Abstract
The early steps of the hepatitis B virus (HBV) life cycle are still poorly understood. Indeed, neither the virus receptor at the cell surface nor the mechanism by which nucleocapsids are delivered to the cytosol of infected cells has been identified. Extensive mutagenesis studies in pre-S1, pre-S2, and most of the S domain of envelope proteins revealed the presence of two regions essential for HBV infectivity: the 77 first residues of the pre-S1 domain and a conformational motif in the antigenic loop of the S domain. In addition, at the N-terminal extremity of the S domain, a putative fusion peptide, partially overlapping the first transmembrane (TM1) domain and preceded by a PEST sequence likely containing several proteolytic cleavage sites, was identified. Since no mutational analysis of these two motifs potentially implicated in the fusion process was performed, we decided to investigate the ability of viruses bearing contiguous deletions or substitutions in the putative fusion peptide and PEST sequence to infect HepaRG cells. By introducing the mutations either in the L and M proteins or in the S protein, we demonstrated the following: (i) that in the TM1 domain of the L protein, three hydrophobic clusters of four residues were necessary for infectivity; (ii) that the same clusters were critical for S protein expression; and, finally, (iii) that the PEST sequence was dispensable for both assembly and infection processes.
The hepatitis B virus (HBV) is the main human pathogen responsible for severe hepatic diseases like cirrhosis and hepatocellular carcinoma. Even though infection can be prevented by immunization with an efficient vaccine, about 2 billion people have been infected worldwide, resulting in 350 million chronic carriers that are prone to develop liver diseases (56). Current treatments consist either of the use of interferon α, which modulates antiviral defenses and controls infection in 30 to 40% of cases, or of the use of viral polymerase inhibitors that allow a stronger response to treatment but require long-term utilization and frequently lead to the outcome of resistant viruses (34, 55). A better understanding of the virus life cycle, and particularly of the mechanism by which the virus enters the cell, could provide background for therapeutics that inhibit the early steps of infection, as recently illustrated with the HBV pre-S1-derived entry inhibitor (25, 45).
HBV belongs to the Hepadnaviridae family whose members infect different species. All viruses of this family share common properties. The capsid containing a partially double-stranded circular DNA genome is surrounded by a lipid envelope, in which two (in avihepadnaviruses infecting birds) or three (in orthohepadnaviruses infecting mammals) envelope proteins are embedded. A single open reading frame bearing several translation initiation sites encodes these surface proteins. Thus, the HBV envelope contains three proteins: S, M, and L that share the same C-terminal extremity corresponding to the small S protein that is crucial for virus assembly (7, 8, 46) and infectivity (1, 31, 53). These proteins are synthesized in the endoplasmic reticulum (ER), assembled, and secreted as particles through the Golgi apparatus (15, 42). The current model for the transmembrane structure of the S domain implies the luminal exposition of both N- and C-terminal extremities and the presence of four transmembrane (TM) domains: the TM1 and TM2 domains, both necessary for cotranslational protein integration into the ER membrane, and the TM3 and TM4 domains, located in the C-terminal third of the S domain (for a review, see reference 6). Among the four predicted TM domains, only the TM2 domain has a defined position between amino acids 80 and 98 of the S domain. The exact localization of the TM1 domain is still unclear, probably because of the relatively low hydrophobicity of its sequence, which contains polar residues and two prolines. The M protein corresponds to the S protein extended by an N-terminal domain of 55 amino acids called pre-S2. Its presence is dispensable for both assembly and infectivity (20, 21, 37). Finally, the L protein corresponds to the M protein extended by an N-terminal domain of 108 amino acids called pre-S1 (genotype D). The pre-S1 and pre-S2 domains of the L protein can be present either at the inner face of viral particles (on the cytoplasmic side of the ER), playing a crucial role in virus assembly (5, 8, 10, 11, 46), or on the outer face (on the luminal side of the ER), available for the interaction with target cells and necessary for viral infectivity (4, 14, 36). The pre-S translocation is independent from the M and S proteins and is driven by the L protein TM2 domain (33). Finally, HBV surface proteins are not only incorporated into virion envelopes but also spontaneously bud from ER-Golgi intermediate compartment membranes (30, 43) to form empty subviral particles (SVPs) that are released from the cell by secretion (8, 40).
One approach to decipher viral entry is to interfere with the function of envelope proteins. Thus, by a mutagenesis approach, two envelope protein domains crucial for HBV infectivity have already been identified: (i) the 77 first amino acids of the pre-S1 domain (4, 36) including the myristic acid at the N-terminal extremity (9, 27) and (ii) possibly a cysteine motif in the luminal loop of the S domain (1, 31). In addition, a putative fusion peptide has been identified at the N-terminal extremity of the S domain due to its sequence homology with other viral fusion peptides (50). This sequence, either N-terminal in the S protein or internal in the L and M proteins, is conserved among the Hepadnaviridae family and shares common structural and functional properties with other fusion peptides (49, 50). Finally, a PEST sequence likely containing several proteolytic cleavage sites has been identified in the L and M proteins upstream of the TM1 domain (39). A cleavage within this sequence could activate the fusion peptide.
In this study, we investigated whether the putative fusion peptide and the PEST sequence were necessary for the infection process. For this purpose, we constructed a set of mutant viruses bearing contiguous deletions in these regions and determined their infectivity using an in vitro infection model based on HepaRG cells (28). The introduction of mutations either in the L and M proteins or in only the S protein allowed us to demonstrate that, in the TM1 domain of L protein, three hydrophobic clusters not essential for viral assembly were crucial for HBV infectivity while their presence in the S protein was critical for envelope protein expression. In addition, we showed that the PEST sequence was clearly dispensable for both assembly and infection processes.
MATERIALS AND METHODS
Plasmids and mutagenesis.
Three different plasmids were necessary for the production of viral particles. The first vector, pHBV L−E−, corresponds to a viral genome competent for viral replication but deficient for envelope protein production. It was derived from the plasmid pHBV L− (37) in which we introduced an opal (UGA) mutation into codon 15 and an amber (UAG) mutation into codon 94 of the S domain without changing the polymerase amino acid sequence. The second plasmid, pSVSX, encodes the wild-type (WT) S protein. It contains the 1,986-bp EcoRI-BglII fragment of WT HBV DNA (subtype ayw; EMBL accession no. X02496) bearing the entire S and X coding regions cloned downstream of the simian virus 40 early promoter in plasmid pSV-SPORT 1 (Life Technology). Finally, the third construct, pSV12SX S−, corresponds to the pSV12SX plasmid that is an expression vector of the three surface proteins (37); in pSV12SX S− the translation initiation codon of the S protein has been changed into a threonine codon, thus preventing the synthesis of S protein. We have verified that this mutation affects neither the synthesis of L protein nor the assembly of infectious viral particles (data not shown). These three plasmids allowed the production of chimeric viruses with WT S protein and mutated L and M proteins and vice versa. Mutations located between amino acids I157 and I191 (see Fig. 2) were introduced by PCR into pSVSX and pSV12SX S−. The first step of the mutagenesis method involved the design of two primers (A and D) flanking two restriction sites present in the sequence of interest. Since the TM1 domain is surrounded by EcoRI and XbaI restriction sites, we used a forward primer, A (AATCGCCAGTCAGGAAGGC), interacting with a sequence upstream of the EcoRI site, and a reverse primer, D (TTGGCCCCCAATACCACATC), interacting with a sequence downstream of the XbaI site. Then, for each mutant we designed a forward primer, B, and a reverse primer, C, bearing the mutation(s) and being complementary to each other. The first independent PCRs generated two fragments resulting from the amplification with the primer pair A and C and the pair B and D, respectively. The complementarities between primers B and C allowed the hybridization between the two amplicons, and the addition of primers A and D in a second PCR step allowed the amplification of the region bearing mutation(s). Finally, amplified fragments of the second PCR step were digested with EcoRI and XbaI and then inserted into EcoRI-XbaI-digested pSVSX and pSV12SX S− plasmids. In the resulting vectors, inserts were sequenced to confirm the presence of the expected mutation(s). The sequences of primers B and C for each mutant are presented in Table 1.
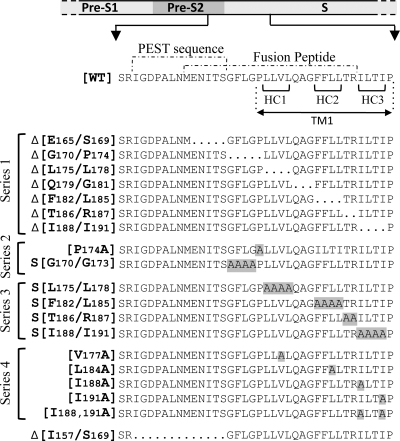
FIG. 2.
