Abstract
The hepatitis C virus (HCV) isolate JFH1 represents the only cloned wild-type sequence capable of efficient replication in cell culture, as well as in chimpanzees. Previous reports have pointed to the viral polymerase NS5B as a major determinant for efficient replication of this isolate. To understand the underlying mechanisms, we expressed and purified NS5B of JFH1 and of the closely related isolate J6, which replicates below the limit of detection in cell culture. The JFH1 enzyme exhibited a 5- to 10-fold-higher specific activity in vitro, consistent with the polymerase activity itself contributing to efficient replication of JFH1. The higher in vitro activity of the JFH1 enzyme was not due to increased RNA binding, elongation rate, or processivity of the polymerase but to higher initiation efficiency. By using homopolymeric and heteropolymeric templates, we found that purified JFH1 NS5B was significantly more efficient in de novo initiation of RNA synthesis than the J6 counterpart, particularly at low GTP concentrations, probably representing an important prerequisite for the rapid replication kinetics of JFH1. Furthermore, we solved the crystal structure of JFH1 NS5B, which displays a very closed conformation that is expected to facilitate de novo initiation. Structural analysis shows that this closed conformation is stabilized by a sprinkle of substitutions that together promote extra hydrophobic interactions between the subdomains “thumb” and “fingers.” These analyses provide deeper insights into the initiation of HCV RNA synthesis and might help to establish more efficient cell culture models for HCV using alternative isolates.
Hepatitis C virus (HCV) is an enveloped positive-strand RNA virus belonging to the genus Hepacivirus in the family Flaviviridae. The genome of HCV encompasses a single ∼9,600-nucleotide (nt)-long RNA molecule carrying one large open reading frame flanked by nontranslated regions (NTRs), which is primarily translated into one polyprotein. The polyprotein precursor is cleaved by cellular and viral proteases into at least 10 different products (for a review, see reference 5). The nonstructural proteins NS3 to NS5B are necessary and sufficient for autonomous RNA replication. They form a membrane-associated replication complex in which NS5B is the RNA-dependent RNA polymerase (RdRp), the key enzyme of viral-RNA replication.
Studies of HCV have long been hampered by the lack of efficient cell culture systems. The advent of subgenomic replicons derived from genotype 1 isolates allowed for the first time a concise analysis of authentic viral-RNA replication (29). However, these isolates required adaptive mutations for efficient propagation in cells (9, 21, 27). Based on these adapted viral sequences, it was not possible to generate infectious virus, since most of these mutations seemed to interfere with virus assembly (38), and only one cell culture-adapted genotype 1 isolate has yet given rise to moderately efficient production of infectious virus (49). This restriction was abolished by a particular genotype 2a isolate designated JFH1, which replicates with outstanding efficiency in cell culture without a requirement for adaptive mutations and thereby efficiently produces virions, which are infectious in vitro and in vivo (19, 46).
The unique features of the JFH1 isolate are not due to its structural proteins; in fact, chimeric genomes harboring the core-NS2 regions of other isolates, even from different genotypes, combined with the NS3-NS5B region of JFH1 are also capable of virion production, some of them even with higher efficiency (24, 37). Therefore, the characteristics of the nonstructural proteins and their mode of RNA synthesis are unique to JFH1. Previous studies have already shown that subgenomic replicons derived from JFH1 exhibit extremely fast RNA synthesis, reaching peak replication levels within 24 h after transfection and exceeding the efficiency of a cell culture-adapted Con1 replicon by orders of magnitude (7). Several lines of evidence point to the polymerase gene as a major determinant of JFH1 RNA replication. An elegant study using a closely related genotype 2a isolate, designated J6, analyzed the replication efficiencies of intragenotypic chimeras with JFH1 (35). HCV J6 has been proven to be infectious in chimpanzees (48) but does not replicate in cell culture at all, thereby providing the ideal requirements to define the regions of the JFH1 genome involved in efficient RNA replication (35). By generating chimeric replicons between these two isolates, Murayama et al. were able to show that the coding regions encompassing the NS3 helicase and NS5B, as well as the 3′ NTR, were the main determinants for efficient JFH1 replication, with NS5B being the most important single factor. In addition, Binder et al. constructed chimeras between JFH1 and the genotype 1b isolate Con1 (7). The Con1 NS5B gene did not give rise to replication-competent chimeric replicons harboring the NS3-5A region of JFH1, in contrast to intergenotypic chimeras, including NS3-5A of Con1 and NS5B of JFH1, again pointing to particular properties residing in the NS5B gene of JFH1. However, a number of different mechanisms could mediate the role of the NS5B coding region, including interactions with other nonstructural proteins or host cell proteins, cis-acting elements residing in the NS5B coding region (16, 50), and the enzymatic activity of the RdRp. Preliminary evidence favoring the last option, without providing mechanistic insight, was provided by the observation that JFH1 RdRp seemed to have a higher specific activity in vitro than Con1 (7). RNA synthesis by NS5B in vitro can be separated into distinct steps, namely, RNA binding, initiation, elongation, and termination. NS5B binds to all kinds of heteropolymeric RNA with ambiguous specificity for virus-derived sequences, while the binding to homopolymers follows a distinct pattern [poly(U) > poly(G) > poly(A) > poly(C)] (28). Purified NS5B can initiate RNA synthesis by a primer-dependent mechanism or de novo (6, 28, 33, 51). De novo initiation at the 3′ end of the viral positive- and negative-strand RNAs is likely to be the physiological mode of initiation of RNA synthesis in infected cells. Once RNA synthesis is initiated, NS5B elongates the nascent RNA by about 200 nt per minute and is capable of processively copying an entire RNA genome in vitro (31, 36). Little is known about termination of RNA synthesis in vitro.
The crystal structure of NS5B was solved initially for the genotype 1 BK isolate (3, 11, 23) and was followed up with many complexes with small molecules. Recently, the structure of a consensus genotype 2a NS5B has been reported (8). The structure of NS5B displays the same organization as and is homologous to other single-chain polymerases. These have been likened to a right hand with a central “palm” domain harboring the catalytic residues and with “fingers” and “thumb” domains on either side (see Fig. 6A, left). In sharp contrast to DNA-dependent polymerases and reverse transcriptases, in which the fingers and thumb move independently of each other during the enzyme's catalytic cycle, the fingers and thumb domains of NS5B are connected by an extension of the fingers (“fingertips”) (see Fig. 6A, right). This feature and the associated “closed-fingers” conformation have since proved to be universal within viral RdRps. Indeed, the subset of RdRps that initiate RNA synthesis de novo (such as NS5B) have been crystallized in a conformation that is nearly suitable for binding of a single-stranded RNA template and the priming nucleotides and thus already nearly competent for de novo initiation. This is exemplified by the φ6 and reovirus RdRps, for which structures of initiation complexes are available that are almost identical to those of the apo RdRps (13, 44). In contrast, a conformational change is clearly required for transitioning from de novo initiation to elongation of the nascent RNA strand. This change is quite small for the reovirus RdRp (44) but is expected to be larger for other RdRps. HCV-NS5B seems to be an extreme case in this respect. First, the extension C terminal to the enzyme (see Fig. 6A, right, where it is labeled “linker”), which in NS5B connects to the transmembrane anchor, folds back toward the catalytic site, and would block off the exit of template RNA. Second, the NS5B thumb is much larger than those of other RdRps and is much too closed in the available structures to allow productive elongation. The so-called beta flap that it harbors (a unique feature of Flaviviridae RdRps) is thus in a position to be engaged in the initiation process, but the whole thumb then has to rotate away to allow egress of the nascent strand, as first proposed by Bressanelli et al. (11).
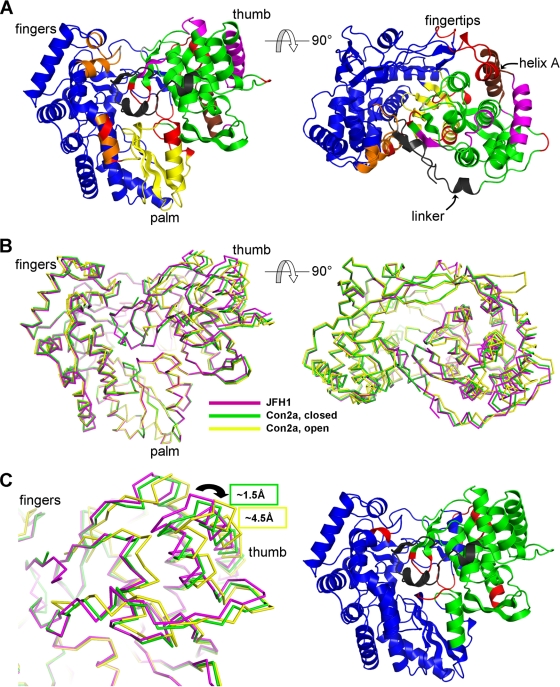
FIG. 6.
