Abstract
Previous studies have demonstrated that c-Src tyrosine kinase interacts specifically with ErbB2, but not with other members of the epidermal growth factor receptor (EGFR) family. To identify the site of interaction, we recently used a chimeric EGFR/ErbB2 receptor approach to show that c-Src requires the kinase region of ErbB2 for binding. Here, we demonstrate that retention of a conserved amino acid motif surrounding tyrosine 877 (referred to here as EGFRYHAD) is sufficient to confer binding to c-Src. Surprisingly the association of c-Src was not dependent on its SH2 or SH3 domain or on the phosphorylation or kinase activity of the receptor. We further show that the chimeric EGFRs that contain the Y877 motif are transforming in vitro and in vivo following ligand stimulation. Transformation was also partially dependent on sustained activation of Stat3. Finally, we demonstrate that EGFRs with mutations in the catalytic domain, originally identified in lung cancer and conferring increased sensitivity to gefitinib and erlotinib, two EGFR kinase inhibitors, gained the capacity to bind c-Src. Moreover, transformation by these EGFR mutants was inhibited by Src inhibitors regardless of their sensitivities to gefitinib and erlotinib. These observations have important implications for understanding the molecular basis for resistance to EGFR inhibitors and implicate c-Src as a critical signaling molecule in EGFR mutant-induced transformation.
The epidermal growth factor receptor (EGFR) family is comprised of four members, EGFR, ErbB2, ErbB3, and ErbB4, with distinct ligand specificities, which, upon homo- or heterodimerization after ligand binding, autophosphorylate and recruit different effector proteins to specific tyrosine residues located in their cytoplasmic tails. These signaling molecules, which are either adapter molecules that recruit other kinases or kinases themselves, mediate diverse functions, such as proliferation, growth, and survival (27). There are now several pieces of evidence demonstrating that these growth factor receptors are mutated or overexpressed in a variety of different cancers, including salivary gland adenocarcinoma (44), breast cancer (47), esophageal squamous carcinoma (22), bladder cancer (58), and lung cancer (57). Accordingly, ErbB2 is overexpressed in 20 to 30% of all human breast cancer, which correlates with poor prognosis, and in 40 to 60% of ductal carcinoma in situ (19). ErbB2 is 100-fold more potent in its transforming ability than ErbB1/EGFR, although the two receptors are 85% homologous (14, 15). Breast carcinoma cells devoid of ErbB2, but not other ErbB receptor family members, are defective in cell invasion upon EGF ligand stimulation (49). In fact, ErbB2 could induce cell migration when overexpressed in cells devoid of any other ErbB receptors. In a three-dimensional cell culture system, overexpression of ErbB2, but not EGFR, disrupts mammary acinus structure by reinitiating cell proliferation, leading to an absence of lumen and disruption of tight junctions and of cell polarity, although the cells still lack invasive properties (31).
Src is a nonreceptor tyrosine kinase implicated in signal transduction pathways downstream of multiple receptors, such as platelet-derived growth factor, insulin receptor, G-coupled receptors, and ErbB family receptors, where it regulates a wide variety of cellular functions that include proliferation, migration, and apoptosis (17). Src tyrosine kinase activity is sporadically increased in many cases of human cancer, including colon and breast cancer (10, 38, 52). Moreover, Src kinase activity is elevated in ErbB2-induced mammary tumors (33). Direct evidence supporting a role in mammary tumor progression derives from observations made in transgenic mice. Constitutive activation of c-Src in mammary epithelia led to frequent mammary epithelial hyperplasias, which occasionally developed into solid tumors (54). Conversely, deletion of c-Src in a mouse mammary tumor virus/polyomavirus middle T-antigen (PyMT) transgenic strain abrogates mammary tumor formation (21).
c-Src is also an important player downstream of the EGFR family. Phosphorylation of several tyrosine residues within the EGFR has been demonstrated to be increased following c-Src overexpression both in vitro and in vivo, suggesting that c-Src is required for full biological response following EGF stimulation (29, 51). In addition to EGFR, c-Src specifically interacts with tyrosine-phosphorylated ErbB2 in ErbB2-induced mammary tumors. This association was further demonstrated to result in enhanced c-Src kinase activity (3, 28, 34, 35). More recently, using chimeric EGF/ErbB2 receptors, we demonstrated that c-Src specifically associates with ErbB2, but not with other ErbB family members. c-Src was demonstrated to specifically associate with the ErbB2 kinase domain (24). Moreover, the chimeric EGFR that contained the c-Src binding site was able to disrupt cell polarity and cell-cell junctions to induce epithelial cell scattering in a three-dimensional cell culture system in a MAPK-dependent manner (24).
Here, we demonstrate that c-Src association with ErbB2 is conformation dependent and that the residues necessary for interaction are centered around Y877 in the kinase domain of ErbB2, an association that is further strengthened by residues located in the amino-terminal part of the kinase domain. This association was not dependent on the SH2 or SH3 domain or the kinase activity of c-Src or ErbB2. We further show that mammary epithelial cells expressing the EGFR/ErbB2 chimeric receptors that have regained the capacity to associate with c-Src have disrupted epithelial polarity that is correlated with enhanced transforming potential, an effect dependent on c-Src kinase activity and Stat3 activation. Finally, we show that mutant EGFRs isolated from lung adenocarcinomas have the capacity to associate with c-Src and that these EGFR mutants require Src kinase activity for transformation.
MATERIALS AND METHODS
Reagents and antibodies.
The following antibodies used in this study were from Cell Signaling unless otherwise specified: phospho-ErbB2 Y877, phospho-ErbB2 Y1221/1222, phospho-ErbB2 Y1248, EGFR, phospho-EGFR Y1173, phospho-EGFR Y845, phospho-MAPK, MAPK, phospho-Stat3 Y705, Stat3, phospho-PTEN, PTEN, phospho-p38, p38, phospho-Akt S473, Akt, ErbB2/Neu (AB-3) (Calbiochem), ErbB2/Neu (AB-4) (Calbiochem), v-Src (AB-1) (Oncogene), c-Src (Upstate), phospho-Src Y416 (Cell Signaling and Calbiochem), actin (Sigma), and EEA1 (Upstate Biotechnology). c-Src constructs were purchased from Upstate Biotechnology (Src cDNA allelic pack). SrcDN251 was a generous gift from P. L. Schwartzberg. PP2 was purchased from Biosource and SKI-1 from Calbiochem. Dominant-negative Stat3 (DN-Stat3) was a generous gift of V. Giguere. Fluorescein-coupled EGF was purchased from Molecular Probes (Invitrogen). Fluorescence-coupled secondary antibodies were from Molecular Probes (Invitrogen).
Chimeric receptor construction and site-directed mutagenesis.
The chimeric receptors EGFRTK, EGFRTK1, and EGFRTK2 were described by Kim et al. (24). EGFRYHAD was created by site-directed mutagenesis. EGFRTK/YHAE and ErbB2YHAE were produced from EGFRTK and activated ErbB2, respectively, by site-directed mutagenesis. The chimeric ErbB2 receptors were constructed by creating a SacI restriction site 5′ of the TK and TK1 regions, an Mfe1 restriction site 3′ of both the TK and TK2 regions, and an FspI restriction site 5′ and 3′ of TK2 and TK1, respectively, in both ErbB2 and EGFR. None of the restriction sites created altered the coding sequences of the receptors. These regions were digested out of EGFR and ligated into the ErbB2 backbone to create ErbB2TK, ErbB2TK1, and ErbB2TK2. To construct a pMSCV-expressing plasmid, chimeric receptors were digested from the pcDNA plasmid with HindIII and EcoRV, blunted, and ligated into the EcoRV restriction site of pMSCV. All constructs were verified by sequencing. c-Src SH2 and SH3 mutants were designed according to the method of Cary et al. (11).
Transfection, immunoprecipitation, and immunoblotting.
293T cells (2 × 106) were plated on 60-mm plates. The cells were transfected 24 h later using Fugene, following the manufacturer's recommendations (Roche). Thirty hours following transfection, the cells were washed twice with ice-cold 1× phosphate-buffered saline (PBS), lysed using TNE lysis buffer (50 mM Tris-HCl, [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 10 mM NaF, 2 mM EDTA, 1 mM sodium orthovanadate, 10 μg/ml leupeptin, and 10 μg/ml aprotinin), incubated on ice for 20 min, and spun down to remove cellular debris. The protein concentration was measured using Bio-Rad's Bradford reagent, and 1 mg was used for overnight immunoprecipitation at 4°C. The next day, protein G-agarose beads (Amersham) were added for 3 h to bind the antibody-protein complexes. The beads were washed five times with lysis buffer and resuspended in 2× loading buffer. Samples were then run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride membranes, and blocked for 2 h at room temperature with 5% milk-TBS (50 mM Tris-HCl and 150 mM NaCl) or 3% bovine serum albumin (BSA)-TBS for phosphoantibodies. The membranes were then incubated overnight at 4°C with the primary antibody. The following day, the membranes were washed four times for 15 min each time with 1× TBS containing 0.3% Tween 20, incubated with the appropriate horseradish peroxidase-coupled secondary antibody for 1 h at room temperature, and finally washed four times for 15 min each time with 1× TBS containing 0.3% Tween 20. The membranes were incubated with enhanced chemiluminescence (Amersham) and exposed on Kodak film. Densitometry was performed using an Odyssey Licor system.
Transformation assay.
