Abstract
MicroRNAs (miRNAs) are endogenous antisense regulators that trigger endonucleolytic mRNA cleavage, translational repression, and/or mRNA decay. miRNA-mediated gene regulation is important for numerous biological pathways, yet the underlying mechanisms are still under rigorous investigation. Here we identify human UPF1 (hUPF1) as a protein that contributes to RNA silencing. When hUPF1 is knocked down, miRNA targets are upregulated. The depletion of hUPF1 also increases the off-target messages of small interfering RNAs (siRNAs), which are imperfectly complementary to transfected siRNAs. Conversely, when overexpressed, wild-type hUPF1 downregulates miRNA targets. The helicase domain mutant of hUPF1 fails to suppress miRNA targets. hUPF1 interacts with human Argonaute 1 (hAGO1) and hAGO2 and colocalizes with hAGO1 and hAGO2 in processing bodies, which are known to be the sites for translational repression and mRNA destruction. We further find that the amounts of target messages bound to hAGO2 are reduced when hUPF1 is depleted. Our data thus suggest that hUPF1 may participate in RNA silencing by facilitating the binding of the RNA-induced silencing complex to the target and by accelerating the decay of the mRNA.
Small RNAs such as microRNAs (miRNAs) and small interfering RNAs (siRNAs) regulate gene expression through processes that are collectively referred to as RNA silencing (4, 36, 59, 69). miRNAs and siRNAs act as the specificity components of the effector complex known as the RNA-induced silencing complex (RISC). In humans, RISC is generated by several proteins, including Argonaute proteins, Dicer, TRBP (human immunodeficiency virus type 1 TAR RNA-binding protein), and PACT (23, 24, 41). The Argonaute proteins are the core components of RISC and directly bind to small RNA. There are four Argonaute proteins in humans (12). Human Argonaute 2 (hAGO2) can directly catalyze target mRNA cleavage (47) and mediate translational repression, while the biochemical roles of other Argonautes remain unclear.
Small RNAs base pair with the target mRNAs, usually resulting in suppressive effects. Complementarity at nucleotide positions 2 to 7 (referred to as “seed” sequences) relative to the 5′ end of small RNA is critical for suppression. Small RNAs can direct endonucleolytic cleavage of a single phosphodiester bond between positions 10 and 11 of the paired bases (relative to the 5′ end of the small RNA) if the complementarity is high throughout the length. If the complementarity between the target and small RNA is low, particularly in the middle of the RNA duplex, the interaction would induce translational repression. The mechanism of this translational repression remains elusive, and other mechanisms acting downstream of the initiation step have been proposed (19, 62).
Studies have also revealed that small RNAs bearing partial mismatches can induce not only translational repression but also mRNA decay. miRNAs, including miR-1, miR-124a, miR-125a, let-7, and miR-430, affect the levels of their mRNA targets (2, 3, 5, 8, 16-18, 22, 46, 53, 64, 72, 73). Furthermore, experimentally introduced siRNAs facilitate the reduction of not only perfectly matching targets (“on-targets”) but also other mRNAs with imperfect complementarity (“off-targets”) (34). The off-target effect induced by siRNA is thought to be mechanistically similar, if not identical, to miRNA-induced mRNA decay (7, 11, 55). The off-target effects are highly problematic in gene knockdown experiments, particularly when their use in clinical applications is contemplated.
Recent studies have highlighted the significance of cytoplasmic foci called processing bodies (P bodies) in RNA silencing (15, 20, 57). P bodies (also termed XRNI foci, GW bodies, or Dcp bodies) contain a number of proteins involved in posttranscriptional regulation: XRN1 (a 5′→3′ exonuclease), GW182/TNRC6 proteins (AIN-1 in nematodes), DCP2 (decapping enzyme), DCP1 (decapping enzyme subunit), Ge-1/Hedls (decapping coactivator), Pat1 (decapping coactivator), Lsm1 to -7 (decapping coactivator complex), RCK/p54 (decapping coactivator and translation regulator), the CCR4-CAF1-NOT complex (deadenylating enzyme), nonsense-mediated decay factors, and the Argonaute proteins. Due to their protein composition, P bodies are thought to be the sites for mRNA destruction and to be involved in RNA silencing (9, 30, 60, 66). Consistently, the presence of miRNA target messages as well as the Argonaute proteins in P bodies is miRNA dependent (49). Moreover, the well-known P body component GW182 was shown to be significant for miRNA-dependent gene silencing (5, 10, 48, 61).
Global mRNA decay factors are generally conserved among eukaryotes, but the actual decay mechanisms of the pathways vary considerably (57). In mammals, global decay of mRNA usually begins with deadenylation mediated by the CCR4-CAF1-NOT complex. Following deadenylation, the mRNA can be rapidly degraded from the 3′ end by the exosome. In addition, deadenylated mRNA molecules are decapped by DCP2 and exonucleolytically degraded in a 5′→3′ direction by XRN1.
Nonsense-mediated mRNA decay (NMD) is a specialized decay process that eliminates aberrant transcripts containing premature termination codons (PTCs), thereby preventing the production of potentially harmful protein fragments (32). NMD is triggered by a premature translation termination event, which leads to the assembly of the surveillance complex on the mRNA. The surveillance complex comprises UPF1, UPF2, UPF3, and four additional NMD factors (SMG1 and SMG5 to SMG7). The detailed mechanism of the assembly of the NMD effectors is not fully understood and varies among species. It is generally thought that, in humans, the UPF complex interacts with the terminating ribosome via eRF1 and eRF3. These interactions are stimulated by the exon junction complex bound downstream of the stop codon (15, 32) or can be inhibited by antagonistic factors such as the cytoplasmic poly(A)-binding protein (PABPC1) (33, 67). The UPF complex then elicits both decapping and deadenylation, but the relative contribution of each pathway has not been estimated (44). Recent studies have also shown that the surveillance complex can induce endonucleolytic cleavage near the PTC and that SMG6 is the endonucleolytic enzyme responsible for this cleavage (14, 29).
Small RNA-induced mRNA decay is another specialized decay pathway. It was indicated that deadenylation takes place during the degradation of miRNA targets, although deadenylation appears not to be absolutely required for silencing (17, 22, 73). Decapping and 5′-to-3′ exonucleolytic activities also contribute to the decay of miRNA targets (3, 63, 73). The factors involved in this pathway were most intensively studied in Drosophila melanogaster. Drosophila AGO1, the deadenylation complex CCR4-NOT, and the decapping complex DCP1-DCP2 are required for miRNA-induced mRNA decay (5), and the requirement for decapping coactivators in this pathway of Drosophila was target specific (18).
In this study, we find that human UPF1 (hUPF1), an NMD factor, is involved in small RNA-induced mRNA decay. The decay of miRNA targets is compromised when hUPF1 is knocked down. When overexpressed, hUPF1 downregulates miRNA targets. Furthermore, hUPF1 interacts with the Argonaute proteins and colocalizes with the Argonaute proteins in P bodies.
MATERIALS AND METHODS
Plasmid construction.
