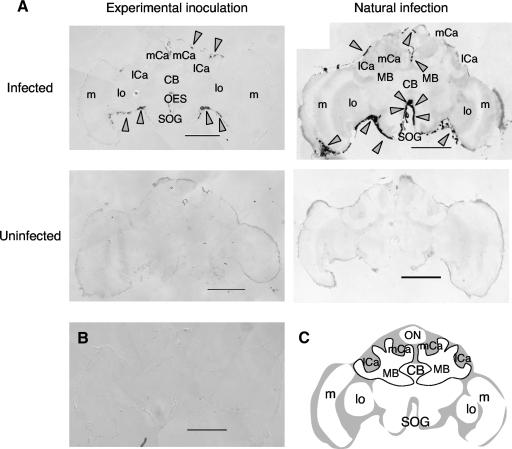
Localization of GB10708 induction with KV infection. In situ hybridization was performed with a GB10708 antisense (A) or sense (B) probe and sections of the brains of worker bees injected with KV-containing homogenate (A, left, and B) or naturally infected with KV (A, right). Representative data from multiple sections of the tested bees are shown (n = 2 for experimental inoculation and n = 2 for natural infection). Arrowheads indicate the positions of the GB10708 signals. (C) Schematic representation of the brain. Gray and white regions indicate neuronal somata and the neuropil, respectively. ON, ocellar nerve; CB, central body; MB, mushroom body; SOG, subesophageal ganglion; OES, esophagus; lo, lobula; m, medulla; mCa, median calyx; lCa, lateral calyx. Bars, 500 μm.
DISCUSSION
In the present study, we examined the distribution of KV in infected worker bees, using in situ hybridization with Kakugo RNA-specific probes. KV infected the somata as well as the axons of various brain regions, in some cases, in naturally KV-infected worker bees, suggesting that KV infection may affect neural function in various brain regions. Although little is known about the brain regions/structures that are involved in aggressive behaviors in insects, octopaminergic neurons in the SOG are important for aggressive behavior in Drosophila (48). KV also infects the SOG, implying that it could affect worker bee aggressive behavior.
In addition, we used the experimental inoculation of KV to compare the progression of the infection to that in naturally infected bees. KV infection was limited to a small number of neuronal somata in the earlier infection phase, whereas it later became detectable in broader regions, such as the somata in the MBs, OLs, central complex, and SOG, the ocellar nerves, and possibly neuronal axons in the MBs. It is possible that changes in KV localization affect various brain functions with the progression of the infection. The broad infection pattern of KV in the brains of experimentally KV-inoculated bees in the later infection phase was similar to that of naturally KV-infected bees, suggesting that the naturally infected bees broadly infected with KV in the brains were in the later phase of infection. The localization of KV in neuronal axons suggests that the virus is transported through the axons, like other picornaviruses (7, 23, 26, 27).
KV infected not only the brains but also the HGs and the fat bodies, suggesting that KV infection is systemic in worker bees. Since the fat body is important for insect metabolism, immunity, and pheromone production (20), KV infection in this tissue possibly influences the physiological states and pheromone production of the worker bees. DWV is also detected in the fat bodies in queen bees, and therefore the influence of KV infection on fat body function may be similar to that of DWV infection (12). In addition, based on the finding of KV infection in the HGs, there may also be an oral transmission route through substance and nutrient exchange that enables horizontal KV infection, similar to the case for sacbrood virus (1). DWV is also possibly transmitted orally, as it is detected in larval food and queen feces (8, 47). Our previous study suggested that KV is transmitted to worker bees partly by Varroa parasitism but also by some unidentified route, because KV was detected in colonies unparasitized by Varroa. Our findings in the present study suggest the likelihood of a horizontal infection route in addition to Varroa parasitization.
In the brains of KV-infected worker bees, the gene expression profile was partially altered, with the induction of GB10708/HP30. Although our inoculation experiments using KV-containing tissue homogenate need to be confirmed using purified KV in future research, the findings that the induction of GB10708/HP30 expression was commonly observed in both naturally KV-infected and experimentally KV-inoculated worker brains and that it was not induced by the injection of homogenate without KV suggest that the change in gene expression profile was due to KV infection. GB10708/HP30 may be a Hymenoptera-specific protein that, based on its primary structure, may interact with other proteins. Because HP30 was identified as a protein induced in the hemolymph after bacterial challenge (33), it may function as a general immune protein rather than being induced specifically by viral (KV) infection. Thus, GB10708/HP30 transcript induction in the periphery of the brain upon KV infection may be involved in the immune reaction around the brain. The roles of GB10708/HP30 need to be clarified by biochemical analysis and identification of the cell types in which the GB10708/HP30 transcript is induced, such as neurons, immune cells, etc.
