Abstract
Ionizing radiation (IR) is a physiologically important stress to which cells respond by the activation of multiple signaling pathways. Using a panel of immortalized and transformed breast epithelial cell lines, we demonstrate that IR regulation of protein synthesis occurs in nontransformed cells and is lost with transformation. In nontransformed cells, IR rapidly activates the MAP kinases ERK1/2, resulting in an early transient increase in cap-dependent mRNA translation that involves mTOR and is radioprotective, enhancing the translation of a subset of mRNAs encoding proteins involved in DNA repair and cell survival. Following a transient increase in translation, IR-sensitive (nontransformed) cells inhibit cap-dependent protein synthesis through a mechanism that involves activation of p53, induction of Sestrin 1 and 2 genes, and stimulation of AMP kinase, inhibiting mTOR and hypophosphorylating 4E-BP1. IR is shown to block proteasome-mediated decay of 4E-BP1, increasing its abundance and the sequestration of eIF4E. The IR signal that impairs mTOR-dependent protein synthesis at late times is assembly of the DNA damage response machinery, consisting of Mre11, Rad50, and NBS1 (MRN); activation of the MRN complex kinase ATM; and p53. These results link genotoxic signaling from the DNA damage response complex to the control of protein synthesis.
Studies have shown that ionizing radiation (IR) alters gene expression much more profoundly at the level of mRNA translation than at transcription (44). This may reflect the fact that up to 40% of the total energy requirement of the cell is invested in protein synthesis (5, 46). Protein synthesis is highly regulated for that reason and is acutely responsive to growth and stress stimuli, thereby coupling mRNA translation activity to the metabolic demands on the cell.
There is very little mechanistic or regulatory understanding of translational control by IR in transformed and nontransformed mammalian cells. High doses of IR have been reported to inhibit overall protein synthesis in highly transformed cells by acting on the cap initiation complex (36) or by reducing levels of initiation factor eIF4G (51), a member of the complex. The cap initiation complex consists of three core proteins: eIF4E, a protein that binds the 7-methyl-GTP (m7GTP) 5′ capped end of the mRNA and promotes assembly of the complex with ribosomes; the molecular scaffold eIF4G, upon which initiation factors and 40S ribosome subunits assemble; and the ATP-dependent RNA helicase eIF4A. Most mRNAs are translated in mammalian cells through a cap-dependent mechanism and are thought to use the cap initiation complex to recruit and assemble scanning ribosomes. IR reportedly disrupts cap initiation complexes by activation of the eIF4E-inhibitory protein 4E-BP1 (36, 51). 4E-BP1 is typically inactivated by phosphorylation carried out by the protein kinase mTOR, but under stress conditions, mTOR activity can be inhibited, thereby blocking cap-dependent mRNA translation. 4E-BP1 and ribosomal S6 kinase (S6K) are direct targets of mTOR, which phosphorylates and inactivates 4E-BP1, phosphorylates and activates S6K, and stimulates cap-dependent mRNA translation (27). 4E-BP1 activity is therefore controlled by mTOR kinase activity, which is in turn regulated by the upstream phosphatidylinositol 3-kinase/Akt pathway (27). The activity of 4E-BP1 can also be controlled by the p38-MAP kinase-extracellular signal-regulated kinase (ERK) pathway (3, 7). Both mTOR and ERK inhibit 4E-BP1 through phosphorylation of its two primary regulatory sites, Thr70 and Ser65 (22, 30).
Studies have not systematically investigated the IR response to and mechanism of action on protein synthesis in nontransformed cells, which are considerably more radioresponsive than highly transformed cells. It has been reported that transformed cells undergo no change in overall protein synthesis with irradiation (63). Notably, transformed cells are thought to be considerably more radioresistant, necessitating the very high doses of IR (20 to 50 Gy) that were used in many studies to achieve measurable inhibition of protein synthesis. Several studies in mouse embryo fibroblast cultures showed that IR can activate mTOR, but the mechanism and impact on protein synthesis were not investigated. This is of particular interest, because mTOR inhibitors, such as rapamycin, have been shown to be effective sensitizers of IR-mediated cancer cell killing and to increase damage to the tumor vasculature (2, 6, 16, 59). Here, we report the surprising finding that irradiation of immortalized breast epithelial cells immediately and transiently stimulates protein synthesis through activation of ERK and increased mTOR activity. We demonstrate that nontransformed cells subsequently inhibit cap-dependent protein synthesis in an IR dose-dependent manner through assembly of the DNA damage response apparatus, comprised of Mre11, Rad50, and NBS1 (the MRN complex), and activation of ATM and p53, resulting in activation of the p53 Sestrin proteins, activation of AMP kinase (AMPK), and inhibition of mTOR. In nontransformed cells, this also results in novel stabilization of the 4E-BP1 protein against proteasome decay, which blocks mRNA translation. These studies elucidate a complex regulatory pathway for the control of protein synthesis that is unique to IR and is lost with transformation in breast cancer cells.