Structural comparison of the three conformationally different available subtype 2a NS5B molecules. Three structures from genotype 2a were analyzed for their conformational flexibility and rigidity using the ESCET program, including the novel JFH1 structure (this report) and the two conformations of a con2a enzyme (8) (PDB codes, 1YUY and 1YVX for the closed and open conformations, respectively). (A) JFH1 NS5B depicted in ribbon representation as two views related by a 90° rotation. With a tolerance threshold of 2.5 sigmas (ESCET lolim parameter), six rigid zones greater than 10 amino acids in size were found and are shown in distinct colors in order of size: blue, green, yellow, orange, magenta, and brown. The red zones indicate more flexible regions or regions that are disordered in one of the structures. The C-terminal linker, a region of lower conformational identity, is colored black for clarity. (B) Three structures of NS5B (JFH1, 1YUY, and 1YVX in magenta, green, and yellow, respectively) superimposed on the palm domain (yellow in panel A). (C) (Left) Close-up of the thumbs in the superposition in panel B. The largest differences occur at the top of the thumb (arrow), with a displacement between the JFH1 enzyme and the con2a closed and open forms of about 1.5 Å and 4.5 Å, respectively (as indicated by the green and yellow boxes). (Right) As in panel A, left, but with a tolerance threshold of 5 sigmas. Only two rigid zones are now found, the thumb and the rest of the enzyme, indicating that the main overall difference between the three molecules is the movement of the thumb.
Our analysis aimed to understand the particular properties of JFH1 polymerase at the molecular level by determining its crystal structure and by comparing the biochemical properties of purified JFH1 NS5B and the closely related J6 enzyme expressed in Escherichia coli. We found that purified JFH1 RdRp indeed has a higher specific activity in vitro than the J6 counterpart. This phenotype could be linked to a more efficient de novo initiation, particularly at low GTP concentrations. The crystal structure revealed a more closed conformation for JFH1 NS5B than for a genotype 2a consensus structure and other available HCV polymerase structures. It is stabilized by the cooperative action of a sprinkle of polymorphisms. This closed conformation is suited for binding single-stranded RNA and 2 nt, thereby promoting efficient de novo initiation.
MATERIALS AND METHODS
Plasmid constructs.
The plasmid encoding NS5B of JFH1 lacking the 21 C-terminal amino acids (ΔC21) and C-terminally fused to His6 in a pET21b vector has been described previously (7). To obtain pET21 NS5BΔC21 J6, the N-terminal coding sequence of the gene was amplified by PCR to add an XbaI restriction site using primers S_pET21Xba (CCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGGCTTCCATGTCATACTCCTGGACC) and A2a8632 (GTGAAGGCTCTCAGGTTCCGCTCG), cut with XbaI and NcoI, and fused to an Nco-MfeI fragment from HC-J6 (48) using XbaI and BlpI restriction sites in pET21 NS5BΔC21 JFH1. In a second step, the C terminus of NS5B J6 was amplified by PCR using primers S2a8913 (GGCTGGGAAACATCATCCAGTATGC) and A_J6deltaC21Hind (ATTAAGCTTAGTGATGGTGATGGTGATGAGATCTGGGTCGGGCACGCGACACGCT GTG) and introduced using an internal MfeI and an artificial HindIII site directly adjacent to the His6 in pET21 NS5BΔC21 JFH1.
Expression and purification of JFH1 and J6 RdRps.
NS5BΔC21 of HCV JFH1 and J6, respectively, C-terminally fused to a hexahistidine tag, were expressed in E. coli BL21(DE3) cells. The bacterial cells were grown to an optical density at 600 nm of 0.8, induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside, incubated for 4 h at room temperature with shaking, and sedimented for 10 min at 6,000 × g. The pellet from 100 ml of culture was lysed in 4 ml lysis buffer I (LBI) (100 mM Tris-HCl, pH 8, 100 mM NaCl, 1 mM MgCl2, 2% Triton X-100, 2 mg/ml Lysozyme [Fluka, Buchs, Switzerland]), and 1 U/ml Benzonase (Merck, Darmstadt, Germany) for 30 min on ice and centrifuged for 15 min at 20,000 × g at 4°C. The supernatant (S1) was removed, the pellet was resuspended in 5 ml of LBII (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.1% β-octylglucopyranoside, 10 mM imidazole, 50% glycerol, 10 mM 2-mercaptoethanol), and the suspension was sonicated on ice five times for 15 s each time at an output control setting of 6 with a Branson 450 Sonifier and a microtip. After a 10-min centrifugation at 20,000 × g, the supernatant (S2) was mixed with 500 μl of a 1:1 Ni-nitrilotriacetic acid (NTA)-agarose slurry (Qiagen, Hilden, Germany), incubated for 30 min at 4°C with mild shaking, and centrifuged for 2 min at 1,000 × g. The supernatant (S3) was discarded, the Ni-NTA-agarose slurry was washed four times with 5 ml of LBII containing 50 mM imidazole, and the bound protein was eluted with 0.5 ml of LBIII containing 250 mM imidazole. Purified NS5B was quantified by the Bradford method and stored in small aliquots at −70°C. A JFH1 NS5BΔC21 variant with an inactivating point mutation at amino acid position D318N, designated GND, was expressed and purified in parallel and served as a negative control for RdRp assays.
In vitro transcription.
To generate templates for RdRp or RNA binding assays, in vitro transcripts of HCV replicons or PCR products were generated using the protocol described recently (7). All replicon and full-length genome constructs used in this study contained the genomic ribozyme of HDV followed by the T7 terminator fused to the HCV 3′ NTR and were transcribed without linearization. The PCR products were used to generate in vitro transcripts corresponding to the 3′ ends of the HCV genome. Primers S/T7/9191 (TTGTAATACGACTCACTATAGGGACCAAGCTCAAACTCACTCCA) and A9678JFH (ACATGATCTGCAGAGAGACC) were used on plasmid pJFH1 or pCV-HC-J6 (48) to generate template DNAs for transcription of 3′(+)JFH and 3′(+)J6, respectively, encompassing nt 9259 to the 3′ end of each genome. Oligonucleotides S_5′G_JFH (GACCTGCCCCTAATAGGGGCG) and A_T7_341 (TTGTAATACGACTCACTATAGGGTGCACGGTCTACGAGACC) or A_T7_2018 (TTGTAATACGACTCACTATAGGGAGTTCATCCACGTGCAGCCGAACC) were used to amplify PCR products to generate the templates 3′(−)341nt and 3′(−)2kb, respectively, from plasmid pJFH1 (46), representing the 3′ end of HCV negative-strand RNA. All PCR products were treated with Klenow enzyme to generate blunt ends, following the instructions of the manufacturer (New England Biolabs). Prior to transcription, DNA was extracted with phenol and chloroform, precipitated with ethanol, and dissolved in RNase-free water. In vitro transcription reactions contained 80 mM HEPES, pH 7.5, 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol, 3.125 mM of each nucleoside triphosphate (NTP), 1 U/μl RNasin (Promega, Mannheim, Germany), 0.05 μg/μl plasmid DNA, and 0.6 U/μl T7 RNA polymerase (Promega). After 2 h at 37°C, an additional 0.3 U/μl T7 RNA polymerase was added, and the reaction mixture was incubated for another 2 h. Transcription was terminated by the addition of 1 U RNase-free DNase (Promega) per μg plasmid DNA and 30 min of incubation at 37°C. After extraction with acidic phenol and chloroform, the RNA was precipitated with isopropanol and dissolved in RNase-free water. The concentration was determined by measurement of the optical density at 260 nm, and RNA integrity was checked by agarose gel electrophoresis. To generate radiolabeled RNAs serving as size markers, in vitro transcriptions were performed as described above but including 10 μCi of [α-32P]CTP (Perkin-Elmer, Rodgau, Germany) and using PCR products corresponding to the 3′ end of the HCV negative strand of ca. 1 kb, 2 kb, 4 kb, and 6 kb as templates. In addition, plasmid DNA pFK_JC1 (37) was used to generate transcripts 9.7 kb in length.
RdRp assays.
All RdRp assays were done in a reaction buffer containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM dithiothreitol, 25 mM KCl, 1 mM EDTA, and 20 U of RNasin (Promega) in a total volume of 25 μl, and the mixtures were incubated for 1.5 h at room temperature unless otherwise stated. Standard reaction mixtures on heteropolymeric templates included 10 μCi of [α-32P]CTP (3,000 Ci/mmol; Perkin-Elmer), 10 μM cold CTP, 1 mM (each) ATP and UTP, and 5 mM GTP; 0.5 μg in vitro-transcribed template RNA; and 0.4 μg of purified J6 or JFH1 polymerase. All reaction components except the nucleotides were preincubated for 30 min at room temperature, and the reaction was started by adding the nucleotide mixture. The reactions were terminated by the addition of 1 volume of 0.5 M EDTA. Processivity and elongation rates were determined in the presence of 1.25 μg of heparin (Braun, Melsungen, Germany) per reaction, which was added together with the nucleotides after 30 min of preincubation of all other reaction components at room temperature. Assays using homopolymeric templates included 2 μg poly(C) (GE Healthcare, Munich, Germany) or 2 μg poly(C) with 20 pmol oligo(G12) primer (MWG, Germany), respectively. In this case, the reaction mixtures were not preincubated and contained 10 μCi of [α-32P]GTP; 50 μM (each) CTP, UTP, and ATP; and total GTP concentrations ranging from 5 to 5,000 μM, as indicated for the respective experiments. To generate reaction products labeled at the 5′ end by de novo initiation, 10 μCi of [γ-32P]GTP was used at various total GTP concentrations in the presence of 50 μM (each) CTP, ATP, and UTP.