The findings of the present study suggest that KV infection may affect some brain functions and/or physiological states in worker honeybees. On the other hand, we usually cannot discriminate KV-infected worker bees from uninfected worker bees in naturally maintained colonies based on behavioral abnormalities, suggesting that KV infection does not severely affect honeybee behavior. Actually, the survival rate of the artificially KV-infected worker bees did not differ significantly from that of PBS-injected worker bees (see Fig. S2 in the supplemental material), suggesting that KV inoculation is not lethal in adult worker bees. This might be why KV was originally identified in the brains of apparently healthy, but aggressive, worker bees (14, 16). In future, KV distribution and gene expression profiles in the brains of aggressive worker bees naturally infected with KV should be examined and compared with the results for experimentally KV-inoculated worker bees to evaluate the similarities and differences between KV infections in aggressive worker bees and nonaggressive worker bees. Understanding the influence of microbial infections on honeybee brain functions will help to elucidate the behavioral effects of microbe-honeybee interactions.
Supplementary Material
Acknowledgments
We thank H. Beier (Würzburg Universität) for instructions on how to handle the honeybee virus and H. Beier and J. Tautz (Würzburg Universität) for collaboration in detecting the bee viruses. We are grateful to K. Shirai, R. K. Paul, Y. Yamazaki, and Y. Uno for technical support in this study.
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN) and by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan, the Terumo Life Science Foundation, and the Naito Foundation. T.F. is the recipient of a grant-in-aid for young scientists from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 2 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bailey, L. 1969. The multiplication and spread of sacbrood virus of bees. Ann. Appl. Biol. 63:483-491. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, L. 1976. Viruses attacking the honey bee. Adv. Virus Res. 20:271-304. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, L., and B. V. Ball. 1991. Honey bee pathology. Academic Press Inc., San Diego, CA.
- 4.Bailey, L., and E. F. W. Fernando. 1972. Effects of sacbrood virus on adult honey-bees. Ann. Appl. Biol. 72:27-35. [Google Scholar]
- 5.Berényi, O., T. Bakonyi, I. Derakhshifar, H. Köglberger, G. Topolska, W. Ritter, H. Pechhacker, and N. Nowotny. 2007. Phylogenetic analysis of deformed wing virus genotypes from diverse geographic origins indicates recent global distribution of the virus. Appl. Environ. Microbiol. 73:3605-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celle, O., P. Blanchard, V. Olivier, F. Schurr, N. Cougoule, J. P. Faucon, and M. Ribiere. 2008. Detection of chronic bee paralysis virus (CBPV) genome and its replicative RNA form in various hosts and possible ways of spread. Virus Res. 133:280-284. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. S., Y. C. Yao, S. C. Lin, Y. P. Lee, Y. F. Wang, J. R. Wang, C. C. Liu, H. Y. Lei, and C. K. Yu. 2007. Retrograde axonal transport: a major transmission route of enterovirus 71 in mice. J. Virol. 81:8996-9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., J. Evans, and M. Feldlaufer. 2006. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebr. Pathol. 92:152-159. [DOI] [PubMed] [Google Scholar]
- 9.Cox-Foster, D. L., S. Conlan, E. C. Holmes, G. Palacios, J. D. Evans, N. A. Moran, P. L. Quan, T. Briese, M. Hornig, D. M. Geiser, V. Martinson, D. vanEngelsdorp, A. L. Kalkstein, A. Drysdale, J. Hui, J. Zhai, L. Cui, S. K. Hutchison, J. F. Simons, M. Egholm, J. S. Pettis, and W. I. Lipkin. 2007. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318:283-287. [DOI] [PubMed] [Google Scholar]
- 10.Eisenhardt, D., A. Fiala, P. Braun, H. Rosenboom, H. Kress, P. R. Ebert, and R. Menzel. 2001. Cloning of a catalytic subunit of cAMP-dependent protein kinase from the honeybee (Apis mellifera) and its localization in the brain. Insect Mol. Biol. 10:173-181. [DOI] [PubMed] [Google Scholar]