Mutations introduced in the S gene. The WT sequence corresponds to the last 9 residues of the pre-S2 domain and to the first 29 residues of the WT S protein (subtype ayw; EMBL accession no. X02496). The position of the TM1 domain was defined between amino acids P174 and P192 according to the TM prediction software programs PHDrhtm (52) and NORSp (38). It is important that the position of the TM1 domain varies depending on the prediction software used and that several positions are found in the literature (2, 3, 6, 13, 18, 47, 48). The dotted lines delimit the putative fusion peptide from amino acids M164 to R187 (50) and the PEST sequence, which spans the last 7 residues of the pre-S2 domain and the first 6 of the S domain (39). Deleted amino acids are replaced by dots in the mutated protein sequences. Alanine substitutions are highlighted in gray. Deletions (Δ) are inclusive, and residues are numbered according to their respective positions relative to the first N-terminal amino acid of the L protein. In series 2 and 3, the S indicates that alanines were substituted for all amino acids in the range specified. For the unique and double substitutions, we used the classical writing convention to indicate the mutation. HC, hydrophobic cluster.
TABLE 1.
Nucleotide sequence of mutagenesis primersa
| Primer B |
Primer C |
||
|---|---|---|---|
| Target | Sequenceb | Target | Sequenceb |
| Δ(E165/S169) | GCTGAACATGGGATTCCTAGGACCC* | Δ(E165/S169) | CTAGGAATCCCATGTTCAGCGCAGG* |
| GCTGAACACGGGATTCCTAGGACCCC** | CTAGGAATCCCGTGTTCAGCGCAG** | ||
| Δ(G170/P174) | CATCACATCACTTCTCGTGTTACAGGC | Δ(G170/P174) | ACACGAGAAGTGATGTGATGTTCTCCATG* |
| ACACGAGAAGTGATGTGATGTTCTCCG** | |||
| Δ(L175/L178) | CCTAGGACCCCAGGCGGGGTTTTTC | Δ(L175/L178) | ACCCCGCCTGGGGTCCTAGGAATCCTG |
| Δ(Q179/G181) | TCTCGTGTTATTTTTCTTGTTGACAAGAATC | Δ(Q179/G181) | ACAAGAAAAATAACACGAGAAGGGGTC |
| Δ(F182/L185) | ACAGGCGGGGACAAGAATCCTCACAATACC | Δ(F182/L185) | GGATCTTGTCCCCGCCTGTAACAC |
| Δ(T186/R187) | TTTCTTGTTGATCCTCACAATACCGC | Δ(T186/R187) | TTGTGAGGATCAACAAGAAAAACCCC |
| Δ(I188/I191) | GTTGACAAGACCGCAGAGTCTAGACTCG | Δ(I188/I191) | GACTCTGCGGTCTTGTCAACAAGAAAAACC |
| Δ(I157/S169) | CTTCTCGAGGGGATTCCTAGGACCCC | Δ(1157/S169) | CTAGGAATCCCCTCGAGAAGATTGACG |
| S(L175/L178) | CCTAGGACCCGCTGCCGCGGCACAGGCGGGGTTTTTC | S(L175/L178) | ACCCCGCCTGTGCCGCGGCAGCGGGTCCTAGGAATCCTG |
| S(F182/L185) | ACAGGCGGGGGCTGCCGCGGCGACAAGAATCCTCACAATACC | S(F182/L185) | GGATTCTTGTCGCCGCGGCAGCCCCCGCCTGTAACAC |
| S(T186/R187) | CTTGTTGGCAGCAATCCTCACAATACCGC | S(T186/R187) | AGGATTGCTGCCAACAAGAAAAACCCC |
| S(I188/I191) | GTTGACAAGAGCCGCCGCAGCACCGCAGAGTCTAGACTCG | S(I188/I191) | GACTCTGCGGTGCTGCGGCGGCTCTTGTCAACAAGAAAAACC |
| V177A | ACCCCTTCTCGCGTTACAGGCGGGG | V177A | CCGCCTGTAACGCGAGAAGGGGTCCTAG |
| L184A | GGGGTTTTTCGCGTTGACAAGAATCCTCAC | L184A | GATTCTTGTCAACGCGAAAAACCCCGCC |
| I188A | GTTGACAAGAGCCCTCACAATACCGCAG | I188A | GTATTGTGAGGGCTCTTGTCAACAAGAAAAACC |
| I191A | GAATCCTCACAGCACCGCAGAGTCTAGACTC | 1191A | GACTCTGCGGTGCTGTGAGGATTCTTGTCAAC |
| I188A I191A | AGAGCCCTCACAGCACCGCAGAGTCTAGACTCG | I188A I191A | CGGTGCTGTGAGGGCTCTTGTCAACAAGAAAAACC |
Primers B and C correspond to forward and reverse primers, respectively (see “Plasmids and mutagenesis” in Materials and Methods). Deletions (Δ) are inclusive, and residues are numbered according to their respective positions relative to the first N-terminal amino acid of the L protein. In mutations preceded by an S, alanines were substituted for all amino acids in the range specified. For the unique and double substitutions, we used the classical writing convention to indicate the mutation.
The same oligonucleotides were used for mutagenesis in pSVSX and pSV12SX S− plasmids with exception of those that include the initiation codon (M164) of the S protein, for which designed primers were used with the pSVSX vector (*) and the pSV12SX S− vector (**).
Virus production.
The cotransfection of the HepG2 hepatoma cell line with the three plasmids (pHBV L−E−, pSVSX, and pSV12SX S−) allowed the production of viral particles in the supernatant of transfected cells. The control L− consisted in the cotransfection of the pHBV L−E− plasmid with only the pSVSX vector, preventing virion production (5, 8). The control Myr− was produced by the cotransfection of pHBV L−E− with pSVSX and an expression vector coding for an unmyristoylated L protein (pSV12SX Myr−), resulting in the production of noninfectious virus (9, 27). Finally, the cotransfection of HepG2 cells with the envelope-defective HBV genome pHBV L−E−, an expression vector of WT or mutated L and M proteins (pSV12SX S−), and an expression vector of WT or mutated S protein (pSVSX) allowed the production of chimeric viruses bearing WT L and M proteins and mutated S proteins or vice versa. The cotransfection was performed by cell electroporation with a unique exponential decrease pulse of 1,800 μF and 230 V in OptiMEM medium supplemented with 10% fetal calf serum (FCS). The absence of significant variations in efficiency of transfection within each experiment was verified by measuring the intracellular hepatitis B secreted core antigen (HBeAg) level in electroporated HepG2 cells (data not shown). The level of this viral antigen directly reflects the efficiency of transfection since its presence depends on the expression of the pHBV L−E− plasmid, which was cotransfected for the production of all mutants and WT viruses. HepG2 cells were then maintained in regular culture medium consisting of William's E medium supplemented with 10% FCS, 2 mM l-glutamine, 100 UI/ml of penicillin, 100 μg/ml of streptomycin, 5 μg/ml of insulin, and 0.5 μM hydrocortisone. The supernatants of cotransfected cells were harvested every 2 days from day 6 to day 12 posttransfection and pooled. After elimination of cellular components by centrifugation at 5,000 × g, viral particles were precipitated from the culture supernatant with 6% polyethylene glycol (PEG) for 12 h at 4°C. After 30 min of centrifugation at 5,000 × g and 4°C, particles aggregated in the pellet were solubilized in phosphate-buffered saline supplemented with 25% FCS to concentrate them 50-fold and stored at −80°C.
Intra- and extracellular HBV envelope protein analysis.