Primer extension reactions to determine elongation rates were run with a radiolabeled G3 RNA oligonucleotide (Sigma-Aldrich) on a poly(C) template. Oligo(G3) (20 pmol) was labeled with 50 μCi [γ-32P]GTP using T4 polynucleotide kinase (NEB) and added to a standard reaction mixture with 2 μg poly(C). After 30 min of precincubation with 2 μg J6 or JFH1 polymerase at room temperature, the reactions were started by adding 0.5 mM (each) ATP, CTP, and UTP; 10 μM GTP; and 1.25 μg of heparin, and the reaction mixtures were incubated for 5 to 20 min at room temperature. The GTP concentration was lowered in these experiments to avoid excessive de novo initiation.
For gel electrophoretic analysis, 10 μl of 3 M Na-acetate and 20 μg glycogen were added to each reaction mixture, samples were extracted with phenol-chloroform, and nucleic acids were precipitated with 0.7 volume of isopropanol. RNAs were separated by denaturing glyoxal-agarose (12) or 6% polyacrylamide-8 M urea gel electrophoresis and analyzed by autoradiography or phosphorimaging after drying of the gel. For quantitative analysis by liquid scintillation counting, samples were precipitated with 10% trichloroacetic acid (TCA) and 0.5% tetrasodium pyrophosphate, passed through GF-C microfilters (GE Healthcare), washed five times with 1% TCA and 0.1% tetrasodium pyrophosphate, and air dried. After the addition of 6 ml of Ultima Gold (Perkin-Elmer), samples were subjected to liquid scintillation counting. All measurements were done at least in duplicate. Specific activities on poly(C) templates were expressed in pmol GTP incorporated per μg of NS5B per hour of reaction time and calculated by determining the fraction of incorporated radioactive GMP, present at 132 nM (ca. 3.3 pmol), in relation to the total GTP concentration. For heteropolymeric templates, the amount of TCA-precipitated radiolabeled CMP was multiplied by 76 to obtain the total amount of incorporated CMP. The total nucleotide monophosphate (NMP) incorporation was calculated by multiplication by 4, assuming a nearly homogeneous distribution of all four NTPs in the newly synthesized RNAs.
Surface plasmon resonance (SPR) biosensor analysis.
A Biacore S51 instrument (GE Healthcare), equilibrated at 25°C, was used to measure the direct interaction between NS5B variants and different RNAs. NS5B JFH1 and J6 (0.02 mg/ml in 50 mM MES [morpholineethanesulfonic acid], pH 6.0) were immobilized by amine coupling to parallel channels of a CM5 biosensor chip (research grade; GE Healthcare), resulting in surface densities of 2,000 to 5,000 response units. Increasing concentrations of the template 3′(+) JFH1 and 3′(+) J6 RNAs (starting from nt 9259) and the negative-strand 3′(−) JFH1 and 3′(−) J6 RNAs, encompassing the terminal 341 nt, were diluted in the running buffer (20 mM Tris, pH 7.5, 50 mM NaCl, 0.005% Tween) and injected for 2 min across the immobilized proteins at a flow rate of 10 μl/min. The surfaces could effectively be regenerated by injections of 2 M NaCl and 2 M MgCl.
Crystal structure determination.
JFH1 NS5B was dialyzed into crystallization buffer (0.2 M ammonium acetate, 5 mM Tris, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol) and concentrated to 5 mg/ml. Initial crystallization screens, set up using robotics (Cartesian MicroSys) with the vapor diffusion method from 200-nl sitting drops, produced spherulites of JFH1 NS5B when mixed with a reservoir solution of 20% polyethylene glycol (PEG) 3,350, 0.2 M NaH2PO4. After optimization of the conditions, the best diffraction quality crystals were grown from a 1:1 mixture of protein solution (4.3 mg/ml) plus 6 to 7% PEG 20,000, 0.2 M NaH2PO4, in 2-μl hanging drops. Before being mounted, the crystals were rapidly swiped through a solution containing 7% PEG 20,000, 0.2 M NaH2PO4, and 30% glycerol. X-ray diffraction data were then collected to 1.9 Å at the Soleil synchrotron (St. Aubin, Gif-sur-Yvette, France) on beamline Proxima 1. Diffraction data were processed with the XDS package (17), and the data were truncated to 2.2 Å due to anisotropy. A test set was chosen to take into account the local C222 symmetry in the asymmetric unit of the crystal with phenix.reflection_file_converter (2). Structure determination was carried out with the CCP4 suite (4). Phases were calculated by the molecular-replacement method with Phaser using the previously published consensus genotype 2a structure, with Protein Data Bank (PDB) code 1YUY (8) as a search model. Rebuilding was carried out with COOT and refinement with Refmac5. Test simulated annealing refinements without NCS restraints were performed at several points during rebuilding with phenix.refine (2) to check whether the four molecules in the asymmetric unit were in identical conformations. As these tests confirmed that there were no significant main-chain differences at this resolution, NCS restraints (tight main chain and loose side chain) were applied in refmac5 throughout. Refinement of torsion, libration, and screw parameters (two blocks per molecule) improved the electron density maps.
Comparative structural analysis.
The PDB was queried for all HCV NS5B polymerase structures based on genotype 2a sequences, and four entries were retrieved. They included two complexes with small-molecule inhibitors and two apo forms (8). It was observed that certain regions of electron density were not optimally modeled, so in order to proceed with an accurate comparative structural analysis, these structures were first subjected to REFMAC5 refinement with torsion, libration, and screw parameters included, which yielded an improvement in the electron density maps and a reduction in the Rfree values of the models.
The program ESCET (43) was employed first to determine which of the genotype 2a structures, including the JFH1 NS5B polymerase structure, had significant conformational differences. The atomic coordinate errors (estimated standard deviations) were first derived for these structures by taking into account the quality of the model based upon its reported resolution, Rfree value, data completeness, and isotropic temperature factors. Next, the invariant and variant regions between the three genotype 2a structures were determined. Error thresholds (lolim parameter) were set at 5.0 or 2.5 sigmas for overall or detailed differences, respectively. Rigid regions having a minimal sequence length of 10 amino acids were specified, and the search was repeated up to 20 times, which was a value in excess of the number of zones present. The variability between the structures was most readily visualized and interpreted by first superimposing them based on the defined invariant region that lies within the palm domain. Figures were generated with PYMOL (the PyMOL Molecular Graphics System [2002] on the World Wide Web [http://www.pymol.org]).
Protein structure accession number.
Coordinates and structure factors for the JFH1_NS5B structure have been deposited in the PDB under accession number 3I5K.
RESULTS
Purification and specific activities of JFH1 and J6 polymerases.
Previous analyses using chimeric replicons revealed that the NS5B gene contributed significantly to the outstanding replication behavior of the HCV isolate JFH1 in cell culture (35). Our study aimed to analyze the potential role of the enzymatic activity and structure of the JFH1 polymerase in this phenotype. Therefore, we expressed NS5B proteins of the closely related genotype 2a isolates JFH1 and J6 in E. coli to compare their properties in vitro. Both enzymes lacked the membrane insertion sequence and could be purified with equal efficiency by affinity chromatography using a C-terminal His6 tag (Fig. 1A). Electrospray mass spectrometry analysis yielded a mass of 64,407 Da (±2 Da), consistent with the expected 64,409 Da for JFH1 NS5B (not shown).
FIG. 1.

Purification of JFH1 and J6 RdRps and determination of specific activities. (A) Expression of NS5B from isolates JFH1 and J6 in E. coli and purification by differential solubilization and affinity chromatography. Both proteins lack 21 C-terminal amino acids and are fused to a His6 tag. T, total bacterial lysate after induction; S1, supernatant after treatment of bacterial cells with LBI and centrifugation; note that NS5B was not soluble under these conditions; S2, supernatant obtained from solubilization of the pellet remaining from S1; P, pellet remaining after centrifugation of S2; FT, flowthrough obtained after incubation of S2 with Ni-NTA-agarose; E1 to E3, eluted proteins. The numbers on the right refer to the masses, in kDa, of reference proteins run on the same gel. For a more detailed explanation, see Materials and Methods. (B) Reaction products generated on a genomic template RNA. Purified NS5B (0.4 μg) from isolate JFH1 or J6 or an inactive point mutant of JFH1 (GND) as indicated above the gel were incubated with in vitro-transcribed RNA corresponding to a subgenomic replicon (I389LucNS3-3′/JFH; 8.9 kb long) (42) in the presence of [α-32P]CTP. Aliquots of each reaction mixture were taken at the indicated time points. The reaction products were separated by denaturing agarose-glyoxal gel electrophoresis, and the gel was dried and subjected to autoradiography. M, radiolabeled marker RNAs of different lengths as indicated on the right. (C) Specific activities of J6 and JFH1 RdRps on a genomic-RNA template. reaction mixtures identical to those in panel B were harvested at the indicated time points and subjected to TCA precipitation and liquid scintillation counting to quantify the amount of radioactivity incorporated into newly synthesized RNA. The data are given in picomoles of incorporated NMP per microgram of enzyme at the given incubation time.