Rat1 fibroblasts (2 × 106 cells) were transfected using 3 μg of pMSCV chimeric receptor and 9 μl of Fugene (Roche) in 60-mm plates following the manufacturer's recommendations. The following day, the cells were split into six 100-mm plates, and puromycin selection (3 μg/ml) was started the next day to control for transfection efficiency. Dulbecco's modified Eagle's medium (DMEM) supplemented with 2% fetal bovine serum (Wisent) and Pen/Strep (Invitrogen) was changed every 3 days. In the case of EGFR and chimeric EGFR, EGF was added daily at a concentration of 10 ng/ml. After 14 days, the plates were washed twice with 1× PBS, fixed with 2% paraformaldehyde, washed three times with 1× PBS, and stained with Giemsa stain.
Immunofluorescence.
Stable Rat1 fibroblasts (1 × 105) were plated on glass coverslips in 24-well plates. The following day, the cells were serum deprived for 48 h with 1× DMEM supplemented with Pen/Strep (Invitrogen). The cells were stimulated with EGF (50 ng/ml) or fluorescein-conjugated EGF (50 ng/ml) for the indicated time. The cells were fixed with 2% paraformaldehyde for 20 min at room temperature, washed three time with 1× PBS, permeabilized with 0.2% Triton X, washed three times with 100 mM glycine, and blocked with immunofluorescence buffer (1× PBS, 0.2% BSA, 0.1% Triton X, 0.05% Tween 20) supplemented with 2% BSA for 30 min. The coverslips were then incubated with the indicated first antibody for 1 h at room temperature, washed three times for 5 min each time with immunofluorescence buffer, incubated with the fluorescence-conjugated secondary antibody for 45 min at room temperature, and washed three times with immunofluorescence buffer. The cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole), and the coverslips were mounted on glass slides with Immunomount (Thermo Scientific). The cells were analyzed using a Zeiss LSM 510 confocal microscope.
Generation of EpH4 stable cell lines.
Using pSV2/EGFR, pJ4/YHAD, and pSV2/TK as templates, 3.8-kbp NotI-DNA fragments of human wild-type EGFR, as well as of the YHAD and TK mutants, were PCR amplified and inserted into the NotI-linearized pβ-actin vector (a gift from Philip Leder, Harvard Medical School [39]). The constructs were linearized and transfected into mouse mammary epithelial EpH4 cells (40) with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After selection with 1 mg/ml G418 (Sigma, St. Louis, MO) for 14 days, stable transfectants were stained with an antibody against the extracellular domain for human EGFR (Oncogene, San Diego, CA; clone 528) and subjected to fluorescence-activated cell sorting (FACS) after secondary-antibody labeling. Sorted, pooled transfectants expressing similar levels of the transgenes were expanded under 250 μg/ml G418, and the presence of the human EGFR extracellular domain was then confirmed by Western blotting, immunostaining, and FACS. The last indicated greater than 90% positivity among the sorted stable transfectants.
Three-dimensional spheroidogenesis assays.
Stable, pooled transfectants of EpH4 cells were seeded onto reconstituted basement membrane gels (Matrigel; BD Biosciences, NJ) at a density of 2 × 105 cells/ml and cultured in serum-free differentiation medium consisting of DMEM/F12 supplemented with 5 μg/ml insulin, 1 μg/ml hydrocortisone, and 5 μg/ml lactogenic prolactin (Sigma). The cultures were maintained for 5 days to allow spheroid formation. The spheroids were then treated with 10 ng/ml EGF (Peprotech Canada, Inc.) for 48 h or left untreated before fixation and staining. For pharmacologic Src inhibition, 5-day-old spheroids were pretreated with 20 μM PP2 (Calbiochem) for 1 hour prior to EGF stimulation for a further 48 h.
At the end of the treatment period, cultures were photographed live by phase-contrast microscopy or were fixed in methanol (−20°C for 15 min), immunostained, and imaged by confocal microscopy. For the latter analysis, the fixed cultures were washed in PBS (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4), blocked with 1% BSA and 10% normal goat serum in PBS, and stained with anti-ZO-1 (Zymed Laboratories Inc., South San Francisco, CA) overnight at 4°C. Primary antibody binding was visualized using species-specific secondary antibodies coupled with Alexa Fluor dyes (Molecular Probes Inc., Eugene, OR), nuclei were stained with 0.5 μg/ml DAPI (Sigma), and the cultures were then mounted with the antifade reagent Prolong (Molecular Probes) and imaged with an Olympus FluoView FV1000 confocal microscope.
Biotin internalization assays.
Biotin internalization assays were performed as described by Grandal et al. (18). Briefly, Rat1 cells were plated on 100-mm dishes and serum deprived for 24 h. The cells were washed several times with ice-cold 1× PBS and then incubated with 0.5 mg/ml of sulfo-NHS-SS-biotin (Pierce, Thermo Scientific) for 30 min at 4°C to label cell surface proteins. The cells were washed several times with 1× PBS to remove nonincorporated biotin and then incubated with DMEM supplemented with 10 ng/ml of EGF for the indicated time. In cases where PP2 (10 μM) was added, the cells were incubated at 37°C for 30 min before the addition of EGF. Internalization was stopped by transferring the cells from 37°C to 4°C and washing them with ice-cold 1× PBS. Noninternalized biotin was stripped off the cell surface by incubating the cells in a reducing buffer (50 mM 2-sodium-2-mercapoethanesulfonate, 100 mM NaCl, 50 mM Tris-HCl, pH 8.7, and 2.5 mM CaCl) two times for 20 min each time at 4°C. Cells used for total cell surface biotin labeling were subjected to the same procedures as other cells except for the reducing step. The cells were lysed, and biotinylated proteins were immunoprecipitated with streptavidin.
Reverse transcription (RT)-PCR.
Rat1 fibroblasts were serum deprived for 48 h, after which the cells were stimulated with EGF (10 ng/ml) for 2 h. In cases where PP2 was used, the cells were pretreated with the inhibitor (10 μM) for 30 min before EGF stimulation. Total RNA was isolated using an RNAeasy kit (Qiagen) following the manufacturer's recommendations. Five micrograms of RNA was reverse transcribed using 800 U of Moloney murine leukemia virus reverse transcriptase and 3 μg of random primers in a 20-μl volume.
RESULTS
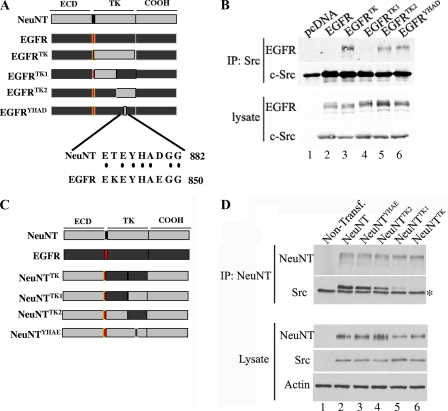
Mutation of EGFR Y845 to ErbB2 Y877 confers c-Src binding to EGFR.
Previous results from our laboratory demonstrated that c-Src specifically interacted with ErbB2 receptor (also referred as NeuNT, the activated rat isoform), but not with EGFR (24, 34). By using a chimeric-receptor approach, in which regions of ErbB2 were swapped into the EGFR, the region binding to c-Src was mapped to the kinase domain of ErbB2. This interaction can be further mapped to the C′-terminal part of the kinase domain (referred as TK2) (Fig. 1A). Previously published results had shown that c-Src association with ErbB2 was phosphotyrosine dependent, as well as SH2 mediated, and closer inspection of the TK2 region revealed the presence of 4 tyrosine residues. Of these 4 residues, only tyrosine (Y) 877 (Y845 in EGFR or Y869 in a different nomenclature) demonstrated a difference in the amino acids surrounding the tyrosine residue (see Fig. S1 in the supplemental material), including the +3 position, which is important for SH2 binding. To test whether residue Y877 was responsible for binding c-Src, the 2 amino acids flanking Y845 in EGFR that differed from those in NeuNT were mutated to the corresponding residues from ErbB2 (K843T and E848D, referred to as EGFRYHAD) (Fig. 1A). This receptor and an EGFR chimeric receptor harboring the TK, TK1, and TK2 regions of ErbB2 were transfected, along with c-Src, into 293T cells. Lysates from these transfected cells were subjected to coimmunoprecipitation using c-Src-specific antibody. Conversion of EGFR Y845 to ErbB2 Y877 was sufficient to confer binding of c-Src to EGFRYHAD (Fig. 1B, lane 6). The association of c-Src with EGFRYHAD was not as stringent as binding to EGFRTK (Fig. 1B, lanes 5 and 3, respectively), suggesting that residues located in the TK1 region might strengthen this association. Therefore, in addition to the YHAD sequence motif, other sequences within the ErbB2 catalytic domain likely contribute to the stability of c-Src binding.
FIG. 1.
Residues surrounding Y877 are responsible for binding c-Src. (A) Schematic representation of chimeric receptors derived from an EGFR backbone, where light gray represents NeuNT, red represents the transmembrane domain and black represents EGFR. The sequence flanking Y877 and Y845 is also depicted to highlight the 2 amino acids changed (no dots) in EGFRYHAD. ECD, extracellular domain. (B) 293T cells were transfected with c-Src and the indicated chimeric receptor. Lysates from these 293T transfected cells were subjected to coimmunoprecipitation using c-Src antibody (clone GD11; Upstate) and separated on SDS-PAGE. These immunoprecipitates (IP) (top) and total cell lysates (bottom) were immunoblotted and probed for the specified proteins. (C) Schematic representation of chimeric receptors derived from a NeuNT backbone. (D) 293T cells were transfected (Transf.) with c-Src and the indicated chimeric receptors. The lysates from the 293T cells transfected with the indicated constructs were subjected to coimmunoprecipitation using an ErbB2 antibody (AB4; Calbiochem) and separated on SDS-PAGE. Both the immunoprecipitates and the total cell lysates were immunoblotted and probed for the specified proteins. The asterisk denotes a nonspecific band associated with the antibody used for immunoprecipitations.