HepG2 cells producing virions were lysed 12 days after cotransfection, and their supernatant was harvested between the day 6 and 12 posttransfection. Cells were lysed in 25 mM Tris-HCl (pH 7.4), 250 mM NaCl, 5 mM EDTA, and 1% NP-40. Nuclei were removed by centrifugation before the immunoblotting analysis, protein concentration was determined with a BC assay for protein quantitation (Uptima), and 20 μg of protein was subjected to electrophoresis. Released viral particles from 1 ml of supernatant were precipitated with 8% PEG 8000, disrupted in loading buffer (Invitrogen), and analyzed as described below. Proteins were analyzed by electrophoresis through NuPAGE Novex 10% Bis-Tris gels from Invitrogen and transferred onto a nitrocellulose filter (Hybond-C; Amersham). Immunoblotting was performed by using enhanced chemiluminescence (SuperSignal West Dura; Pierce) with a primary horse polyclonal antibody (ab9193; Abcam) targeting HBV surface antigen (Ad/Ay) (29) at a dilution of 1:1,000 and a secondary rabbit polyclonal antibody to horse immunoglobulin G ([IgG] ab6921-1; Abcam), linked to horseradish peroxidase, at a dilution of 1:10,000 (29). To verify the homogeneity of the intracellular protein load, we used an anti-α-tubulin antibody (Sigma) at a dilution of 1:25,000 and a secondary goat polyclonal antibody to mouse immunoglobulin G (Dako) linked to horseradish peroxidase at a dilution of 1:5,000. For extracellular proteins, the load was assessed with Ponceau S red staining of the nitrocellulose membranes.
HBsAg detection.
The secretion of envelope proteins as SVPs or complete viral particles in the culture supernatant of transfected HepG2 cells was assessed by a commercial enzyme-linked immunosorbent assay (ELISA) (Monolisa AgHBs Ultra; catalog no. 72346) from Bio-Rad. The concentration of hepatitis B surface antigen (HBsAg) in the medium was determined by comparison with a purified solution of HBsAg of known concentration. The HBsAg concentration in supernatants of HepG2 cells producing WT virions was between 20 and 160 ng/ml, depending on the experiment.
Virus titration.
To measure the amount of virus produced with WT or mutated envelope proteins, particles from 50 μl of concentrated inocula were fixed on 96-well plates coated with a monoclonal anti pre-S1 antibody (MA18/7; a generous gift from W. H. Gerlich), and viral DNA was quantified by quantitative PCR (Q-PCR). The coating with anti-pre-S1 antibody (4 μg/ml) was performed overnight at 4°C in a bicarbonate buffer at pH 9.6. Then, after saturation with a solution of phosphate-buffered saline supplemented with 3% bovine serum albumin, viral particles from inocula were fixed on the coated plates overnight at room temperature. The viral DNA was released from particles by a proteinase K treatment in the presence of sodium dodecyl sulfate, which destroys virions, and purified by the classical method of phenol-chloroform extraction and isopropyl alcohol precipitation. Finally, the number of genome equivalents (GEq) per milliliter of inoculum was determined by Q-PCR with primers that amplified a sequence in the core gene (12). The virion concentration in WT inocula was between 8 × 108 and 4.7 × 109 GEq/ml, depending on the experiment.
Infection.
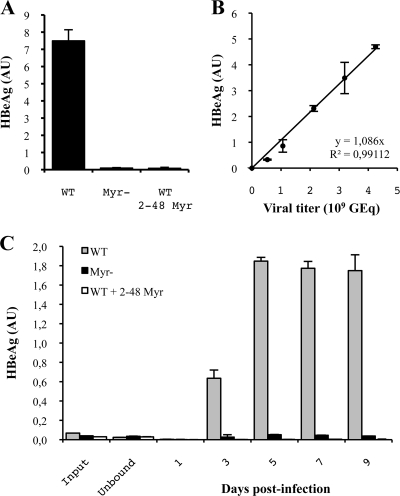
We performed infections with WT and mutant virus on HepaRG cells with 4% PEG, an enhancer of viral infection (26, 28). The level of infection was measured by quantification of HBeAg in the culture supernatant of infected cells with the Bio-Rad kit Monolisa HBe Ag/Ab Plus (catalog number 72396). This marker of HBV infection has previously been proven to be sensitive and reliable in our system and others by comparison with intracellular HBV RNA quantification and HBsAg secretion (19, 22, 35, 53, 54). We further demonstrated its specificity by observing that neither the well-assembled noninfectious control Myr− (9, 27) nor WT virions incubated with the entry inhibitor peptide preS/2-48myr (a myristoylated peptide consisting of residues 2 to 48 of pre-S) (25) led to the production of HBeAg (Fig. 1A). Moreover, we demonstrated that there was a linear and proportional relationship between the level of HBeAg produced by infected HepaRG cells and the amount of virus used to infect them (Fig. 1B). Finally, the analysis of the HBeAg secretion kinetics in the culture supernatant of infected cells demonstrated that while almost no HBeAg could be detected in the viral input and in the unbound fraction, a sustained production of HBeAg starting 3 days after infection was observed for the WT virus (Fig. 1C). This production corresponds to a de novo synthesis since no HBeAg was detected for the two negative controls. In agreement with these results, in a review published in 2007 and focusing on the hepadnavirus entry mechanism (23), Dieter Glebe and Stephan Urban described the HBeAg as a reliable marker of infection that offered the advantage of being undetectable in viral inocula at dilutions used for infection. Importantly, since the HBeAg level was proportional to the viral input used for infection, infectivity results were expressed as a ratio between the level of HBeAg and the amount of virus used for infection to take into account slight variations between different inocula. For the infection of the HepaRG cell line, highly confluent cells (4.75 × 105 cells per 1.9-cm2 well), differentiated in the presence of 2% dimethyl sulfoxide, were covered with 250 μl of serum-free culture medium containing 25 μl of inoculum and 25 μl of PEG 40% (28). After infection, cells were washed three times, and the medium consisting of William's E medium supplemented with 5% FCS, 2% dimethyl sulfoxide, 2 mM l-glutamine, 100 UI/ml of penicillin, 100 μg/ml of streptomycin, 5 μg/ml of insulin, and 50 μM hydrocortisone was renewed every 2 days. We assessed infection 10 days postinfection by measuring the HBeAg level.
FIG. 1.
Characterization of the HBeAg infection marker. (A) The specificity of the HBeAg as an infection marker was assessed by infecting HepaRG cells with either WT virions, noninfectious Myr− virions, or WT viruses incubated with the entry inhibitor preS/2-48myr (25). (B) The linear relationship between the HBeAg level and the amount of virus used to infect cells was demonstrated by infecting HepaRG cells with serial dilutions of a WT inoculum. The black line corresponds to the experimental linear regression calculated at the indicated data points. (C) The kinetics of HBeAg secretion in the culture supernatant of infected cells was determined. The input corresponds to the culture medium containing viruses, which were prepared to infect HepaRG cells, before the incubation with cells, and unbound corresponds to the same medium after overnight incubation with cells. For all experiments, standard deviations were calculated by the analysis of three experiments. AU, arbitrary units.
RESULTS
S protein expression is reduced by deletion of hydrophobic residues in the putative fusion peptide.
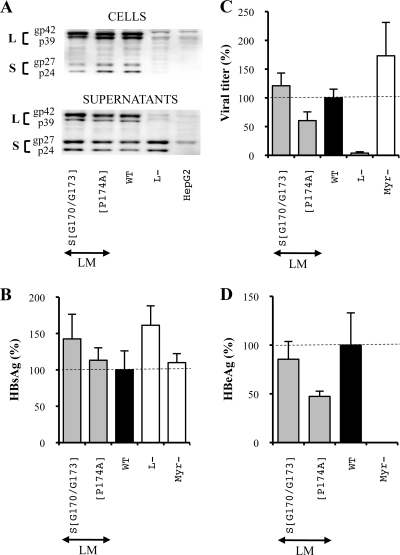
To evaluate the role of the S protein N-terminal extremity including the putative fusion peptide during the infection process, we constructed mutant viruses in which hydrophobic or neutral/polar residues of the region were deleted only in the S protein (Fig. 2, series 1). For each mutant, we assessed intra- and extracellular mutated envelope protein expression, HBsAg secretion, virus production, and infectivity.
For all deletion mutants, the intracellular level of S protein was notably decreased, and for three mutants [S protein with a deletion of the residues between L175 and L178 [SΔ(L175/L178)], SΔ(F182/L185), and SΔ(I188/I191)] it was barely detectable (Fig. 3A). Interestingly, each of these mutants harbored a deletion of one hydrophobic cluster of the TM1 domain. Otherwise, the level of WT L protein was often reduced even though mutations were introduced in only the S protein, suggesting that the expression of the L protein was favored by the coexpression of the S protein. Even though the anti-S antibody should recognize all HBV envelope proteins, the M protein was barely detectable, probably as a consequence of a lower expression level (Fig. 3A). Indeed, when the same extracts were analyzed with a monoclonal anti-Pre-S2 antibody that gave a stronger signal, the M protein was detected in the WT control and in most mutant extracts (data not shown). However, since its presence is dispensable for both assembly and infection processes, we describe only the L and S protein expression level variations.