To compare the specific activities of the RdRps, we first used a positive-strand replicon RNA derived from JFH1 as a template and analyzed the reaction products obtained at different time points by denaturing agarose gel electrophoresis and autoradiography (Fig. 1B). The average lengths and amounts of reaction products linearly increased over time for both RdRps, but the JFH1 enzyme significantly synthesized more reaction products than J6 at each time point, indicating a higher specific activity. We repeated the experiment and analyzed the number of incorporated radioactive nucleotides by TCA precipitation and liquid scintillation counting and observed a specific activity of JFH1 four- to fivefold higher than that of J6 polymerase on this template (Fig. 1C and Table 1, genomic RNA). This difference was reproducible for many different batches of enzymes and also with template RNAs derived from other isolates, indicating that the apparently higher specific activity was not due to specific interactions with cis-acting elements (data not shown). Since these data were consistent with our previous observation that JFH1 polymerase had a higher activity than its Con1 (genotype 1b) counterpart (7), we speculated that JFH1 RdRp might have special enzymatic properties, which might contribute to its outstanding RNA replication capability in cell culture. Therefore, by analyzing different distinct steps of RNA synthesis, we wanted to understand the mechanistic basis underlying the higher specific activity of JFH1 than of J6 RdRp, in particular regarding the elongation rate, RNA binding properties, and initiation efficiency.
TABLE 1.
Specific activities of JFH1 and J6 RdRps on different templates
| Template | cGTP (μM) | Sp act (pmol NTP/μg/h)a |
|
|---|---|---|---|
| JFH | J6 | ||
| Poly(C) | 1 | ND | ND |
| 5 | 1.3 ± 0.5 | 0.2 ± 0.1 | |
| 5,000 | 931 ± 348 | 258 ± 26 | |
| Poly(C)/oligo(G) | 1 | 0.5 ± 0.2 | 0.4 ± 0.2 |
| 5 | 3.7 ± 0.9 | 1.6 ± 0.5 | |
| 5,000 | 525 ± 40 | 392 ± 47 | |
| Genomic RNA | 5,000 | 6.0 ± 0.8 | 1.6 ± 0.6 |
| Antigenomic RNA | 5,000 | 4.1 ± 0.8 | 1.4 ± 0.5 |
ND, not detectable.
Analysis of the elongation rates and processivities of JFH1 and J6 polymerases.
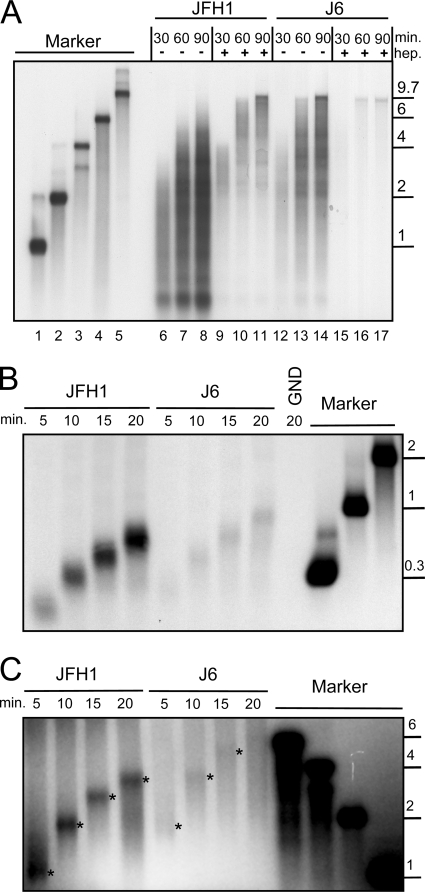
A standard RdRp assay like that shown in Fig. 1B does not allow any conclusions about the elongation rate or processivity of the polymerase, since individual polymerase molecules might fall off the template or reinitiate RNA synthesis de novo or in a primer-dependent mode at any time during the incubation period. To exclude reinitiation, we preincubated a positive-strand template RNA encompassing the entire HCV genome with purified NS5B and started the reaction by adding the nucleotides in the presence of excess amounts of heparin. This polyanion traps all RdRp molecules that are not involved in RNA synthesis at the start of the experiment or that fall off the template during elongation. In the absence of heparin, the reaction products were similar to those of the previous experiment, being very heterogeneous in size, including a large number of short RNAs even after a 90-min incubation time (Fig. 2A, lanes 6 to 8 and 12 to 14). In the presence of heparin, the overall amount of reaction products was reduced, as expected, but most newly synthesized RNAs steadily increased in length over time, reaching template length after 60 to 90 min (Fig. 2A, lanes 9 to 11 and 15 to 17). The maximum length of JFH1 reaction products was about 4 kb after 30 min (lane 9), and some reached template length at about 60 min (lane 10) in the presence of heparin, indicating an elongation rate of ca. 100 to 150 bases per minute. RNAs synthesized by the J6 enzyme had already reached 6 kb in length after 30 min (lane 15), and most of the reaction products had gained the template length of 9.7 kb after 60 min, corresponding to a polymerization velocity of ca. 150 to 200 bases per minute. Along with the lower elongation rate, JFH1 polymerase also appeared to be less processive than the J6 enzyme. This was indicated by a significant amount of reaction products 2 to 6 kb in length remaining at the same size in the presence of heparin at 60 versus 90 min of incubation time (Fig. 2, compare lanes 10 and 11) and by the steadily increasing amounts of very short RNA products (below 1 kb) in the absence of heparin (compare lanes 7 and 8), which was observed only in the case of JFH1 RdRp.
FIG. 2.
Elongation rates and processivity of JFH1 and J6 polymerases on different RNA templates. (A) RNA synthesis of purified JFH1 and J6 RdRps in the presence and absence of heparin on a full-length genomic-RNA template. JFH1 or J6 polymerase, as shown above the gel, was preincubated for 30 min with a genomic template RNA (JC1, 9.7 kb in length) in the presence of all reaction components except nucleotides and heparin (hep.). Reactions were started by adding a nucleotide mixture with or without heparin, as indicated above the gel, and incubated for the indicated time in minutes. (B) Elongation rate on an antigenomic template RNA. A 2-kb antigenomic-RNA template, as shown schematically in Fig. 5A [3′(−)2kb] was incubated with JFH1 or J6 polymerase for the indicated time in the presence of heparin. GND, purified JFH1 polymerase with an inactivating point mutation at D318N. (C) Extension of a radiolabeled oligo(G3) primer on a homopolymeric poly(C) template in the presence of heparin by JFH1 or J6 RdRp. The main reaction product at each time point is marked by an asterisk. All reaction products were analyzed by denaturing agarose-glyoxal gel electrophoresis and autoradiography. Marker, radiolabeled RNAs, with sizes in kilobases on the right, were generated by in vitro transcription. min., incubation time in minutes.
To substantiate our findings, we repeated the experiment in the presence of heparin on a shorter template corresponding to the 3′ portion of the JFH1 negative-strand RNA encompassing 2 kb [3′(−)2kb] (see Fig. 5A for a schematic representation). The average length of the reaction products again constantly grew within the time scale of the experiment (Fig. 2B). Interestingly, elongation was much slower for both polymerases on this template, at ca. 30 nt per minute for JFH1 and ca. 50 nt per minute for J6 polymerase. However, J6 polymerase was still slightly faster than JFH1 on this template RNA.
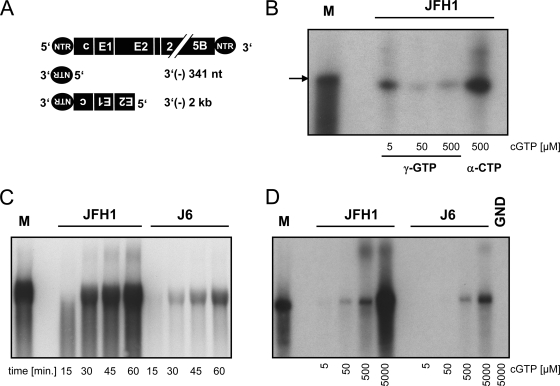
FIG. 5.
De novo initiation efficiency on template RNAs corresponding to the 3′ end of the HCV antigenome. (A) Schematic representation of the template RNAs used for in vitro RdRp assays. Sequences corresponding to the 3′ terminus of the negative strand are shown upside down and in 3′-to-5′ orientation. (B) RNAs synthesized by JFH1 RdRp in the presence of [γ-32P]GTP or [α-32P]CTP. The 341-nt template was used at various concentrations of cold GTP (cGTP) as indicated below the gel with the indicated radiolabeled nucleotides. Reaction products obtained after 1 hour of incubation time were analyzed by denaturing gel electrophoresis on a polyacrylamide-urea gel and autoradiography. The arrow points to the size of the template RNA. (C and D) De novo initiation efficiency of JFH1 and J6 RdRps on the 2-kb template RNA schematically shown in panel A. (C) Time course of RNA synthesis. One microgram each of purified JFH1 and J6 RdRp, as indicated, was incubated for 15 to 60 min as shown below the gel in the presence of 5 mM cold GTP to allow optimal de novo initiation. (D) Impact of the GTP concentration. RdRp assay mixtures with JFH1 or J6 polymerase or an inactive point mutant of JFH1 (GND) as indicated were incubated with the 2-kb template RNA for 1 hour at different GTP concentrations as shown below the gel. The reaction products were analyzed by denaturing agarose-glyoxal gel electrophoresis and autoradiography. M, radiolabeled in vitro transcript corresponding to the template size.
