Abstract
Rhinoviruses are prevalent human pathogens that are associated with life-threatening acute asthma exacerbations. The innate immune response to rhinovirus infection, which may play an important role in virus-induced asthma induction, has not been comprehensively investigated. We examined the innate immune response in cells infected with human rhinovirus 1a (HRV1a). Beta interferon (IFN-β) mRNA was induced in HRV1a-infected cells at levels significantly lower than in cells infected with Sendai virus. To understand the basis for this observation, we determined whether components of the pathway leading to IFN-β induction were altered during infection. Dimerization of the transcription factor IRF-3, which is required for synthesis of IFN-β mRNA, is not observed in cells infected with HRV1a. Beginning at 7 h postinfection, IPS-1, a protein that is essential for cytosolic sensing of viral RNA, is degraded in HRV1a-infected cells. Induction of apoptosis by puromycin led to the cleavage of IPS-1, but treatment of HRV1a-infected cells with the pan-caspase inhibitor, zVAD, did not block cleavage of IPS-1. IPS-1 is cleaved in vitro by caspase-3 and by the picornaviral proteinases 2Apro and 3Cpro. Expression of HRV1a and polioviral 2Apro and 3Cpro led to degradation of IPS-1 in cells. These results suggest that IPS-1 is cleaved during HRV1a infection by three different proteases. Cleavage of IPS-1 may be a mechanism for evasion of the type I IFN response, leading to a more robust infection.
Human rhinoviruses, positive-stranded RNA viruses of the Picornaviridae family, account for greater than 50% of all upper respiratory tract infections (28). Although usually mild and self-limiting, viral upper respiratory tract infections are one of the most common illnesses in humans, with 500 million cases and an economic burden estimated at $40 billion annually in the United States (8). Furthermore, rhinovirus is implicated in causing severe exacerbation of other diseases including chronic bronchitis, sinusitis, and asthma (22).
The innate immune response to rhinovirus infection may play an important role in rhinovirus-induced asthma induction. Viral infection leads to a signaling cascade that stimulates the antiviral innate immune response, limiting viral replication and initiating the adaptive immune system (14). Two sensors for viral RNA have been described, retinoic acid-inducible gene (RIG-I) and melanoma differentiation associated gene-5 (MDA-5) (12, 17, 37). Each RNA sensor comprises a DEXD/H box RNA helicase domain and a caspase recruitment and activation domain (36). MDA-5 and RIG-I appear to sense infections by different viruses. RIG-I, which detects double-stranded RNA or single-stranded RNA with a 5′-triphosphate, is responsible for alpha/beta interferon (IFN-α/β) induction by paramyxoviruses, flaviviruses, influenza viruses, and Japanese encephalitis virus including Newcastle disease virus and Sendai virus (SeV) (11, 31, 35-37). MDA-5 recognizes double-stranded RNA and is the critical sensor of infection by encephalomyocarditis virus and mengovirus, two members of the picornavirus family (10, 13).
Both MDA-5 and RIG-I interact with the caspase recruitment and activation domain-containing adaptor protein, IFN-β promoter stimulator 1 (IPS-1) (also known as MAVS, VISA, or Cardiff) (15, 24, 26, 32). IPS-1 is localized to the outer mitochondrial matrix and activates the kinases IΚΚɛ (inhibitor of nuclear factor kappa B[NF-κB] kinase) and TBΚ1 (TANK-binding kinase), which are required for the phosphorylation of IFN regulatory factor 3 (IRF-3) (9). Phosphorylated IRF-3 dimerizes and translocates to the nucleus where it recruits p300/CREB binding protein (20, 33). IRF-3 cooperatively binds with the transcription factors NF-κB and AFT-2/c-Jun to form an enhancesome on the IFN-β promoter (23). This complex leads to the activation of the IFN-β gene and synthesis of type I IFNs.
Numerous mechanisms for circumvention of the innate immune response pathway have been revealed in virus-infected cells. The study of these strategies has illuminated the function of innate sensing pathways (reviewed in reference 21). For example, the proteases of hepatitis C, GB, and A viruses cleave IPS-1 (6, 34), and both MDA-5 and RIG-I are degraded in cells infected with picornaviruses (2, 3).
The innate response to rhinovirus infection has not been comprehensively investigated. We therefore examined induction of IFN-β during infection with HRV1a. IFN-β mRNA was detected in HRV1a-infected cells at levels significantly lower than in cells infected with SeV. To understand the basis for this observation, we determined whether components of the pathway leading to IFN-β induction were altered during infection. Dimerization of the transcription factor IRF-3, which is required for synthesis of IFN-β mRNA, is not observed in cells infected with HRV1a. When IRF-3-depleted cells were infected with SeV or HRV1a, small amounts of IFN-β mRNA were produced. The level of IFN-β mRNA was similar to that observed during HRV1a infection of the parental HeLa cell line. IRF-3 depletion did not affect HRV1a growth kinetics. Beginning at 7 h postinfection, IPS-1, a protein that is essential for cytosolic sensing of viral RNA, was degraded in HRV1a-infected cells. Induction of apoptosis by puromycin also led to the cleavage of IPS-1, but treatment of HRV1a-infected cells with the pan-caspase inhibitor zVAD did not protect IPS-1 from cleavage. IPS-1 was cleaved in vitro by caspase-3 and by the picornaviral proteinases 2Apro and 3Cpro. Expression of picornaviral proteinases 2Apro and 3ABC in cell lines led to cleavage of IPS-1. These results suggest that IPS-1 is cleaved during HRV1a infection by viral proteinases 2Apro, 3C, and activated caspase-3. Cleavage of IPS-1 during HRV1a infection may be a mechanism for evasion of the type I IFN response, leading to a more robust infection.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
S3 HeLa, 293T, and BSR T7/5 cells were grown in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA), 10% fetal calf serum (HyClone, Logan, UT), and 1% penicillin-streptomycin (Invitrogen). BSR T7/5 cells, a BHK-derived cell line stably expressing T7 RNA polymerase, were generously provided by Klaus Conzelmann, Ludwig Maximilians Universitat Munich, Germany (4). Selection of BSR T7/5 was maintained by the addition of G418 (1 μg/ml) at every other passage. Stocks of HRV1a were obtained from the ATCC (Manassas, VA) and were propagated in HeLa cells. Rhinovirus plaque assays were carried out using HeLa cells grown in Dulbecco's modified Eagle medium (Specialty Media, Philipsburg, NJ), 0.05% NaHCO3, 2% heat-inactivated bovine calf serum, 1% penicillin-streptomycin, and 1% type VII low-gelling temperature agarose (Sigma-Aldrich, St. Louis, MO). Cells were incubated for 72 h and developed using 10% trichloroacetic acid and crystal violet. The Cantell strain of SeV was a generous gift from Adolfo Garcia-Sastre, Mount Sinai School of Medicine, New York, NY. pFLAG-IPS-1 was a generous gift from Zhijian Chen, University of Texas Southwestern Medical Center, Dallas, TX.