The cDNAs of Argonaute genes were amplified by PCR and subcloned into the Flag-pCK vector at the EcoRI and NotI sites. Primer sequences used for PCR are as follows: hAGO1, 5′-GAA TTC TGA TGG AAG CGG GAC CCT CGG-3′ (forward) and 5′-GCG GCC GCT CAA GCG AAG TAC ATG GTG CGC-3′ (reverse); hAGO2, 5′-GAA TTC GCA TGG GTG TTC TCT CTG C-3′ (forward) and 5′-GCG GCC GCT CAA GCA AAG TAC ATG GTG CG-3′ (reverse). The 3′ untranslated region (UTR) fragments that include miR-124a target sites were cloned into the XbaI site of the pGL3-CMV vector to generate the luciferase expression plasmids FL-IQGAP1 (also designated pGL3-CMV-IQGAP1-3′ UTR) and FL-CD164 (also designated pGL3-CMV-CD164-3′ UTR). The primers used to amplify the UTRs are as follows: IQGAP1-3′ UTR, 5′-TCT AGA CAA TTC ACT CCA TCC CTA TGG C-3′ (forward) and 5′-TCT AGA GGC TCA GCA GCA TGA TTT CA-3′ (reverse); CD164-3′ UTR, 5′-TCT AGA AAT AAG ATG CCA CAC AAG GAA CTA CA-3′ (forward) and 5′-TCT AGA AAT GTG AGG TAA ACC GAC CTT TCC-3′ (reverse).
Cell culture and transfection.
HeLa cells and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (WelGENE) supplemented with 10% fetal bovine serum (WelGENE). Transfections of HEK293T cells were carried out using the calcium phosphate method. Plasmid transfections of HeLa cells for quantitative reverse transcription-PCR (qRT-PCR) were performed twice during the first 2 days (on the first day and the second day) using Metafectene (Biontex), and total RNAs were isolated on the fourth day.
RNAi.
HeLa cells were cotransfected with 20 nM of miR-124a RNA duplex or siRNA against green fluorescent protein (siGFP) and with 60 nM siGFP, siUPF1, or siUPF2. Seventy-two hours later, Northern blot analysis was performed. For analysis of endogenous miRNA targets, HeLa cells were treated with siGFP, siUPF1, or siAGO2 on the first day and the third day and the cells were harvested on the fourth day. Lipofectamine 2000 (Invitrogen) was used for the transfections. RNA interference (RNAi) for Argonaute proteins was carried out as follows. HeLa cells were transfected with 100 nM concentrations of siRNA against luciferase (siLuc), siAGO1, siAGO2, siAGO3, or siAGO4. Thirty hours later, cells were retransfected with 30 nM of miR-124a RNA duplex. Northern blot analysis was performed after an additional 40 h. RNAiFect (Qiagen) was used for transfection. miRNA mimics and siRNAs were synthesized by Samchully Pharmaceuticals. The miR-124a and miR-124a-m RNA duplex sequences, which were annealed, are as follows: miR-124a, 5′-UUA AGG CAC GCG GUG AAU GCC A-3′ (guide) and 5′-GCA UUC ACC GCG UGC CUU AAU U-3′ (passenger); miR-124a-m, 5′-UAA GCG ACG CGG UGA AUG CCA-3′ (guide) and 5′-GCA UUC ACC GCG UCG CUU AAU U-3′ (passenger). Sequences of the functional strands of siRNAs are 5′-UCG AAG UAC UCA GCG UAA G-3′ (siLuc), 5′-UUC UUG AGC ACC UCU UCU CUU-3′ (siAGO1), 5′-CAU CAG AAU GUU CAA GGC U-3′ (siAGO2), 5′-UUA CCA AUC UGC UAA UUU C-3′ (siAGO3), 5′-AUU GCU AUU AGU UCU GGC C-3′ (siAGO4), 5′-UGA AUU AGA UGG CGA UGU U-3′ (siGFP), 5′-UGG AGC GGA ACU GCA UCU U-3′ (siUPF1-1), 5′-GGC UUU UGU CCC AGC CAU C-3′ (siUPF2-1), 5′-AAU GGA GCG GAA CUG CAU C-3′ (siUPF1-3), 5′-UUC AGA UGG ACU UCC GUG C-3′ (siAGO2-1), 5′-AAC CGC AGU UCU CUG UAG G-3′ (siRNA duplex against mitogen-activated protein kinase 14 [siMAPK14]), 5′-UUU UUG GAA CAG UCU UUC C-3′ (siPPIB-1), and 5′-UUU GUA GCC AAA UCC UUU C-3′ (siPPIB-2). An additional set of RNAs (stealth siRNAs) were designed to target different sites on the transcripts of hUPF1, hUPF2, and hUPF3b and were purchased from Invitrogen. The sequences of the functional strands are 5′-UUG CCC ACA AUG AUG ACG CCA UAC C-3′ (siUPF1-2), 5′-AAU AAC AGC UGA UUC CAC CAU UCG G-3′ (siUPF2-2), and 5′-AAU UCU ACU AUA GCG GGA UAU UCC U-3′ (siUPF3b-2). The negative control (siCtrl) was the Stealth RNAi-negative control duplex (Invitrogen; Medium GC duplex).
Northern blot analysis of mRNAs.
Total RNA was isolated from transfected HeLa cells using TRIzol reagent (Invitrogen), separated on denaturing formaldehyde agarose gels (13 μg per well), and blotted onto Zeta-probe membranes (Bio-Rad). 32P-labeled probes were generated by the Prime-It random primer labeling kit (Stratagene) using PCR products as the templates. After the membrane was incubated for 40 min at 68°C in prehybridization solution (Express hybridization solution; Clontech), hybridization was carried out for 80 min at 68°C. Membranes were washed twice for 45 min at room temperature in washing solution Ι (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.05% sodium dodecyl sulfate) and then twice for 30 min at 50°C in washing solution II (0.1× SSC, 0.1% sodium dodecyl sulfate).
Luciferase assay.
HeLa cells cultured in 12-well plates were cotransfected with 100 ng of pCMV-firefly luciferase reporter (pGL3-CMV-IQGAP1 3′ UTR or pGL3-CMV-CD164 3′ UTR), 20 ng of pRL-CMV Renilla luciferase, 10 nM of miRNA duplex, and 40 nM of siUPF1, siUPF2, siUPF3b, or siCtrl (Invitrogen). Lipofectamine 2000 (Invitrogen) was used for the transfections, which were performed in duplicate. Forty-eight hours later, cells were lysed and assayed for luciferase activity using the Dual-Luciferase reporter system (Promega). All firefly luciferase activities were normalized against Renilla luciferase activities.
qRT-PCR.
The comparative threshold cycle method with SYBR green was conducted with the 7300 real-time PCR system (Applied Biosystems). For mRNA detection, the following primers were used: p27, 5′-TCC GGC TAA CTC TGA GGA CAC-3′ (forward) and 5′-TGT TTT GAG TAG AAG AAT CGT CGG T-3′ (reverse); Hmga2, 5′-CTG CTA TAC ACA AGC AAT GCA AG-3′ (forward) and 5′-GTA AGG AGA TTG CTT CTT TAA CTG-3′ (reverse); DnaJb11, 5′-AAT TTG TCC ATT TGC ATT CG-3′ (forward) and 5′-TTT GCT CCT GGA GTC CTC TT-3′ (reverse); GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-ACC CAG AAG ACT GTG GAT GG-3′ (forward) and 5′-CAG TGA GCT TCC CGT TCA G-3′ (reverse). qRT-PCR was performed on technical triplicates and biological triplicates.
Isolation of hAGO2-associated RNA.
HeLa cells grown in 10-cm dishes were treated twice with siGFP or siUPF1 on the first day and the third day. The cells were harvested on the fifth day. Plasmids were transfected twice into HeLa cells (on the first day and the second day) using Metafectene (Biontex), and the cells were harvested on the fourth day. The harvested cells were lysed and immunoprecipitated as previously described (6, 71). Anti-hAGO2 antibody (4G8) was used for immunoprecipitation (IP). RNA was isolated using TRIzol reagent (Invitrogen) from the immunoprecipitates or 5% of the lysates used for IP (as input). The purified RNA was analyzed by qRT-PCR. IP was performed in duplicate.
IP and Western blot analysis.