Finally, we measured the elongation of a radiolabeled oligo(G3) primer on a homopolymeric poly(C) template in the presence of heparin (Fig. 2C). The average size of the reaction products constantly increased over time for JFH1 polymerase and reached almost 4 kb after 20 min. The sizes of reaction products generated by J6 polymerase were a little more heterogeneous, but at 5, 10, and 15 min, some distinct product lengths of ca. 2 kb, 4 kb, and 6 kb, respectively, could be determined. Therefore, it seemed that RNA synthesis was fastest on a poly(C) template, reaching ca. 200 nt per minute for JFH1 and almost 400 nt per minute for J6 polymerase.
In summary, we found different elongation rates of the HCV polymerase on different template RNAs, but in all cases, the JFH1 enzyme exhibited a lower elongation rate and was less processive than its J6 counterpart. Therefore, the elongation rate and processivity cannot account for the increased specific activity of JFH1 polymerase.
RNA binding.
The initial binding between NS5B and an RNA template could be measured using an SPR biosensor and was intended to provide a qualitative and comparative measure for the two NS5B variants. The polymerase molecules were immobilized, and substrate RNAs derived from the 3′ end of the positive-strand genome, including an important cis-acting RNA element in NS5B (16, 50) (Fig. 3A), as well as the negative strands [3′(−) JFH1 or J6, respectively] (Fig. 3B), were injected. We tested RNAs derived from both isolates to exclude subtle differences in binding affinity due to potential isolate-specific cis-acting signals. Titration with increasing concentrations of RNA revealed a stoichiometry of only about 0.05 for the protein-RNA complex (data not shown), which compares to previously reported low values (20% [25]). The possibility of multiple NS5B molecules on the biosensor surface binding to the same RNA strand might account for the lower binding capacity determined here. Similar SPR sensorgrams obtained with 3′ JFH1 RNA and 3′ J6 RNA, positive or negative strand, showed that NS5B JFH1 and NS5B J6 were indistinguishable in their abilities to bind RNA. The same was true for binding to the genomic replicon RNA template used in Fig. 1A and poly(U) and poly(C) RNAs (data not shown).
FIG. 3.
RNA binding of JFH1 and J6 NS5B measured by an SPR biosensor. (A) Interaction between immobilized NS5B variants (JFH1, black; J6, gray) and template RNA [3.3 μg/ml 3′(+) JFH1 RNA; 0.86 μg/ml 3′(+) J6 RNA], corresponding to the 3′ region of the respective positive-strand genome. (B) Interaction between immobilized NS5B variants (JFH1, black; J6, gray) and the 3′ portion of the JFH1 or J6 negative-strand RNA [50 μg/ml 3′(−) JFH1 RNA; 3.1 μg/ml 3′(−) J6 RNA], each 341 nt in length [3′(−)341nt] (Fig. 5A). The SPR biosensor response scale on the y axis was normalized so that 1 corresponds to a 1:1 stoichiometry (RNA-NS5B).
Although we could not find any differences in RNA binding of the two enzymes, differences in the apparent binding capacities for the different RNAs were observed, with 3′(+) RNA > 3′(−) RNA and poly(U) > poly(C); the latter showed almost no binding to NS5B, as previously described (28). However, these data are difficult to interpret, since an RNA strand has many sites that can interact with NS5B, and therefore, the actual concentration of functional RNA binding sites in this experimental setup was not clearly defined. The apparent binding capacity might also vary for RNAs of different lengths due to the possibility of multiple binding to several protein molecules. Therefore, affinity constants could not be determined with this experimental setup, but only comparative data sets.
Still, our data clearly demonstrate that the higher in vitro activity of the JFH1 enzyme did not originate from a higher affinity for RNA or a higher ratio of RNA binding-competent enzyme in the protein preparation.
De novo and primer-dependent initiation on homopolymeric template RNA.
HCV NS5B can initiate RNA synthesis de novo (33, 51) or in a primer-dependent manner by so called copy-back initiation (6). Due to the complexity of reaction products synthesized on templates corresponding to viral genomes and the potential presence of short RNAs in these in vitro transcripts that could act as primers, we focused on homopolymeric templates with and without primers to differentiate between these two initiation mechanisms. Since poly(C) was the only homopolymeric template giving rise to efficient RNA synthesis (28, 33), we used either poly(C) for analysis of de novo initiation or poly(C) and an oligo(G12) primer for primer-dependent initiation of RNA synthesis.
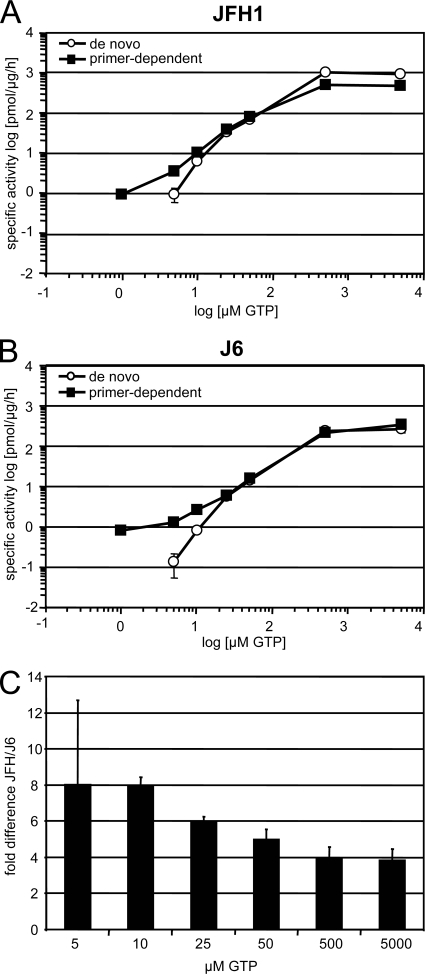
At a total GTP concentration of 1 μM, no primer-independent incorporation of radioactivity could be observed, indicating that de novo initiation did not occur under these conditions (Fig. 4 and Table 1). This is in line with previous results showing that de novo initiation requires a certain threshold concentration of GTP (33). In contrast, considerable amounts of radioactive products were generated in a primer-dependent mode by both polymerases. Interestingly, the two enzymes exhibited almost identical specific activities under these conditions, thereby demonstrating that the higher specific activity of JFH1 NS5B was not due to primer-dependent initiation of RNA synthesis. When GTP concentrations were raised to 5 μM, both polymerases gave rise to significant de novo initiation of RNA synthesis; however, JFH1 polymerase was on average 6- to 10-fold more efficient than J6 under these conditions, demonstrating that the increased in vitro activity of JFH1 NS5B was due to efficient de novo initiation.
FIG. 4.
De novo and primer-dependent initiation of RNA synthesis on homopolymeric templates. Standard RdRp assay mixtures using 1 μg of JFH1 (A) or J6 (B) RdRp were incubated for 1 hour with a poly(C) (de novo) or a poly(C)/oligo(G12) (primer-dependent) template with 10 μCi [α-32P]GTP and various concentrations of cold GTP, as shown on the x axis. Incorporation of radioactivity was quantified by TCA precipitation and liquid scintillation counting. After background subtraction, the picomoles of incorporated GMP per microgram of enzyme per hour were calculated based on the ratio of labeled to unlabeled GTP. (C) Difference in de novo initiation efficiencies between JFH1 and J6 RdRp at various GTP concentrations. The data represent the ratio of specific activities of JFH1 versus J6 polymerase on poly(C) templates, as shown in panels A and B at the indicated GTP concentrations.
Previous studies have shown that high concentrations of GTP dramatically stimulate the enzymatic activity of NS5B in vitro (30), most probably by stimulating de novo initiation of RNA synthesis (33) by still barely defined mechanisms. We analyzed the impact of increasing GTP concentrations on the de novo initiation efficiencies of JFH1 and J6 RdRps to evaluate whether the difference in specific activity remained constant (Fig. 4A and B and Table 1). Enzymatic activity was dramatically stimulated by increasing concentrations of GTP, reaching a plateau at 500 μM for both polymerases. Starting from 25 μM, de novo initiation remained the predominant mode of initiation on both templates, since addition of an oligo(G) primer did not further enhance the overall specific activity. Oligo(G12) even had a slight inhibitory effect, particularly on JFH1 RdRp at very high GTP concentrations (Fig. 4A, 500 and 5,000 μM GTP). The specific activity of JFH1 remained higher at all analyzed GTP concentrations. However, when we calculated the difference in specific activity between JFH and J6, we found an eightfold difference at low concentrations of GTP and only a fourfold difference at high concentrations of GTP (Fig. 4C), indicating that high GTP concentrations did compensate to some extent for the lower capability of J6 polymerase to initiate de novo and showing that JFH1 was more efficient, particularly in de novo initiation at low GTP concentrations, than J6.