Generation of stable knockdown cell lines.
Depletion of IRF-3 was achieved by using retroviral vectors encoding microRNAs (miRNAs) generously provided by Jeremy Luban, University of Geneva, Switzerland. Virus stocks were generated in 293T cells by cotransfection using Fugene 6 (Roche, Indianapolis, IN) with either pAPM-IRF-3 (targeting human IRF-3) or pAPM-L1221 (control miRNA targeting luciferase), psPAX2, and pMD.G. Viruses were harvested 24 h posttransfection and filtered (0.45-mm-pore-size filter; Pall Corp., East Hills, NY). To generate cell lines stably expressing miRNAs, HeLa cells were infected with virus stocks, and 48 h posttransduction cells were subjected to puromycin selection with increasing dosages of puromycin every 2 days for 10 days from 1 μM to 5 μM. Depletion of IRF-3 was confirmed by Western blot analysis.
Reagents.
Rabbit polyclonal IRF-3 and mouse monoclonal IRF-3 were purchased from Sigma-Aldrich. The mouse monoclonal β-actin antibody was purchased from Sigma-Aldrich. Rabbit anti-IPS-1 antibody, chicken anti-SeV antibody, and mouse monoclonal anti-poly(A)-binding protein (PABP) were purchased from AbCam (Cambridge, MA). Rabbit polyclonal anti-poly(ADP-ribose) polymerase (PARP) was purchased from Cell Signaling Technology, Danvers, MA. The general caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-(OMe) fluoromethylketone (Z-VAD-FMK) was purchased from R&D Systems, Minneapolis, MN. DNA-mediated transformation of BSR T7/5 cells was performed using Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. Cell extracts were prepared as described below for Western blot analysis.
Virus replication in cultured cells.
Adherent cell monolayers were grown in 3.5-cm plates and infected with virus at a multiplicity of infection (MOI) of 10 or 0.1 PFU per cell. Virus was absorbed for 45 min at 37°C, and then 2 ml of culture medium was added. Cells were incubated at 33°C for the times indicated in the figure legends. Cells were then scraped into the medium and subjected to two freeze-thaw cycles, and cellular debris was removed by centrifugation. Virus titer was determined by plaque assay as described above.
Infections and Western blot analysis.
Confluent monolayers of HeLa cells were infected with HRV1a or SeV for the times indicated in the figure legends. Cells were scraped into the culture medium and pelleted by centrifugation. The cell pellet was washed once with phosphate-buffered saline (PBS), centrifuged, and lysed for 15 min on ice in NP-40 buffer (50 mM Tris-Cl, pH 8.0, 1% Nonidet P-40, 150 mM NaCl) containing complete protease inhibitor cocktail (Roche). Total protein was determined by Bradford assay (Bio-Rad, Hercules, CA), and 40 μg of each sample was mixed with sodium dodecyl sulfate (SDS) loading dye. Proteins were separated on a 10% SDS-polyacrylamide gel and electrotransferred overnight at 30 V onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). Membranes were incubated in blocking buffer (PBS containing 0.1% Tween and 5% nonfat milk) for 1 h at room temperature and subsequently incubated overnight at 4°C with antibodies diluted in blocking buffer, as indicated in the figure legends. Membranes were washed three times for 15 min in PBS-0.1% Tween and incubated for 1 h with horseradish peroxidase-conjugated secondary antibody (1:1,000 anti-goat and 1:3,000 anti-rabbit; Dako, Carpinteria, CA). Proteins were visualized using the ECL or ECL+ chemiluminescence system (Amersham Biosciences, Piscataway, NJ).
Native polyacrylamide gel electrophoresis (PAGE) and Western blot analysis were performed as described previously (25), with the following changes: 10% gels were made with ProSieve 50 gel solution (Cambrex, Rockland, ME). Thirty micrograms of protein was loaded and wet transferred to PVDF membrane overnight at 30 V. Membranes were probed for IRF-3 overnight at 4°C with a rabbit polyclonal antibody, FL-425 (Sigma-Aldrich), or mouse monoclonal SC-12 (Sigma-Aldrich). Membranes were developed with the ECL+ system (Amersham Biosciences).
Quantitative real-time PCR.