MATERIALS AND METHODS
Cell culture.
Human breast cancer cell lines were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). The cell lines consisted of MCF10A immortalized breast epithelial cells, UACC-893 cells derived from a primary stage 2 invasive ductal carcinoma (IDC) (grade 3; Her2/neu+ p53+ estrogen receptor [ER]/PR−); HCC70 cells derived from a stage 3A primary IDC (grade 3; Her2/neu− p53+ ER+/PR−), HCC1395 cells derived from a stage 1 primary 1DC (grade 3; Her2/neu− p53+ ER+/PR−), and BT474 cells derived from a stage 4 IDC (grade 3; p53+ ER+/PR−). The cells were grown under the guidelines of the ATCC in their recommended media. The cells were transfected with DNA plasmids using Fugene reagent as described by the manufacturer (Roche).
Irradiation, hypoxia, and etoposide treatments.
Hypoxic culture conditions (0.5% O2, 5% CO2) for up to 24 h were used, and all experiments were performed as described previously (8). The cells were irradiated with a Varian Linac 2300 linear accelerator at a dose rate of 22 Gy/min at room temperature for the doses shown. Mock-treated cells were handled identically, except for irradiation. Etoposide was added at 34 μM to the cultures for the times indicated in the text.
[35S]methionine incorporation assay.
Cells were labeled with 50 μCi of [35S]methionine/cysteine per ml (Easytag Express Protein Labeling Mix; Dupont/NEN) in methionine/cysteine-free Dulbecco's modified Eagle's medium for 1 h, and lysates were prepared as described previously (8). The specific activity of 35S incorporation was determined by trichloroacetic acid precipitation onto GF/C filters and liquid scintillation counting.
Antibodies and immunoblot analysis.
Rabbit polyclonal antiserum to eIF4G was described previously (12). Mouse monoclonal anti-eIF4A antibody was provided by W. Merrick (Case Western Reserve University, Cleveland, OH). Other antisera—rabbit polyclonal anti-eIF2αP (Ser51) antibody (Biosource) and rabbit polyclonal anti-eIF2α antibody (Santa Cruz Biotechnology)—were from commercial sources. All other antibodies were from Cell Signaling Technology. An enhanced-chemiluminescence system (Amersham) was used for detection. Following treatments, the cells were washed twice in ice-cold phosphate-buffered saline, lysed in 0.5% NP-40 lysis buffer at 4°C, and clarified by centrifugation at 13,000 × g for 10 min. Protein concentrations were determined for each sample by Bradford assay (Bio-Rad, Hercules, CA). To determine the total levels and phosphorylation statuses of specific proteins, equal amounts of protein from NP-40 lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by protein immunoblotting with specific antibodies. The phosphorylation status of 4E-BP1 using electrophoretic migration was determined by electrophoresis in SDS-15% PAGE, whereas 8% gels were used to determine total 4E-BP1 levels. The phosphorylation statuses of most proteins were determined by first immunoblotting membranes with phosphospecific antibody and then stripping the membranes using Restore Western blot stripping buffer (Pierce), followed by reprobing the membranes with nonphosphospecific antibodies.
Analysis of eIF4E and 4E-BP1 interaction.
Equal amounts of protein from NP-40 cell lysates were incubated with m7GTP-Sepharose 4B (30 μl of settled bed volume) for 2 h at 4°C. The pelleted beads were washed and eluted as described previously (8). The pellet was solubilized in 1× SDS sample buffer and heated to 37°C for 20 min before being boiled and analyzed by SDS-PAGE and immunoblotting.
Retroviral and lentivirus expression studies.
Constitutively active Flag-GADD34 C-terminal protein fragment (A1) cloned into pBABE-puro, a retrovirus expression vector, was provided by D. Ron (NYU Medical School, New York, NY) (49). Cloning of 4E-BP1 expression vector into the pBABE vector and transformation of cells with vectors were previously described (8). Interfering RNAs were delivered by transduction of cells with lentivirus short hairpin RNA (shRNA) expression vectors. Double-stranded shRNAs for cloning into lentivirus vectors were directed to either the 5′ or 3′ untranslated regions of mRNAs targeted for gene silencing. To produce virus containing the shRNA-generating cassette, 293GP cells were transfected with 4 μg each of pCI-VSV-G, pCMVD8.2R′, and pLK0.1 with the use of Fugene (Roche). Target cells were infected in the presence of Polybrene (4 mg/ml) and selected with puromycin for 48 h. Retrovirus-containing supernatants were harvested after 48 h, passed through a 0.45-μm filter, and frozen at −80°C until they were used. Target cells were subsequently infected by the addition of retrovirus-containing supernatants to the medium, along with Polybrene.