These data clearly demonstrate that the higher specific activity of JFH1 polymerase was due to its increased capability to initiate RNA synthesis de novo, whereas primer-dependent initiation was equally efficient for both polymerases.
De novo initiation of RNA synthesis on an authentic part of the HCV genome.
Next, we aimed to analyze the characteristics found on homopolymeric RNAs on more authentic RNA templates. Since templates containing the 3′ end of the positive strand always gave rise to very heterogeneous reaction products (Fig. 1B) (7), we used RNAs derived from the 3′ end of the negative strand [3′(−)341 nt] (Fig. 5A), which predominantly yielded reaction products of template size in previous studies (7). By using [γ-32P]GTP and a 341-nt template corresponding to the 3′ end of the HCV antigenome, we first wanted to clarify the nature of the reaction products synthesized on these templates. In the presence of [γ-32P]GTP, only those reaction products incorporating the triphosphorylated nucleotide at the 5′ end are radiolabeled, and this happens only in cases of de novo initiation. As shown in Fig. 5B, the predominant reaction product generated by JFH1 polymerase in the presence of [γ-32P]GTP was identical in size to the template RNA and to those RNAs labeled with [α-32P]CTP. Therefore, the majority of newly synthesized RNA on this template was most likely de novo initiated at the very 3′ end, which represents the way RNA synthesis is expected to take place in vivo. The apparent decrease in band intensity with increasing concentrations of GTP was due to the higher dilution of the radiolabeled [γ-32P]GTP, which was not fully compensated for by the expected higher initiation efficiency at 50 μM and 500 μM GTP.
For the comparative analysis, we chose a 2-kb template RNA corresponding to the 3′ end of the HCV negative strand [(3′(−)2 kb)] (Fig. 5A) and analyzed the activities of JFH1 and J6 polymerases at high GTP concentrations (5 mM) in a time course experiment (Fig. 5C). Again, the predominant reaction product was a template-size RNA concordant with the expectation of RNAs initiated de novo at the very 3′ end of the template. As for the homopolymeric template, JFH1 was also clearly more efficient in de novo initiation than J6 using this authentic RNA substrate obtained from the HCV genome, arguing for the physiological relevance of our results. We also analyzed the GTP dependence of de novo initiation on this template RNA (Fig. 5D). The stimulating effect of increasing GTP concentrations on RNA synthesis was obvious for both enzymes, but JFH1 was again more efficient at all GTP concentrations. At 5 μM and 50 μM GTP, J6 RdRp did not synthesize any detectable amounts of RNA, whereas JFH1 RdRp produced small but significant amounts of reaction products, substantiating the results obtained with poly(C). At high GTP concentrations, JFH1 polymerase was four- to fivefold more efficient than J6, as determined by phosphorimaging. Similar results were obtained for the 341-nt template (data not shown).
In summary, the higher specific activity of JFH1 NS5B could clearly be attributed to its better capability to also initiate RNA synthesis de novo on authentic template RNAs, and this might contribute to its unique properties in cell culture.
Structural analysis of JFH1 polymerase.
To gain deeper insight into the structural basis of the special features of JFH1 polymerase, we solved the crystal structure of the soluble construct used in the activity assays (Table 2 shows crystallographic details). JFH1 NS5B was crystallized in a slightly but significantly more closed conformation than previously reported for other HCV strains (all four JFH1 NS5B molecules in the crystal asymmetric unit have the same conformation within experimental error). This is most readily shown by comparing its structure to the available NS5B structure with the highest sequence identity, that of the consensus sequence 2a polymerase (hereafter called con2a NS5B) (8). Compared with JFH1ΔC21, 28 residues are mutated in J6 NS5B and 20 residues in con2a NS5B. Eighteen of the latter are identical in con2a NS5B and J6 NS5B. The structure of con2a NS5B was reported in two crystal forms: one structure displayed a closed conformation (form I; PDB code 1YUY), while in the other the molecule's conformation was more open (form II; PDB codes 1YVX, 1YVZ, and 1YV2). We used the program ESCET (43) to objectively assess significant movements and rigid domains between JFH1 NS5B and the closed and open forms of con2a NS5B. ESCET was developed to objectively identify rigid domains in proteins, accounting for coordinate errors in X-ray crystal structures. These errors are estimated for each atom and are used to test the identity of all pairs of three-dimensional models considered. Models are thus characterized as identical or nonidentical within a given error threshold (typically 2 to 3 sigmas, but higher levels can be chosen if one wishes to retain only large differences; see the legend to Table 3). Nonidentical models were further analyzed to iteratively yield a division in rigid (identical within error) subdomains. Table 3 shows that all three structures considered are conformationally different from each other when all significant differences are considered (leftmost Table 3 values; lolim = 2.0). Indeed, JFH1 NS5B is more closed than even the closed form of con2a (Fig. 6B), although this difference is sufficiently small that it becomes blurred when one considers only larger differences (rightmost Table 3 values; lolim = 5.0). At this crude level of analysis, ESCET delineates two major moving blocks, the smaller being the thumb and the larger the palm and fingers (Fig. 6C, right), as had been previously reported for subtype 1b NS5B by Adachi et al. (1). At a finer analysis level, ESCET further breaks up the NS5B structure (Fig. 6A and B). There are three large (>50-residue) blocks, making up most of the fingers, thumb, and palm. Three smaller blocks (about 20 residues each) were also highlighted by the ESCET analysis. Strikingly, they all pinpoint zones of polymorphism between JFH1 and other HCV 2a strains.
TABLE 2.
Crystallographic detailsa
| Parameter | Value |
|---|---|
| Data collection and processing | |
| Unit cell parameters (Å; degrees) | a = 130.60, b = 115.81, |
| c = 133.24; β = 107.19 | |
| Space group | P21 |
| X-ray source; wavelength | Soleil synchrotron, Proxima 1; 0.975 Å |
| Resolution range (Å) | 30-2.2 (2.3-2.2) |
| No. of unique reflections | 191, 348 |
| Redundancy | 3.8 (3.9) |
| Completeness (%) | 99.7 (99.9) |
| Rmerge (%) | 11.7 (62.0) |
| I/sigma(I) | 8.65 (2.13) |
| No. of Molecules in asymmetric unit | 4 |
| Refined model | |
| R factors | |
| Rwork (%) | 19.1 |
| Rfree (%) | 22.6 |
| Rmsd, bond lengths (Å) | 0.013 |
| Rmsd, bond angles (degrees) | 1.36 |
| No. of non-H atoms | 19,472 |
Numbers in parentheses refer to the highest resolution shell. Rmerge=ΣhΣj|Ih,j−<Ih>|/ΣhΣjIh,j, where <Ih> is the average intensity of the reflection of unique index h with j observations (j>1). Rwork=Σh||Fobs|−k|Fcalc||/Σh|Fobs| for the 187,370 reflections used in refinement; Rfree was computed with the same formula for the 3,969 reflections not used in refinement.
TABLE 3.
Comparison of three conformationally different subtype 2a NS5B structures
| Parametera | Valueb |
||
|---|---|---|---|
| JFH1_B | 1YUY (con2a, closed) | 1YVX (con2a, open) | |
| <esd> (Å) | 0.14 | 0.10 | 0.12 |
| JFH1_B | 90.4/94.9/99.9 | 57.7/63.5/80.5 | |
| 1YUY | 58.9/64.3/80.8 | ||
JFH1_B, most precisely refined JFH1 NS5B in the four-molecule asymmetric unit. <esd>, mean coordinate uncertainty for alpha carbons.
The values are the percentages of residues whose alpha carbons are not in significantly different positions given a tolerance of 2.0, 2.5, and 5.0 sigmas (ESCET lolim parameter). Two models can be considered to be in identical conformations if this value is larger than 98% (43).