RNA was harvested from HeLa cells or from cell lines producing miRNAs targeting luciferase or IRF-3 (mi-L1221 or mi-IRF-3, respectively) infected with HRV1a or SeV using an RNeasy minikit (Qiagen, Valencia, CA) at the times indicated in the figure legends. RNA concentrations were determined by an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Contaminating genomic DNA was removed by treating 8 μg of each sample with a Turbo DNA-free kit (Ambion, Austin, TX). cDNA was generated using the SuperScript III First-Strand Synthesis SuperMix for quantitative reverse transcription-PCR (qRT-PCR) according to the manufacturer's protocol (Invitrogen), and quantitative PCR mix was made with 5× Fast-Start SYBR green master mix containing 6-carboxy-X-rhodamine (Roche Diagnostics, Indianapolis, IN) with a final concentration of 400 nM of each primer. Triplicate reactions were performed on the Prism 7500 real-time PCR system in a 96-well optical plate (Applied Biosystems, Foster City, CA) with a final volume of 50 μl. The primer sequences were as follows: for IFN-β forward, CGA CAC TGT TCG TGT TGT CA; IFN-β reverse, GAA GCA CAA CAG GAG AGC AA; porphobilinogen deaminase (PBDG) forward, CTG GTA ACG GCA ATG CGG CT; and PBDG reverse, CGA GAT GGC TCC GAT GGT GA.
In vitro translation.
Coupled transcription-translation experiments were performed using a TNT quick-coupled transcription/translation system (Promega Corp, Madison, WI) in the presence of [35S]methionine (Amersham BioSciences, Piscataway, NJ). One microgram of DNA was added to each 50-μl reaction mixture, and the manufacturer's protocol was followed. The in vitro translated IPS-1 was added to cleavage buffer containing 100 mM NaCl2, 5 mM MgCl2, and 10 mM HEPES-KOH, pH 7.4, in the presence or absence of poliovirus 3Cpro (provided by B. Semler, University of California, Irvine, CA), coxsackievirus B3 2Apro (provided by R. Lloyd, Baylor College of Medicine, Houston, TX), or activated human caspase-3 (Calbiochem, La Jolla, CA). The cleavage reaction mixtures were incubated at 37°C for 18 h. The products were separated by 10% SDS-PAGE, transferred to a PVDF membrane overnight at 30 V, and exposed to a phosphorimager plate or subjected to Western blot analysis.
RESULTS
HRV1a infection does not induce robust IFN-β transcription.
The results of previous studies have indicated that members of the Picornaviridae family antagonize the innate immune system at multiple levels. In a previous investigation we showed that MDA-5 and RIG-I, innate cytoplasmic sensors of RNA, are degraded in cells infected with picornaviruses (2, 3). Furthermore, IPS-1 is cleaved in cells infected with hepatitis A virus, leading to a block in IFN-β synthesis (34). Therefore, we investigated IFN-β expression during HRV1a infection. HeLa cells were infected with HRV1a or SeV. We used the Cantell strain of SeV, which is sensed by RIG-I and induces high levels of IFN-β (27). At different times after infection, total cellular RNA was harvested and reverse transcribed into cDNA, and IFN-β transcript levels were assessed by quantitative real-time PCR.
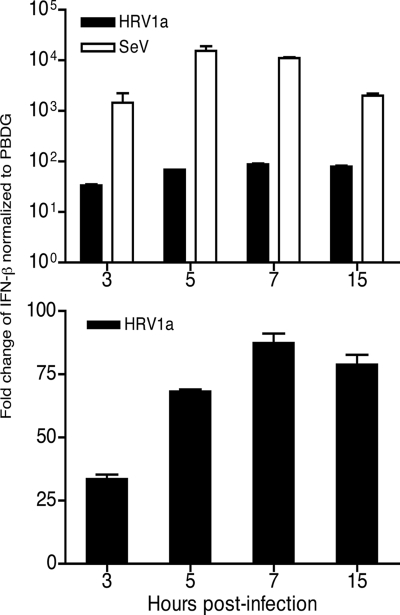
SeV induced high levels of IFN-β mRNA as early as 1 h after infection, peaking just after 7 h and falling slightly by 15 h (Fig. 1A). In contrast, although HRV1a did induce IFN-β compared to levels in mock-treated cells (Fig. 1B), induction was 100- to 1,000-fold lower than that observed during SeV infection (Fig. 1A). The low level of IFN-β mRNA suggests that HRV1a infection interferes with innate sensing of viral RNA.
FIG. 1.
Induction of IFN-β mRNA during HRV1a infection. HeLa cells were infected with HRV1a or SeV, total cellular RNA was harvested at the times indicated, and IFN-β mRNA was detected by SYBR green qRT-PCR. Results were normalized to PBDG mRNA copy number and displayed as the change in induction relative to uninfected cells using the ΔΔCT (where CT is threshold cycle) method. (Top) Induction of IFN-β in cells infected with either HRV1a or SeV. (Bottom) Induction of IFN-β during HRV1a infection. The HRV1a results were plotted separately in panel B to highlight the induction of IFN-β mRNA synthesis.
Dimerization of IRF-3 is blocked during infection with HRV1a.
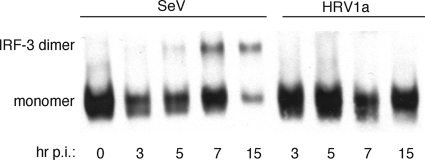
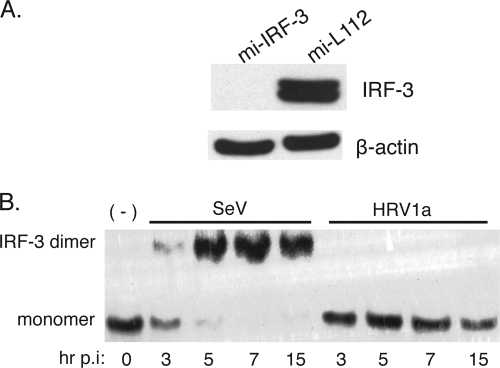
To provide an explanation for the low levels of IFN-β mRNA observed during HRV1a infection, we examined dimerization of the transcriptional activator, IRF-3, by native PAGE. In cells infected with SeV, IRF-3 dimers were first detected at 3 h postinfection (Fig. 2). Levels of IRF-3 dimers peaked by 7 h and fell slightly by 15 h. IRF-3 dimers were not observed in HRV1a-infected cells.