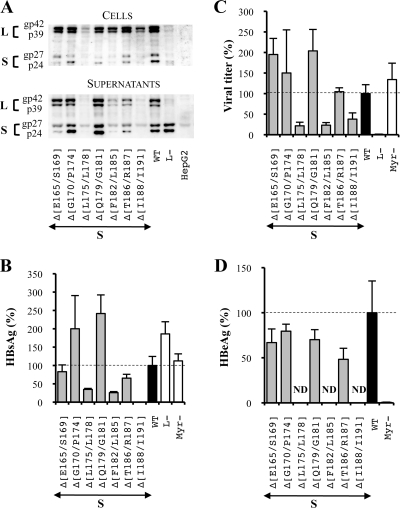
FIG. 3.
Three hydrophobic clusters of four amino acids are crucial in the TM1 domain of the S protein for its expression. S means that mutations were introduced in the sole S protein. For the three graphics, values were expressed as percentages of the WT condition, and sample standard deviations were determined by the analysis of three sets of experiments. The horizontal dotted lines indicate the level of the WT condition. L− and Myr− were used as controls. (A) Analysis of intra- and extracellular mutated protein expression. Cellular proteins (cells), harvested 12 days posttransfection, were probed with an anti-S antibody (1:1,000). Pools of culture supernatants that were collected between days 6 and 12 posttransfection were precipitated with 8% PEG, disrupted in sample buffer (Invitrogen), and analyzed as described above. HepG2 and L− were used as negative controls. Molecular sizes of glycosylated (gp) and unglycosylated (p) HBV envelope proteins are indicated. (B) HBsAg secretion. The effect of deletions on the secretion ability of envelope proteins was assessed by measuring the HBsAg level in pools of culture supernatants, collected between days 6 and 12 posttransfection, of HepG2 cells producing virions. The HBsAg concentration was determined with a commercial ELISA (Bio-Rad). (C) Virus titers. The number of complete particles in inocula was determined by Q-PCR analysis of viral DNA extracted from immuno-captured virus. (D) Infectivity. Infection was assessed by measuring the level of HBeAg in the culture supernatant of infected HepaRG cells 10 days postinfection with a commercial ELISA (Bio-Rad). The infectivity was expressed as a ratio between the level of HBeAg and the number of GEq used for infection.
With respect to the extracellular envelope protein level as assessed by Western blot analysis and ELISA, when a significant amount of intracellular S protein was detected, it was either increased or slightly decreased while it was barely detectable for the three mutants whose intracellular S protein levels were strongly reduced (Fig. 3A and B). Overall, both intra- and extracellular envelope protein levels were strongly reduced by the deletion of hydrophobic clusters in the TM1 domain of the S protein.
Then, we assessed the ability of deleted proteins to complement an envelope protein-defective genome for viral particle secretion. In the L− control, the L and M protein expression vector was omitted, thus preventing virus production. As expected, no virus was detected in this control. In addition, we showed that in the S protein, the deletion of any of the TM1 domain hydrophobic clusters inhibited the assembly process by 60 to 80% while other deletions did not alter virus production (Fig. 3C).
Finally, we evaluated the infectious ability of the mutants whose assembly was not impaired by measuring the amount of HBeAg in the culture supernatant of infected HepaRG cells. This specific marker of HBV infection has been previously proven to be sensitive and reliable in our system and in others by comparison with intracellular HBV RNA quantification and HBsAg secretion (19, 22, 35, 53, 54) (Fig. 1). To take into account the slight variations between the different inocula, infectivity results were expressed as a ratio between the level of HBeAg and the amount of virus used for infection. First, we verified that no HBeAg was detected in the noninfectious assembly-competent Myr− control (9, 27), thus confirming our infection assay specificity. Then, we observed that the analyzed mutants remained fully infectious (Fig. 3D).
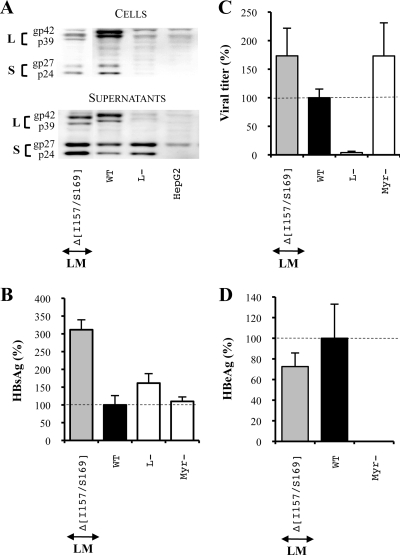
Deletions in the N-terminal extremity of the L and M protein S domain evidence the important role of the TM1 domain for infectivity.
Since deletions exclusively introduced in the S protein either prevent viral assembly or have no effect on viral infectivity, we tested the impact of the same mutations when introduced in only the L and M proteins using the pSV12SX S− vector. This vector does not produce any S protein owing to the mutation of the S protein initiation codon into a threonine codon (see Materials and Methods). This time, Western blot analysis showed that intracellular mutated L protein and WT S protein levels were either comparable to the WT condition or reduced [L and M proteins with a deletion of residues F182 to L185 [LMΔ(F182/L185)], LMΔ(T186/R187) and LMΔ(I188/I191)] while their extracellular levels were never decreased (Fig. 4A). Thus, all deletions, and notably the ones that suppress hydrophobic clusters, allowed sustained envelope protein expression and secretion. As a further support of this conclusion, the SVP secretion level measured by an HBsAg-specific ELISA tended to be increased and even sometimes exceeded the secretion level in the L− control (Fig. 4B). In this control, the absence of the L protein resulted in increased S protein secretion because this protein was no longer retained by the L protein and thus became barely detectable within cells (Fig. 4A). Indeed, it is well known that the M and S surface proteins are spontaneously secreted in the form of SVPs while the L protein is not self-competent for secretion and retained the S protein in the ER in a dose-dependent manner by virtue of its association with this protein (32, 41, 44). Thus, since the mutated L proteins were present in supernatants, it is likely that the interaction between the L and S proteins was preserved and that the observed increase of secretion for all the deletions resulted from a diminution of L protein intracellular retention.
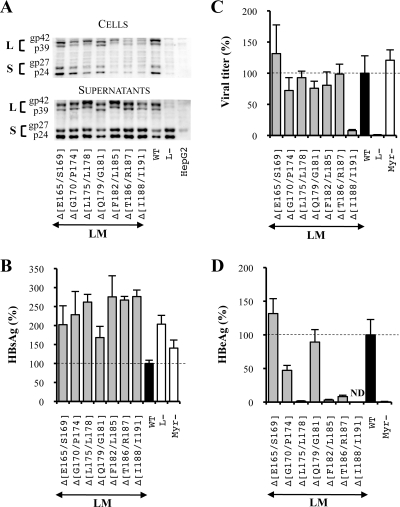
FIG. 4.
Effect of deletions at the N-terminal extremity of the L and M protein S domains on the HBV viral cycle. For the three graphics, values were expressed as percentages of the WT condition, and sample standard deviations were determined by the analysis of three sets of experiments. The horizontal dotted lines indicate the level of the WT condition. L− and Myr− were used as controls. (A) Analysis of intra- and extracellular mutated protein expression. Cellular proteins (cells), harvested 12 days posttransfection, were probed with an anti-S antibody (1:1,000). Pools of culture supernatants that were collected between days 6 and 12 posttransfection were precipitated with 8% PEG, disrupted in sample buffer (Invitrogen), and analyzed as described above. HepG2 and L− were used as negative controls. Molecular sizes of glycosylated (gp) and unglycosylated (p) HBV envelope proteins are indicated. (B) HBsAg secretion. The effect of deletions on the secretion ability of envelope proteins was assessed by measuring the HBsAg level in pools of culture supernatants, collected between days 6 and 12 posttransfection, of HepG2 cells producing virions. HBsAg concentration was determined with a commercial ELISA (Bio-Rad). (C) Virus titers. The number of complete particles in inocula was determined by Q-PCR analysis of viral DNA extracted from immuno-captured virus. (D) Infectivity. Infection was assessed by measuring the level of HBeAg in the culture supernatant of infected HepaRG cells 10 days postinfection with a commercial ELISA (Bio-Rad). The infectivity was expressed as a ratio between the level of HBeAg and the number of GEq used for infection.