Residues within the TK1 region of ErbB2/NeuNT are involved in stabilization of c-Src binding.
To further identify the critical sequences within the ErbB2 catalytic domain responsible for stabilizing c-Src binding, we derived activated ErbB2 (NeuNT) chimeric receptors that contained the comparable regions of the EGFR catalytic domain. These chimeric receptors carried either the entire catalytic domain of EGFR (NeuNTTK), the N′-terminal region of the EGFR catalytic domain (NeuNTTK1), the C′-terminal domain of EGFR (NeuNTTK2), or the YHAE motif found surrounding Y845 in EGFR (NeuNTYHAE) (Fig. 1C). To assess whether the replacement of these regions in the ErbB2 catalytic domain affected recruitment of c-Src, we performed coimmunoprecipitation analyses with ErbB2/Neu- and c-Src-specific antibodies. The results revealed that substitution of either the TK1 region or the entire TK region of the ErbB2 catalytic domain resulted in a dramatic reduction in the capacity of c-Src to bind NeuNT (Fig. 1D, lanes 5 and 6).
To address whether the biological transforming activities of these chimeric receptors were altered as a consequence of the abrogation of c-Src binding, we tested the transforming potentials of the activated ErbB2 chimeric receptors on Rat1 fibroblasts. NeuNTTK demonstrated a twofold decrease in transformation efficiency (Table 1), which is significant given the strong transforming potential of NeuNT. On the other hand, NeuNTTK1 slightly increased transformation, which is reminiscent of results presented by Xu et al. (55), which demonstrated that mutating the HSP90 binding site (referred to as the M5 loop) in the TK1 region regulated phosphorylation of the C′-terminal tyrosine residues. Accordingly, phosphorylation of these residues was increased in the NeuNTTK1 construct (data not shown). Taken together, these observations show that sequences located within the Neu TK1 region participate in c-Src binding and further demonstrate that recruitment of c-Src to activated ErbB2 facilitates the morphological transformation of Rat1 fibroblasts.
TABLE 1.
Relative transformation abilities of ErbB2-derived constructs in Rat1 fibroblasts
| Construct | Focus assay 1 |
Focus assay 2 |
Relative transformation ability | ||
|---|---|---|---|---|---|
| Mean no. of foci ± SD | % Transformation of NeuNT | Mean no. of foci ± SD | % Transformation of NeuNT | ||
| NeuNT | 221 ± 15 | 100 | 289 ± 11 | 100 | 100 |
| NeuNTYHAE | 206 ± 13 | 93 | 277 ± 15 | 96 | 95 |
| NeuNTTK2 | 212 ± 17 | 96 | 295 ± 33 | 102 | 99 |
| NeuNTTK1 | 287 ± 28 | 130 | 354 ± 44 | 122 | 126 |
| NeuNTTK | 103 ± 19 | 47 | 169 ± 24 | 58 | 53 |
| NeuNTY882F | 211 ± 12 | 96 | 287 ± 21 | 99 | 98 |
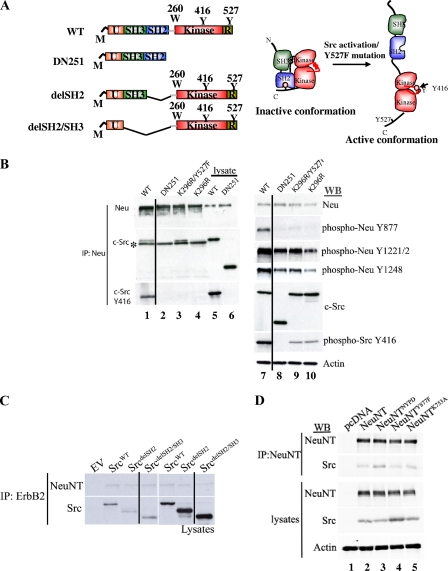
c-Src requires an open conformation to bind ErbB2 through its kinase domain.
Whereas the above-mentioned data demonstrate that the ErbB2 catalytic domain plays an instrumental role in mediating the association of c-Src, the sequence requirements within c-Src that are required for association with ErbB2 are unclear. To address this issue, we used a number of well-characterized c-Src mutations to identify the regions within c-Src that are required for binding to ErbB2. They include the SrcDN251 mutant, which is missing the kinase domain, as well as the inhibitory C′-terminal tail, but retains the SH2 and SH3 domain; the kinase-inactive c-SrcK296R mutant; and the DN c-SrcK296R/Y527F mutant, which is in an active conformation but is kinase inactive (Fig. 2A). To assess whether these mutations could impact c-Src capacity to bind ErbB2, we again performed coimmunoprecipitation analyses with both c-Src- and ErbB2-specific antisera. Despite the retention of SH3 and SH2, SrcDN251 was unable to interact with activated ErbB2 (Fig. 2B, lane 2). Conversely, c-Src mutants containing mutations that modified the specificity of c-Src SH2, SH3, or both domains still retained the ability to bind ErbB2 (see Fig. S2 in the supplemental material). Moreover, c-Src mutants in which the SH2 or SH2 and SH3 domains were removed (Fig. 2A) still retained the capacity to bind to NeuNT (Fig. 2C). Although the kinase-inactive c-SrcK296R mutant was not capable of associating with ErbB2 (Fig. 2B, lane 4), the c-SrcK296R/Y527F mutant was able to associate with ErbB2 (Fig. 2B, lane 3). Interestingly, tyrosine phosphorylation of Y877 in ErbB2 was prevented by these three mutated c-Src constructs, including SrcK296R/Y527F (Fig. 2B, lanes 8 to 10). Therefore, while residues surrounding Y877 mediated part of the binding to c-Src (Fig. 1B), the residue was not required to be phosphorylated to promote interaction, excluding an SH2-mediated interaction (Fig. 2B, lane 3, and C). Given that the c-SrcK296R/Y527F mutant is also kinase dead but possesses an open conformation, these results demonstrate that the association of c-Src with ErbB2 requires c-Src in an open conformation.
FIG. 2.
c-Src association with ErbB2 is likely mediated through the kinase domain. (A) Schematic representation of some of the c-Src constructs derived from the wild-type (WT) c-Src (right). The conformations of inactive and active c-Src are depicted on the left. M, myristylation; U, unique region; R, repression domain. (B) Several c-Src constructs were transfected along with activated ErbB2 in 293T cells. Lysates from these transfected cells were probed with the specified antibodies. A line was added to the blots to indicate that an extra lane was removed from the original image. The asterisk denotes a nonspecific band. (C) c-Src with SH2 or SH2/SH3 deleted (as depicted in panel A) was transfected with NeuNT, and the lysates were subjected to coimmunoprecipitation (IP) and probed with the indicated antibody. A line was added to the blots to indicate that an extra lane was removed from the original image. WB, Western blot. (D) NeuNT and kinase-inactive NeuNTK753A, NeuNTNYPD, and NeuNTY877F receptors were transfected in 293T cells along with c-Src. The lysates were subjected to coimmunoprecipitation and probed with the indicated antibodies.
Consistent with this model, mutation of tyrosine 877 within the YHAD motif in the NeuNT catalytic domain (NeuNTY877F) or a lysine residue within the ATP binding domain (NeuNTK753A) was unable to disrupt the ability of c-Src to bind NeuNT (Fig. 2D, lanes 4 and 5). Moreover, mutation of the five major tyrosine phosphorylation sites within the NeuNT C′-terminal domain (NeuNTNPYD) (Fig. 2D, lane 3) and a C′-terminally truncated NeuNT in which the five major tyrosine phosphorylation sites were deleted (data not shown) retained their capacities to associate with c-Src. Together with data from a c-Src SH2, a c-SrcdelSH2, and a c-SrcK296R/Y527F mutant, these observations argue that c-Src association with ErbB2 occurs independently of an SH2-phosphotyrosine interaction.
EGFRYHAD transforms Rat1 fibroblasts and Eph4 cells in two-dimensional and three-dimensional cell cultures.
To address whether the ability of EGFRYHAD to interact with c-Src provided transformation potential to EGFR, we performed a Rat1 focus assay. Rat1 fibroblasts were transfected with EGFR, EGFRTK, or EGFRYHAD and tested for the ability to form foci in the absence or presence of EGF (10 ng/ml). As depicted in Table 2, no foci were seen in the absence of EGF for the three receptors. In the presence of EGF, the transformation potential of EGFRTK and of EGFRYHAD was 24-fold and 4-fold, respectively, higher than for EGFR alone. This result suggests that the strength of c-Src association with the receptor is correlated with the ability to transform Rat1 fibroblasts. These observations contrast with the data showing that disruption of c-Src association from a constitutively activated ErbB2 receptor has a relatively modest effect on the specific transforming activity of activated ErbB2. This difference in behavior likely reflects the fact that the chimeric EGFRs are dependent on transient ligand activation whereas the chimeric activated ErbB2 receptors are constitutively activated due to mutation in the ErbB2 transmembrane domain.
TABLE 2.
Relative transformation abilities of EGFR-derived constructs in Rat1 fibroblasts
| Construct | Focus assay 1 |
Focus assay 2 |
Relative transformation ability | ||
|---|---|---|---|---|---|
| Mean no. of foci ± SD | Fold increase over EGFR | Mean no. of foci ± SD | Fold increase over EGFR | ||
| No EGF | |||||
| EGFR | 0 | NAa | 0 | NA | NA |
| EGFRTK | 0 | NA | 0 | NA | NA |
| EGFRYHAD | 0 | NA | 0 | NA | NA |
| EGF | |||||
| EGFR | 4 ± 1 | 1 | 3 ± 1 | 1 | 1 |
| EGFRTK | 75 ± 5 | 18.75 | 90 ± 8 | 30 | 24.38 |
| EGFRYHAD | 14 ± 3 | 3.5 | 14 ± 2 | 4.67 | 4.09 |
NA, not applicable.