FIG. 2.
HRV1a infection does not induce IRF-3 homodimers. HeLa cells were infected with either SeV or HRV1a and harvested at the times indicated. Cell extracts were prepared and fractionated by native PAGE, and IRF-3 was detected by Western blot analysis using polyclonal rabbit antiserum against IRF-3, which strongly recognizes both the monomeric and dimeric forms of the protein. Monomeric and dimeric forms of IRF-3 are labeled. p.i., postinfection.
Inhibition of SeV-induced IRF-3 activation by HRV1a.
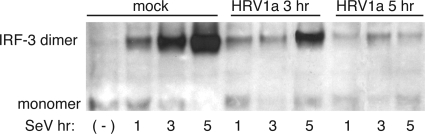
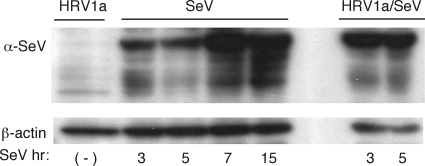
The low levels of IFN-β observed during HRV1a infection may be a consequence of poor sensing of the viral RNA or of virus-induced cleavage of one or more cell proteins in the sensing pathway. To address these possibilities, we determined whether IRF-3 activation induced by SeV was altered during HRV1a infection. HeLa cells were mock infected or infected with HRV1a at an MOI of 10 for 3 or 5 h and then superinfected with SeV. Cell lysates were harvested at different times postinfection with SeV, and IRF-3 dimers were examined by native PAGE. Infection with HRV1a caused a dramatic loss in the ability of SeV infection to induce dimerization of IRF-3 (Fig. 3). SeV proteins accumulated during HRV1a infection, indicating that the inability of SeV to induce dimerization of IRF-3 is not due to inhibition of viral replication by HRV1a (Fig. 4). These results suggest that HRV1a infection disrupts the innate immune sensing pathway.
FIG. 3.
HRV1a infection inhibits SeV-induced homodimerization of IRF-3. HeLa cells were either mock treated or infected with HRV1a for 3 h or 5 h and then superinfected with SeV for the times indicated. Cell extracts were prepared and fractionated by native PAGE, and IRF-3 was detected by Western blot analysis using a mouse monoclonal antibody. Monomeric and dimeric forms of IRF-3 are labeled. (-), no SeV added.
FIG. 4.
Accumulation of SeV proteins during coinfection with HRV1a. HeLa cells were infected with HRV1a for 8 h (lane 1); with SeV for 3, 5, 7, or 15 h (lanes 3 to 5); or with HRV1a for 5 h followed by SeV for 3 or 5 h (lanes 7 and 8). No sample was loaded in lane 6. Cell extracts were prepared and fractionated by SDS-PAGE, and SeV proteins were detected by Western blot analysis using anti-SeV antiserum. α, anti.
IPS-1 is degraded in HRV1a-infected cells.
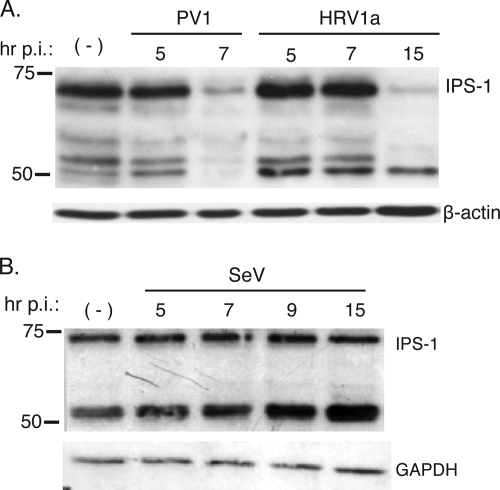
The dimerization of IRF-3 requires phosphorylation by the kinases TBK and TANK-1, which are selectively activated through IPS-1 (9). Because IPS-1 is an essential protein of the pathway leading to IFN-β induction, we determined if it was altered during rhinovirus infection. HeLa cells were infected with HRV1a, and at different times after infection, IPS-1 protein was visualized by Western blot analysis. By 15 h postinfection, IPS-1 protein was barely detectable (Fig. 5A). Degradation of IPS-1 was also observed by 5 h after infection with another member of the picornavirus family, poliovirus. IPS-1 remained intact in SeV-infected cells (Fig. 5B), demonstrating that degradation of the protein is not a general cellular response to viral infection.
FIG. 5.
Cleavage of IPS-1 in cells infected with HRV1a or poliovirus. Monolayers of HeLa cells were infected with HRV1a, poliovirus, or SeV. Cell extracts were prepared after infection at the times indicated and fractionated by SDS-PAGE, and IPS-1 was detected by Western blot analysis. −, mock-infected cells. A separate bottom panel shows detection of β-actin (A) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (B) to ensure that equal amounts of protein were applied to each lane. p.i., postinfection.
Picornavirus infection can lead to the inhibition of host cell translation (19, 29). To ensure that IPS-1 degradation was not due to aggregate cell protein shutoff, cells were treated with cycloheximide (5 μg/ml) or infected with HRV1a, and IPS-1 levels were determined by Western blot analysis. IPS-1 levels remained stable during treatment with cycloheximide for 24 h, whereas HRV1a infection led to the degradation of IPS-1 (unpublished data). This observation suggests that the decrease in IPS-1 during an HRV1a infection is not a consequence of the rapid turnover of the protein.