- 11.Fiala, A., U. Müller, and R. Menzel. 1999. Reversible downregulation of protein kinase A during olfactory learning using antisense technique impairs long-term memory formation in the honeybee, Apis mellifera. J. Neurosci. 19:10125-10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fievet, J., D. Tentcheva, L. Gauthier, J. de Miranda, F. Cousserans, M. E. Colin, and M. Bergoin. 2006. Localization of deformed wing virus infection in queen and drone Apis mellifera L. Virol. J. 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foley, E., and P. H. O'Farrell. 2003. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 17:115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiyuki, T., S. Ohka, H. Takeuchi, M. Ono, A. Nomoto, and T. Kubo. 2006. Prevalence and phylogeny of Kakugo virus, a novel insect picorna-like virus that infects the honeybee (Apis mellifera L.), under various colony conditions. J. Virol. 80:11528-11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiyuki, T., H. Takeuchi, M. Ono, S. Ohka, T. Sasaki, A. Nomoto, and T. Kubo. 2005. Kakugo virus from brains of aggressive worker honeybees. Adv. Virus Res. 65:1-27. [DOI] [PubMed] [Google Scholar]
- 16.Fujiyuki, T., H. Takeuchi, M. Ono, S. Ohka, T. Sasaki, A. Nomoto, and T. Kubo. 2004. Novel insect picorna-like virus identified in the brains of aggressive worker honeybees. J. Virol. 78:1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez, E., D. Wiggins, B. Fielding, and A. P. Gould. 2007. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445:275-280. [DOI] [PubMed] [Google Scholar]
- 18.Kamikouchi, A., H. Takeuchi, M. Sawata, S. Natori, and T. Kubo. 2000. Concentrated expression of Ca2+/ calmodulin-dependent protein kinase II and protein kinase C in the mushroom bodies of the brain of the honeybee Apis mellifera L. J. Comp. Neurol. 417:501-510. [DOI] [PubMed] [Google Scholar]
- 19.Kamikouchi, A., H. Takeuchi, M. Sawata, K. Ohashi, S. Natori, and T. Kubo. 1998. Preferential expression of the gene for a putative inositol 1,4,5-trisphosphate receptor homologue in the mushroom bodies of the brain of the worker honeybee Apis mellifera L. Biochem. Biophys. Res. Commun. 242:181-186. [DOI] [PubMed] [Google Scholar]
- 20.Krupp, J. J., C. Kent, J. C. Billeter, R. Azanchi, A. K. C. So, J. A. Schonfeld, B. P. Smith, C. Lucas, and J. D. Levine. 2008. Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster. Curr. Biol. 18:1373-1383. [DOI] [PubMed] [Google Scholar]
- 21.Kubo, T., M. Sasaki, J. Nakamura, H. Sasagawa, K. Ohashi, H. Takeuchi, and S. Natori. 1996. Change in the expression of hypopharyngeal-gland proteins of the worker honeybees (Apis mellifera L.) with age and/or role. J. Biochem. 119:291-295. [DOI] [PubMed] [Google Scholar]
- 22.Lemaitre, B., and J. Hoffmann. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697-743. [DOI] [PubMed] [Google Scholar]
- 23.Martinat, C., N. Jarousse, M. C. Prévost, and M. Brahic. 1999. The GDVII strain of Theiler's virus spreads via axonal transport. J. Virol. 73:6093-6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller, U. 1996. Inhibition of nitric oxide synthase impairs a distinct form of long-term memory in the honeybee, Apis mellifera. Neuron 16:541-549. [DOI] [PubMed] [Google Scholar]
- 25.Müller, U. 2000. Prolonged activation of cAMP-dependent protein kinase during conditioning induces long-term memory in honeybees. Neuron 27:159-168. [DOI] [PubMed] [Google Scholar]
- 26.Ohka, S., N. Matsuda, K. Tohyama, T. Oda, M. Morikawa, S. Kuge, and A. Nomoto. 2004. Receptor (CD155)-dependent endocytosis of poliovirus and retrograde axonal transport of the endosome. J. Virol. 78:7186-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohka, S., W. X. Yang, E. Terada, K. Iwasaki, and A. Nomoto. 1998. Retrograde transport of intact poliovirus through the axon via the fast transport system. Virology 250:67-75. [DOI] [PubMed] [Google Scholar]
- 28.Olivier, V., I. Massou, O. Celle, P. Blanchard, F. Schurr, M. Ribière, and M. Gauthier. 2008. In situ hybridization assays for localization of the chronic bee paralysis virus in the honey bee (Apis mellifera) brain. J. Virol. Methods 153:232-237. [DOI] [PubMed] [Google Scholar]