Virus production analysis demonstrated that deletions in the N-terminal part of the L protein S domain did not affect particle assembly except for one mutation, LMΔ(I188/I191), which prevented complete viral particle secretion (Fig. 4C). Since the protocol used for measuring the virus titer depends on the immuno-capture of virus particles with an anti-pre-S1 antibody (MA18/7), it is possible that the LMΔ(I188/I191) mutation blocks the appearance on the virus surface of the pre-S domain of the L protein normally occurring during the topology switch of the L protein and prevents the detection of virions. However, it seems unlikely that small deletions in the TM1 region affected the pre-S1 domain translocation since it was demonstrated that a larger deletion spanning the totality of the TM1 domain (between amino acids 9 and 32 of the S domain) did not affect the translocation and that the TM2 domain was necessary and sufficient for the pre-S translocation (33). Importantly, an interesting feature of our titration protocol is that when a mutation does not affect the viral titer but inhibits infectivity, this result cannot be attributed to a lack of pre-S translocation.
Finally, we analyzed the infectious ability of this series of mutated virions. First, we observed a moderate decrease of infectivity for the mutant LMΔ(G170/P174) (Fig. 4D). The presence of a proline in this deletion led us to investigate whether its sole substitution could similarly impact viral infectivity. Indeed, the same level of inhibition was observed when the P174 residue was replaced by an alanine while the substitution of all other amino acids of this group including the two highly hydrophobic residues (F171 and L172) (Fig. 2, series 2) had no effect on infectivity (Fig. 5). More importantly, we showed that the deletion of two hydrophobic clusters, residues L175 to L178 (L175/L178) and F182/L185, and of a group of two polar amino acids, T186 and R187, in the L and M proteins reduced viral infectivity by more than 90% (Fig. 4D).
FIG. 5.
The replacement of the proline 174 by an alanine reduced HBV infectivity. For the three graphics, values were expressed as percentages of the WT condition, and sample standard deviations were determined by the analysis of three sets of experiments. The horizontal dotted lines indicate the level of the WT condition. L− and Myr− were used as controls. (A) Analysis of intra- and extracellular mutated protein expression. Cellular proteins (cells), harvested 12 days posttransfection, were probed with an anti-S antibody (1:1,000). Pools of culture supernatants that were collected between days 6 and 12 posttransfection were precipitated with 8% PEG, disrupted in sample buffer (Invitrogen), and analyzed as described above. HepG2 and L− were used as negative controls. Molecular sizes of glycosylated (gp) and unglycosylated (p) HBV envelope proteins are indicated. (B) HBsAg secretion. The effect of mutations on the secretion ability of envelope proteins was assessed by measuring the HBsAg level in pools of culture supernatants, collected between days 6 and 12 posttransfection, of HepG2 cells producing virions. The HBsAg concentration was determined with a commercial ELISA (Bio-Rad). (C) Virus titers. The number of complete particles in inocula was determined by Q-PCR analysis of viral DNA extracted from immuno-captured virus. (D) Infectivity. Infection was assessed by measuring the HBeAg level in the culture supernatant of infected HepaRG cells 10 days postinfection with a commercial ELISA (Bio-Rad). The infectivity was expressed as a ratio between the level of HBeAg and the number of GEq used for infection.
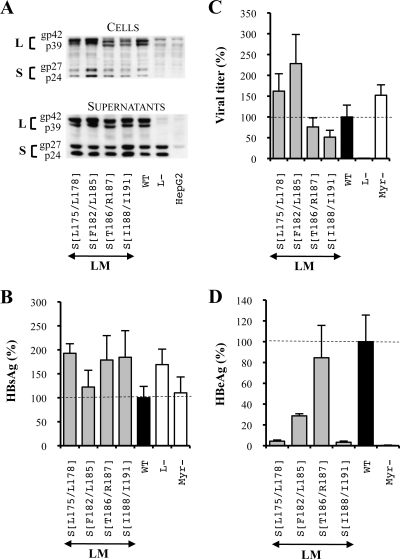
Multiple alanine substitutions reveal a third hydrophobic cluster essential for infectivity.
Since deletions within proteins may result in artifacts, we decided to confirm by a multiple-alanine substitution approach the role of residues whose deletion in the L and M proteins inhibited infectivity or assembly processes. Our results showed that the intracellular level of viral envelope proteins bearing the substitutions described in Fig. 2 (series 3) was either slightly reduced or remained unaffected (Fig. 6A) while the SVP secretion as well as the extracellular protein level was not impaired (Fig. 6A and B). Then, we observed that none of the mutations strongly inhibited virus production (Fig. 6C). Interestingly, while the deletion of the third hydrophobic cluster in the L and M proteins drastically inhibited viral particle assembly (Fig. 4C), the corresponding replacement by alanines decreased the viral titer by no more than 50% (Fig. 6C). Finally, we confirmed the importance of the two first hydrophobic clusters L175/L178 and F182/L185 in the infection process since their mutation inhibited infectivity by 95 and 70%, respectively (Fig. 6D), and we observed that substitutions in the third hydrophobic cluster, I188/I191, inhibited infectivity by more than 95%. Thus, we demonstrated that the three TM1 domain hydrophobic clusters play a major role in HBV infectivity. Moreover, we demonstrated that it was possible to simultaneously mutate the T186 and R187 residues without disturbing the infectious process (Fig. 6D). Since their deletion in the L and M proteins had a pronounced inhibitory effect on infectivity (Fig. 4D), we may speculate that this short hydrophobic sequence plays only a role of spacer between two adjacent hydrophobic stretches. All in all, these results identified three hydrophobic clusters of four amino acids at the N-terminal extremity of the L and M protein S domain that are crucial for HBV infectivity.
FIG. 6.
Multiple alanine substitutions in the L and M protein TM1 domain revealed the presence of three hydrophobic clusters crucial for HBV infectivity. For the three graphics, values were expressed as percentages of the WT condition, and sample standard deviations were determined by the analysis of three sets of experiments. The horizontal dotted lines indicate the level of the WT condition. L− and Myr− were used as controls. (A) Analysis of intra- and extracellular mutated protein expression. Cellular proteins (cells), harvested 12 days posttransfection, were probed with an anti-S antibody (1:1,000). Pools of culture supernatants that were collected between days 6 and 12 posttransfection were precipitated with 8% PEG, disrupted in sample buffer (Invitrogen), and analyzed as described above. HepG2 and L− were used as negative controls. Molecular sizes of glycosylated (gp) and unglycosylated (p) HBV envelope proteins are indicated. (B) HBsAg secretion. The effect of substitutions on the secretion ability of envelope proteins was assessed by measuring the HBsAg level in pools of culture supernatants, collected between days 6 and 12 posttransfection, of HepG2 cells producing virions. The HBsAg concentration was determined with a commercial ELISA (Bio-Rad). (C) Virus titers. The number of complete particles in inocula was determined by Q-PCR analysis of viral DNA extracted from immuno-captured virus. (D) Infectivity. Infection was assessed by measuring the level of HBeAg in the culture supernatant of infected HepaRG cells 10 days postinfection with a commercial ELISA (Bio-Rad). The infectivity was expressed as a ratio between the level of HBeAg and the number of GEq used for infection.
The integrity of the TM1 domain heptad repeat structure is dispensable for infectivity.
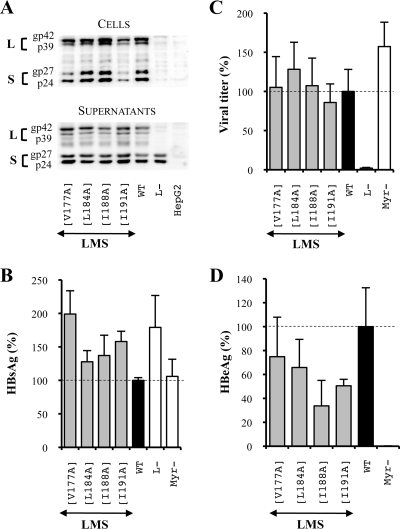
To evaluate whether the hydrophobic residues that were crucial for infectivity were components of a heptad repeat motif that may be important for fusion activity (50), we constructed new mutant viruses whose hydrophobic amino acids, constitutive of the most hydrophobic side of the heptad repeat helical structure, were replaced by alanines (Fig. 2, series 4). In the duck hepatitis B virus (DHBV) putative fusion peptide, the mutation of such amino acids, also located at the N-terminal extremity of the S domain, was able to disrupt infectivity (13). First, we made single substitutions, and then, since no strong inhibition of infectivity was observed even when mutations were introduced in the three envelope proteins (Fig. 7), we decided to combine up to four mutations either in the L and M proteins or in the S protein to finally identify two isoleucines, I188 and I191, located at the C-terminal extremity of the TM1 domain (Fig. 2, series 4), that were crucial for infectivity in the L and M proteins. The analysis of intra- and extracellular mutated protein expression levels revealed an increase in the extracellular protein level and HBsAg secretion for the mutant L and M proteins with alanine substitutions for residues I188 and I191 [LM(I188A I191A)] (Fig. 8A and B), probably because of reduced L protein intracellular retention, as observed when the whole hydrophobic cluster was replaced by alanines. Then, the analysis of virus production demonstrated that neither the S(I188A I191A) nor the LM(I188A I191A) mutant affected viral particle assembly (Fig. 8C). Finally we demonstrated that in the L and M proteins, isoleucines of the third hydrophobic cluster (I188 and I191) were crucial for the HBV infection process. Indeed, when these residues were both mutated in the L and M proteins, HBV infectivity was reduced by 90% (Fig. 8D). By contrast, when the same mutation was introduced in the S protein, it had no significant effect on the infection process. Since the M protein is dispensable for virus infectivity, we can assume that these isoleucines are crucial for HBV infectivity in the sole L protein.