Induction of IFN-β during rhinovirus infection in cells depleted of IRF-3.
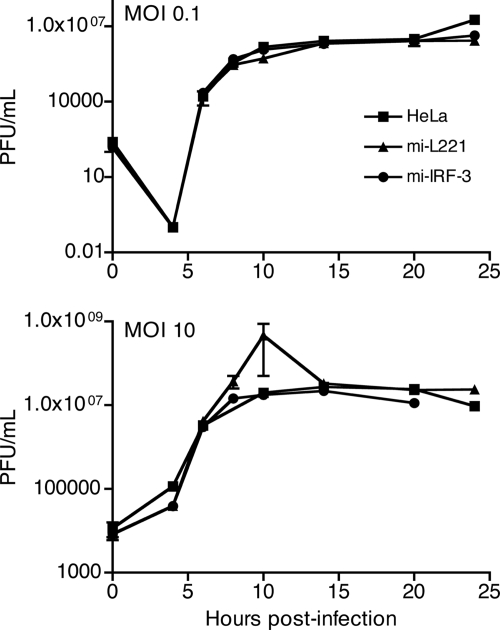
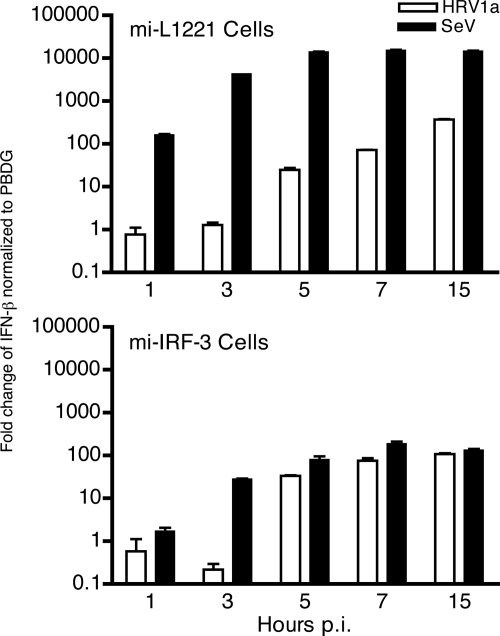
If cleavage of IPS-1 accounts for the low level of IFN-β observed during HRV1a infection, then infected cells depleted of IRF-3 should exhibit similar levels of the cytokine. Efficient knockdown of IRF-3 was achieved in stable cell lines that express an IRF-3-specific miRNA (mi-L1221), compared with control stable cell lines expressing a luciferase-specific miRNA (mi-IRF-3) (Fig. 6A). As expected, IRF-3 dimers were observed during SeV but not HRV1a infection in the control cell line (Fig. 6B). Absence of IRF-3 did not affect replication of HRV1a at low or high MOI (Fig. 7). Induction of IFN-β by SeV or HRV1a in the control and parental HeLa cell lines was similar (Fig. 1 and 7). Low levels of IFN-β were observed after SeV and HRV1a infection of cells depleted of IRF-3 (Fig. 8). The amounts of IFN-β mRNA resembled those observed during HRV1a infection of HeLa cells (Fig. 1).
FIG. 6.
Knockdown of IRF-3 in HeLa cells. Stable cells lines mi-IRF-3 and mi-L1221 were produced. Cell extracts were prepared and analyzed for IRF-3 protein levels by SDS-PAGE and Western blot analysis (A). Dimerization of IRF-3 was examined by native PAGE in cells infected with HRV1a or SeV (B). p.i., postinfection.
FIG. 7.
Effect of IRF-3 knockdown on HRV1a replication. HeLa, mi-IRF-3, and mi-L1221 cells were infected with HRV1a at an MOI of 0.1 or 10. At the indicated times after infection, virus titers were determined by plaque assay on HeLa cell monolayers.
FIG. 8.
Induction of IFN-β mRNA during HRV1a infection in cells with reduced IRF-3. mi-L1221 or mi-IRF-3 cells were infected with HRV1a or SeV, total cellular RNA was harvested at the times indicated, and IFN-β mRNA was detected by SYBR green qRT-PCR. Results were normalized to PBDG mRNA copy number and are displayed as the change in induction relative to uninfected cells using the ΔΔCT method (where CT is threshold cycle). p.i., postinfection.
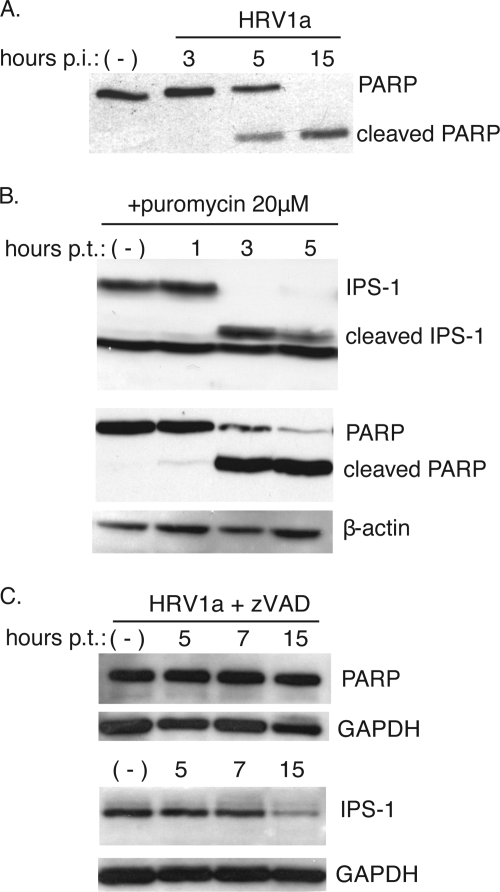
Cleavage of IPS-1 during apoptosis.