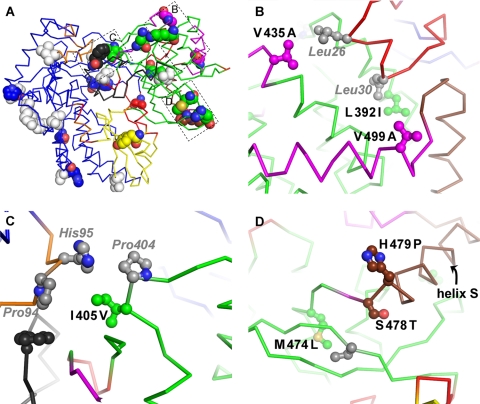
First, a region at the top of the thumb harbors V435A and V499A (Fig. 7B) (we used JFH1 as the prototype subtype 2a sequence and hence denote the polymorphisms as mutations of con2a/J6 from the JFH1 sequence). V435A and the nearby L392I strengthen the interaction of the thumb with helix A of the fingertips. V435, at the base of the beta flap, makes hydrophobic contacts with L26 in helix A of the fingertips, an interaction that is not made by A435 even in the con2a closed form I. In the open con2a form II, L26 actually moves to occupy the hydrophobic pocket lined with I392 and occupied by L30 in the closed forms. L392 in JFH1 NS5B leads to a slight bulge in this pocket, likely stabilizing the interaction with L30 that tethers helix A to the thumb in the closed forms and making the replacement by L26 seen in con2a form II less favorable. V499 is part of the GTP surface site (10) and may have a possible indirect influence on helix A interaction. The second smaller moving block (Fig. 6 and 7) is located in part at the top of the fingers. In this region (Fig. 7C), the unusual JFH1 I405V polymorphism helps buttress the interaction between H95 and P404 that bridges the gap between the thumb and the top of the fingers in the closed forms. The extra methyl group in I405 indeed makes direct contact with the H95 side chain, as well as extra contacts with the beta flap. When the differences between JFH1 NS5B and con2a form II are considered, residues 405 and 95 move apart by ∼2.5 Å, opening the access to the catalytic cleft. In the closed forms, the linker is ordered and residue 561 participates in interactions with both the fingers (through contacts with P94) and the beta flap, although the F561Y polymorphism entails one hydrogen bond less between the linker and the beta flap in JFH1 NS5B. The third smaller moving block (Fig. 6 and 7) is, unexpectedly, helix S of the thumb. This helix thus moves independently of the rest of the thumb, and the loop at its top that is displaced by this movement is part of both the interaction site with helix A and the surface GTP binding site. Another unusual JFH1 polymorphism, H479P, lies at the base of helix S, together with the more common S478T (Fig. 7D). In this region, M474L modifies the hinge around which the main thumb rotation (Fig. 6C) occurs, although the effect (if any) of this is uncertain.
FIG. 7.
JFH1 polymorphisms contributing to the stabilization of the closed thumb conformation. (A) View of JFH1 NS5B as in Fig. 5A, left, but in alpha carbon representation. The 28 polymorphisms of JFH1 compared to J6 are displayed as all-atom spheres. The 18 that are identical in J6 and con2a are colored by atom types for oxygens (red), nitrogens (blue), and sulfurs (yellow) and by rigid zones for carbons. The 10 polymorphisms that are identical in JFH1 and con2a, and on the effect (if any) of which we consequently have no structural information (S42T, A62S, S79T, L85M, Q131E, Q241R, R250H, V309I, V421A, and S450A), are colored white. The boxed areas correspond to the locations of panels B, C, and D. (B, C, and D) Close-ups of the three smaller rigid zones (in magenta, orange, and brown, respectively). Polymorphisms are labeled in black, colored as in panel A, and displayed as ball and stick diagrams. Some of the (conserved) residues interacting with them are labeled in gray italics, colored gray, and displayed as ball and stick diagrams.
This accounts for 8 of the 18 residues that differ when JFH1 is compared to con2a/J6. The other 10 polymorphisms are sprinkled over the enzyme's surface. We could not discern any influence of these on NS5B conformation. We note that some are in regions that have been reported as important for in vitro activity, e.g., positions 150 (22), 57 and 63 (close to positions 345) (28), and 65 (32). Others, like N455S and K517R, induce a remodeling of the surface of NS5B. These two mutations lead to a large change in the pattern of hydrogen bonding at the base of the beta flap. Remarkably, this does not lead to any conformational change in this part of the structure.
DISCUSSION
The present study provides evidence that the polymerase of the HCV isolate JFH1 contains special structural and functional properties that might contribute to its outstanding replication efficiency in cell culture. By comparing the enzymatic activities of NS5B from the isolates JFH1 and J6, we could show that the JFH1 RdRp exerts a significantly higher specific activity, which is due to its greater capability to initiate RNA synthesis de novo.
The increased de novo initiation efficiency of the polymerase fits well with the replication kinetics of JFH1 in living cells. Previous studies have shown extremely fast replication kinetics of subgenomic replicons derived from this isolate (7). After a short lag phase of 4 h, the first synthesis of the negative strand could be detected, which exponentially amplified in the following 20 hours and thereafter reached a peak level that was most likely limited by host cell determinants; this was in striking contrast to replicons derived from other isolates that gave rise to a much slower linear amplification of RNA (21, 26). The lag phase might represent the time required for excessive translation of the viral RNA (39) and the generation of the membrane alterations containing the viral replication complexes and is similar in length for cell culture-adapted Con1 replicons (V. Lohmann, unpublished data). The next step, amplification of the RNA, is much faster for JFH1 and was most likely determined by the ability of a replication complex to initiate RNA synthesis. Some evidence that the initiation step of RNA synthesis is indeed rate limiting in cell culture is provided by the strong impact of chimeric 5′- and 3′-NTR sequences specifically on the efficiency of positive- and negative-strand synthesis of JFH1 (7). Our results now indicate that the competence of NS5B to initiate RNA synthesis might be an important limiting determinant in this process.
Currently it is not clear to what extent the increased de novo initiation efficiency of the viral RdRp contributes to the overall replication competence of JFH1. Given the genetic evidence provided by Murayama et al. (35), it is certain that the enzymatic activity of NS5B is not the sole determinant defining the particular properties of JFH1 but that additional factors reside in the region encoding helicase and in the 3′ NTR. In addition, cis-acting elements within the NS5B coding region (16, 50) or functions in the C-terminal membrane insertion sequence (34), which was excluded from our analysis, might also be significant. In essence, the requirements for efficient JFH1 replication substantially overlap with the cis- and trans-acting factors that are directly involved in initiation of RNA synthesis, i.e., helicase, NS5A, NS5B, and the NTRs (7). Therefore, the outstanding efficiency of the JFH1 nonstructural proteins might be due to the particular architecture of the entire replicase and the corresponding cis-acting elements. A subtle analysis of mutant purified NS5B polymerases and replicons in parallel will be necessary to clarify the contribution of RdRp activity in this complex. These analyses are beyond the scope of this study, but the structural analysis of the JFH1 polymerase will now allow valuable predictions of how to modulate the conformation of NS5B to facilitate de novo initiation and to analyze the impact of these mutations on replication efficiency in cell culture.
To this end, a very recent study confirmed that JFH1 polymerase also had a substantially higher specific activity in vitro than a genotype 1b polymerase (47), which is in line with our previously published results (7). By converting 11 individual residues in the genotype 1b enzyme to their JFH1 counterparts, Weng et al. found some enhanced enzymatic activity for residues 377, 450, 455, and 561, all of which are located in the thumb domain. These results provide some experimental evidence for the contribution of the thumb domain to higher JFH1 RdRp activity, as suggested by our structural analysis. Still, it will be most critical to analyze whether the same mutations that increase polymerase activity in vitro really contribute to efficient RNA replication in cell culture.
Although our structural analysis compares con2a NS5B with JFH1 NS5B (while our biochemical data compare J6 NS5B with JFH1 NS5B), we surmise that our conclusions likely also hold for a JFH1/J6 structural comparison. Indeed, J6 and con2a NS5Bs are closer (12 substitutions in the Δ21 forms, all of them common polymorphisms in 2a strains) than either is to JFH1 NS5B (28 and 20 substitutions, respectively, 18 of which are the same in con2a NS5B and J6 NS5B). Of these 18 substitutions, several are very unusual polymorphisms of JFH1 (e.g., L392, I405, V435, M474, H479, and V499 are virtually unknown in other 2a strains). The structure of JFH1 NS5B revealed the closest conformation of all HCV RdRps published so far. In fact, all available NS5B structures represent closed conformations in the sense that they are suited to hold a single-stranded template and the nucleotides required for de novo initiation of RNA synthesis but too closed for egress of a double-stranded RNA and thus do not allow productive elongation. This suggests that the closed conformation should be competent for de novo initiation and that the transition from initiation to processive elongation requires a conformational change of the polymerase. Biochemical analyses showing that a single-stranded portion at the 3′ end of the template RNA is indeed required for de novo initiation support this assumption (18). The small difference we have observed for the JFH1 structure could be dismissed as the result of crystal-packing forces, but our structural and biochemical analysis points to an involvement in de novo initiation. First, the four JFH1 NS5B molecules in the crystal asymmetric unit make different crystal contacts but are in identical main chain conformations. Second, the polymorphisms stabilizing the JFH1 NS5B closed conformation, though mostly conservative substitutions, bridge gaps that would be left by the less bulky J6 residues. Given the higher de novo initiation rate of the JFH1 enzyme we found in vitro, this subtle conformational difference actually seems to make the difference between being closed enough to initiate often (JFH1 NS5B) and being closed enough to initiate sometimes, which seems to account for all other NS5B structures.