FIG. 7.
The unique substitution of amino acids constitutive of the TM1 domain heptad repeat does not affect the HBV viral cycle. LMS means that mutations were introduced in the three envelope proteins. For this purpose, the mutagenesis was performed in the pSV12SX and pSVSX plasmids. For the three graphics, values were expressed as percentages of the WT condition, and sample standard deviations were determined by the analysis of three sets of experiments. The horizontal dotted lines indicate the level of the WT condition. L− and Myr− were used as controls. (A) Analysis of intra- and extracellular mutated protein expression. Cellular proteins (cells), harvested 12 days posttransfection, were probed with an anti-S antibody (1:1,000). Pools of culture supernatants that were collected between days 6 and 12 posttransfection were precipitated with 8% PEG, disrupted in sample buffer (Invitrogen), and analyzed as described above. HepG2 and L− were used as negative controls. Molecular sizes of glycosylated (gp) and unglycosylated (p) HBV envelope proteins are indicated. (B) HBsAg secretion. The effect of mutations on the secretion ability of envelope proteins was assessed by measuring the HBsAg level in pools of culture supernatants, collected between days 6 and 12 posttransfection, of HepG2 cells producing virions. The HBsAg concentration was determined with a commercial ELISA (Bio-Rad). (C) Virus titers. The number of complete particles in inocula was determined by Q-PCR analysis of viral DNA extracted from immuno-captured virus. (D) Infectivity. Infection was assessed by measuring the HBeAg level in the culture supernatant of infected HepaRG cells 10 days postinfection with a commercial ELISA (Bio-Rad). The infectivity was expressed as a ratio between the level of HBeAg and the number of GEq used for infection.
FIG. 8.
Two isoleucines of the third hydrophobic cluster are crucial in L and M proteins for infectivity. For the three graphics, values were expressed as percentages of the WT condition, and sample standard deviations were determined by the analysis of three sets of experiments. The horizontal dotted lines indicate the level of the WT condition. L− and Myr− were used as controls (A) Western Blot analysis of intra- and extracellular mutated protein expression. Cellular proteins (cells), harvested 12 days posttransfection, were probed with an anti-S antibody (1:1,000). The pools of culture supernatants that were collected between days 6 and 12 posttransfection were precipitated with 8% PEG, disrupted in sample buffer (Invitrogen), and analyzed as described above. HepG2 and L− were used as negative controls. Molecular sizes of glycosylated (gp) and unglycosylated (p) HBV envelope proteins are indicated. (B) HBsAg secretion. The effect of the mutation on the secretion ability of envelope proteins was assessed by measuring the HBsAg level in pools of culture supernatants, collected between days 6 and 12 posttransfection, of HepG2 cells producing virions. The HBsAg concentration was determined with a commercial ELISA (Bio-Rad). (C) Virus titers. The number of complete particles in inocula was determined by Q-PCR analysis of viral DNA extracted from immuno-captured virus. D: Infectivity. Infection was assessed by measuring the HBeAg level in the culture supernatant of infected HepaRG cells 10 days postinfection with a commercial ELISA (Bio-Rad). The infectivity was expressed as a ratio between the level of HBeAg and the number of GEq used for infection.
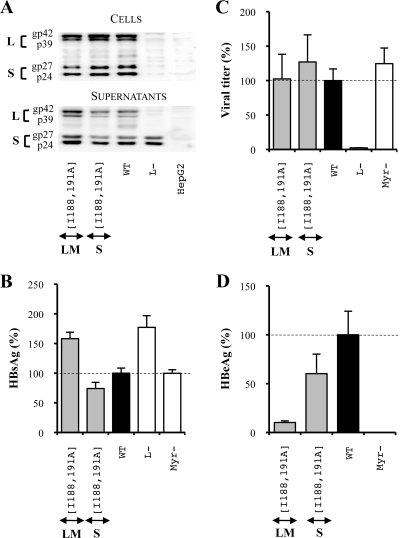
The PEST sequence upstream of the L and M protein TM1 domains is dispensable for HBV infectivity.
To interact with target membranes as fusion loops or fusion peptides do, the TM1 domain of the L protein should either form a loop at the surface of viral particles or be exposed at the N-terminal extremity of the protein by a proteolytic cleavage. Since Lu et al. identified a PEST sequence likely containing several proteolytic cleavage sites upstream of the TM1 domain (39), we asked whether this sequence was necessary for infectivity. To answer this question, we constructed an additional mutant in which the L and M protein PEST sequences were deleted (Fig. 2). First, our results showed that the PEST sequence deletion led to a decrease of intracellular viral envelope proteins and to a great increase in HBsAg secretion and extracellular protein levels (Fig. 9A and B), probably again as a consequence of a reduced L protein intracellular retention capacity. Then, we observed that neither the assembly nor the infection process was affected by the deletion (Fig. 9C and D), meaning that the PEST sequence was clearly dispensable for infectivity.
FIG. 9.
The PEST sequence is dispensable for HBV assembly and infection processes. For the three graphics, values were expressed as percentages of the WT condition, and sample standard deviations were determined by the analysis of three sets of experiments. The horizontal dotted lines indicate the level of the WT condition. L− and Myr− were used as controls. (A) Analysis of intra- and extracellular mutated protein expression. Cellular proteins (cells), harvested 12 days posttransfection, were probed with an anti-S antibody (1:1,000). Pools of culture supernatants that were collected between days 6 and 12 posttransfection were precipitated with 8% PEG, disrupted in sample buffer (Invitrogen), and analyzed as described above. HepG2 and L− were used as negative controls. Molecular sizes of glycosylated (gp) and unglycosylated (p) HBV envelope proteins are indicated. (B) HBsAg secretion. The effect of the deletion on the secretion ability of envelope proteins was assessed by measuring the HBsAg level in pools of culture supernatants, collected between days 6 and 12 posttransfection, of HepG2 cells producing virions. The HBsAg concentration was determined with a commercial ELISA (Bio-Rad). (C) Virus titers. The number of complete particles in inocula was determined by Q-PCR analysis of viral DNA extracted from immuno-captured virus. (D) Infectivity. Infection was assessed by measuring the HBeAg level in the culture supernatant of infected HepaRG cells 10 days postinfection with a commercial ELISA (Bio-Rad). The infectivity was expressed as a ratio between the level of HBeAg and the number of GEq used for infection.
DISCUSSION
In this work, we evaluated the role during HBV entry of two motifs, possibly important for HBV infectivity, which were identified at the N-terminal extremity of the S domain by sequence homology but were not demonstrated to be necessary for the infection process. The first motif corresponds to a putative fusion peptide that has been identified by comparison with other N-terminal viral fusion peptides (50) even though HBV envelope protein structure, characterized by the presence of four TM-spanning domains, strikingly differs from known viral fusion proteins. The second motif, a PEST sequence likely containing several proteolytic cleavage sites, was suggested to be implicated in the proteolytic exposure of the L and M protein putative fusion peptide (39).