HEK293T cells grown in 10-cm dishes were collected in 500 μl of ice-cold buffer D-K'200 (20 mM Tris, pH 8.0, 200 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) at 48 h posttransfection. The cells were then sonicated on ice and centrifuged at 12,000 rpm for 10 min at 4°C. The supernatant was incubated with 10 μl of anti-Flag antibody conjugated to agarose beads (anti-Flag M2 affinity gel; Sigma) with constant rotation for 60 min at 4°C. The beads were then washed five times in buffer D-K'200, drained, and used for Western blot analysis.
For IP of endogenous proteins, HeLa cells were sonicated in buffer D-K'80 (20 mM Tris, pH 8.0, 80 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) and centrifuged at 10,000 rpm for 10 min at 4°C. The supernatant was precleared and then rotated in the presence of either M2 monoclonal anti-Flag antibody (Sigma), monoclonal anti-hAGO antibody (a gift from Zissimos Mourelatos), goat polyclonal anti-PERK antibody (Santa Cruz Biotechnology), or goat polyclonal anti-hUPF1 antibody (Merck) at 4°C overnight. Subsequently, 30 μl of protein A-Sepharose beads and protein G-Sepharose beads (1:1 mixture) was added to the immune complex for 90 min at 4°C. The beads were washed five times with buffer D-K'80 and used for Western blot analysis.
Primary antibodies used in Western blot analysis are rabbit anti-Dicer antibody, rabbit anti-TRBP antibody, mouse anti-Flag M2 antibody (Sigma), mouse anti-V5 antibody (Invitrogen), mouse anti-myc antibody (9E10), mouse anti-Y14 antibody (4C4), mouse anti-hnRNP A1 antibody (4B10), mouse anti-hAGO antibody (a gift from Zissimos Mourelatos), and goat anti-hUpf1 antibody (Merck).
Immunofluorescence.
For immunofluorescence analysis, HeLa cells were grown on glass coverslips in six-well plates and cotransfected with a myc-hUPF1 expression plasmid and a V5-tagged Argonaute protein expression plasmid using Lipofectamine 2000. Forty-eight hours after transfection, the cells were fixed using 2% formaldehyde in phosphate-buffered saline (PBS) for 30 min, washed with PBS, and permeabilized in PBS containing 0.1% Triton X-100 for a further 15 min. After several washes, the cells were saturated with 4% bovine serum albumin in PBS for 30 min and then immunostained for 2 h with rabbit anti-V5 tag antibody (1:500 dilution; Sigma), human anti-GW182 serum (18033 serum; 1:600 dilution), and mouse anti-myc antibody 9E10 (1:40 dilution) in PBS containing 4% bovine serum albumin as a blocking agent. Subsequently, the cells were washed with PBS and incubated for 1 h with secondary antibodies Alexa Fluor 647-goat anti-human immunoglobulin G (IgG) (1:400 dilution), Alexa Fluor 488-goat anti-mouse IgG (1:400 dilution), and Alexa Fluor 594-goat anti-rabbit IgG (1:400 dilution). Cells were visualized using the 40× objective of a confocal microscope.
Northern blot analysis of miRNAs.
RNA was resolved on a 12.5% urea-polyacrylamide gel and transferred electronically to a Zeta-probe membrane (Bio-Rad) as previously described (40). An oligonucleotide complementary to miR-222 or let-7a was end labeled with [γ-32P]ATP and used as the probe.
RESULTS
Knockdown of hUPF1 upregulates the targets of a miR-124a mimic.
In an attempt to identify the factors required for small RNA-induced mRNA decay, we depleted various mRNA decay factors by transfecting siRNAs into HeLa cells. The siRNAs were cotransfected with miR-124a, which is known to reduce the CD164 and IQGAP1 mRNAs (46). The 3′ UTRs of CD164 and IQGAP1 contain multiple sites that can partially base pair with miR-124a (see Fig. S1A in the supplemental material). Following transfection, the levels of mRNAs were determined by Northern blotting. Consistent with a previous report (46), both the CD164 and IQGAP1 mRNAs were reduced specifically when an miR-124a RNA duplex was transfected (Fig. 1A, lanes 1 and 2). When an RNA duplex (miR-124a-m) that carries mutations in the seed region of the miRNA was used as a negative control, no significant effect on the target mRNA levels was observed (see Fig. S1A in the supplemental material).
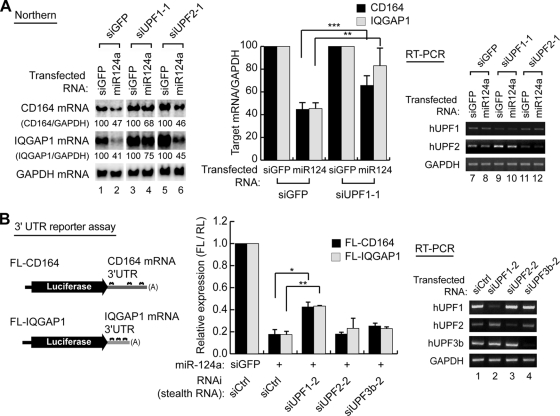
FIG. 1.
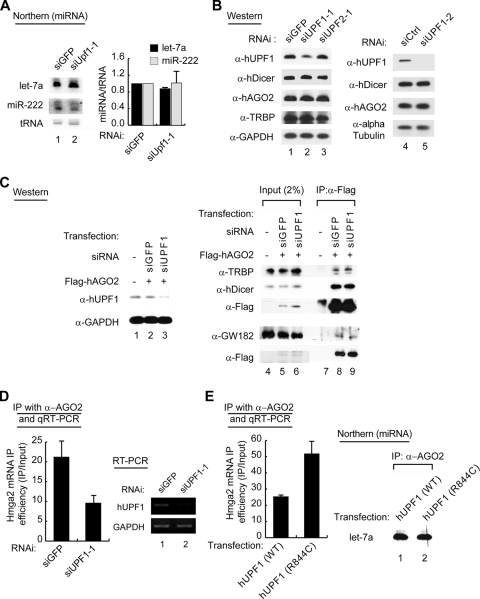
hUPF1 depletion derepresses miRNA target messages. (A) Northern blotting (lanes 1 to 6) shows that hUPF1 is required for efficient mRNA downregulation induced by miR-124a. HeLa cells were cotransfected with control siRNA (siGFP) or the miR-124a duplex along with siGFP, siUPF1-1, or siUPF2-1. The graph shows quantification of the relative levels of target mRNAs. Relative target mRNA levels from Northern blotting (left) were normalized against that of GAPDH mRNA (n = 3; means ± standard deviations [SD]). RT-PCR (lanes 7 to 12) visualizes the efficiency of knockdown. (B, left) Schematic representation of reporter mRNAs. The 3′ UTR fragment from either IQGAP1 or CD164 mRNA was inserted downstream of firefly luciferase (FL) coding sequences to generate the reporter mRNAs FL-IQGAP1 and FL-CD164, respectively. (Middle) Luciferase assay using the UTR reporter constructs. HeLa cells were cotransfected with the FL reporter construct, the Renilla luciferase (RL) plasmid, and siUPF1-2, siUPF2-2, siUPF3b-2, or siCtrl together with either miR-124a or miR-124a-m. FL activity was normalized against RL activity. The relative expression in the presence of miR-124a was again normalized against the activity determined in the presence of miR-124a-m (n = 2; means ± SD). (Right) RT-PCR to confirm the knockdown of UPF1, UPF2, and UPF3. A paired one-tailed t test was used to calculate the P values for Northern blotting and the luciferase assay (***, P < 0.01; **, P < 0.05; *, P < 0.1).
