Abstract
The P, V, and C proteins of measles virus are encoded in overlapping reading frames of the P gene, which makes it difficult to analyze the functions of the individual proteins in the context of virus infection. We established a system to analyze the C protein independently from the P and V proteins by placing its gene in an additional transcription unit between the H and L genes. Analyses with this system indicated that a highly attenuated Edmonston lineage vaccine strain encodes a fully functional C protein, and the P and/or V protein is involved in the attenuated phenotype.
Measles is a highly contagious disease characterized by high fever and a maculopapular rash. About 4% of deaths in children aged under 5 years are caused by measles worldwide (9). The causative agent, measles virus (MV), belongs to the genus Morbillivirus in the family Paramyxoviridae. The genome of MV is a nonsegmented negative-strand RNA of ∼16 kb in length and contains six genes. Each gene encodes a single structural protein, namely the nucleocapsid (N), phospho (P), matrix (M), fusion (F), hemagglutinin (H), and large (L) proteins (17). The genome forms a helical ribonucleoprotein complex with the N protein and viral RNA-dependent RNA polymerase composed of the P and L proteins. The P protein acts as an essential cofactor of RNA-dependent RNA polymerase and tethers the L protein onto the nucleocapsid template (20). The P gene encodes additional gene products, the V and C proteins, by the processes of RNA editing and alternative translational initiation in a different reading frame, respectively (17). Although the V and C proteins are nonessential for MV replication (29, 35), they act as important virulence factors in vivo (11, 28, 44-46). Many lines of evidence have indicated that the C and V proteins of MV antagonize the host interferon (IFN) responses (10, 16, 22, 23, 25, 26, 37, 42, 47). The V protein directly interferes with pathways of IFN induction (1) and IFN signaling (25, 26, 42), while the C protein contributes to circumvention of IFN induction by controlling the levels of viral RNA synthesis (22, 23, 31). Direct interference with IFN signaling by the C protein has also been reported, although its effects are weaker than those of the V protein (16, 37).
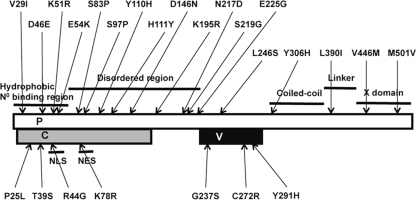
In a recent study, we showed that a recombinant IC-B strain possessing the P gene of the attenuated Edmonston tag strain (IC/EdP) replicates less efficiently than the parental IC-B strain (a virulent strain) (40). The Edmonston tag strain is a recombinant MV derived from the Edmonston B vaccine strain, which has been passaged numerous times in various cultured cells (30). There are many amino acid differences between the P gene products of the IC-B and Edmonston tag strains (Fig. 1) (27, 30, 43). Most of the changes found in the Edmonston tag strain are common to the Edmonston lineage MV strains (27). However, owing to two amino acid substitutions at positions 110 and 272, which are not conserved among the Edmonston lineage MV strains (25-27), the V protein of the Edmonston tag strain is defective in counteracting IFN signaling (10, 12, 16, 25). The changes at these two positions are also found in other lineages of MV vaccine strains as well as cultured, cell-adapted MV strains (2, 8, 40). Therefore, we consider the P gene of the Edmonston tag strain to be a representative P gene encoded in attenuated MV strains.
FIG. 1.
Amino acid differences between the P, C, and V proteins of the Edmonston tag strain and those of the IC-B strain. The white, gray, and black boxes indicate the reading frames for the P protein and the C protein and the unique carboxyl terminus of the V protein of the Edmonston tag strain, respectively. Substitutions in the P protein compared with the IC-B strain are shown above the boxes, while those in the C and V proteins are shown below the boxes. The functional domains of the P protein (20) are shown as black lines above the white box, while those of the C protein (24) are shown below the gray box. NLS, nuclear localization signal; NES, nuclear export signal.
The C protein expressed from the newly created C gene supports MV replication efficiently.