- 29.Ongus, J. R., D. Peters, J. M. Bonmatin, E. Bengsch, J. M. Vlak, and M. M. van Oers. 2004. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 85:3747-3755. [DOI] [PubMed] [Google Scholar]
- 30.Paul, R. K., H. Takeuchi, and T. Kubo. 2006. Expression of two ecdysteroid-regulated genes, broad-complex and E75, in the brain and ovary of the honeybee (Apis mellifera L.). Zool. Sci. 23:1085-1092. [DOI] [PubMed] [Google Scholar]
- 31.Quackenbush, J. 2002. Microarray data normalization and transformation. Nat. Genet. 32(Suppl.):496-501. [DOI] [PubMed] [Google Scholar]
- 32.Racaniello, V. R. 2007. Picornaviridae: the viruses and their replication, p.796-838. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, vol. 1, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 33.Randolt, K., O. Gimple, J. Geissendörfer, J. Reinders, C. Prusko, M. J. Mueller, S. Albert, J. Tautz, and H. Beier. 2008. Immune-related proteins induced in the hemolymph after aseptic and septic injury differ in honey bee larvae and adults. Arch. Insect Biochem. Physiol. 69:155-167. [DOI] [PubMed] [Google Scholar]
- 34.Roossinck, M. 2005. Symbiosis versus competition in plant virus evolution. Nat. Rev. Microbiol. 3:917-924. [DOI] [PubMed] [Google Scholar]
- 35.Rortais, A., D. Tentcheva, A. Papachristoforou, L. Gauthier, G. Arnold, M. E. Colin, and M. Bergoin. 2006. Deformed wing virus is not related to honey bees' aggressiveness. Virol. J. 3:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith-Gill, S. J. 1991. Protein-protein interactions: structural motifs and molecular recognition. Curr. Opin. Biotechnol. 2:568-575. [DOI] [PubMed] [Google Scholar]
- 37.Snodgrass, R. E. 1956. Anatomy of the honey bee. Cornell University Press, London, United Kingdom.
- 38.Takeuchi, H., T. Fujiyuki, K. Shirai, Y. Matsuo, A. Kamikouchi, Y. Fujinawa, A. Kato, A. Tsujimoto, and T. Kubo. 2002. Identification of genes expressed preferentially in the honeybee mushroom bodies by combination of differential display and cDNA microarray. FEBS Lett. 513:230-234. [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi, H., E. Kage, M. Sawata, A. Kamikouchi, K. Ohashi, M. Ohara, T. Fujiyuki, T. Kunieda, K. Sekimizu, S. Natori, and T. Kubo. 2001. Identification of a novel gene, Mblk-1, that encodes a putative transcription factor expressed preferentially in the large-type Kenyon cells of the honey bee brain. Insect Mol. Biol. 10:487-494. [DOI] [PubMed] [Google Scholar]
- 40.Terio, V., V. Martella, M. Camero, N. Decaro, G. Testini, E. Bonerba, G. Tantillo, and C. Buonavoglia. 2008. Detection of a honeybee iflavirus with intermediate characteristics between Kakugo virus and deformed wing virus. New Microbiol. 31:439-444. [PubMed] [Google Scholar]
- 41.Uno, Y., T. Fujiyuki, M. Morioka, H. Takeuchi, and T. Kubo. 2007. Identification of proteins whose expression is up- or down-regulated in the mushroom bodies in the honeybee brain using proteomics. FEBS Lett. 581:97-101. [DOI] [PubMed] [Google Scholar]
- 42.Wang, D.-I., and F. E. Moeller. 1970. The division of labor and queen attendance behavior of Nosema-infected worker honey bees. J. Econ. Entomol. 63:1539-1541. [Google Scholar]
- 43.Watanabe, T., M. Kikuchi, D. Hatakeyama, T. Shiga, T. Yamamoto, H. Aonuma, M. Takahata, N. Suzuki, and E. Ito. 2007. Gaseous neuromodulator-related genes expressed in the brain of honeybee Apis mellifera. Dev. Neurobiol. 67:456-473. [DOI] [PubMed] [Google Scholar]
- 44.White, G. F. 1917. Sacbrood. USDA Bull. 431:1-55. [Google Scholar]
- 45.Winston, M. L. 1987. The biology of the honeybee. Harvard University Press, Cambridge, MA.
- 46.Yamazaki, Y., K. Shirai, R. K. Paul, T. Fujiyuki, A. Wakamoto, H. Takeuchi, and T. Kubo. 2006. Differential expression of HR38 in the mushroom bodies of the honeybee brain depends on the caste and division of labor. FEBS Lett. 580:2667-2670. [DOI] [PubMed] [Google Scholar]
- 47.Yue, C., and E. Genersch. 2005. RT-PCR analysis of deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 86:3419-3424. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, C., Y. Rao, and Y. Rao. 2008. A subset of octopaminergic neurons are important for Drosophila aggression. Nat. Neurosci. 11:1059-1067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.