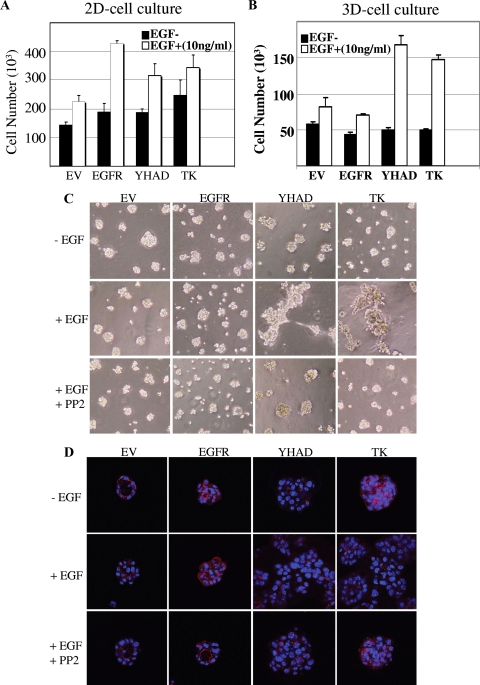
To identify whether the chimeric receptor EGFRYHAD was also able to promote transformation in a more physiological setting, we next examined whether expression of these chimeric EGFRs could perturb the ability of EpH4 mammary epithelial cells to form size-restricted spheroids on reconstituted basement membrane gels (Matrigel) (36). To accomplish this, EpH4 cells were transfected with EGFR, EGFRTK, or EGFRYHAD; selected with G418; and sorted using FACS against cell surface expression of the receptors to isolate a highly expressing cell population for each receptor (see Fig. S3 in the supplemental material). The sorted cells were then plated either as a monolayer in cell culture plates (two dimensional) or as spheroids on basement membrane gels (three dimensional) and tested for their proliferative capacities following EGF stimulation for 3 days. In two-dimensional cell culture, EGF was able to efficiently induce proliferation of EpH4 cells overexpressing EGFR, EGFRTK, or EGFRYHAD to a similar level (Fig. 3A). This increase in proliferation was lost when EGFR-expressing EpH4 cells were plated on Matrigel and allowed to form spheroid structures (Fig. 3B). In contrast, EpH4-EGFRTK and EpH4-EGFRYHAD cells displayed similar proliferative capacities following EGF stimulation when plated on Matrigel, suggesting that recruitment of c-Src to these receptors enabled them to maintain their EGF-dependent proliferative status under these three-dimensional cell culture conditions.
FIG. 3.
(A and B) Sorted EpH4 cell transfectants were plated as monolayers on tissue culture plastic (A) or as spheroids on basement membrane gels (B) and maintained in differentiation medium without or with 10 ng/ml EGF for 72 h. The monolayers and spheroids were then enzymatically dissociated, and single viable cells were counted (n = 3 for each condition; representative of two separate experiments). The error bars indicate standard deviations. EV, empty vector. (C and D) Cells were plated on Matrigel in differentiation medium, and spheroids were allowed to form for 5 days before they were treated with EGF (10 ng/ml) (+) or left untreated (−) in the absence or presence of the Src inhibitor PP2 (20 μM) for a further 48 h. The cultures were then imaged live by phase-contrast microscopy to assess morphology (C) or were immunostained for ZO-1 (red), followed by nuclear marking with DAPI (blue), and imaged by confocal microscopy (D).
The recruitment of c-Src to activated receptors also had profound effects on spheroid morphology in three-dimensional culture. Normally, EpH4 cells form small, cohesive, size-restricted spheroids of 20 to 40 cells on Matrigel, a process that was not affected by EGF treatment of either the vector control or EGFR-expressing cells (Fig. 3C and D). In contrast, in both EpH4-EGFRYHAD and EpH4-EGFRTK cells, EGF-mediated receptor activation led to the formation of large, disorganized clusters. This morphogenic effect was not associated with the total loss of cell-cell adhesion, as demonstrated by the membrane localization of the tight-junction-associated molecule ZO-1 under all conditions. However, the localization of ZO-1 became extremely disorganized after receptor activation in EpH4-EGFRTK and EpH4-EGFRYHAD cells (Fig. 3D), indicating a loss of epithelial polarity (2). The morphogenic effect of receptor activation in EpH4-EGFRTK and EpH4-EGFRYHAD cells was abrogated by the Src inhibitor PP2 (Fig. 3C and Fig. D), indicating that recruitment of c-Src played a causal role in the loss of epithelial polarity.
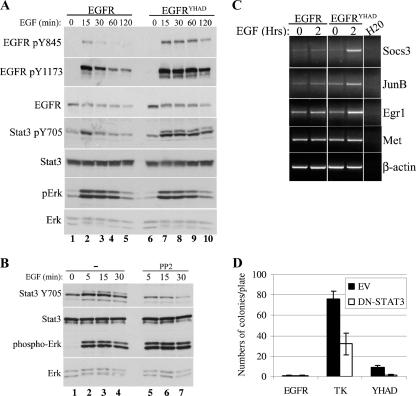
The increased transformation potential of EGFRYHAD is mediated by sustained EGFR signaling and specific activation of Stat3.
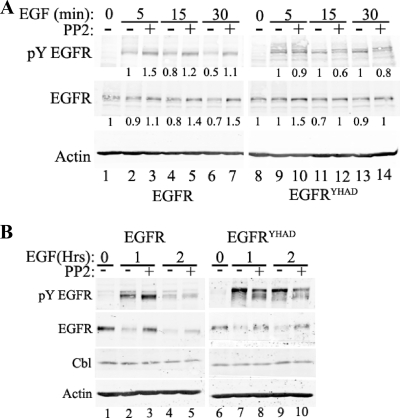
To identify the molecular mechanism responsible for transformation following EGF stimulation downstream of the EGFR chimeric receptor, we performed a series of immunoblot analyses with antibodies directed against a number of downstream signaling molecules on lysates derived from Rat1 cells expressing the different chimeric EGFRs. Examination of the phosphorylation status of EGFR at Y845 revealed that EGF stimulation of EGFR and EGFRYHAD led to a rapid increase in phosphorylation of Y845 (Fig. 4A, lanes 1 to 10). However, in contrast to wild-type EGFR, where tyrosine phosphorylation was rapidly downregulated (Fig. 4A, lanes 3 to 5), phosphorylation of this tyrosine residue in EGFRYHAD-expressing cells was sustained (Fig. 4A, lanes 8 to 10, and 5C, lane 9). To identify the signaling pathways differently activated downstream of EGFRYHAD, several of the known effector proteins activated downstream of EGFR and c-Src were probed using phosphospecific antibodies in a pooled population of EGFR- and EGFRYHAD-expressing Rat1 cells. The results revealed that EGFRYHAD-expressing cells possessed elevated levels of tyrosine phosphorylation on two of the EGFR autophosphorylation sites (Y845 and Y1173). In addition, increased tyrosine phosphorylation of Stat3 (Y705) was noted on both Stat3α (86-kDa) and Stat3β (79-kDa) isoforms (for example, Fig. 4A, compare lane 4 and lane 9). Examination of other key signaling molecules revealed that the phosphorylation levels of Erk (Fig. 4A), p38, PTEN, and Akt were comparable in the EGFR and EGFRYHAD cell lines (see Fig. S4A in the supplemental material). Because c-Src had previously been implicated in Stat3 activation (6, 9), we next assessed whether inhibition of Src with the Src family inhibitor PP2 would impact Stat3 phosphorylation (Fig. 4B). The results showed that PP2 inhibition led to a dramatic inhibition of Stat3α (and, to a lesser extent, Stat3β) phosphorylation (Fig. 4B, lane 3 and lane 6). This inhibition was highly specific to Stat3, since no difference in phosphorylation was seen for Erk (Fig. 4B). To confirm that an increase in phospho-Stat3 led to transcriptional activation of Stat3 target genes, RT-PCR was performed on total RNA isolated from EGFR- and EGFRYHAD-expressing cells 2 h post-EGF stimulation. Consistent with elevated levels of Stat3, an increase in the levels of the Stat3 Socs3, JunB, and Egr1 target gene mRNAs was noted in EGFRYHAD-expressing cells compared to the parental EGFR-expressing cells (Fig. 4C). Consistent with these transcriptional analyses, phospho-Stat3 was localized in the nuclei of EGFRYHAD-expressing cells (data not shown).
FIG. 4.
EGFRYHAD specifically activates Stat3 in Rat1 cells. (A) Cells were serum deprived for 48 h and stimulated with EGF (10 ng/ml) for the indicated times. The lysates from EGFR- and EGFRYHAD-expressing Rat1 fibroblasts were subjected to Western blotting and probed for the specified proteins. (B) Rat1 fibroblasts were serum deprived for 48 h and stimulated with EGF for the indicated times. In the c-Src inhibition experiment, cells were pretreated with PP2 (10 μM) for 30 min prior to EGF stimulation. (C) RT-PCR analysis of EGFR- and EGFRYHAD-expressing Rat1 fibroblasts. Rat1 fibroblasts were serum deprived for 48 h and stimulated with EGF for the indicated times. RNA was extracted using Qiagen RNeasy following the manufacturer's instructions and subjected to RT-PCR as described in Materials and Methods. (D) EGFR-, EGFRTK-, and EGFRYHAD-expressing Rat1 fibroblasts were subjected to a focus transformation assay as described in Materials and Methods. The cells were cotransfected with DN-Stat3, as well as the indicated construct. The cells were provided with 10 ng/ml EGF daily. The error bars indicate standard deviations. EV, empty vector.