Previous studies using expression plasmids have suggested that functional differences in the C protein are possibly involved in the attenuated phenotypes of the Edmonston and vaccine strains of MV (3, 24, 31). These observations motivated us to compare the C protein functions between the IC-B and Edmonston tag strains in the context of virus infection. It should be noted that the licensed, highly attenuated Zagreb vaccine strain encodes a C protein with an amino acid sequence identical to that of the Edmonston tag strain (4, 27). However, analyses of the C protein using infectious MVs pose the problem that mutations in the C protein are often accompanied by mutations in the P and V proteins because they are encoded in overlapping reading frames. In the present study, we established a reverse-genetics system to analyze the C protein independently from the P and V proteins by placing its gene in an additionally created transcription unit (termed the C gene) between the H and L genes (Fig. 2A). The expression levels of MV mRNAs decrease progressively from the 3′ end to the 5′ end of the virus genome. Although the C protein is expressed from the P gene (the second locus of the virus genome) in the original virus genome, the expression level is relatively low. It is because the C protein is translated using the second AUG codon in the P gene transcripts via a leaky scanning mechanism. We, therefore, placed the C gene at a downstream locus (between the H and L genes) of the virus genome in order to achieve an expression level of the C protein similar to that from the P gene. The original reading frame of the C protein in the P gene was knocked out by introducing nonsense mutations into the frame, as reported previously (22, 44), and the mutated P gene was termed PΔC. A recombinant MV IC-B strain possessing a genome with these alterations was generated by reverse-genetics techniques (39, 41) and designated ICΔC-add[IC-C] (the C protein of the IC-B strain was termed IC-C). Another recombinant MV, designated ICΔC-add[ΔC], was also generated. This second recombinant MV possessed the C gene but lacked C protein expression owing to introduced nonsense mutations. A third recombinant MV, ICΔC, which lacks C protein expression from the P gene and does not have an additional C gene, has already been reported (22, 44). All the recombinant MV strains analyzed in the present study were derived from IC323-EGFP (18) (termed the wild-type [wt] virus in this study), which was engineered to express enhanced green fluorescent protein (EGFP) from another additional transcriptional unit at the first locus of the genome of the IC-B strain (Fig. 2A) (18). A pulse-labeling experiment revealed that the expression levels of the C protein from the newly created C gene at 24 h postinfection (p.i.) in Vero/hSLAM and A549/hSLAM cells were similar to those from the original P gene (Fig. 2B).
FIG. 2.
The C protein expressed from the newly created C gene is fully functional in supporting MV replication. (A) Insertion of an additional transcription unit (the C gene) between the H and L genes. Transcription regulatory regions (gene end [GE], intergenic, and gene start [GS] sequences) and the coding sequence for the C protein (C-ORF) were inserted at the junction between the H and L genes using appropriate restriction enzyme recognition sites (SpeI, FseI, and RsrII). The recombinant MV genome also possesses a transcription unit for EGFP (green). Le and Tr indicate leader and trailer sequence, respectively. (B) Vero/hSLAM and A549/hSLAM cells were infected with wt and ICΔC-add[IC-C] viruses at a multiplicity of infection (MOI) of 0.5. At 12 and 24 h p.i., the cells were pulse-labeled with [35S]methionine-cysteine. The C proteins were immunoprecipitated with an anti-C protein monoclonal antibody (2D10) (23), subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detected using a Fuji BioImager 1000 (Fuji, Tokyo, Japan). (C) Indirect immunofluorescence assay for IRF3. A549/hSLAM cells were infected with recombinant MVs (wt, ICΔC, ICΔC-add[IC-C], and ICΔC-add[ΔC]), which express EGFP (18), at an MOI of 0.5 and incubated in medium containing a fusion-blocking peptide (32). At 36 h p.i., IRF3 was detected by an indirect immunofluorescence assay. Green and red fluorescence indicate EGFP encoded in the recombinant MV genome and IRF3, respectively. (D) Expression of viral proteins. A549/hSLAM cells were infected with recombinant MVs at an MOI of 0.5. At 24 h p.i., the N, P, V, C, and H proteins in the cells were detected by SDS-PAGE and Western blotting using appropriate primary and secondary antibodies. (E) Growth kinetics. Monolayers of gA549/hSLAM cells were infected with recombinant MVs (□, wt; ×, ICΔC; ○, ICΔC-add[IC-C];  , ICΔC-add[ΔC]) at an MOI of 0.01. At various time intervals, the infectious virus titers were determined by plaque assays. Data represent the means ± standard deviations (SD) of results from triplicate samples.