In this small-scale screening for mRNA decay factors, we found that the CD164 and IQGAP1 mRNAs accumulate when siUPF1-1 was transfected (Fig. 1A, lane 4; see the right panel for quantitation). siUPF2, siY14, siXRN1, siCCR4, siPARN, and siTTP did not significantly affect the decay of the IQGAP1 and CD164 mRNAs (Fig. 1A; see Fig. S1B in the supplemental material; data not shown). The knockdown of hUPF1 and hUPF2 was confirmed by RT-PCR (Fig. 1A, lanes 7 to 12). We observed similar effects on the IQGAP1 and CD164 mRNAs when we used a separate set of siRNAs that are complementary to distinct sites of hUPF1 and hUPF2 mRNAs (siUPF1-2 and siUPF2-2) (data not shown). Notably, knockdown of hUPF2 failed to derepress miRNA targets (Fig. 1A; see Fig. S1B in the supplemental material), suggesting that hUPF1, but not hUPF2, plays a role in small RNA-induced mRNA decay.
To exclude the possibility that inefficient knockdown of hUPF2 explains this observation, we examined whether knockdown of hUPF2 increases a known NMD substrate. For this, we transfected siRNAs and a well-known NMD reporter (Gl-Norm or Gl-Ter) (37, 74) into HeLa cells and then measured mRNA levels by Northern blotting (see Fig. S1C in the supplemental material). MUP was used as an internal control (37). We found that both siUPF1 and siUPF2 can efficiently upregulate the nonsense-containing NMD reporter Gl-Ter (39Ter). This indicates that hUPF2 knockdown in our experiment was sufficient to suppress NMD but not small RNA-induced mRNA decay. Thus, the observed effect on the miRNA targets, CD164 and IQGAP1 (Fig. 1A), is unlikely to be a consequence of NMD.
If hUPF1 is directly involved in the small RNA pathway, it is expected that hUPF1 would be dependent on miRNA target sites for the regulation of the mRNAs. To test this possibility, we generated firefly luciferase reporters containing the 3′ UTRs of CD164 or IQGAP1 and determined the expression of the reporter genes relative to that for control groups lacking the 3′ UTRs of the targets (Fig. 1B; see Fig. S2A in the supplemental material). HeLa cells were transfected with these reporters along with either miR-124a or control RNA (miR-124a-m) (Fig. 1B). A Renilla luciferase construct was cotransfected as an internal control. Depletion of hUPF1 compromised miR-124a-induced inhibition of luciferase expression (Fig. 1B). This set of siUPF1-2, siUPF2-2, and siUPF3b-2 was effective, as examined by RT-PCR (Fig. 1B, lanes 1 to 4). Similar results were obtained when different siRNAs (siUPF1-1 and siUPF2-1) were used instead in this experiment (data not shown). Knockdown of hUPF1 did not affect the expression of a reporter lacking the target sites (firefly luciferase) (see Fig. S2B in the supplemental material). Moreover, we could observe the upregulation of the miRNA reporter specifically when wild-type miR-124a, but not the mutant miR-124a (miR-124a-m), was introduced (see Fig. S2C in the supplemental material). Taken together, these data indicate that hUPF1 directly participates in miRNA-induced mRNA decay through the miRNA target sites.
hUPF1 is involved in the silencing of siRNA off-targets.
In order to test whether hUPF1 has a general role in RNA silencing, we next employed siRNA instead of miRNA. siMAPK14 was transfected into HeLa cells, and Northern blot analysis was performed to detect the MAPK14 mRNA (containing one perfect target site) and the RPA2 mRNA (harboring two imperfect target sites) (34). siMAPK14 could downregulate both MAPK14 mRNA (on-target) and the RPA2 mRNA (off-target) (Fig. 2A, compare lanes 1 and 2). When hUPF1 was depleted, the RPA2 mRNA was derepressed while the MAPK14 mRNA was only slightly increased (Fig. 2A, lane 3).
FIG. 2.
Effects of hUPF1 depletion on siRNA-induced mRNA decay. (A) Northern blotting (lanes 1 to 3) shows that hUPF1 is required for silencing of the off-targets (RPA2) but not on-targets (MAPK14). HeLa cells were transfected with siUPF1-1 (lane 3) or siGFP (lanes 1 and 2). After 30 h, the cells were transfected with siMAPK14 (lanes 2 and 3) or siGFP (lane 1) and incubated for 40 h before harvest. RT-PCR (lanes 4 to 6) confirms the knockdown of hUPF1. (B) Northern blot analysis and RT-PCR of mRNA. The experiment was done under the same conditions as that for Fig. 1A.
To further confirm the effect of hUPF1 depletion on RNAi, we transfected two kinds of siRNAs, both of which perfectly match the PPIB mRNA (Fig. 2B). We found that the knockdown of hUPF1 did not affect the PPIB mRNA (on-target), although hAGO2 knockdown impaired RNAi under the same condition (Fig. 2B). This result suggests that hUPF1 is required for the decay of imperfectly matching off-targets but may not be involved in endonucleolytic cleavage of the on-target of siRNA.
hUPF1 is required for the downregulation of endogenous miRNA targets.
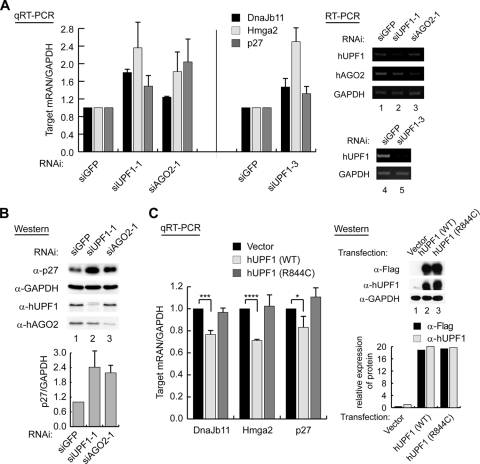
Because exogenous small RNAs were used in the experiments above, we next asked if hUPF1 is necessary for endogenous miRNAs to suppress their targets. For this, we monitored the levels of known miRNA targets in hUPF1-depleted HeLa cells. siAGO2 was used as a positive control. qRT-PCR was carried out to determine the levels of the Hmga2, p27, and DnaJb11 mRNAs, which are known to be targeted by endogenous miRNAs let-7, miR-222, and miR-29b, respectively (6, 39, 42, 45, 51). We found that these targets were increased when hUPF1 or hAGO2 was knocked down (Fig. 3A, left). Similar results were obtained when a different siUPF1, siUPF1-3, was used (Fig. 3A, middle). RT-PCR confirmed the knockdown of hUPF1 and hAGO2 (Fig. 3A, lanes 1 to 5). To exclude the possibility that the accumulated mRNAs contained PTCs, we determined the level of p27, a target of miR-222. Western blotting shows that knockdown of hUPF1 increases the level of the full-length p27 protein (Fig. 3B). This result indicates that the accumulated mRNAs are functional transcripts, not PTC-containing NMD substrates. Interestingly, we found that the level of p27 is upregulated by ∼2.4-fold (Fig. 3B), while the level of p27 mRNA is increased by ∼1.5-fold (Fig. 3A). This suggests that hUPF1 may also contribute to translational repression to some extent. This is consistent with recent findings that hUPF1 is an inhibitor of translational initiation (31) and termination (33).
FIG. 3.