The genomes of HRV1a and poliovirus encode two viral proteinases, 2Apro and 3Cpro, that process the viral polyprotein and also degrade cellular proteins. Picornaviral proteinases are known to induce apoptosis, a process that occurs in cells infected with certain rhinoviruses (5, 7, 30). Caspases are activated during apoptosis, leading to cleavage of cellular proteins such as PARP. In cells infected with HRV1a, PARP cleavage is observed starting at 5 h postinfection, suggesting that HRV1a induces apoptosis (Fig. 9A). Furthermore, activated caspase-3 was observed in cells infected with HRV1a by 5 h postinfection (unpublished data). To address whether induction of apoptosis leads to cleavage of IPS-1, cells were treated with puromycin, and IPS-1 was examined by Western blot analysis. Puromycin treatment for 3 and 5 h induced apoptosis, as shown by cleavage of PARP (Fig. 9B). In the same cells IPS-1 was degraded, and a ∼50-kDa putative cleavage product was observed (Fig. 9B).
FIG. 9.
Cleavage of IPS-1 and PARP during apoptosis. Monolayers of HeLa cells were infected with HRV1a (MOI of 10) (A), treated with puromycin (20 μM) to induce apoptosis (B), or infected with HRV1a (MOI of 10) and treated with zVAD (20 μM) (C). At different times after infection or after the addition of puromycin or zVAD to the culture medium, cell extracts were prepared and fractionated by SDS-PAGE, and PARP and IPS-1 were detected by Western blot analysis. Separate bottom panels show detection of β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to ensure that equal amounts of protein were applied to each lane. p.i., postinfection; p.t., posttreatment; −, no treatment.
Activated caspases are not necessary for the HRV1a-induced IPS-1 degradation.
If caspases are the only mediators of IPS-1 cleavage during an HRV1a infection, then inhibiting activated caspases should prevent IPS-1 degradation. To test this hypothesis, the effect of zVAD, a pan-caspase inhibitor, on IPS-1 cleavage was determined. HeLa cells were infected with HRV1a in the presence or absence of inhibitor. The levels of IPS-1 at different times after infection were determined by Western blot analysis. Degradation of IPS-1 was not affected by zVAD (Fig. 9C). As expected, the inhibitor prevented PARP cleavage (Fig. 9C). Furthermore, the yield of HRV1a was not altered by zVAD treatment, indicating that the drug does not inhibit viral proteinases 2Apro or 3Cpro (unpublished data). Taken together, these results suggest that HRV1a-induced degradation of IPS-1 is mediated not only by caspases but also by other cellular or even viral proteases.
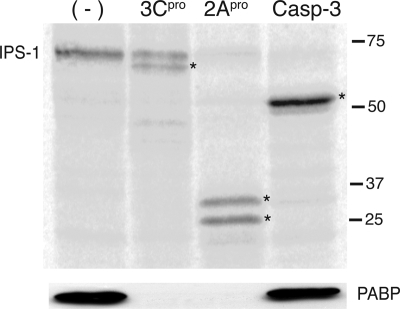
IPS-1 is cleaved by 3Cpro, 2Apro, and caspase-3.
We determined whether IPS-1 could be cleaved in vitro by purified poliovirus 3Cpro, coxsackievirus 2Apro, or activated human caspase-3. The poliovirus and coxsackievirus enzymes were used because the HRV1a proteins were not available. IPS-1 was produced by in vitro translation in a reticulocyte lysate in the presence of [35S]methionine and incubated with each enzyme. Both viral proteinases were active in the reticulocyte lysate, as shown by cleavage of PABP (Fig. 10). IPS-1 was cleaved into fragments of different sizes by all three enzymes (Fig. 10).
FIG. 10.
Effect of 3Cpro, 2Apro, and caspase-3 on IPS-1 in vitro. IPS-1 was produced by in vitro translation in a reticulocyte lysate in the presence of [35S]methionine. The lysate was subsequently incubated with purified 3CDpro, 2Apro, or caspase-3 and fractionated by SDS-PAGE, and [35S]methionine-labeled proteins were detected by phosphorimaging. PABP (to confirm enzyme activity) and β-actin (loading control [data not shown]) were detected by Western blot analysis. Asterisks indicate putative cleavage products of IPS-1. −, no treatment.
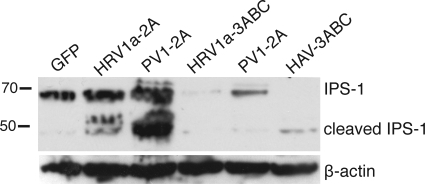
To determine if 2Apro and 3Cpro proteins of HRV1a can cleave IPS-1 in cells, plasmids encoding the proteins were introduced into cells by DNA-mediated transformation. It has been shown that the hepatitis A virus proteinase 3Cpro must be synthesized as part of the precursor protein 3ABC to properly target the proteinase to mitochondria (34). Therefore, plasmids encoding epitope-tagged 3ABC proteins of HRV1 and poliovirus were produced. However, we failed to detect the proteinases by Western blot analysis when these plasmids were introduced into different cell lines by a variety of methods.
To increase the expression levels of the viral proteinases, plasmids were introduced into a BHK cell line (BSR T7/5) which stably produces T7 RNA polymerase. BSR cells were cotransformed with a plasmid encoding IPS-1 and either enhanced green fluorescent protein or a viral proteinase. Twenty-four hours later cells were harvested for Western blot analysis. As expected, synthesis of hepatitis A virus 3ABC protein led to cleavage of IPS-1 (Fig. 11). Expression of 2Apro and 3ABC proteins of HRV1a and poliovirus 3ABC led to degradation of IPS-1 (Fig. 11). These results show that degradation of IPS-1 observed during HRV1a and poliovirus infection is likely a consequence of the activity of both viral proteinases and caspases.