, ICΔC-add[ΔC]) at an MOI of 0.01. At various time intervals, the infectious virus titers were determined by plaque assays. Data represent the means ± standard deviations (SD) of results from triplicate samples.
Our previous studies indicated that IFN regulatory factor 3 (IRF3) is activated in cells infected with ICΔC, leading to the production of IFN (22, 23). Furthermore, protein translation is inhibited in these cells through phosphorylation of the eukaryotic translation initiation factor eIF-2α (22). Consequently, ICΔC replicates poorly in these cells possessing a functional IFN system but replicates fairly well in cells with a defective IFN system (22). As observed in ICΔC-infected cells, IRF3 was translocated into the nucleus in ICΔC-add[ΔC]-infected cells, whereas it was hardly translocated into the nucleus in ICΔC-add[IC-C]-infected cells (Fig. 2C). Synthesis of viral proteins was restored in ICΔC-add[IC-C]-infected cells (Fig. 2D). Furthermore, this Western blot analysis reconfirmed the similar expression levels of the C protein for the wt and ICΔC-add[IC-C] viruses at 24 h p.i. (Fig. 2D). ICΔC-add[IC-C] replicated efficiently, and its maximum virus titer was as high as that of the wt virus (Fig. 2E). However, the virus titer of ICΔC-add[IC-C] dropped off more rapidly than that of the wt virus (Fig. 2E), revealing some differences between the growth kinetics of the wt and ICΔC-add[IC-C] viruses at the late stage of virus infection. The rapid decrease in the virus titer of ICΔC-add[IC-C] may thus suggest some difference in the expression levels of the C protein between ICΔC-add[IC-C] and wt viruses after 48 h p.i. Nevertheless, all the data indicate that the C protein expressed from the newly created C gene efficiently supports virus replication similar to that of the C protein expressed from the original P gene.
The C protein of the Edmonston tag strain supports MV replication as efficiently as the wt C protein.
Next, the function of the C protein of the Edmonston tag strain (Ed-C) was compared with that of IC-C using the newly developed recombinant virus system with the inserted C gene. Recombinant ICΔC-add[Ed-C], which encodes the C gene of Ed-C, was generated. Synthesis of viral RNAs in either ICΔC-add[IC-C]- or ICΔC-add[Ed-C]-infected cells was controlled to a level similar to that in wt virus-infected cells, whereas it was accelerated in ICΔC-add[ΔC]-infected cells, as observed in ICΔC-infected cells (Fig. 3A). No significant differences were found between the intracellular distribution patterns of Ed-C and IC-C in virus-infected cells (Fig. 3B). IRF3 was hardly translocated into the nucleus in ICΔC-add[Ed-C]-infected cells (Fig. 3C). In addition, ICΔC-add[Ed-C] produced levels of viral proteins similar to those of ICΔC-add[IC-C] (Fig. 3D). Consequently, ICΔC-add[Ed-C] replicated as efficiently as ICΔC-add[IC-C] (Fig. 3E). These data indicate that Ed-C is fully functional in supporting virus replication and circumventing host IFN responses, similar to IC-C. These results are consistent with our previous observation that the ability of the C protein to inhibit viral RNA synthesis is correlated with the abilities of MV to circumvent IFN induction and support virus growth in IFN-competent cells (23). Many studies regarding the roles of the MV C protein in virus replication and pathogenesis have been carried out using the Edmonston tag strain (7, 15, 28, 29, 46). Our present study helps to validate the knowledge regarding the functions of the C protein obtained using the Edmonston tag strain.
FIG. 3.