To further assess the functional importance of Stat3 in EGFR-mediated transformation, we next determined whether expression of a DN mutant of Stat3 (DN-Stat3) could impact the transforming capacities of the chimeric EGFRs. The results showed that stable expression of DN-Stat3 resulted in 50% and 75% reduction in the number of foci for EGFRTK and EGFRYHAD, respectively (Fig. 4D). Last, we performed coimmunoprecipitation experiments to assess whether preferential activation of Stat3 by EGFRYHAD was caused by a difference in association with the receptor. No difference in association was seen between EGFR and EGFRYHAD (see Fig. S4B in the supplemental material). Collectively, these results demonstrate that EGFRYHAD does not specifically activate Stat3 through increased binding and that this activation is partly responsible for the increased transformation potential of the receptor.
One possible explanation for the sustained phosphorylation of EGFRYHAD is that, unlike the parental EGFR, which is rapidly degraded following EGF stimulation, the recruitment of c-Src stabilizes EGFRYHAD. To directly test whether sustained EGFR signaling in EGFRYHAD was due to prolonged stability of the mutant EGFRYHAD, we examined both the levels and state of activation of the EGFR mutant over a 2-h time course (Fig. 5). Consistent with the short-term EGF stimulation, Y845 phosphorylation levels of EGFRYHAD were enhanced in comparison to the wild-type EGFR (Fig. 5A, lane 6 versus lane 13, and B, lanes 4 and 9). Levels of EGFR were decreased by 50% after 30 min of EGF stimulation compared to no significant decrease for EGFRYHAD. Accordingly, the phosphorylation kinetics of Y845 was correlated with total levels for both receptors for up to 30 min (Fig. 5A).
FIG. 5.
EGFRYHAD promotes sustained signaling. (A and B) Stable Rat1 fibroblast populations expressing the indicated constructs were serum deprived for 48 h. The cells were then stimulated with 10 ng/ml of EGF for the indicated times. The lysates were subjected to immunoblotting and probed with the indicated antibodies. The numbers below the EGFR immunoblots are quantifications performed with the Licor Odyssey system and are averages of three different Western blots. In the case of total EGFR, serum deprivation (time zero) was set as a reference for the EGF stimulation time point, and 5 min of EGF stimulation was set as a reference for pY EGFR (Y845 phosphorylation). In the cases where PP2 was used, the cells were pretreated for 30 min with 10 μM of the inhibitor.
To evaluate whether the observed stabilization of EGFRYHAD involved activation of c-Src, we next assessed whether inhibition of c-Src activity could impact receptor stabilization. Surprisingly, the results showed that inhibition of Src activity with PP2 inhibitor actually increased the stability of both EGFR and EGFRYHAD (Fig. 5B). Despite the increase in levels of EGFRYHAD, tyrosine phosphorylation of residue 845 was suppressed by PP2 administration (Fig. 5B, lanes 8 and 10). These data demonstrate that the increase in tyrosine phosphorylation of residue 845 in EGFRYHAD was not due to the impact of c-Src on receptor stability. However, phosphorylation of Y845 in EGFR was not affected by PP2 (the slight increase in phosphorylation [Fig. 5B, lanes 2 and 3 versus lanes 7 and 8] was due to an increase in the receptor) while phosphorylation was greatly reduced by PP2 in EGFRYHAD, suggesting that in that receptor, but not in EGFR, Y845 might be phosphorylated by Src.
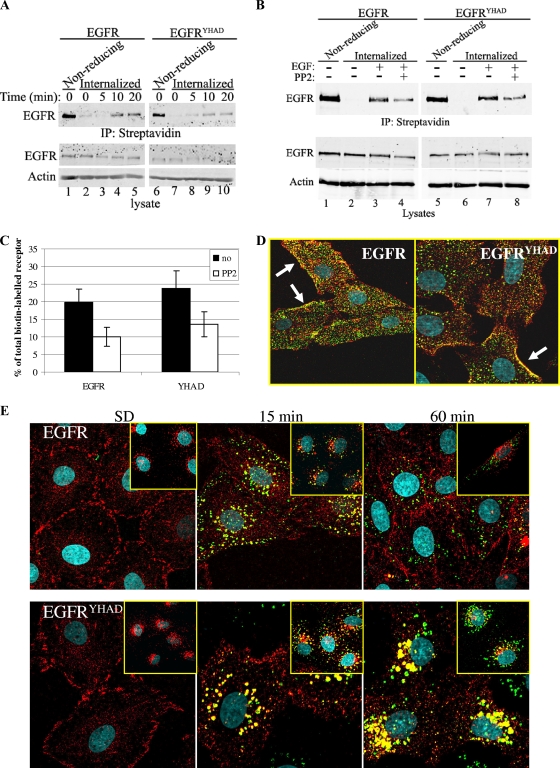
Another possible explanation for the increase in long-term signaling by EGFRYHAD is a difference in receptor internalization. To test the receptor internalization rate, we performed a biotin internalization assay in which cell surface EGFR was labeled with biotin and allowed to internalize following EGF stimulation. The results showed that the internalization rates of EGFR and EGFRYHAD did not differ significantly, as roughly 20% of the total biotinylated receptors were internalized following 20 min of EGF stimulation (Fig. 6A and C). Inhibition of Src kinase activity using PP2 decreased internalization by roughly 50% for both receptors (Fig. 6B and C). To further evaluate the relationship between activation of c-Src and receptor internalization, we followed the kinetics of receptor internalization and activation of c-Src with fluorescein-labeled EGF and phosphospecific antibodies directed against activated c-Src (Fig. 6D and E). Shortly after growth factor administration, these immunofluorescence studies revealed the presence of activated Src and EGF/EGFR complex at the plasma membrane in both EGFR- and EGFRYHAD-expressing cells (Fig. 6D). However, these immunofluorescence studies revealed a sustained complex of EGF/EGFR and activated c-Src within the perinuclear region of EGFRYHAD-expressing cells that was absent in the parental EGFR-expressing cells (Fig. 6E, 60-min time point). Although both activated c-Src/EGFR and c-Src/EGFRYHAD complexes were found in endosomes shortly after EGF stimulation (15 min), the perinuclear compartment where these complexes are found in EGFRYHAD-expressing cells appears to be distinct from the endosomal compartment, as determined by EEA1 staining (Fig. 6E, insets). Taken together, these data argue that the sustained signaling observed in EGFRYHAD-expressing cells is due to the stabilization of the c-Src/EGFRYHAD complex in a perinuclear compartment and not to a defect in receptor internalization.
FIG. 6.
(A, B, and C) Cells were subjected to a biotin internalization assay as described in Materials and Methods. Internalized biotinylated proteins were immunoprecipitated (IP) with streptavidin and probed with EGFR antibody. (B) PP2 (5 μM) was added 30 min before EGF stimulation, and the cells were stimulated for 20 min. (C) Percentages of internalized EGFR following 20 min of EGF stimulation. The numbers represent averages of three independent experiments, and the error bars indicate standard deviations. no, vehicle treated. (D and E) Stablly EGFR- and EGFRYHAD-expressing Rat1 fibroblasts were subjected to immunofluorescence assays with the indicated antibodies. (D) The arrows indicate activated Src and EGF/EGFR complex at the plasma membrane. (E) The cells were serum deprived (SD) for 48 h and stimulated with 50 ng/ml of fluorescein-coupled EGF for the indicated times. Red staining corresponds to phospho-Src Y416. (Insets) Fluorescein-conjugated EGF (green) and EEA1 (red) staining, an early endosome marker. Images were taken using a Zeiss LSM confocal microscope.
Mutants of EGFR identified in lung cancer interact with c-Src.
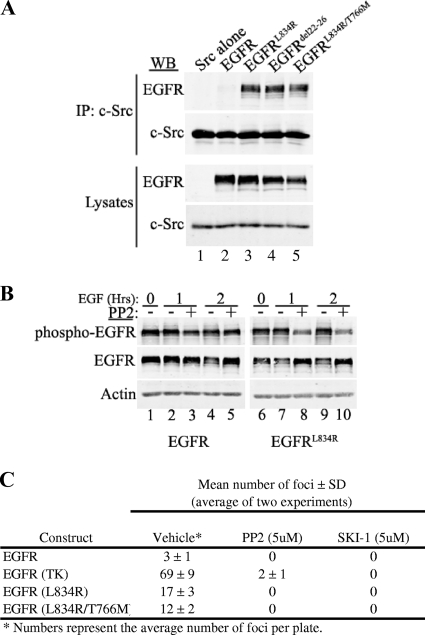
Recently, several somatic mutations have been identified in human lung adenocarcinomas. Of these mutations, L834R and Δ722-726 account for 80 to 90% of all mutations (these mutations are also referred to as L858R and Δ746-50 in a different numbering scheme) (45). These mutations are associated with greater sensitivity to gefitinib and erlotinib, two EGFR tyrosine kinase inhibitors (45). These EGFR mutants usually become resistant to the two drugs by acquiring another somatic mutation, T766M (also referred as T790M in a different numbering scheme), in more than 50% of resistant cases. Since these EGFR mutants shared several proprieties associated with EGFRYHAD activation, such as an increase in transformation potential, Stat3 activation, and a defect in degradation (1, 46), we next assessed whether these naturally occurring EGFR mutants had also acquired the capacity to associate with c-Src. To test this possibility, the mutations were engineered in an EGFR expression cassette, and these constructs were transfected into 293T cells and tested by coimmunoprecipitation analyses for the capacity to associate with c-Src. In contrast to parental EGFR, which could not associate with c-Src (Fig. 7A, lane 2), but like EGFRYHAD, these naturally occurring EGFR mutants had regained the capacity to associate with c-Src (Fig. 7A, lanes 3 to 5). The differences in c-Src binding were not due to levels of EGFR, as all of the cells expressed levels of mutant EGFR comparable to those of the wild-type receptor (Fig. 7A). We then tested whether, like EGFRYHAD, the phosphorylation of Y845 in the EGFRL834R mutant was dependent on Src activity by using PP2. Cells (293T) were transfected with EGFR or EGFRL834L, serum deprived for 16 h, and stimulated with 10 ng/ml of EGF. Cells expressing EGFRL834R and pretreated with PP2 demonstrated a significant decrease in Y845 phosphorylation after 1 and 2 h of EGF stimulation (Fig. 7B, lanes 8 and 10). No difference was seen for EGFR (Fig. 7B, lanes 3 and 5). Again, for EGFRL834R as for EGFRYHAD, Src inhibition slightly prevented receptor degradation (Fig. 7B, lanes 8 and 10). Collectively, these data imply that the EGFR variants have acquired the ability to recruit and that their activities are dependent on c-Src tyrosine kinase.