Single-step real-time qRT-PCR.
Quantitative reverse transcription (qRT)-PCR analysis of mRNAs was performed using the SYBR green QRT-PCR Kit (Sigma) in a Lightcycler instrument (Roche). RT was carried out at 61°C for 20 min and denaturation at 95°C for 30 s, followed by 45 cycles of amplification: 95°C for 2 s for denaturation, 59°C for 5 s for annealing, and 72°C for 10 s for amplification and acquisition. The primer sets employed for specific gene quantification were designed using Roche's RT-PCR primer design tool.
Statistical analysis.
Statistical analyses used the two-tailed Student t test, with a P value of <0.05 taken as significant.
RESULTS
Nontransformed breast epithelial cells exhibit a scalable biphasic translational response to IR.
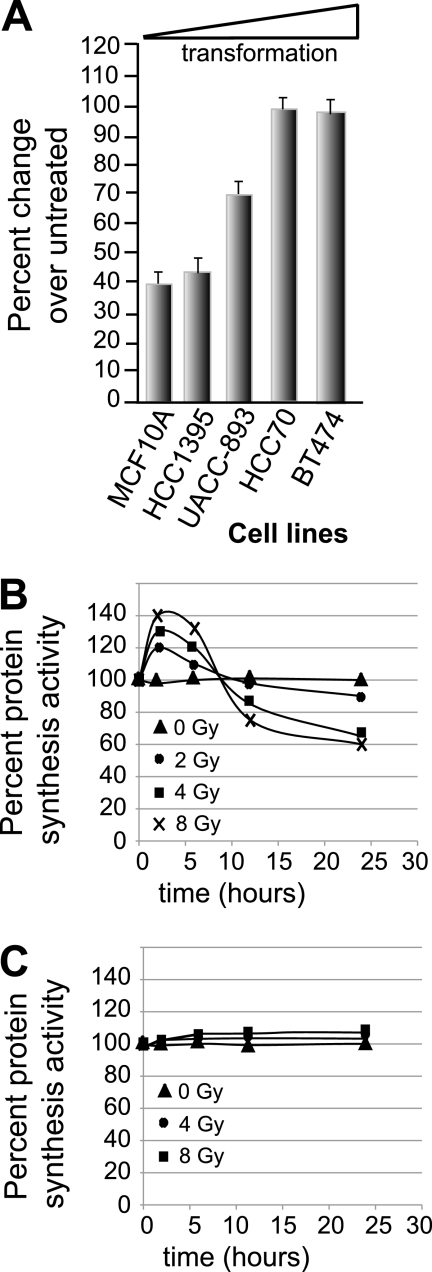
We investigated the effect of IR on overall protein synthesis activity and its relationship to increasing transformation of breast cancer cell lines. Breast cancer cells that were previously shown to comprise a panel of increasingly transformed cell lines (8) were chosen. They consisted of MCF10A cells (immortalized human breast epithelial cells), HCC1395 cells (ER+, wild-type p53, early-stage, intermediate-grade human ductal carcinoma), UACC-893 cells (ER−, wild-type p53, intermediate-stage and -grade ductal carcinoma), HCC70 cells (ER−, p53 mutated, intermediate-stage and -grade ductal carcinoma), and BT474 cells (ER+, p53 mutated, high-stage and -grade ductal carcinoma). Transformation was previously assessed based on a panel of in vitro criteria, i.e., growth in soft agar and the ability to form tumors in nude mice, among other standard features of transformation. The cells were treated with a single fraction of 8 Gy IR, followed 24 h later by metabolic labeling with [35S]methionine/cysteine to determine the overall protein synthesis activity of the viable fraction of cells. Nontransformed, immortalized breast epithelial MCF10A cells demonstrated >60% reduction in protein synthesis by 24 h following treatment with 8 Gy IR, whereas highly transformed BT474 cells were resistant (Fig. 1A). Cell lines, such as HCC1395 and UACC-893, that were more transformed than MCF10A cells but less so than BT474 cells demonstrated intermediately decreased IR-mediated inhibition of protein synthesis (Fig. 1A). This was also observed in other cell lines in similar states of transformation (data not shown).
FIG. 1.