The C protein of the Edmonston tag strain is equivalent in functionality to the C protein of the IC-B strain. (A) Quantification of viral mRNAs. Vero/hSLAM cells were infected with recombinant MVs (red, wt; light blue, ICΔC; purple, ICΔC-add[IC-C]; blue, ICΔC-add[ΔC]; yellow, ICΔC-add[Ed-C]) at an MOI of 0.5. At 36 h p.i., the viral mRNA levels in the cells were analyzed by reverse transcription-quantitative PCR as previously described (23). Data represent the means ± SD of results from triplicate samples. (B) Intracellular distribution of the C protein. Vero/hSLAM cells were infected with recombinant MVs expressing EGFP (18) at an MOI of 0.5 in the presence of a fusion-blocking peptide (32). At 36 h p.i., the intracellular distribution of the C protein was analyzed by an indirect immunofluorescence assay using appropriate primary and secondary antibodies (23). Green and red fluorescence indicate EGFP encoded in the recombinant MV genome and the C protein, respectively. (C) Indirect immunofluorescence assay for IRF3. ICΔC-add[Ed-C]-infected A549/hSLAM cells were subjected to an indirect immunofluorescence assay for IRF3 using the same procedures described in the legend for Fig. 2C. (D) Expression of viral proteins. Viral proteins in recombinant MV-infected A549/hSLAM cells were detected using the same procedures described in the legend for Fig. 2D. (E) Growth kinetics. Monolayers of A549/hSLAM, HeLa/hSLAM, and H358 cells were infected with recombinant MVs (○, ICΔC-add[IC-C];  , ICΔC-add[ΔC]; ▵, ICΔC-add[Ed-C]) at an MOI of 0.01. At various time intervals, the infectious virus titers were determined by plaque assays. Data represent the means ± SD of results from triplicate samples.
, ICΔC-add[ΔC]; ▵, ICΔC-add[Ed-C]) at an MOI of 0.01. At various time intervals, the infectious virus titers were determined by plaque assays. Data represent the means ± SD of results from triplicate samples.
The P/V protein of the Edmonston tag strain is responsible for the attenuated phenotype.
None of the data showed any functional differences between IC-C and Ed-C. Consequently, the reduction in virus growth observed for IC/EdP (40) should be caused by the P and/or V (P/V) protein of the Edmonston tag strain. The P gene of the Edmonston tag strain in which C protein expression was knocked out (EdPΔC) was introduced into the ICΔC-add[IC-C] and ICΔC-add[Ed-C] genomes to replace PΔC. The generated viruses were termed IC/EdPΔC-add[IC-C] and IC/EdPΔC-add[Ed-C], respectively. Recombinant MVs possessing EdPΔC (IC/EdPΔC-add[IC-C] and IC/EdPΔC-add[Ed-C]) synthesized smaller amounts of viral RNAs (Fig. 4A) and replicated less efficiently (Fig. 4B) than those possessing PΔC (ICΔC-add[IC-C] and ICΔC-add[Ed-C]). In addition, MVs possessing EdPΔC, including the Edmonston strain and IC/EdP, produced smaller plaques than those possessing PΔC (Fig. 4C) (40). These data confirm that the attenuated growth of IC/EdP is caused by the P/V protein of the Edmonston tag strain and not by the C protein. It is unlikely that the inability of the V protein of the Edmonston tag strain to counteract IFN signaling (10, 12, 16, 25) is associated with the growth attenuation of MV possessing EdPΔC, because we used Vero cells that are defective in the IFN system (14, 21) in these analyses.
FIG. 4.
The P/V protein, but not the C protein, attenuates MV RNA synthesis and growth. (A) Quantification of viral mRNAs. Monolayers of Vero/hSLAM cells were infected with recombinant MVs (light gray, ICΔC-add[IC-C]; black, ICΔC-add[Ed-C]; dark gray, IC/EdPΔC-add[IC-C]; white, IC/EdPΔC-add[Ed-C]) at an MOI of 0.01 in the presence of a fusion-blocking peptide (32). At 18 h p.i., mRNAs were purified from the cells, and the viral mRNA levels were determined by reverse transcription-quantitative PCR as previously described (40). Data represent the means ± SD of results from triplicate samples. (B) Growth kinetics. Monolayers of Vero/hSLAM cells were infected with recombinant MVs (○, ICΔC-add[IC-C]; ▵, ICΔC-add[Ed-C]; •, IC/EdPΔC-add[IC-C]; ▴, IC/EdPΔC-add[Ed-C]) at an MOI of 0.01. At various time intervals, the infectious virus titers were determined by plaque assays. Data represent the means ± SD of results from triplicate samples. (C) Plaque assays. Monolayers of Vero/hSLAM cells on 12-well cluster plates were infected with recombinant viruses and overlaid with Dulbecco modified Eagle medium containing 2% fetal bovine serum and 1% methylcellulose. At 6 days p.i., the cells were stained with the RTU Vectastain Elite ABC reagent (Vector Laboratories) using anti-MV H protein monoclonal antibodies and a biotinylated secondary antibody.