FIG. 7.
(A) EGFR somatic mutants identified in lung adenocarcinoma associate with c-Src. 293T cells were transfected with the indicated EGFR mutants, as well as wild-type c-Src. The lysates from these cells were subjected to coimmunoprecipitation (IP) experiments and probed for the indicated proteins. WB, Western blot. (B) 293T cells were transfected with EGFRTK or EGFRTK1, as well as c-Src. The cells were serum deprived for 24 h and stimulated with EGF (10 ng/ml) for the indicated time (0, 1, or 2 h). To inhibit Src activity, the cells were pretreated for 30 min with 10 μM of PP2 before EGF stimulation. Phospho-EGFR corresponds to phospho-Y845. (C) Rat1 transformation assay. The numbers are averages of two independent assays. Medium with or without PP2 or SKI-1 supplemented with 10 ng/ml of EGF was changed every 3 days. SD, standard deviation.
To assess the role of Src in the transformation potential of these EGFR mutants, we performed a Rat1 focus assay. Cells were transfected with EGFRTK, EGFRL834R, and EGFRL834R/T766M and allowed to grow in the presence of EGF (10 ng/ml) for 14 days with or without the Src inhibitor PP2 (5 μM). SKI-1 (5 μM), a more selective Src inhibitor, was also used to ensure that transformation inhibition was not mediated through a nonselective PP2 substrate. Both EGFRL834R and EGFRL834R/T766M induced Rat1 transformation, although the extent of transformation was much less than for EGFRTK (Fig. 7C). Both PP2 and SKI-1 completely inhibited Rat1 transformation by EGFRL834R and EGFRL834R/T766M, demonstrating that transformation by these EGFR mutants is Src dependent. This result has important therapeutic implications, since EGFRL834R/T766M demonstrates resistance to erlotinib and gefetinib treatment.
DISCUSSION
The biochemical data presented in this paper, as well as previously published results, suggest that association of c-Src with ErbB2 is conformation dependent. The kinase region of ErbB2 was previously demonstrated to be sufficient for c-Src binding to a chimeric EGFR containing the ErbB2 kinase domain (24). This catalytic domain likely contains all the residues required for this association, since a chimeric ErbB2 receptor containing the EGFR kinase region was unable to interact with c-Src (Fig. 1D, lane 6). Upon generation of chimeric EGFR containing either the N′-terminal (TK1) or the C′-terminal (TK2) portion of the ErbB2 kinase domain, association of c-Src was restricted to the TK2 region. EGFR and ErbB2 share over 95% homology at the amino acid level in this catalytic region, having only 16 divergent amino acids between the two receptors. Two of these residues flank the tyrosine residues Y877 in ErbB2 and Y845 in EGFR. Mutating these two residues in an EGFR background to mimic ErbB2 is sufficient to confer binding of c-Src to EGFR, although this association was weak compared to a chimeric EGFR containing the full kinase domain of ErbB2 (EGFRTK), suggesting that residues located in the TK1 part of the kinase domain are also important for association with c-Src. Surprisingly, while substituting the NeuNT TK region for the EGFR TK region completely abrogated Src binding, substituting only the TK2 region slightly reduced binding of c-Src. This result is puzzling because it suggests that in the context of the NeuNT backbone the TK1 region is sufficient for Src binding. At this point, the basis for this potential discrepancy is unknown.
The mutations identified in lung cancer, as well as the “c-Src binding” site identified in this paper by mimicking ErbB2 in the EGFR backbone, surround the region delineated by the activation loop and the αC helix making up the ATP binding pocket (7, 45). Recent structural studies have demonstrated that the L834R and G695S somatic mutations in EGFR also likely shift EGFR from an “inactive” to an “active” conformation equilibrium. Our results suggest that the combined lysine-to-threonine (K843T) and glutamic acid-to-aspartic acid (G848E) mutations also induce a similar opening of the ATP binding pocket, measured by an increase in Y845 phosphorylation. While c-Src activity seems to be required for phosphorylation of Y877 in ErbB2 (Fig. 2B) (55) and in EGFRYHAD (Fig. 5A and B), phosphorylation of Y845 in EGFR was not dependent on c-Src activity following EGF stimulation (Fig. 5A and B), which contrasts with previously reported results (4). Interestingly, phosphorylation of Y845 in EGFRL834R was also dependent on Src activity, raising the possibility that ErbB2, EGFRYHAD, and EGFRL834R are Src substrates. The association between c-Src and ErbB2 or EGFR mutants (EGFRYHAD and lung cancer mutants) is reminiscent of either a kinase binding to one of its substrates through a docking peptide interaction or two kinase dimers undergoing autophosphorylation through a mechanism known as activation segment exchange (37, 41). These models have never been described for c-Src, although Csk was reported to use a docking peptide interaction to phosphorylate c-Src Y527 (26). On the other hand, the activation segment exchange would explain how c-Src can phosphorylate a tyrosine residue (Y877, which would be located in the exchanged activation loop) that does not correspond to its consensus substrate sequence. However, this model requires a dimerization domain that allows the two kinases to orient and come into close proximity for the exchange to occur. Given our results, this would likely be in the TK1 region, since this domain greatly enhances binding between c-Src and chimeric EGFR or ErbB2. Future studies should allow this hypothesis to be validated.
c-Src comprises several well-described domains implicated in signal transduction: an SH2 domain, which binds phosphotyrosine residues; an SH3 domain, which binds proline-rich sequences; a highly conserved kinase domain; and a short cytoplasmic tail that harbors a negative regulatory tyrosine residue when phosphorylated (30). c-Src associates mostly with other kinases or adapter molecules involved in signal transduction through its SH2 and/or SH3 domain. In this study, the specificities of these two domains seem irrelevant in mediating the association of c-Src with ErbB2 or EGFRYHAD, since mutations that modified either SH2 or SH3 binding specificity (D99N and T215W) (11) were insufficient to alter binding. Moreover, removing the entire SH2 and SH3 domains did not affect the association of c-Src with ErbB2 (Fig. 2C). These results suggest that c-Src associates with ErbB2 independently of these two domains. Using various kinase-dead and DN mutants of c-Src, it seems that c-Src kinase activity itself is also not required for association with ErbB2 or EGFRYHAD. Surprisingly, while a kinase-dead mutant could not associate with ErbB2 or EGFRYHAD (K296R), a DN mutant (K296R/Y527F) could associate with these receptors. Based on the current structural model for c-Src activation, mutation of Y527F would release the looping of the C′-terminal tail and binding of that tail to the SH2 domain, opening up c-Src in an extended conformation and exposing its kinase domain (Fig. 2A) (5, 13). The fact that DN251, a c-Src construct that lacks the kinase domain but retains both the SH3 and SH2 domains, did not interact with these receptors strongly suggests that only the kinase domain is required for association with ErbB2 or EGFRYHAD, independently of its kinase activity. Moreover, since the K296R/Y527F Src mutant can productively associate with ErbB2 in the absence of phosphorylation of Y877 within ErbB2, this suggests that engagement of the phosphotyrosine is dispensable for the formation of the c-Src/ErbB2 complex. Although our group and others have previously reported that the c-Src SH2 domain can interact with phosphorylated ErbB2/NeuNT (28, 32, 34), these studies were mostly done in vitro with glutathione S-transferase-SH2, not with the full protein. Moreover, although these results claimed that c-Src binds ErbB2 through its SH2 domain, the specific tyrosine residue responsible for this association was never identified. In fact, mutating all the major autophosphorylation tyrosine residues located in the C′-terminal tail (Fig. 2D) or deleting the C′-terminal tail (data not shown) of ErbB2 does not abolish the association of c-Src with ErbB2, suggesting that an unidentified tyrosine residue promotes the binding of c-Src or that c-Src mainly binds ErbB2 through a non-SH2-mediated association (24). However, it is possible that c-Src interacts with ErbB2 through its SH2 domain, an interaction masked by a more stringent association through its kinase domain. The concept that c-Src can interact with ErbB family members or with one of its substrate proteins through its kinase domain has been noted previously. For example, it has been demonstrated that c-Src could associate with EGFR through its kinase domain (42, 43). c-Src has also been demonstrated to interact with PyMT through its kinase domain. In fact, the association of c-Src with PyMT is strikingly similar to its association with ErbB2, requiring an open conformation and its kinase domain to interact with PyMT (16). Overall, the results presented in this paper, along with previously published papers on the structure of EGFR and ErbB2, demonstrate that c-Src requires the kinase region of ErbB2 and mutated EGFRs for interaction.