Effects of knockdown or overexpression of hUPF1 on endogenous miRNA targets. (A) qRT-PCR shows that the targets of endogenous miRNAs are upregulated when hUPF1 or hAGO2 was knocked down in HeLa cells (n = 3; means ± standard deviations [SD]). RT-PCR (lanes 1 to 5) was carried out to confirm the knockdown of hUPF1 and hAGO2. (B) Cells were prepared as for panel A and were used for Western blotting. The graph shows the normalized levels of the p27 protein from three independent experiments (n = 3, means ± SD). α, anti. (C, left) qRT-PCR shows that the overexpression of hUPF1 decreases miRNA targets. A Flag-tagged wild-type hUPF1 (WT) or mutant hUPF1 (R844C) expression plasmid was transfected into HeLa cells, and the target mRNA levels were determined by qRT-PCR (n = 3; means ± SD). A paired one-tailed t test was used to calculate the P values for qRT-PCR (****, P < 0.001; ***, P < 0.01; *, P < 0.1). (Right) Expression levels of the Flag-hUPF1 proteins were measured by Western blotting.
We next asked if overexpression of hUPF1 has any effects on the downregulation of target mRNAs. For this, the hUPF1 expression plasmid was transfected into HeLa cells and the Hmga2, p27, and DnaJb11 mRNA levels were measured by qRT-PCR (Fig. 3C). As expected, the expression of hUPF1 resulted in the reduction of the miRNA targets, again in support of a role of hUPF1 in RNA silencing. We also expressed a hUPF1 mutant harboring a mutation in the helicase domain (R844C) to a level comparable to that of the wild type (68). The R844C mutant did not significantly change the target mRNA levels (Fig. 3C). Therefore, the helicase domain may be required for the function of hUPF1 in the downregulation of miRNA targets. The requirement for the active helicase domain of hUPF1 suggests an active role for hUPF1 in the mRNA decay process.
hUPF1 interacts with the Argonaute proteins and localizes in P bodies.
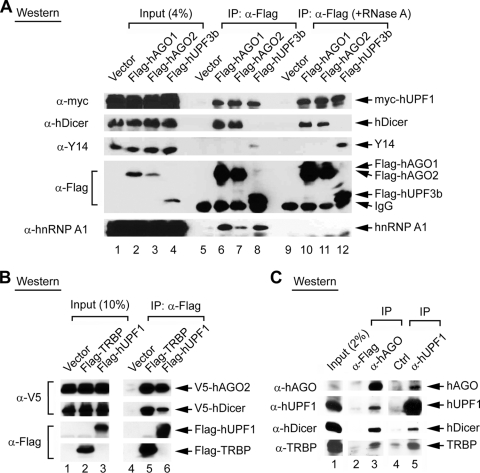
In an attempt to understand how hUPF1 may act in the miRNA pathway, we examined whether hUPF1 interacts with the Argonaute proteins. Flag-tagged hAGO1 or hAGO2 was coexpressed with myc-tagged hUPF1 in HEK293T cells and was immunoprecipitated with anti-Flag antibody. Western blotting showed that hUPF1 was coprecipitated with both hAGO1 and hAGO2 (Fig. 4A, lanes 6 and 7). Flag-tagged hUPF3b was used as a positive control for interaction with hUPF1 because hUPF1 and hUPF3b are well-known binding partners in the NMD pathway (Fig. 4A, lane 8). hAGO1 and hAGO2 interacted with hUPF1 even in the presence of RNase A (Fig. 4A, lanes 10 to 12), whereas the interaction with hnRNP A1 was disrupted by RNase A treatment. Thus, the Argonaute proteins may bind to hUPF1 through a protein-protein interaction. Notably, hAGO1 and hAGO2 were not precipitated with Y14, the key component of the exon junction complex, which interacts with NMD factors (Fig. 4A, lanes 10 and 11). hUPF3b is associated with Y14 (Fig. 4A, lane 12), while no significant interaction between hUPF3b and the RISC components was found (Fig. 4A, lanes 8 and 12, and data not shown). These results indicate that the hUPF1-RISC complex may constitute a distinct protein interaction module that is separable from the module involved in the NMD pathway, which consists of hUPF1, hUPF3, and Y14. When the tagging was changed to express Flag-hUPF1 and V5-hAGO2, Flag-hUPF1 still interacted with a V5-tagged hAGO2 (Fig. 4B, lane 6). Flag-tagged TRBP was used as a control for the interaction with hAGO2 and hDicer (Fig. 4B, lane 5).
FIG. 4.
Interaction of hUPF1 with Argonaute proteins. IP was followed by Western blotting. (A) The myc-hUPF1 protein was coexpressed with Flag-hAGO1, Flag-hAGO2, or Flag-hUPF3b. IP was carried out using anti-Flag antibody in the absence or presence of RNase A. Western blotting was carried out using anti-myc (α-myc) antibody (9E10), anti-hDicer antibody, anti-Y14 antibody (4C4), anti-Flag antibody, or anti-hnRNP A1 antibody (4B10). (B) Flag-tagged proteins were coexpressed with V5-hAGO2 and V5-hDicer. IP was carried out using anti-Flag antibody, and the proteins were visualized using either anti-V5 or anti-Flag antibody. (C) IP of endogenous complexes using anti-hAGO antibody (lane 3). Anti-Flag antibody was used as a negative control (lane 2). Lane 5, goat anti-hUpf1 antibody; lane 4, goat anti-PERK antibody (control).
We further tested the interactions between endogenous RNA decay factors by precipitating endogenous complexes using a monoclonal anti-Argonaute antibody and a goat polyclonal anti-hUPF1 antibody. The anti-Argonaute antibody precipitated hUPF1 as well as hDicer and TRBP (Fig. 4C, lane 3). The anti-hUPF1 antibody precipitated the Argonautes and, to a lesser extent, hDicer and TRBP (Fig. 4C, lane 5). These data suggest that hUPF1 may interact with RISC components to modulate RNA silencing.
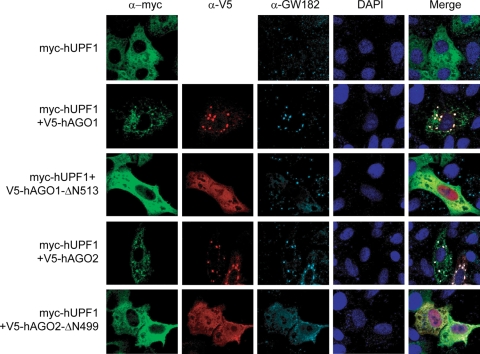
Since Argonaute proteins have been shown to localize to P bodies (49, 65) and hUPF1 was also found in cytoplasmic bodies when coexpressed with other NMD factors such as SMG-5 and SMG-7 (21, 70), we examined the localization of hUPF1 in comparison with that of the Argonaute proteins. When expressed alone in HeLa cells, hUPF1 was dispersed in the cytoplasm (Fig. 5, top row) as previously observed by others (21, 70). However, when coexpressed with hAGO1 or hAGO2, hUPF1 colocalized with hAGO1 or hAGO2 in P bodies (Fig. 5), suggesting that the RISC may recruit hUPF1 into P bodies. We then expressed hUPF1 along with a mutant hAGO1 (hAGO1-ΔN513) missing the N-terminal half. This mutant failed to enter P bodies because it lacks the PAZ domain, which is required for P body localization (49). This mutant was unable to direct hUPF1 to P bodies (Fig. 5), although it can still interact with hUPF1 (see Fig. S3 in the supplemental material). Coexpression of the hAGO2 N terminus mutant (hAGO2-ΔN499) also failed to localize hUPF1 into P bodies (Fig. 5). These results suggest that hUPF1 localization in P bodies may, at least in part, be dependent on AGO proteins. Given that the localization required AGO overexpression (Fig. 5) although the effect on changes in mRNA did not require AGO overexpression (Fig. 1 to 3), it is unclear at this point whether the observed P bodies are functional sites where RNA silencing takes place. Because P bodies are known to dynamically change in size and number depending on the condition, it is plausible that endogenous hUPF1 and AGO may normally function in smaller microscopically invisible P bodies (15).
FIG. 5.