Initiation of RNA synthesis in vitro can be further subdivided into distinct steps (25): binding of the RNA, formation of a productive RNA-enzyme complex, and synthesis of a trinucleotide primer, followed by the transition from initiation to elongation, which probably requires a conformational change of the enzyme. In accordance with published data (25), we found only a subfraction of JFH1 and J6 NS5B bound to RNAs corresponding to the 3′ ends of the genomes of both isolates, but with identical affinities and kinetics for both enzymes and templates. However, our binding assay, as well as most of those used by others, cannot distinguish between nonproductive and productive binding of the polymerase to RNA. Previous results indicated that the formation of a productive RNA-enzyme complex was the slowest step in the catalysis of RNA synthesis by NS5B on a heteropolymeric template, which can take hours to days (25). This might be due to the necessity to thread a single-stranded template into the closed conformation of the enzyme (14, 18, 20). Enhancement of this critical step might therefore represent the major difference between JFH1 and J6 polymerases, since the majority of RNA-protein binding events measured in the SPR might represent nonproductive binding events, thereby hiding a critical difference in productive binding. Still, increased productive binding of RNA seems not to be very likely for de novo initiation on poly(C), since our current and previous analyses clearly showed that NS5B does not stably bind to this RNA at all (28). Alternatively, formation of the initial trinucleotide or the switch from initiation to elongation might be enhanced for JFH1 RdRp, since these steps sum up to the apparent de novo initiation efficiency determined in our assays. We favor a model in which the closed conformation stabilizes the ternary complex of enzyme-template and priming nucleotides, thereby facilitating the formation of the first di-/trinucleotide. This conclusion is borne out by the fact that the pattern of polymorphisms stabilizing the closed JFH1 NS5B conformation should facilitate this very first step of RNA synthesis. By the same argument, it should also somewhat hinder the transition to elongation, which requires opening of NS5B. However, since the extra interactions involved are hydrophobic in nature (Fig. 7B and C), and therefore short range, they vanish as soon as NS5B starts to open. The tradeoff is therefore small and obviously in favor of the more efficiently initiating enzyme.
The difference in de novo initiation efficiency between JFH1 and J6 RdRps decreased with increasing GTP concentrations. The mechanism underlying the stimulating effect of high GTP concentrations on HCV and also pestiviral RdRps is still somewhat enigmatic (30). Several studies have already shown that elevated concentrations of the initiating nucleotide, GTP or ATP, are required to allow de novo initiation (33, 41). A recent study showed that in fact high concentrations of nucleotide triphosphates corresponding to all of the first three incorporated nucleotides are required for efficient de novo initiation, with apparent Km values in the range of 100 to 400 μM (15). This might explain at least some of the stimulating effect of GTP, since both templates used for de novo initiation analysis in our study depended on GTP as the initiating nucleotide. In addition, high concentrations of GTP have been shown to actively suppress primer-dependent initiation of RNA synthesis, most probably by stabilizing the closed, de novo initiation-competent conformation of the enzyme (14, 40). This model best fits our results, since primer-dependent initiation indeed does not significantly contribute to RNA synthesis at GTP concentrations above 100 μM (Fig. 4A and B and Table 1) and the difference in de novo initiation efficiency between JFH1 and J6 polymerases decreased at high GTP concentrations (Fig. 4C). An extensive mutagenic analysis focusing on the polymorphisms between JFH1 and J6 and analyzing their impacts on the stimulating effect of GTP on de novo initiation will probably help us to further understand the underlying mechanism. Beyond this mechanistic insight, it is hard to predict whether the reduced GTP dependence of JFH1 RdRp has any physiological relevance for replication in cell culture, since intracellular GTP concentrations are typically in the range of 500 μM (45).
In conclusion, we found that the polymerase of the HCV isolate JFH1 has a very closed conformation, stabilized by hydrophobic interactions not present in other HCV isolates, and efficiently supports de novo initiation of RNA synthesis in vitro, which might contribute to the outstanding properties of the isolate. Understanding the molecular basis of efficient JFH1 replication in cell culture might help to modulate the replication competence of other viral isolates in a way that does not interfere with the generation of infectious particles and thereby widens the availability of HCV variants accessible for studies of the entire viral life cycle in vitro.
Acknowledgments
We are grateful to T. Wakita and J. Bukh for the generous gift of plasmids, I.-H. Shih for helpful discussions, and R. Bartenschlager for critically reading the manuscript. We thank the Structural Biology and Proteomics pole of the IMAGIF integrated platform (https://www.imagif.cnrs.fr/?nlang=en) for access to crystallization and mass spectrometry services and Synchrotrons SOLEIL (beamline PROXIMA 1) and ESRF (European Synchrotron Radiation Facility) for generous allocation of beam time.
This work was funded by Deutsche Forschungsgemeinschaft (DFG) grant Lo 1556/1-1 to V.L. and by grants from the European Community (VIRGIL Network of Excellence, grant LSHM-CT-2004-503359) and l'Agence Nationale de Recherches sur le SIDA et les Hépatites Virales (ANRS) to S.B. P.S. acknowledges an ANRS postdoctoral fellowship.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 3.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 6.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 7.Binder, M., D. Quinkert, O. Bochkarova, R. Klein, N. Kezmic, R. Bartenschlager, and V. Lohmann. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J. Virol. 81:5270-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswal, B. K., M. M. Cherney, M. Wang, L. Chan, C. G. Yannopoulos, D. Bilimoria, O. Nicolas, J. Bedard, and M. N. James. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202-18210. [DOI] [PubMed] [Google Scholar]
- 9.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 10.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, T., K. Mackey, and T. Du. 2004. Analysis of RNA by Northern and slot blot hybridization. Curr. Protoc. Mol. Biol. Chapter 4, unit 4.9. [DOI] [PubMed]
- 13.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 14.Chinnaswamy, S., I. Yarbrough, S. Palaninathan, C. T. Kumar, V. Vijayaraghavan, B. Demeler, S. M. Lemon, J. C. Sacchettini, and C. C. Kao. 2008. A locking mechanism regulates RNA synthesis and host protein interaction by the hepatitis C virus polymerase. J. Biol. Chem. 283:20535-20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrari, E., Z. He, R. E. Palermo, and H. C. Huang. 2008. Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation. J. Biol. Chem. 283:33893-33901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 18.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 20.Kim, Y. C., W. K. Russell, C. T. Ranjith-Kumar, M. Thomson, D. H. Russell, and C. C. Kao. 2005. Functional analysis of RNA binding by the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 280:38011-38019. [DOI] [PubMed] [Google Scholar]
- 21.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labonte, P., V. Axelrod, A. Agarwal, A. Aulabaugh, A. Amin, and P. Mak. 2002. Modulation of hepatitis C virus RNA-dependent RNA polymerase activity by structure-based site-directed mutagenesis. J. Biol. Chem. 277:38838-38846. [DOI] [PubMed] [Google Scholar]
- 23.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 24.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Y., W. W. Jiang, J. Pratt, T. Rockway, K. Harris, S. Vasavanonda, R. Tripathi, R. Pithawalla, and W. M. Kati. 2006. Mechanistic study of HCV polymerase inhibitors at individual steps of the polymerization reaction. Biochemistry 45:11312-11323. [DOI] [PubMed] [Google Scholar]
- 26.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 30.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 31.Lohmann, V., A. Roos, F. Körner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 32.Lou, H., Y. H. Choi, J. E. LaVoy, M. E. Major, and C. H. Hagedorn. 2003. Analysis of mutant NS5B proteins encoded by isolates from chimpanzees chronically infected following clonal HCV RNA inoculation. Virology 317:65-72. [DOI] [PubMed] [Google Scholar]
- 33.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moradpour, D., V. Brass, E. Bieck, P. Friebe, R. Gosert, H. E. Blum, R. Bartenschlager, F. Penin, and V. Lohmann. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 78:13278-13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murayama, A., T. Date, K. Morikawa, D. Akazawa, M. Miyamoto, M. Kaga, K. Ishii, T. Suzuki, T. Kato, M. Mizokami, and T. Wakita. 2007. The NS3 helicase and NS5B-to-3′X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells. J. Virol. 81:8030-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh, J. W., T. Ito, and M. C. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietschmann, T., M. Zayas, P. Meuleman, G. Long, N. Appel, G. Koutsoudakis, S. Kallis, G. Leroux-Roels, V. Lohmann, and R. Bartenschlager. 2009. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS. Pathog. 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranjith-Kumar, C. T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330:675-685. [DOI] [PubMed] [Google Scholar]
- 41.Ranjith-Kumar, C. T., Y. C. Kim, L. Gutshall, C. Silverman, S. Khandekar, R. T. Sarisky, and C. C. Kao. 2002. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 76:12513-12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaller, T., N. Appel, G. Koutsoudakis, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider, T. R. 2004. Domain identification by iterative analysis of error-scaled difference distance matrices. Acta Crystallogr. D 60:2269-2275. [DOI] [PubMed] [Google Scholar]
- 44.Tao, Y., D. L. Farsetta, M. L. Nibert, and S. C. Harrison. 2002. RNA synthesis in a cage—structural studies of reovirus polymerase λ3. Cell 111:733-745. [DOI] [PubMed] [Google Scholar]
- 45.Traut, T. W. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]
- 46.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng, L., J. Du, J. Zhou, J. Ding, T. Wakita, M. Kohara, and T. Toyoda. 2009. Modification of hepatitis C virus 1b RNA polymerase to make a highly active JFH1-type polymerase by mutation of the thumb domain. Arch. Virol. 154:765-773. [DOI] [PubMed] [Google Scholar]
- 48.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 49.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]