FIG. 11.
Expression of viral proteinases in cells. BSR T7/5 cells were cotransformed with plasmids encoding IPS-1 and either enhanced green fluorescent protein (GFP) or proteinases of HRV1a, poliovirus (PV1), or hepatitis A virus (HAV). After 24 h cell extracts were prepared and fractionated by SDS-PAGE, and IPS-1 was detected by Western blot analysis. The separate bottom panel shows detection of β-actin to ensure that equal amounts of protein were applied to each lane.
DISCUSSION
HRV1a induced expression of IFN-β mRNA in HeLa cells but at levels far lower than those observed during infection with SeV. Consistent with this observation, we found that IRF-3 dimers did not form during HRV1a infection. Phosphorylation of IRF-3 is mediated by protein kinases whose activation depends upon signals received from IPS-1 and, in turn, MDA-5 or RIG-I. IPS-1 is cleaved in cells infected with HRV1a, which could explain the absence of IRF-3 dimers. It is also possible that the kinases IΚΚɛ and TBΚ1 are also modified during infection, but degradation of IPS-1 would be sufficient to prevent dimerization of IRF-3. MDA-5 is cleaved in HRV1a-infected cells (3), but the kinetics and extent of cleavage are not consistent with the complete absence of IRF-3 dimers observed here. To prove that cleavage of IPS-1 is a mechanism to abrogate IFN-β synthesis will require production of IPS-1 that is not cleaved in HRV1a-infected cells.
Our results show that cleavage of IPS-1 during HRV1a infection may be carried out by either of the two viral proteinases, 2Apro or 3Cpro. These proteinases process the viral polyprotein and, for some picornaviruses, have been shown to cleave a variety of cell proteins such as eukaryotic initiation factor 4G (18) (cleaved by 2Apro) and PABP (19) (cleaved by both proteinases). Previously, it had not been demonstrated that 2Apro and 3Cpro of HRV1a cleaved any cellular proteins. In cells infected with hepatitis A virus, IPS-1 is cleaved by 3Cpro produced from a 3ABC precursor (34).
Our findings also imply that cellular caspases are involved in cleavage of IPS-1 during HRV1a infection. It was previously demonstrated that apoptosis is induced during infection with certain rhinoviruses. HeLa cells infected with human rhinovirus 14, a major group rhinovirus, exhibited typical apoptotic cellular alterations including cell contraction, nuclear condensation, and activation of caspase-9 and caspase-3 (7). Induction of apoptosis has also been demonstrated in cells infected with the minor group rhinovirus 1B, which is highly related to the serotype used in these studies, HRV1a (30). We have found that apoptosis is also induced in HRV1a-infected cells, as indicated by activation of caspase-3 and cleavage of PARP, a known capsase-3 substrate, within 5 h of infection. Activated caspase-3 can cleave IPS-1 in vitro to yield a ∼50-kDa protein. Induction of apoptosis during HRV1a infection may be a unique viral strategy to cleave IPS-1 and limit IFN-β mRNA levels.
Induction of apoptosis by puromycin treatment of cells caused cleavage of IPS-1. The ∼50-kDa putative cleavage product is similar in size to the fragment produced when IPS-1 is cleaved by caspase-3 in vitro. However, treating cells with the pan-caspase inhibitor zVAD did not block IPS-1 degradation. This observation indicates that other proteases are involved in the degradation of the protein. Surprisingly, IPS-1 was cleaved in a rabbit reticulocyte lysate by poliovirus 3Cpro and coxsackievirus B3 2Apro, and specific products were formed. In contrast, no cleavage products of IPS-1 are observed in HRV1a-infected cells or in cells producing 3ABC from plasmid vectors. The combined action of the viral proteinases and caspase-3 on IPS-1 in vivo may make the cleavage products unstable.
Although IPS-1 is degraded in cells upon expression of 3ABC or 2Apro, it is not known if IPS-1 is a direct substrate of the viral proteinases. Synthesis of picornaviral 2Apro or 3Cpro in cells induces apoptosis (5, 7, 30), which may lead to degradation of IPS-1 through activated caspase-3. However, two of our observations suggest a role for the viral proteinases in direct cleavage of the protein: the failure to block degradation of IPS-1 by treatment of HRV1a-infected cells with the caspase inhibitor zVAD and the direct cleavage of IPS-1 by purified viral proteinases in vitro.
There are two predicted caspase-3 cleavage sites in IPS-1 protein, at amino acids 86 and 429. Cleavage only at amino acid 429 would produce the ∼50-kDa putative cleavage product observed in cells treated with puromycin, in reticulocyte lysates incubated with caspase-3, and in cells expressing HRV1a and poliovirus 2Apro. There are several potential cleavage sites for 2Apro and 3Cpro within IPS-1 that would, if utilized, produce the putative cleavage products observed in vitro. Experiments are currently in progress to identify the cleavage sites for each protease.
IPS-1 undergoes lysine 48-linked polyubiquitination by the E3 ubiquitin ligase RNF125 and is likely subject to proteasomal degradation (1). Degradation of IPS-1 during HRV1a infection could be due in part to proteasome degradation. To test this hypothesis, we treated cells with the proteasome inhibitor MG132 and infected cells with HRV1a. IPS-1 protein levels decreased within 15 h postinfection; however, the protein was partially protected from degradation when protein levels were compared to cells not treated with MG132 (data not shown). These data suggest a role for the proteasome in IPS-1 degradation. We also observed some inhibition of the viral proteinases in cells treated with MG132, which could lead to protection of IPS-1. Additional experiments are needed to determine the role of the proteasome in IPS-1 degradation during HRV1a infection.