Implications.
The mechanisms involved in the attenuation of the Edmonston and vaccine strains of MV remain to be elucidated. MV can be attenuated through adaptation to growth in cultured cells (19), and the Edmonston and vaccine strains have been generated through large numbers of passages in various cultured cells (34). Mutations in the P gene are often observed during the adaptation process of MV (2, 13, 27, 38, 43) as well as in related viruses (5, 6, 33). The C protein was shown to be dispensable for virus growth in some cultured cells (29) but acts as an important virulence factor in vivo (11, 28, 44). Therefore, mutations in the C protein are possibly responsible for the attenuated phenotypes of the Edmonston and vaccine strains of MV. However, recent studies have indicated that the C protein plays important roles in circumventing the host innate immune responses (16, 22, 23, 37, 47) and is therefore dispensable for virus growth in cultured cells only when the cells have a defective IFN system (15, 22, 44). Our data indicate that Ed-C is fully functional in supporting virus growth in cells possessing a functional IFN system. Preservation of a functional C protein by the Edmonston tag and vaccine strains of MV would be reasonable, since they have to grow in chicken embryo fibroblasts (34), which possess an intact IFN system (36).
It has long been believed that the Edmonston strain and its derivative vaccines have acquired mutations that may promote viral RNA synthesis in cultured cells. However, this is unlikely to be the case. Our present data suggest that the P/V protein of the Edmonston tag strain attenuates MV growth by reducing the level of viral RNA synthesis. We speculate that this may allow the virus to circumvent the host innate immune responses effectively, thereby leading to better survival in various cultured cells. Our present data provide clues toward understanding why MV vaccine strains have become attenuated during the process of adaptation to growth in unnatural host cells and which viral proteins have contributed to this attenuation.
Acknowledgments
We thank M. A. Billeter for providing the p(+)MV2A plasmid encoding the genome of the Edmonston tag strain.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare of Japan.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bankamp, B., G. Hodge, M. B. McChesney, W. J. Bellini, and P. A. Rota. 2008. Genetic changes that affect the virulence of measles virus in a rhesus macaque model. Virology 373:39-50. [DOI] [PubMed] [Google Scholar]
- 3.Bankamp, B., J. Wilson, W. J. Bellini, and P. A. Rota. 2005. Identification of naturally occurring amino acid variations that affect the ability of the measles virus C protein to regulate genome replication and transcription. Virology 336:120-129. [DOI] [PubMed] [Google Scholar]
- 4.Baricevic, M., D. Forcic, T. K. Gulija, R. Jug, and R. Mazuran. 2005. Determination of the coding and non-coding nucleotide sequences of genuine Edmonston-Zagreb master seed and current working seed lot. Vaccine 23:1072-1078. [DOI] [PubMed] [Google Scholar]
- 5.Baron, M. D., A. C. Banyard, S. Parida, and T. Barrett. 2005. The Plowright vaccine strain of Rinderpest virus has attenuating mutations in most genes. J. Gen. Virol. 86:1093-1101. [DOI] [PubMed] [Google Scholar]
- 6.Baron, M. D., Y. Kamata, V. Barras, L. Goatley, and T. Barrett. 1996. The genome sequence of the virulent Kabete “O” strain of rinderpest virus: comparison with the derived vaccine. J. Gen. Virol. 77:3041-3046. [DOI] [PubMed] [Google Scholar]
- 7.Billeter, M. A., H. Y. Naim, and S. A. Udem. 2009. Reverse genetics of measles virus and resulting multivalent recombinant vaccines: applications of recombinant measles viruses. Curr. Top. Microbiol. Immunol. 329:129-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borges, M. B., E. Caride, A. V. Jabor, J. M. Malachias, M. S. Freire, A. Homma, and R. Galler. 2008. Study of the genetic stability of measles virus CAM-70 vaccine strain after serial passages in chicken embryo fibroblasts primary cultures. Virus Genes 36:35-44. [DOI] [PubMed] [Google Scholar]