Subcellular localization of hUPF1. HeLa cells were transiently transfected with plasmids expressing hUPF1 and/or Argonaute proteins and stained with anti-myc (9E10), anti-V5, and anti-GW182 antibodies. The transfected plasmids are indicated on the left. Alexa Fluor 488-goat anti-mouse IgG, Alexa Fluor 594-goat anti-rabbit IgG, and Alexa Fluor 647-goat anti-human IgG were used as the secondary antibodies.
We next tested which Argonaute proteins are required for miRNA-induced mRNA degradation. We introduced siAGO1, siAGO2, siAGO3, or siAGO4 together with the miR-124a duplex into HeLa cells (see Fig. S4A in the supplemental material). When hAGO1 and hAGO2 were depleted, the miR-124a target mRNAs were stabilized, indicating that hAGO1 and hAGO2 may be involved in miRNA-induced mRNA decay. On the other hand, knockdown of hAGO3 and hAGO4 did not significantly affect target repression, although these RNAi data do not exclude the possibility that hAGO3 and hAGO4 are significant in other conditions and cell types. When hAGO1 and hAGO2 were depleted by RNAi, expression of an miR-124a reporter that contains the IQGAP1 3′ UTR was also derepressed (see Fig. S4C in the supplemental material). This result provides further evidence that hAGO1 and hAGO2 are mainly responsible for small RNA-induced mRNA decay, at least in HeLa cells.
hUPF1 may assist RISC to bind to the target mRNA and accelerate mRNA degradation.
We postulated and tested several possible mechanisms for the action of hUPF1 (see Fig. 7 for the model). First, it is conceivable that hUPF1 may be involved in miRNA biogenesis. To test this possibility, we measured miRNA levels in cells depleted of hUPF1 (Fig. 6A), but we did not detect significant changes in the levels of let-7a and miR-222. It was also noted that the small RNA duplexes used for our initial experiments were mimics of Dicer products (Fig. 1 and 2). Therefore, our results indicate that hUPF1 is unlikely to affect miRNA processing, and it may act downstream of miRNA processing.
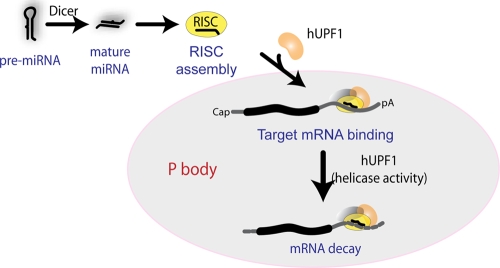
FIG. 7.
A model for the action mechanism of hUPF1 in small RNA-induced mRNA silencing. hUPF1 may bind to hAGO2 after RISC assembly and colocalize to P bodies. hUPF1 may facilitate the binding of RISC to the target mRNA and accelerate the degradation of the mRNA.
FIG. 6.
Effects of hUPF1 on hAGO2-associated mRNAs. (A) Northern blotting of miRNAs using total RNA. The graph shows the mean values (± standard deviations [SD]) of relative miRNA levels measured in two independent experiments (n = 2). (B) Western blotting of RNA silencing factors. The levels of Dicer, hAGO2, and TRBP proteins are not affected by hUPF1 knockdown. GAPDH or alpha-tubulin was used as the loading control. α, anti. (C) IP followed by Western blotting shows that the interactions between RISC components are not affected by knockdown of hUPF1. HeLa cells were cotransfected with Flag-tagged hAGO2 plasmids and siUPF1 or siGFP. IP was carried out using anti-Flag antibody in the presence of RNase A. (D) Knockdown of hUPF1 reduces the Hmga2 mRNA associated with hAGO2. hUpf1 depleted-HeLa cells were lysed and used for IP with antibody against hAGO2. Coprecipitated mRNA was extracted and quantified by qRT-PCR. The Hmga2 mRNA levels were normalized against the GAPDH mRNA levels in the same sample. The IP efficiency was then calculated by dividing the normalized Hmga2 mRNA level from IP by that for the input (n = 2; means ± SD). (E) qRT-PCR shows that overexpression of mutant hUPF1 (R844C) increases the level of hAGO2-associated Hmga2 mRNA, compared to overexpression of wild-type hUPF1 (WT). A Flag-tagged wild-type hUPF1 (WT) or mutant hUPF1 expression plasmid was transfected into HeLa cells. The cells were lysed and used for IP with antibody against hAGO2. IP efficiency was calculated as for panel D (n = 2; means ± SD). Northern blotting of miRNA (right) shows that hAGO2-associated let-7 miRNA was not affected by overexpression of mutant hUPF1 compared to overexpression of the wild type.
Second, we tested the possibility that hUPF1 may upregulate the levels of RISC components, which would indirectly influence RNA silencing (Fig. 6B). Depletion of hUPF1 using two different siRNAs did not alter the levels of hAGO2, hDicer, and TRBP, as examined by Western blotting (Fig. 6B), excluding the possibility that hUPF1 changes the levels of the RISC components.
We then asked if hUPF1 regulates the interactions between RISC components. A plasmid expressing Flag-hAGO2 was cotransfected with siUPF1 or siGFP into HeLa cells (Fig. 6C). Flag-hAGO2 was then immunoprecipitated, and the associated proteins were visualized by Western blotting. The binding of hAGO2 to hDicer, TRBP, or GW182 was not affected by depletion of hUPF1 (Fig. 6C, lanes 8 and 9). Comparable results were obtained when hAGO1 was used instead of hAGO2 (see Fig. S5 in the supplemental material). Therefore, it is unlikely that the interactions between RISC components are affected by the depletion of hUPF1.
To further understand the action mechanisms of hUPF1, we next determined the levels of mRNAs bound to hAGO2. It was recently reported that the Hmga2 mRNA, the target of let-7, is enriched in the hAGO2 complex in HeLa cells (71). Consistent with this report, more Hmga2 mRNA was precipitated with a monoclonal antibody against hAGO2 (4G8) than with the control antibody (SP2/0), while similar levels of GAPDH mRNA were immunoprecipitated with both antibodies (see Fig. S6 in the supplemental material). We depleted hUPF1 in HeLa cells by RNAi, immunoprecipitated the hAGO2 complex using anti-hAGO2 antibody, and measured the mRNA levels by qRT-PCR. When hUPF1 was knocked down, less Hmga2 mRNA was associated with hAGO2 compared to the control (Fig. 6D). This result suggests that hUPF1 increases the binding of hAGO2 to the miRNA targets.
Because it is plausible that hUPF1 may act at a downstream process(es) as well, we examined whether hUPF1 is required for the downstream decay step. For this, we overexpressed either wild-type hUPF1 or the R844C helicase mutant and determined the hAGO2-associated mRNA. If the helicase activity of hUPF1 functions only at the mRNA binding step, not at the mRNA decay step, it is expected that less mRNA would be bound to hAGO2 when the R844C mutant is expressed because this mutant was shown to be inactive in silencing (Fig. 3C). However, our data show that more Hmga2 mRNA remains associated with the hAGO2 complex in the presence of R844C mutant than in the presence of the wild type (Fig. 6E), suggesting that the RISC-bound mRNA accumulates without getting degraded if the helicase activity of hUPF1 is not provided. Northern blotting of the hAGO2-bound RNA showed that comparable amounts of let-7a were associated with hAGO2 in the presence of R844C mutant and the wild type, indicating that hUPF1 helicase activity is not required for the interaction between hAGO2 and miRNA (Fig. 6E). Thus, hUPF1 may act at the mRNA decay step as well as the mRNA binding step. The helicase activity of hUPF1 may be required at the decay step.