The presence of the putative fusion peptide at different positions in envelope proteins, either N-terminal in the S protein or internal in the L and M proteins, led us to introduce deletions separately in these proteins. This strategy allowed us to observe that the deletion of any of the three TM1 domain hydrophobic clusters of the S protein strongly reduced its intracellular level, in agreement with a previous study (47) which reported that S protein expression was altered by deletions in the C-terminal part of the TM1 domain. Since all other S protein mutants showed sustained expression and secretion and normal viral assembly and infectivity, this suggested that the main function of the S protein TM1 domain is to ensure the expression of this protein. By contrast, when mutations were introduced in the L and M proteins, they did not affect the synthesis or stability of envelope proteins and tended to increase HBsAg secretion. As it was established that the first 32 residues of the S domain act as an uncleaved signal peptide allowing the cotranslational insertion of the S protein TM1 domain in the ER membrane (16, 18), it was not surprising that interference with this essential function affected S protein synthesis or stability. On the contrary, it was logical that the expression of the L protein was not altered since in the pre-S form of this protein, which is present at the inner face of viral particles, the TM1 domain is cytoplasmically exposed, and thus its cotranslational translocation function is not required. Moreover, the topogenic signal overlapping the TM1 domain was implicated in the ER translocation of upstream sequences and was then supposed to be necessary for the translocation of the M protein pre-S2 domain (17). Therefore, the deletions of hydrophobic clusters that disturbed S protein expression could have prevented the translocation of the M protein pre-S2 domain when the deletions were introduced in the L and M proteins, leading to the suppression of glycosylation of the pre-S2 domain asparagine. Consequently, we verified by Western blot analysis performed with an anti-pre-S2 antibody the presence of the glycosylated forms (gp33 and ggp36) of the M protein. Since we observed no alteration of the glycosylation profile of the deleted M proteins (data not shown), we concluded that small deletions in this topogenic signal of the M protein did not affect its pre-S2 domain translocation.
Surprisingly, we observed that a drop in the mutated S protein intracellular level in some mutants triggered a strong decrease in the WT L protein level, suggesting that its expression was favored by the coexpression of the S protein. We confirmed this hypothesis by an additional experiment in which we noticed that the intra- and extracellular levels of a WT L protein produced without S protein were strongly reduced compared to the levels of the same WT L protein produced with S protein (data not shown). It is well known that the L protein is not secreted in the absence of the S protein. We had therefore anticipated that reduced S protein expression would lead to intracellular accumulation of L protein. On the contrary, we observed that not only extracellular but also intracellular L protein levels were reduced when S protein levels were decreased. We may speculate that in the absence of the S protein, the L protein may accumulate in a misfolded conformation, which is progressively degraded within the culture time.
Concerning the assembly process, we observed that none of the deletions introduced in the L and M proteins affected virus production with the exception of one mutant, LMΔ(I188/I191), for which no complete particles were detected. Nevertheless, the replacement of these four residues by alanines only slightly reduced complete particle production, suggesting that these amino acids act as spacers.
Finally, infectivity analysis of mutant viruses bearing contiguous deletions or substitutions in the N-terminal extremity of the L and M protein S domain allowed the identification of three hydrophobic clusters in the TM1 domain (HC1, HC2, and HC3) that are crucial for HBV entry. Whether these residues crucial for infectivity are involved in the binding or in the fusion step of viral entry remained to be determined. Indeed, since the L protein is presumably involved in binding to host cells and since no binding test for the virus receptor is yet available, we cannot rule out that our mutations affect this initial step of viral entry. Nevertheless, previous studies (described below) and the hydrophobic nature of these clusters suggest that they may be implicated in a direct interaction with the external lipid leaflet of a target cellular membrane during a fusion process. Indeed, in agreement with this view, previous work showed that synthetic peptides covering amino acids M164 to Q179 of the putative fusion peptide, in which the first TM1 domain hydrophobic cluster is included, can destabilize liposomes (51). Moreover, Berting et al. demonstrated that the influenza virus hemagglutinin fusion protein whose fusion peptide sequence was replaced by L protein amino acids G170 to Q179, which includes the first hydrophobic cluster, can induce a hemifusion process (2). Otherwise, we noticed that of the three described hydrophobic clusters, the first and third strikingly resemble each other and seem to have a more important role for infectivity than the second one since their deletion, as well as their replacement by alanines, inhibited infectivity by more than 95%. Thus, even though a synthetic fusion peptide including only the first hydrophobic cluster was sufficient in a peptide fusion assay to destabilize lipids (51), we postulate that the third cluster may be necessary for inducing fusion in the natural infection process. Interestingly, the two isoleucines present in this cluster were dispensable for infectivity in the S protein while they were essential in the L protein. We may then suggest that all TM1 domain hydrophobic clusters, even though they are critical for S protein synthesis and their effect on infectivity cannot be directly assessed, play a role in viral entry only when they are present in the L and M proteins. Accordingly, in the DHBV infection model, Chojnacki et al., by introducing similar mutations in the TM1 domain of DHBV envelope proteins, showed that the sole L protein was implicated in the fusion step of the entry process (13).
The infectivity determinant that we identified in the L and M protein TM1 domains, if involved in the fusion process, would act either as an internal fusion loop or as an N-terminal fusion peptide. Given the internal position of this determinant, its N-terminal exposure would require proteolytic cleavage upstream of its sequence. Since the deletion of a PEST sequence identified upstream of the TM1 domain and likely containing several proteolytic cleavage sites did not inhibit viral infectivity, we concluded that the L protein TM1 domain was not likely to support the function of N-terminal fusion peptide and, rather, that it acts as a fusion loop.
To directly participate in the entry process, the TM1 domain should be exposed at the surface of viral particles. However, current models propose that in the L protein this domain is either inside viral particles or embedded within the lipid bilayer of virions. Since the TM1 domain has a relatively low hydrophobicity, this dual topology is not surprising. Ultimately, its mild hydrophobic character could also allow its full translocation outside the viral particle. In agreement with this hypothesis, Grgacic and Schaller observed in the DHBV model that low pH could trigger the translocation and exposure of the L protein TM1 domain at the surface of viral particles (24). Thus, these data suggested that the infectivity determinant of the HBV TM1 domain could directly interact with the targeted host cell membrane. It remains to be determined, however, whether the HBV L protein TM1 domain is also exposed during the infection process either as a consequence of exposure to low pH, disulfide bridge isomerization, or binding to a cellular receptor. In addition, it should be very informative to establish an in vitro fusion assay based on viral particles and liposomes for directly measuring the fusion activity of TM1 hydrophobic cluster mutants.
Acknowledgments
This work was supported by funding from INSERM (Institut National de la Santé et de la Recherche Médicale), ARC (Association pour la Recherche sur le Cancer), and ANRS (Agence Nationale de Recherche contre le Sida et les hépatites virales). C. Lepère-Douard and M. Trotard are recipients of fellowships, respectively, from the Ministère de l'Education Nationale de la Recherche et de la Technologie and from the Région Bretagne.