Induction of IFN-β mRNA was examined in a stable cell line depleted of IRF-3. After infection with SeV, cytokine levels were significantly reduced as expected. However, during HRV1a infection, IFN-β levels were similar whether IRF-3 was present or not. This observation suggests that other transcription factors, such as IRF-7 and NF-κB, may allow synthesis of IFN-β when IRF-3 is not present or inactive.
Infection by the major group rhinovirus HRV14 leads to very low levels of IFN-β mRNA, which correlated with impairment of IRF-3 activation (16). This observation is consistent with our finding that IFN-β mRNA synthesis is impaired during HRV1a infection, concomitant with IPS-1 cleavage and inhibition of IRF-3 activation. Cleavage of IPS-1 by both viral proteases and cellular caspase-3 may be a strategy to ensure effective abrogation of the innate antiviral response.
Acknowledgments
This work was supported in part by Public Health Service grants AI50754 and T32AI07161 from the National Institute of Allergy and Infectious Diseases.
We thank Adolfo Garcia-Sastre for the Cantell strain of SeV, Richard Lloyd for the gift of 2Apro and 3Cpro, Jeremy Luban for the lentiviral vectors, Klaus Conzelmann for BSR T7/5 cells, and Zhijian Chen for pFLAG-IPS-1.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Arimoto, K., H. Takahashi, T. Hishiki, H. Konishi, T. Fujita, and K. Shimotohno. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. USA 104:7500-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barral, P., D. Sarkar, P. Fisher, and V. Racaniello. 2009. RIG-I is cleaved during picornavirus infection. Virology 391:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barral, P. M., J. M. Morrison, J. Drahos, P. Gupta, D. Sarkar, P. B. Fisher, and V. R. Racaniello. 2007. MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 81:3677-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calandria, C., A. Irurzun, A. Barco, and L. Carrasco. 2004. Individual expression of poliovirus 2Apro and 3Cpro induces activation of caspase-3 and PARP cleavage in HeLa cells. Virus Res. 104:39-49. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Z., Y. Benureau, R. Rijnbrand, J. Yi, T. Wang, L. Warter, R. E. Lanford, S. A. Weinman, S. M. Lemon, A. Martin, and K. Li. 2007. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J. Virol. 81:964-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deszcz, L., E. Gaudernak, E. Kuechler, and J. Seipelt. 2005. Apoptotic events induced by human rhinovirus infection. J. Gen. Virol. 86:1379-1389. [DOI] [PubMed] [Google Scholar]
- 8.Fendrick, A. M., A. S. Monto, B. Nightengale, and M. Sarnes. 2003. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch. Intern. Med. 163:487-494. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 10.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 103:8459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 12.Kang, D. C., R. V. Gopalkrishnan, L. Lin, A. Randolph, K. Valerie, S. Pestka, and P. B. Fisher. 2004. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene 23:1789-1800. [DOI] [PubMed] [Google Scholar]
- 13.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 14.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 15.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 16.Kotla, S., T. Peng, R. E. Bumgarner, and K. E. Gustin. 2008. Attenuation of the type I interferon response in cells infected with human rhinovirus. Virology 374:399-410. [DOI] [PubMed] [Google Scholar]
- 17.Kovacsovics, M., F. Martinon, O. Micheau, J. L. Bodmer, K. Hofmann, and J. Tschopp. 2002. Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr. Biol. 12:838-843. [DOI] [PubMed] [Google Scholar]
- 18.Krausslich, H. G., M. J. H. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eukaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuyumcu-Martinez, N. M., M. E. Van Eden, P. Younan, and R. E. Lloyd. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo, Y. M., and M. Gale, Jr. 2007. Viral regulation and evasion of the host response. Curr. Top. Microbiol. Immunol. 316:295-313. [DOI] [PubMed] [Google Scholar]
- 22.Mallia, P., and J. S. 2006. How viral infections cause exacerbation of airway diseases. Chest 130:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 24.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 25.Mori, M., M. Yoneyama, T. Ito, K. Takahashi, F. Inagaki, and T. Fujita. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 279:9698-9702. [DOI] [PubMed] [Google Scholar]
- 26.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 27.Strahle, L., J. B. Marq, A. Brini, S. Hausmann, D. Kolakofsky, and D. Garcin. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J. Virol. 81:12227-12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss, J. H., and E. G. Strauss. 2002. Viruses and human disease. Academic Press, San Diego, CA.
- 29.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taimen, P., H. Berghall, R. Vainionpaa, and M. Kallajoki. 2004. NuMA and nuclear lamins are cleaved during viral infection-inhibition of caspase activity prevents cleavage and rescues HeLa cells from measles virus-induced but not from rhinovirus 1B-induced cell death. Virology 320:85-98. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi, O., and S. Akira. 2007. Recognition of viruses by innate immunity. Immunol. Rev. 220:214-224. [DOI] [PubMed] [Google Scholar]
- 32.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 33.Yang, H., G. Ma, C. H. Lin, M. Orr, and M. G. Wathelet. 2004. Mechanism for transcriptional synergy between interferon regulatory factor (IRF)-3 and IRF-7 in activation of the interferon-beta gene promoter. Eur. J. Biochem. 271:3693-3703. [DOI] [PubMed] [Google Scholar]
- 34.Yang, Y., Y. Liang, L. Qu, Z. Chen, M. Yi, K. Li, and S. M. Lemon. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. USA 104:7253-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoneyama, M., and T. Fujita. 2007. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 282:15315-15318. [DOI] [PubMed] [Google Scholar]
- 36.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DEXD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 37.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]