DISCUSSION
Our study suggests a novel role for hUPF1 in modulating RNA silencing in humans. The precise mechanism of action remains to be determined. And we cannot fully rule out the possibility that hUPF1 acts indirectly by regulating some unknown factor that is involved in RNA silencing. However, the simplest way of interpreting our data is that hUPF1 is recruited to RISC after miRNA processing and RISC assembly (Fig. 7 shows a model). hUPF1 then helps RISC to bind to the target mRNA and brings in nucleases to induce mRNA decay. Because hUPF1 is known to interact with the DCP1/DCP2 complex, exosome, and deadenylation factors in Saccharomyces cerevisiae and humans (25, 44, 50, 54), it is conceivable that hUPF1 may mediate the interaction between the RISC and mRNA decay factors. We can also speculate that hUPF1 remodels the messenger ribonucleoprotein complex (mRNP) via its ATP-dependent RNA helicase domain to facilitate the recruitment of RISC and mRNA decay factors. Alternatively, but not mutually exclusively, it is plausible that hUPF1 may act through aberrant translation. In support of this, hUPF1 was reported to interfere with efficient termination (33), and miRNAs are known to inhibit protein synthesis from polysome-associated mRNAs at a postinitiation step(s) (56, 58). These hypotheses remain to be tested.
Hock and colleagues recently showed that hAGO1 and hAGO2 form three distinct mRNPs (complexes I to III) and that hUPF1 is associated with the largest complex (complex III) (26). It will be of interest to find out whether hAGO2 directly recruits hUPF1 or indirectly associates with hUPF1 through an as yet unknown molecular link(s).
Although UPF1 is best known as a central effector of NMD, previous studies have revealed additional functions of UPF1 outside the NMD pathway (28, 37). Staufen, which binds to specific sites in the 3′ UTRs of certain mRNAs, interacts with hUPF1 to recruit decay factors and induces specific degradation of a Staufen-binding transcript (37, 38). Furthermore, hUPF1 and the protein kinase ATR are involved in the degradation of replication-dependent histone mRNAs at the end of S phase through an unknown mechanism (35). In nematodes, SMG2, the UPF1 homologue, is required for the maintenance of double-stranded RNA-induced RNAi (13), although the mechanism underlying this phenomenon remains obscure. Knockdown of the fly UPF1 homolog did not affect miRNA-dependent suppression of reporter genes in Drosophila S2 cells (61). In Arabidopsis thaliana, gene silencing by an inverted repeat transgene is impaired in upf1 mutants, suggesting a role for UPF1 in RNA silencing in plants (1). Plant miRNA targets usually have nearly perfect matches to miRNAs and they were unaffected in this study (1), suggesting that plant UPF1 may not be required for the miRNA pathway but may instead participate in another small RNA pathway(s). Taken together, these results imply functional divergence of UPF1 homologues among eukaryotic organisms. In fact, UPF1 null yeast and nematode mutants are viable (27, 43), whereas the knockout of upf1 in mice is lethal (52). It is currently unclear whether these seemingly unrelated observations can be explained by a unified mechanistic model. But what becomes increasingly clear is that hUPF1 interacts with multiple RNAs and proteins and thereby acts as a central player in diverse mRNA decay processes.
We find that hUPF1 is required for efficient silencing of siRNA off-targets while it is not important for suppression of on-targets. This result demonstrates that target mRNA decay and endonucleolytic cleavage are two separate processes that involve distinct sets of protein factors. hUPF1 is a factor that is specifically involved in the decay of the off-target transcripts and the natural targets of endogenous miRNAs. The off-target effect is a major hurdle in RNAi application because it compromises the specificity of RNAi. Many off-target effects in RNAi experiments are due to siRNAs acting like miRNAs on unintended targets (7). Our current finding may provide a basis for further studies to overcome off-target effects.
Supplementary Material
Acknowledgments
We specially thank Lynne Maquat and Yoon Ki Kim for hUPF1 cDNA and NMD reporters. We also thank Elisa Izaurralde and Matthias Hentze for critical comments, Zissimos Mourelatos for Argonaute cDNAs, Gideon Dreyfuss for anti-Y14 and anti-hnRNP A1 antibodies, Tom Tuschl for Argonaute cDNAs, J. Lykke-Andersen and Mikiko C. Siomi for antibodies, Marvin J. Fritzler for anti-GW182 antibody, Hyunsook Lee for anti-myc antibody, and Young-Kook Kim for discussion and technical help.
This work was supported by Creative Research Initiatives (R16-2007-073-01000-0). H.J., J.H., and K.-H.Y. were supported by the BK21 studentship. H.J. was supported by the Seoul Science Fellowship.
Footnotes
Published ahead of print on 24 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arciga-Reyes, L., L. Wootton, M. Kieffer, and B. Davies. 2006. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47:480-489. [DOI] [PubMed] [Google Scholar]
- 2.Baek, D., J. Villen, C. Shin, F. D. Camargo, S. P. Gygi, and D. P. Bartel. 2008. The impact of microRNAs on protein output. Nature 455:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagga, S., J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus, and A. E. Pasquinelli. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122:553-563. [DOI] [PubMed] [Google Scholar]
- 4.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 5.Behm-Ansmant, I., J. Rehwinkel, T. Doerks, A. Stark, P. Bork, and E. Izaurralde. 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beitzinger, M., L. Peters, J. Y. Zhu, E. Kremmer, and G. Meister. 2007. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 4:76-84. [DOI] [PubMed] [Google Scholar]
- 7.Birmingham, A., E. M. Anderson, A. Reynolds, D. Ilsley-Tyree, D. Leake, Y. Fedorov, S. Baskerville, E. Maksimova, K. Robinson, J. Karpilow, W. S. Marshall, and A. Khvorova. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3:199-204. [DOI] [PubMed] [Google Scholar]
- 8.Chendrimada, T. P., K. J. Finn, X. Ji, D. Baillat, R. I. Gregory, S. A. Liebhaber, A. E. Pasquinelli, and R. Shiekhattar. 2007. MicroRNA silencing through RISC recruitment of eIF6. Nature 447:823-828. [DOI] [PubMed] [Google Scholar]
- 9.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding, L., A. Spencer, K. Morita, and M. Han. 2005. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 19:437-447. [DOI] [PubMed] [Google Scholar]
- 11.Doench, J. G., C. P. Petersen, and P. A. Sharp. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi, N., S. Zenno, R. Ueda, H. Ohki-Hamazaki, K. Ui-Tei, and K. Saigo. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13:41-46. [DOI] [PubMed] [Google Scholar]
- 13.Domeier, M. E., D. P. Morse, S. W. Knight, M. Portereiko, B. L. Bass, and S. E. Mango. 2000. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289:1928-1931. [DOI] [PubMed] [Google Scholar]
- 14.Eberle, A. B., S. Lykke-Andersen, O. Muhlemann, and T. H. Jensen. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 16:49-55. [DOI] [PubMed] [Google Scholar]
- 15.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 16.Eulalio, A., E. Huntzinger, and E. Izaurralde. 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15:346-353. [DOI] [PubMed] [Google Scholar]
- 17.Eulalio, A., E. Huntzinger, T. Nishihara, J. Rehwinkel, M. Fauser, and E. Izaurralde. 2009. Deadenylation is a widespread effect of miRNA regulation. RNA 15:21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eulalio, A., J. Rehwinkel, M. Stricker, E. Huntzinger, S. F. Yang, T. Doerks, S. Dorner, P. Bork, M. Boutros, and E. Izaurralde. 2007. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 21:2558-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filipowicz, W., S. N. Bhattacharyya, and N. Sonenberg. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9:102-114. [DOI] [PubMed] [Google Scholar]
- 20.Fillman, C., and J. Lykke-Andersen. 2005. RNA decapping inside and outside of processing bodies. Curr. Opin. Cell Biol. 17:326-331. [DOI] [PubMed] [Google Scholar]
- 21.Fukuhara, N., J. Ebert, L. Unterholzner, D. Lindner, E. Izaurralde, and E. Conti. 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 17:537-547. [DOI] [PubMed] [Google Scholar]