We gratefully acknowledge M. Samson for critical reading of the manuscript.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Abou-Jaoude, G., and C. Sureau. 2007. Entry of hepatitis delta virus requires the conserved cysteine residues of the hepatitis B virus envelope protein antigenic loop and is blocked by inhibitors of thiol-disulfide exchange. J. Virol. 81:13057-13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berting, A., C. Fischer, S. Schaefer, W. Garten, H. D. Klenk, and W. H. Gerlich. 2000. Hemifusion activity of a chimeric influenza virus hemagglutinin with a putative fusion peptide from hepatitis B virus. Virus Res. 68:35-49. [DOI] [PubMed] [Google Scholar]
- 3.Blanchet, M., and C. Sureau. 2006. Analysis of the cytosolic domains of the hepatitis B virus envelope proteins for their function in viral particle assembly and infectivity. J. Virol. 80:11935-11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchet, M., and C. Sureau. 2007. Infectivity determinants of the hepatitis B virus pre-S domain are confined to the N-terminal 75 amino acid residues. J. Virol. 81:5841-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 71:9350-9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss, V. 2007. Hepatitis B virus morphogenesis. World J. Gastroenterol. 13:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss, V., and D. Ganem. 1991. Mutational analysis of hepatitis B surface antigen particle assembly and secretion. J. Virol. 65:3813-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruss, V., J. Hagelsten, E. Gerhardt, and P. R. Galle. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218:396-399. [DOI] [PubMed] [Google Scholar]
- 10.Bruss, V., and R. Thomssen. 1994. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J. Virol. 68:1643-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruss, V., and K. Vieluf. 1995. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J. Virol. 69:6652-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, R. W., H. Piiparinen, M. Seppanen, P. Koskela, S. Sarna, and M. Lappalainen. 2001. Real-time PCR for detection and quantitation of hepatitis B virus DNA. J. Med. Virol. 65:250-256. [DOI] [PubMed] [Google Scholar]
- 13.Chojnacki, J., D. A. Anderson, and E. V. Grgacic. 2005. A hydrophobic domain in the large envelope protein is essential for fusion of duck hepatitis B virus at the late endosome. J. Virol. 79:14945-14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chouteau, P., J. Le Seyec, I. Cannie, M. Nassal, C. Guguen-Guillouzo, and P. Gripon. 2001. A short N-proximal region in the large envelope protein harbors a determinant that contributes to the species specificity of human hepatitis B virus. J. Virol. 75:11565-11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubois, M. F., C. Pourcel, S. Rousset, C. Chany, and P. Tiollais. 1980. Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc. Natl. Acad. Sci. USA 77:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eble, B. E., V. R. Lingappa, and D. Ganem. 1986. Hepatitis B surface antigen: an unusual secreted protein initially synthesized as a transmembrane polypeptide. Mol. Cell. Biol. 6:1454-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eble, B. E., V. R. Lingappa, and D. Ganem. 1990. The N-terminal (pre-S2) domain of a hepatitis B virus surface glycoprotein is translocated across membranes by downstream signal sequences. J. Virol. 64:1414-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eble, B. E., D. R. MacRae, V. R. Lingappa, and D. Ganem. 1987. Multiple topogenic sequences determine the transmembrane orientation of the hepatitis B surface antigen. Mol. Cell. Biol. 7:3591-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelke, M., K. Mills, S. Seitz, P. Simon, P. Gripon, M. Schnolzer, and S. Urban. 2006. Characterization of a hepatitis B and hepatitis delta virus receptor binding site. Hepatology 43:750-760. [DOI] [PubMed] [Google Scholar]
- 20.Fernholz, D., P. R. Galle, M. Stemler, M. Brunetto, F. Bonino, and H. Will. 1993. Infectious hepatitis-B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology 194:137-148. [DOI] [PubMed] [Google Scholar]
- 21.Fernholz, D., M. Stemler, M. Brunetto, F. Bonino, and H. Will. 1991. Replicating and virion secreting hepatitis B mutant virus unable to produce preS2 protein. J. Hepatol. 13(Suppl. 4):S102-S104. [DOI] [PubMed] [Google Scholar]
- 22.Glebe, D., M. Aliakbari, P. Krass, E. V. Knoop, K. P. Valerius, and W. H. Gerlich. 2003. Pre-s1 antigen-dependent infection of Tupaia hepatocyte cultures with human hepatitis B virus. J. Virol. 77:9511-9521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glebe, D., and S. Urban. 2007. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 13:22-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grgacic, E. V., and H. Schaller. 2000. A metastable form of the large envelope protein of duck hepatitis B virus: low-pH release results in a transition to a hydrophobic, potentially fusogenic conformation. J. Virol. 74:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gripon, P., I. Cannie, and S. Urban. 2005. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 79:1613-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gripon, P., C. Diot, and C. Guguen-Guillouzo. 1993. Reproducible high level infection of cultured adult human hepatocytes by hepatitis-B virus: effect of polyethylene glycol on adsorption and penetration. Virology 192:534-540. [DOI] [PubMed] [Google Scholar]
- 27.Gripon, P., J. Le Seyec, S. Rumin, and C. Guguen-Guillouzo. 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213:292-299. [DOI] [PubMed] [Google Scholar]
- 28.Gripon, P., S. Rumin, S. Urban, J. Le Seyec, D. Glaise, I. Cannie, C. Guyomard, J. Lucas, C. Trepo, and C. Guguen-Guillouzo. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 99:15655-15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guarnieri, M., K. H. Kim, G. Bang, J. Li, Y. Zhou, X. Tang, J. Wands, and S. Tong. 2006. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J. Virol. 80:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huovila, A. P., A. M. Eder, and S. D. Fuller. 1992. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J. Cell Biol. 118:1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaoude, G. A., and C. Sureau. 2005. Role of the antigenic loop of the hepatitis B virus envelope proteins in infectivity of hepatitis delta virus. J. Virol. 79:10460-10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroki, K., R. Russnak, and D. Ganem. 1989. Novel N-terminal amino acid sequence required for retention of a hepatitis B virus glycoprotein in the endoplasmic reticulum. Mol. Cell. Biol. 9:4459-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert, C., and R. Prange. 2001. Dual topology of the hepatitis B virus large envelope protein: determinants influencing post-translational pre-S translocation. J. Biol. Chem. 276:22265-22272. [DOI] [PubMed] [Google Scholar]
- 34.Leemans, W. F., H. L. Janssen, and R. A. de Man. 2007. Future prospectives for the management of chronic hepatitis B. World J. Gastroenterol. 13:2554-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lepere, C., M. Regeard, J. Le Seyec, and P. Gripon. 2007. The translocation motif of hepatitis B virus envelope proteins is dispensable for infectivity. J. Virol. 81:7816-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 73:2052-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 72:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, J., and B. Rost. 2003. NORSp: predictions of long regions without regular secondary structure. Nucleic Acids Res. 31:3833-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, X., T. M. Block, and W. H. Gerlich. 1996. Protease-induced infectivity of hepatitis B virus for a human hepatoblastoma cell line. J. Virol. 70:2277-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nassal, M. 1996. Hepatitis B virus morphogenesis. Curr. Top. Microbiol. Immunol. 214:297-337. [DOI] [PubMed] [Google Scholar]
- 41.Ou, J. H., and W. J. Rutter. 1987. Regulation of secretion of the hepatitis B virus major surface antigen by the preS-1 protein. J. Virol. 61:782-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patient, R., C. Hourioux, P. Y. Sizaret, S. Trassard, C. Sureau, and P. Roingeard. 2007. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J. Virol. 81:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patzer, E. J., G. R. Nakamura, C. C. Simonsen, A. D. Levinson, and R. Brands. 1986. Intracellular assembly and packaging of hepatitis B surface antigen particles occur in the endoplasmic reticulum. J. Virol. 58:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persing, D. H., H. E. Varmus, and D. Ganem. 1986. Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science 234:1388-1391. [DOI] [PubMed] [Google Scholar]
- 45.Petersen, J., M. Dandri, W. Mier, M. Lutgehetmann, T. Volz, F. von Weizsacker, U. Haberkorn, L. Fischer, J. M. Pollok, B. Erbes, S. Seitz, and S. Urban. 2008. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat. Biotechnol. 26:335-341. [DOI] [PubMed] [Google Scholar]
- 46.Poisson, F., A. Severac, C. Hourioux, A. Goudeau, and P. Roingeard. 1997. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology 228:115-120. [DOI] [PubMed] [Google Scholar]
- 47.Prange, R., R. Nagel, and R. E. Streeck. 1992. Deletions in the hepatitis B virus small envelope protein: effect on assembly and secretion of surface antigen particles. J. Virol. 66:5832-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez-Crespo, I., J. Gomez-Gutierrez, J. A. Encinar, J. M. Gonzalez-Ros, J. P. Albar, D. L. Peterson, and F. Gavilanes. 1996. Structural properties of the putative fusion peptide of hepatitis B virus upon interaction with phospholipids. Circular dichroism and Fourier-transform infrared spectroscopy studies. Eur. J. Biochem. 242:243-248. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez-Crespo, I., J. Gomez-Gutierrez, M. Nieto, D. L. Peterson, and F. Gavilanes. 1994. Prediction of a putative fusion peptide in the S protein of hepatitis B virus. J. Gen. Virol. 75:637-639. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Crespo, I., E. Nunez, J. Gomez-Gutierrez, B. Yelamos, J. P. Albar, D. L. Peterson, and F. Gavilanes. 1995. Phospholipid interactions of the putative fusion peptide of hepatitis B virus surface antigen S protein. J. Gen. Virol. 76:301-308. [DOI] [PubMed] [Google Scholar]
- 52.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5:1704-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salisse, J., and C. Sureau. 2009. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J. Virol. 83:9321-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulze, A., P. Gripon, and S. Urban. 2007. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 46:1759-1768. [DOI] [PubMed] [Google Scholar]
- 55.Tillmann, H. L. 2007. Antiviral therapy and resistance with hepatitis B virus infection. World J. Gastroenterol. 13:125-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.World Health Organization. 2002. Hepatitis B. Department of Communicable Diseases Surveillance and Response, World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/hepatitis/HepatitisB_whocdscsrlyo2002_2.pdf.