- 9.Bryce, J., C. Boschi-Pinto, K. Shibuya, and R. E. Black. 2005. W. H. O. estimates of the causes of death in children. Lancet 365:1147-1152. [DOI] [PubMed] [Google Scholar]
- 10.Caignard, G., M. Guerbois, J. L. Labernardiere, Y. Jacob, L. M. Jones, F. Wild, F. Tangy, and P. O. Vidalain. 2007. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology 368:351-362. [DOI] [PubMed] [Google Scholar]
- 11.Devaux, P., G. Hodge, M. B. McChesney, and R. Cattaneo. 2008. Attenuation of V- or C-defective measles viruses: infection control by the inflammatory and interferon responses of rhesus monkeys. J. Virol. 82:5359-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devaux, P., V. von Messling, W. Songsungthong, C. Springfeld, and R. Cattaneo. 2007. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology 360:72-83. [DOI] [PubMed] [Google Scholar]
- 13.Druelle, J., C. I. Sellin, D. Waku-Kouomou, B. Horvat, and F. T. Wild. 2008. Wild type measles virus attenuation independent of type I IFN. Virol. J. 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 15.Escoffier, C., S. Manié, S. Vincent, C. P. Muller, M. Billeter, and D. Gerlier. 1999. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J. Virol. 73:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontana, J. M., B. Bankamp, W. J. Bellini, and P. A. Rota. 2008. Regulation of interferon signaling by the C and V proteins from attenuated and wild-type strains of measles virus. Virology 374:71-81. [DOI] [PubMed] [Google Scholar]
- 17.Griffin, D. E. 2007. Measles virus, p. 1551-1585. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 18.Hashimoto, K., N. Ono, H. Tatsuo, H. Minagawa, M. Takeda, K. Takeuchi, and Y. Yanagi. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76:6743-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobune, F., H. Sakata, and A. Sugiura. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64:700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longhi, S. 2009. Nucleocapsid structure and function. Curr. Top. Microbiol. Immunol. 329:103-128. [DOI] [PubMed] [Google Scholar]
- 21.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakatsu, Y., M. Takeda, S. Ohno, R. Koga, and Y. Yanagi. 2006. Translational inhibition and increased interferon induction in cells infected with C protein-deficient measles virus. J. Virol. 80:11861-11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakatsu, Y., M. Takeda, S. Ohno, Y. Shirogane, M. Iwasaki, and Y. Yanagi. 2008. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J. Virol. 82:8296-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishie, T., K. Nagata, and K. Takeuchi. 2007. The C protein of wild-type measles virus has the ability to shuttle between the nucleus and the cytoplasm. Microbes Infect. 9:344-354. [DOI] [PubMed] [Google Scholar]
- 25.Ohno, S., N. Ono, M. Takeda, K. Takeuchi, and Y. Yanagi. 2004. Dissection of measles virus V protein in relation to its ability to block alpha/beta interferon signal transduction. J. Gen. Virol. 85:2991-2999. [DOI] [PubMed] [Google Scholar]
- 26.Palosaari, H., J.-P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks, C. L., R. A. Lerch, P. Walpita, H.-P. Wang, M. S. Sidhu, and S. A. Udem. 2001. Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J. Virol. 75:910-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patterson, J. B., D. Thomas, H. Lewicki, M. A. Billeter, and M. B. Oldstone. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80-89. [DOI] [PubMed] [Google Scholar]
- 29.Radecke, F., and M. A. Billeter. 1996. The nonstructural C protein is not essential for multiplication of Edmonston B strain measles virus in cultured cells. Virology 217:418-421. [DOI] [PubMed] [Google Scholar]