Our data also suggest that the recruitment of c-Src by ErbB2 through the interaction of catalytic domains plays a critical role in its transforming activity. First, conversion of the 2 residues surrounding Y845 in EGFR to mimic Y877, the sequence context in ErbB2, was sufficient to confer ligand-dependent transforming activity (Table 2). Replacement of the EGFR catalytic domain with that of ErbB2 further enhanced the ligand-dependent transforming potential of EGFR. Significantly, this elevated transforming activity was further correlated with the stronger association of c-Src with the chimeric receptor and was dependent on Src activity (Fig. 7C). On the other hand, exchanging the TK domain of ErbB2/NeuNT for the EGFR TK domain was sufficient to decrease the transformation potential by 50%. Given that NeuNT is a very potent oncogene and that this chimeric NeuNTTK receptor can still recruit other adapters/kinases through its C′-terminal tyrosine residue and/or dimerize with other ErbB family members (Rat1 cells express EGFR), this 50% reduction in transformation potential is significant. It suggests that the full transformation potential of NeuNT is not uniquely dependent on c-Src binding. Consistent with the morphological transforming data, EpH4 mammary epithelial cells that expressed the chimeric EGFRs that were competent for c-Src binding resulted in ligand-dependent loss of epithelial cell polarity (Fig. 3C and D), an effect that was dependent on Src activity. The ligand-dependent loss of epithelial polarity in EpH4 cells was further associated with uncontrolled epithelial cell proliferation in cells plated on Matrigel (Fig. 3B). Taken together, these observations argue that recruitment of c-Src to chimeric EGFRs can modulate the transforming potential of these receptors.
While these studies provide compelling evidence that recruitment of c-Src can dramatically modulate the transforming potential of EGFR, the molecular mechanism by which this occurs remains to be elucidated. One potential clue to how this might occur derives from the differences in the trafficking of the chimeric receptor and the parental EGFR. Following ligand stimulation, EGFR is internalized via clathrin-dependent endocytosis into early endosomes, an effect mediated by the binding of c-Cbl to phosphorylated Y1045 and subsequent multiple monoubiquitination of the receptor (48). Deregulated c-Src activity promotes c-Cbl degradation, resulting in stabilization of EGFR at the cell surface (25). Surprisingly, in our hands, Src activity seemed responsible for degradation of both EGFR and EGFRYHAD (Fig. 5A and B). This function of Src activity has been reported before for other cell types (53). Moreover, our studies suggest that while recruitment of c-Src to EGFR results in receptor activation, the internalization rates were similar in EGFR and EGFRYHAD (Fig. 6A to E). In addition, molecular analyses have revealed that despite being rapidly internalized, like the parental EGFR, the chimeric receptors are still productively tyrosine phosphorylated and likely still signal (Fig. 6). The capacity of c-Src to associate with the chimeric receptor is likely responsible for this enhanced tyrosine phosphorylation, since inhibiting c-Src activity decreased receptor phosphorylation (Fig. 5A and B). In addition, c-Src has been reported to be enriched on endosomal membranes (23) and EGFR has been reported to signal from endosomes (8), suggesting that both EGFRYHAD and c-Src could still signal from that compartment.
Another important signaling difference between EGFR and EGFRYHAD is that the latter resulted in preferential and sustained tyrosine phosphorylation of Stat3 (Fig. 4A). Stat3 activation was mediated by c-Src, and this activation was required for transformation by EGFRYHAD (Fig. 4B to D). Preferential activation of Stat3 was not caused by a selective association with the chimeric receptor, since EGFR and EGFRYHAD bound Stat3 to the same extent (see Fig. S4B in the supplemental material). Consistent with the importance of Stat3 in oncogenic signaling by these chimeric receptors, expression of DN-Stat3 can dramatically impair their transforming potentials (Fig. 4D). These data are consistent with previous studies that suggested that ErbB2-induced tumor maintenance is dependent on Stat3 activation (20, 50). Sustained signaling does not explain the preferential activation of Stat3, since this event occurs as early as 5 min. At this point, the mechanism responsible for this preferential activation remains unresolved.
The observation that EGFRYHAD was able to preferentially activate Stat3 and had increased phosphorylation following activation is strikingly similar to the behavior of EGFR somatic mutants found in non-small-cell lung carcinoma (1, 46). In all of these cases, any mutation near the activation loop of EGFR that favors the active conformation seems to promote the association of c-Src with the receptor (56). It is conceivable that the ability of these mutant EGFRs to recruit c-Src is in part responsible for their tumorigenic potential, since pharmacological inhibitors of Src prevented transformation by these mutants (Fig. 7C) (12). Strikingly, inhibition of Src activity also prevented transformation by an erlotinib- and gefitinib-resistant T766M EGFR mutant (Fig. 7C), suggesting that a potential strategy to treat EGFR mutant-driven lung cancers might benefit from Src kinase activity inhibition. Identification of the molecular basis for this phenomenon could lead to the development of improved therapeutic strategies that would potentially include Src inhibitors.
Supplementary Material
Acknowledgments
We acknowledge support from CIHR/CBCRA grant 015420. In addition, we acknowledge support from a Terry Fox Foundation Team Grant, the NIH Mouse Models of Human Cancer Consortium, and the DOD Centers of Excellence. W.J.M. was supported by a CRC Chair in Molecular Oncology.
Footnotes
Published ahead of print on 24 August 2009.
Supplemental material for this article can be found at http://mcb.asm.org/.
REFERENCES
- 1.Alvarez, J. V., H. Greulich, W. R. Sellers, M. Meyerson, and D. A. Frank. 2006. Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor. Cancer Res. 66:3162-3168. [DOI] [PubMed] [Google Scholar]
- 2.Arakaki, N., T. Nagao, R. Niki, A. Toyofuku, H. Tanaka, Y. Kuramoto, Y. Emoto, H. Shibata, K. Magota, and T. Higuti. 2003. Possible role of cell surface H+-ATP synthase in the extracellular ATP synthesis and proliferation of human umbilical vein endothelial cells. Mol. Cancer Res. 1:931-939. [PubMed] [Google Scholar]
- 3.Belsches, A. P., M. D. Haskell, and S. J. Parsons. 1997. Role of c-Src tyrosine kinase in EGF-induced mitogenesis. Front. Biosci. 2:d501-d518. [DOI] [PubMed] [Google Scholar]
- 4.Biscardi, J. S., M. C. Maa, D. A. Tice, M. E. Cox, T. H. Leu, and S. J. Parsons. 1999. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J. Biol. Chem. 274:8335-8343. [DOI] [PubMed] [Google Scholar]
- 5.Boggon, T. J., and M. J. Eck. 2004. Structure and regulation of Src family kinases. Oncogene 23:7918-7927. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg, J. F., C. M. Horvath, D. Besser, W. W. Lathem, and J. E. Darnell, Jr. 1998. Stat3 activation is required for cellular transformation by v-src. Mol. Cell Biol. 18:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess, A. W., H. S. Cho, C. Eigenbrot, K. M. Ferguson, T. P. Garrett, D. J. Leahy, M. A. Lemmon, M. X. Sliwkowski, C. W. Ward, and S. Yokoyama. 2003. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell 12:541-552. [DOI] [PubMed] [Google Scholar]
- 8.Burke, P., K. Schooler, and H. S. Wiley. 2001. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell 12:1897-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, X., A. Tay, G. R. Guy, and Y. H. Tan. 1996. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol. Cell Biol. 16:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cartwright, C. A., M. P. Kamps, A. I. Meisler, J. M. Pipas, and W. Eckhart. 1989. pp60c-src activation in human colon carcinoma. J. Clin. Investig. 83:2025-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cary, L. A., R. A. Klinghoffer, C. Sachsenmaier, and J. A. Cooper. 2002. SRC catalytic but not scaffolding function is needed for integrin-regulated tyrosine phosphorylation, cell migration, and cell spreading. Mol. Cell. Biol. 22:2427-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung, B. M., M. Dimri, M. George, A. L. Reddi, G. Chen, V. Band, and H. Band. 2009. The role of cooperativity with Src in oncogenic transformation mediated by non-small cell lung cancer-associated EGF receptor mutants. Oncogene 28:1821-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowan-Jacob, S. W., G. Fendrich, P. W. Manley, W. Jahnke, D. Fabbro, J. Liebetanz, and T. Meyer. 2005. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure 13:861-871. [DOI] [PubMed] [Google Scholar]
- 14.Di Fiore, P. P., J. H. Pierce, T. P. Fleming, R. Hazan, A. Ullrich, C. R. King, J. Schlessinger, and S. A. Aaronson. 1987. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell 51:1063-1070. [DOI] [PubMed] [Google Scholar]
- 15.Di Fiore, P. P., J. H. Pierce, M. H. Kraus, O. Segatto, C. R. King, and S. A. Aaronson. 1987. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science 237:178-182. [DOI] [PubMed] [Google Scholar]
- 16.Dunant, N. M., M. Senften, and K. Ballmer-Hofer. 1996. Polyomavirus middle-T antigen associates with the kinase domain of Src-related tyrosine kinases. J. Virol. 70:1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frame, M. C. 2004. Newest findings on the oldest oncogene; how activated src does it. J. Cell Sci. 117:989-998. [DOI] [PubMed] [Google Scholar]