- 22.Giraldez, A. J., Y. Mishima, J. Rihel, R. J. Grocock, S. Van Dongen, K. Inoue, A. J. Enright, and A. F. Schier. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312:75-79. [DOI] [PubMed] [Google Scholar]
- 23.Gregory, R. I., T. P. Chendrimada, N. Cooch, and R. Shiekhattar. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123:631-640. [DOI] [PubMed] [Google Scholar]
- 24.Haase, A. D., L. Jaskiewicz, H. Zhang, S. Laine, R. Sack, A. Gatignol, and W. Filipowicz. 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6:961-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, F., and A. Jacobson. 2001. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 21:1515-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hock, J., L. Weinmann, C. Ender, S. Rudel, E. Kremmer, M. Raabe, H. Urlaub, and G. Meister. 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 8:1052-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodgkin, J., A. Papp, R. Pulak, V. Ambros, and P. Anderson. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123:301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holbrook, J. A., G. Neu-Yilik, M. W. Hentze, and A. E. Kulozik. 2004. Nonsense-mediated decay approaches the clinic. Nat. Genet. 36:801-808. [DOI] [PubMed] [Google Scholar]
- 29.Huntzinger, E., I. Kashima, M. Fauser, J. Sauliere, and E. Izaurralde. 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoans. RNA 14:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingelfinger, D., D. J. Arndt-Jovin, R. Luhrmann, and T. Achsel. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489-1501. [PMC free article] [PubMed] [Google Scholar]
- 31.Isken, O., Y. K. Kim, N. Hosoda, G. L. Mayeur, J. W. Hershey, and L. E. Maquat. 2008. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133:314-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isken, O., and L. E. Maquat. 2007. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21:1833-1856. [DOI] [PubMed] [Google Scholar]
- 33.Ivanov, P. V., N. H. Gehring, J. B. Kunz, M. W. Hentze, and A. E. Kulozik. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635-637. [DOI] [PubMed] [Google Scholar]
- 35.Kaygun, H., and W. F. Marzluff. 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12:794-800. [DOI] [PubMed] [Google Scholar]
- 36.Kim, V. N. 2005. Small RNAs: classification, biogenesis, and function. Mol. Cells 19:1-15. [PubMed] [Google Scholar]
- 37.Kim, Y. K., L. Furic, L. Desgroseillers, and L. E. Maquat. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120:195-208. [DOI] [PubMed] [Google Scholar]
- 38.Kim, Y. K., L. Furic, M. Parisien, F. Major, L. DesGroseillers, and L. E. Maquat. 2007. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 26:2670-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, Y. K., J. Yu, T. S. Han, S. Y. Park, B. Namkoong, D. H. Kim, K. Hur, M. W. Yoo, H. J. Lee, H. K. Yang, and V. N. Kim. 2009. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 37:1672-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 41.Lee, Y., I. Hur, S. Y. Park, Y. K. Kim, M. R. Suh, and V. N. Kim. 2006. The role of PACT in the RNA silencing pathway. EMBO J. 25:522-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, Y. S., and A. Dutta. 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 21:1025-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 44.Lejeune, F., X. Li, and L. E. Maquat. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675-687. [DOI] [PubMed] [Google Scholar]
- 45.le Sage, C., R. Nagel, D. A. Egan, M. Schrier, E. Mesman, A. Mangiola, C. Anile, G. Maira, N. Mercatelli, S. A. Ciafre, M. G. Farace, and R. Agami. 2007. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 26:3699-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim, L. P., N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley, and J. M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769-773. [DOI] [PubMed] [Google Scholar]
- 47.Liu, J., M. A. Carmell, F. V. Rivas, C. G. Marsden, J. M. Thomson, J. J. Song, S. M. Hammond, L. Joshua-Tor, and G. J. Hannon. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437-1441. [DOI] [PubMed] [Google Scholar]
- 48.Liu, J., F. V. Rivas, J. Wohlschlegel, J. R. Yates III, R. Parker, and G. J. Hannon. 2005. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7:1261-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, J., M. A. Valencia-Sanchez, G. J. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22:8114-8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayr, C., M. T. Hemann, and D. P. Bartel. 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315:1576-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medghalchi, S. M., P. A. Frischmeyer, J. T. Mendell, A. G. Kelly, A. M. Lawler, and H. C. Dietz. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10:99-105. [DOI] [PubMed] [Google Scholar]
- 53.Mishima, Y., A. J. Giraldez, Y. Takeda, T. Fujiwara, H. Sakamoto, A. F. Schier, and K. Inoue. 2006. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 16:2135-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitchell, P., and D. Tollervey. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11:1405-1413. [DOI] [PubMed] [Google Scholar]
- 55.Nielsen, C. B., N. Shomron, R. Sandberg, E. Hornstein, J. Kitzman, and C. B. Burge. 2007. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 13:1894-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nottrott, S., M. J. Simard, and J. D. Richter. 2006. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 13:1108-1114. [DOI] [PubMed] [Google Scholar]
- 57.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121-127. [DOI] [PubMed] [Google Scholar]
- 58.Petersen, C. P., M. E. Bordeleau, J. Pelletier, and P. A. Sharp. 2006. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 21:533-542. [DOI] [PubMed] [Google Scholar]
- 59.Pillai, R. S. 2005. MicroRNA function: multiple mechanisms for a tiny RNA? RNA 11:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pillai, R. S., S. N. Bhattacharyya, C. G. Artus, T. Zoller, N. Cougot, E. Basyuk, E. Bertrand, and W. Filipowicz. 2005. Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309:1573-1576. [DOI] [PubMed] [Google Scholar]
- 61.Rehwinkel, J., I. Behm-Ansmant, D. Gatfield, and E. Izaurralde. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11:1640-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richter, J. D. 2008. Think you know how miRNAs work? Think again. Nat. Struct. Mol. Biol. 15:334-336. [DOI] [PubMed] [Google Scholar]
- 63.Schmitter, D., J. Filkowski, A. Sewer, R. S. Pillai, E. J. Oakeley, M. Zavolan, P. Svoboda, and W. Filipowicz. 2006. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 34:4801-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selbach, M., B. Schwanhausser, N. Thierfelder, Z. Fang, R. Khanin, and N. Rajewsky. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58-63. [DOI] [PubMed] [Google Scholar]
- 65.Sen, G. L., and H. M. Blau. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633-636. [DOI] [PubMed] [Google Scholar]
- 66.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh, G., I. Rebbapragada, and J. Lykke-Andersen. 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 6:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun, X., H. A. Perlick, H. C. Dietz, and L. E. Maquat. 1998. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 95:10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomari, Y., and P. D. Zamore. 2005. Perspective: machines for RNAi. Genes Dev. 19:517-529. [DOI] [PubMed] [Google Scholar]
- 70.Unterholzner, L., and E. Izaurralde. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16:587-596. [DOI] [PubMed] [Google Scholar]
- 71.Weinmann, L., J. Hock, T. Ivacevic, T. Ohrt, J. Mutze, P. Schwille, E. Kremmer, V. Benes, H. Urlaub, and G. Meister. 2009. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136:496-507. [DOI] [PubMed] [Google Scholar]
- 72.Wu, L., and J. G. Belasco. 2005. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 25:9198-9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu, L., J. Fan, and J. G. Belasco. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA 103:4034-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, J., X. Sun, Y. Qian, and L. E. Maquat. 1998. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4:801-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.