- 30.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reutter, G. L., C. Cortese-Grogan, J. Wilson, and S. A. Moyer. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285:100-109. [DOI] [PubMed] [Google Scholar]
- 32.Richardson, C. D., A. Scheid, and P. W. Choppin. 1980. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology 105:205-222. [DOI] [PubMed] [Google Scholar]
- 33.Rivals, J. P., P. Plattet, C. Currat-Zweifel, A. Zurbriggen, and R. Wittek. 2007. Adaptation of canine distemper virus to canine footpad keratinocytes modifies polymerase activity and fusogenicity through amino acid substitutions in the P/V/C and H proteins. Virology 359:6-18. [DOI] [PubMed] [Google Scholar]
- 34.Rota, J. S., Z. D. Wang, P. A. Rota, and W. J. Bellini. 1994. Comparison of sequences of the H, F, and N coding genes of measles virus vaccine strains. Virus Res. 31:317-330. [DOI] [PubMed] [Google Scholar]
- 35.Schneider, H., K. Kaelin, and M. A. Billeter. 1997. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology 227:314-322. [DOI] [PubMed] [Google Scholar]
- 36.Sekellick, M. J., W. J. Biggers, and P. I. Marcus. 1990. Development of the interferon system. I. In chicken cells development in ovo continues on time in vitro. In Vitro Cell. Dev. Biol. 26:997-1003. [DOI] [PubMed] [Google Scholar]
- 37.Shaffer, J. A., W. J. Bellini, and P. A. Rota. 2003. The C protein of measles virus inhibits the type I interferon response. Virology 315:389-397. [DOI] [PubMed] [Google Scholar]
- 38.Takeda, M., A. Kato, F. Kobune, H. Sakata, Y. Li, T. Shioda, Y. Sakai, M. Asakawa, and Y. Nagai. 1998. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J. Virol. 72:8690-8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeda, M., S. Ohno, F. Seki, K. Hashimoto, N. Miyajima, K. Takeuchi, and Y. Yanagi. 2005. Efficient rescue of measles virus from cloned cDNA using SLAM-expressing Chinese hamster ovary cells. Virus Res. 108:161-165. [DOI] [PubMed] [Google Scholar]
- 40.Takeda, M., S. Ohno, M. Tahara, H. Takeuchi, Y. Shirogane, H. Ohmura, T. Nakamura, and Y. Yanagi. 2008. Measles viruses possessing the polymerase protein genes of the Edmonston vaccine strain exhibit attenuated gene expression and growth in cultured cells and SLAM knock-in mice. J. Virol. 82:11979-11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda, M., K. Takeuchi, N. Miyajima, F. Kobune, Y. Ami, N. Nagata, Y. Suzaki, Y. Nagai, and M. Tashiro. 2000. Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74:6643-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi, K., S. I. Kadota, M. Takeda, N. Miyajima, and K. Nagata. 2003. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett. 545:177-182. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi, K., N. Miyajima, F. Kobune, and M. Tashiro. 2000. Comparative nucleotide sequence analysis of the entire genomes of B95a cell-isolated and Vero cell-isolated measles viruses from the same patient. Virus Genes 20:253-257. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi, K., M. Takeda, N. Miyajima, Y. Ami, N. Nagata, Y. Suzaki, J. Shahnewaz, S. Kadota, and K. Nagata. 2005. Stringent requirement for the C protein of wild-type measles virus for growth both in vitro and in macaques. J. Virol. 79:7838-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. D. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valsamakis, A., H. Schneider, P. G. Auwaerter, H. Kaneshima, M. A. Billeter, and D. E. Griffin. 1998. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J. Virol. 72:7754-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokota, S., H. Saito, T. Kubota, N. Yokosawa, K. Amano, and N. Fujii. 2003. Measles virus suppresses interferon-alpha signaling pathway: suppression of Jak1 phosphorylation and association of viral accessory proteins, C and V, with interferon-alpha receptor complex. Virology 306:135-146. [DOI] [PubMed] [Google Scholar]