- 18.Grandal, M. V., R. Zandi, M. W. Pedersen, B. M. Willumsen, B. van Deurs, and H. S. Poulsen. 2007. EGFRvIII escapes down-regulation due to impaired internalization and sorting to lysosomes. Carcinogenesis 28:1408-1417. [DOI] [PubMed] [Google Scholar]
- 19.Gullick, W. J. 2002. A new model for ductal carcinoma in situ suggests strategies for treatment. Breast Cancer Res. 4:176-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, W., Y. Pylayeva, A. Pepe, T. Yoshioka, W. J. Muller, G. Inghirami, and F. G. Giancotti. 2006. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell 126:489-502. [DOI] [PubMed] [Google Scholar]
- 21.Guy, C. T., S. K. Muthuswamy, R. D. Cardiff, P. Soriano, and W. J. Muller. 1994. Activation of the c-Src tyrosine kinase is required for the induction of mammary tumors in transgenic mice. Genes Dev. 8:23-32. [DOI] [PubMed] [Google Scholar]
- 22.Hollstein, M. C., A. M. Smits, C. Galiana, H. Yamasaki, J. L. Bos, A. Mandard, C. Partensky, and R. Montesano. 1988. Amplification of epidermal growth factor receptor gene but no evidence of ras mutations in primary human esophageal cancers. Cancer Res. 48:5119-5123. [PubMed] [Google Scholar]
- 23.Kaplan, K. B., J. R. Swedlow, H. E. Varmus, and D. O. Morgan. 1992. Association of p60c-src with endosomal membranes in mammalian fibroblasts. J. Cell Biol. 118:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, H., R. Chan, D. L. Dankort, D. Zuo, M. Najoukas, M. Park, and W. J. Muller. 2005. The c-Src tyrosine kinase associates with the catalytic domain of ErbB-2: implications for ErbB-2 mediated signaling and transformation. Oncogene 24:7599-7607. [DOI] [PubMed] [Google Scholar]
- 25.Kirisits, A., D. Pils, and M. Krainer. 2007. Epidermal growth factor receptor degradation: an alternative view of oncogenic pathways. Int. J. Biochem. Cell Biol. 39:2173-2182. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S., X. Lin, N. H. Nam, K. Parang, and G. Sun. 2003. Determination of the substrate-docking site of protein tyrosine kinase C-terminal Src kinase. Proc. Natl. Acad. Sci. USA 100:14707-14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linggi, B., and G. Carpenter. 2006. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 16:649-656. [DOI] [PubMed] [Google Scholar]
- 28.Luttrell, D. K., A. Lee, T. J. Lansing, R. M. Crosby, K. D. Jung, D. Willard, M. Luther, M. Rodriguez, J. Berman, and T. M. Gilmer. 1994. Involvement of pp60c-src with two major signaling pathways in human breast cancer. Proc. Natl. Acad. Sci. USA 91:83-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maa, M. C., T. H. Leu, D. J. McCarley, R. C. Schatzman, and S. J. Parsons. 1995. Potentiation of epidermal growth factor receptor-mediated oncogenesis by c-Src: implications for the etiology of multiple human cancers. Proc. Natl. Acad. Sci. USA 92:6981-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, G. S. 2004. The road to Src. Oncogene 23:7910-7917. [DOI] [PubMed] [Google Scholar]
- 31.Muthuswamy, S. K., D. Li, S. Lelievre, M. J. Bissell, and J. S. Brugge. 2001. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat. Cell Biol. 3:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muthuswamy, S. K., and W. J. Muller. 1995. Activation of Src family kinases in Neu-induced mammary tumors correlates with their association with distinct sets of tyrosine phosphorylated proteins in vivo. Oncogene 11:1801-1810. [PubMed] [Google Scholar]
- 33.Muthuswamy, S. K., and W. J. Muller. 1994. Activation of the Src family of tyrosine kinases in mammary tumorigenesis. Adv. Cancer Res. 64:111-123. [DOI] [PubMed] [Google Scholar]
- 34.Muthuswamy, S. K., and W. J. Muller. 1995. Direct and specific interaction of c-Src with Neu is involved in signaling by the epidermal growth factor receptor. Oncogene 11:271-279. [PubMed] [Google Scholar]
- 35.Muthuswamy, S. K., P. M. Siegel, D. L. Dankort, M. A. Webster, and W. J. Muller. 1994. Mammary tumors expressing the neu proto-oncogene possess elevated c-Src tyrosine kinase activity. Mol. Cell. Biol. 14:735-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niemann, C., V. Brinkmann, E. Spitzer, G. Hartmann, M. Sachs, H. Naundorf, and W. Birchmeier. 1998. Reconstitution of mammary gland development in vitro: requirement of c-met and c-erbB2 signaling for branching and alveolar morphogenesis. J. Cell Biol. 143:533-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver, A. W., S. Knapp, and L. H. Pearl. 2007. Activation segment exchange: a common mechanism of kinase autophosphorylation? Trends Biochem. Sci. 32:351-356. [DOI] [PubMed] [Google Scholar]
- 38.Ottenhoff-Kalff, A. E., G. Rijksen, E. A. van Beurden, A. Hennipman, A. A. Michels, and G. E. Staal. 1992. Characterization of protein tyrosine kinases from human breast cancer: involvement of the c-src oncogene product. Cancer Res. 52:4773-4778. [PubMed] [Google Scholar]
- 39.Pinkas, J., and P. Leder. 2002. MEK1 signaling mediates transformation and metastasis of EpH4 mammary epithelial cells independent of an epithelial to mesenchymal transition. Cancer Res. 62:4781-4790. [PubMed] [Google Scholar]
- 40.Reichmann, E., R. Ball, B. Groner, and R. R. Friis. 1989. New mammary epithelial and fibroblastic cell clones in coculture form structures competent to differentiate functionally. J. Cell Biol. 108:1127-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remenyi, A., M. C. Good, and W. A. Lim. 2006. Docking interactions in protein kinase and phosphatase networks. Curr. Opin. Struct. Biol. 16:676-685. [DOI] [PubMed] [Google Scholar]
- 42.Sato, K., A. Sato, M. Aoto, and Y. Fukami. 1995. c-Src phosphorylates epidermal growth factor receptor on tyrosine 845. Biochem. Biophys. Res. Commun. 215:1078-1087. [DOI] [PubMed] [Google Scholar]
- 43.Sato, K., A. Sato, M. Aoto, and Y. Fukami. 1995. Site-specific association of c-Src with epidermal growth factor receptor in A431 cells. Biochem. Biophys. Res. Commun. 210:844-851. [DOI] [PubMed] [Google Scholar]
- 44.Semba, K., N. Kamata, K. Toyoshima, and T. Yamamoto. 1985. A v-erbB-related protooncogene, c-erbB-2, is distinct from the c-erbB-1/epidermal growth factor-receptor gene and is amplified in a human salivary gland adenocarcinoma. Proc. Natl. Acad. Sci. USA 82:6497-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma, S. V., D. W. Bell, J. Settleman, and D. A. Haber. 2007. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7:169-181. [DOI] [PubMed] [Google Scholar]
- 46.Shtiegman, K., B. S. Kochupurakkal, Y. Zwang, G. Pines, A. Starr, A. Vexler, A. Citri, M. Katz, S. Lavi, Y. Ben-Basat, S. Benjamin, S. Corso, J. Gan, R. B. Yosef, S. Giordano, and Y. Yarden. 2007. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene 26:6968-6978. [DOI] [PubMed] [Google Scholar]
- 47.Slamon, D. J., G. M. Clark, S. G. Wong, W. J. Levin, A. Ullrich, and W. L. McGuire. 1987. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235:177-182. [DOI] [PubMed] [Google Scholar]
- 48.Sorkin, A., and L. K. Goh. 2008. Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 314:3093-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spencer, K. S., D. Graus-Porta, J. Leng, N. E. Hynes, and R. L. Klemke. 2000. ErbB2 is necessary for induction of carcinoma cell invasion by ErbB family receptor tyrosine kinases. J. Cell Biol. 148:385-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan, M., K. H. Lan, J. Yao, C. H. Lu, M. Sun, C. L. Neal, J. Lu, and D. Yu. 2006. Selective inhibition of ErbB2-overexpressing breast cancer in vivo by a novel TAT-based ErbB2-targeting signal transducers and activators of transcription 3-blocking peptide. Cancer Res. 66:3764-3772. [DOI] [PubMed] [Google Scholar]
- 51.Tice, D. A., J. S. Biscardi, A. L. Nickles, and S. J. Parsons. 1999. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc. Natl. Acad. Sci. USA 96:1415-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verbeek, B. S., T. M. Vroom, S. S. Adriaansen-Slot, A. E. Ottenhoff-Kalff, J. G. Geertzema, A. Hennipman, and G. Rijksen. 1996. c-Src protein expression is increased in human breast cancer. An immunohistochemical and biochemical analysis. J. Pathol. 180:383-388. [DOI] [PubMed] [Google Scholar]
- 53.Ware, M. F., D. A. Tice, S. J. Parsons, and D. A. Lauffenburger. 1997. Overexpression of cellular Src in fibroblasts enhances endocytic internalization of epidermal growth factor receptor. J. Biol. Chem. 272:30185-30190. [DOI] [PubMed] [Google Scholar]
- 54.Webster, M. A., R. D. Cardiff, and W. J. Muller. 1995. Induction of mammary epithelial hyperplasias and mammary tumors in transgenic mice expressing a murine mammary tumor virus/activated c-src fusion gene. Proc. Natl. Acad. Sci. USA 92:7849-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, W., X. Yuan, K. Beebe, Z. Xiang, and L. Neckers. 2007. Loss of Hsp90 association up-regulates Src-dependent ErbB2 activity. Mol. Cell. Biol. 27:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, S., K. Park, J. Turkson, and C. L. Arteaga. 2008. Ligand-independent phosphorylation of Y869 (Y845) links mutant EGFR signaling to stat-mediated gene expression. Exp. Cell Res. 314:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yatabe, Y., and T. Mitsudomi. 2007. Epidermal growth factor receptor mutations in lung cancers. Pathol. Int. 57:233-244. [DOI] [PubMed] [Google Scholar]
- 58.Zhau, H. Y., R. J. Babaian, and S. J. Hong. 1990. A new 180 kDa urine protein marker associated with bladder cancer. J. Urol. 144:47-52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.