IR differentially regulates protein synthesis in transformed and nontransformed cells. (A) Established cell lines with increasing transformation, from immortalized breast epithelial cells (MCF10A) to highly transformed breast cancer cells (BT474), were propagated under identical conditions and either left untreated or treated with 8 Gy IR (the highest tolerated dose for MCF10A cells), and protein-synthetic activity was measured 24 h later in the surviving fraction by [35S]methionine/cysteine incorporation and determination of protein specific activity. The data are presented as the ratio of treated to untreated specific activities of protein labeling plus standard errors of the mean. (B and C) Protein synthesis rates of MCF10A cells (B) and BT474 cells (C) with increasing IR doses. The rates were measured by [35S]methionine/cysteine incorporation and determination of protein specific activity. The data presented were derived from the mean of at least three independent experiments.
The kinetics of the protein synthesis response revealed an unexpected dose-dependent early increase in protein synthesis by as much as 30 to 40% at 4 to 8 Gy IR in radioresponsive MCF10A cells (Fig. 1B) (P < 0.05). The early increase in protein synthesis immediately following IR was not observed in transformed BT-474 cells (Fig. 1C) (P < 0.05) or other highly transformed cells (data not shown). The maximum increase in protein synthesis by IR occurred most strongly at 4 to 8 Gy but was still pronounced at 2 Gy and occurred 2 to 6 h posttreatment. There was also a correlation between an increased IR dose level and later inhibition of protein synthesis in MCF10A cells (Fig. 1B). IR doses greater than 8 Gy showed even more substantial inhibition of protein synthesis but were associated with considerable cytotoxicity at 24 h (data not shown). Collectively, these data demonstrate a dose-dependent biphasic response in global protein-synthetic rates following IR in radioresponsive immortalized breast epithelial cells. They also suggest that with frank transformation cells become resistant to inhibition of protein synthesis by IR.
Rapid, early translation increase promotes accumulation of radioprotective proteins.
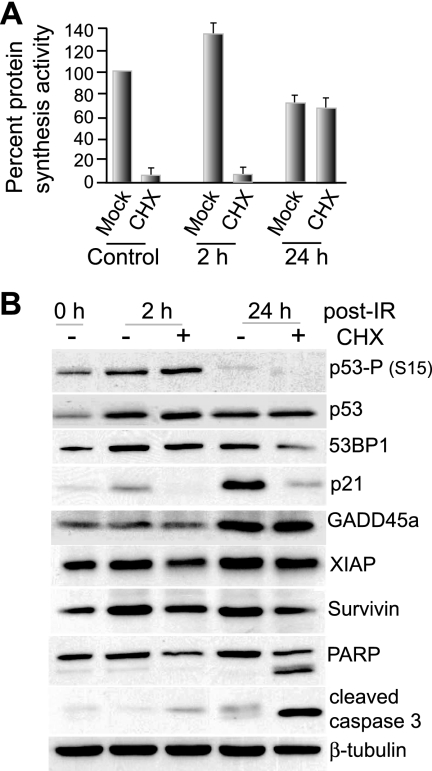
It was previously proposed that maintenance or increased protein synthesis following IR might serve as a protective stress response (44) or, in dendritic cells, as part of an IR-induced inflammatory signal required for immune functions and cell survival (39). We therefore transiently blocked the early translation phase by treatment of cells with the protein synthesis inhibitor cycloheximide for 1 h immediately following treatment with 8 Gy IR. The 1-h cycloheximide treatment blocked the initial IR-mediated increase in protein synthesis at 4 to 6 h in MCF10A cells, but not the late inhibition at 24 h (Fig. 2A). As the DNA damage response is associated with induction of coordinated programs of cell cycle arrest, DNA repair, and modulation of apoptosis, we examined MCF10A cells for factors associated with these functions (Fig. 2B). Immunoblot analysis of IR-treated cells showed induction of 53BP1 and p21, which are involved in p53 activation, DNA repair, and G2/M arrest, all of which were decreased in the cycloheximide-treated group at 24 h. Of note, XIAP and survivin, which suppress apoptosis, were blocked in induction following IR treatment by cycloheximide treatment, which was also associated with induction of apoptosis, as indicated by cleavage of caspase 3 and PARP. These data suggest, but do not prove, that the IR-mediated transient increase in protein synthesis may be critical for the expression of multiple factors that protect the cell against IR stress.
FIG. 2.
Effect of early-phase protein-synthetic activity on gene expression and viability. (A) MCF10A cells were left untreated (0 h) or treated with 8 Gy IR and then, at 2 or 24 h, treated with 10 μg/ml cycloheximide (CHX) or mock treated, and the protein-synthetic rates were determined by [35S]methionine incorporation for 1 h. (B) Immunoblot analysis of cellular lysates from cells treated in parallel with those in panel A. Equal amounts of total protein were resolved by 10% SDS-PAGE and identified by specific antibodies as indicated. The immunoblots are representative of typical results from at least three studies. All data presented were derived from the mean of at least three independent experiments, with standard errors of the mean shown.
IR stimulates protein synthesis transiently by acting on 4E-BP1.
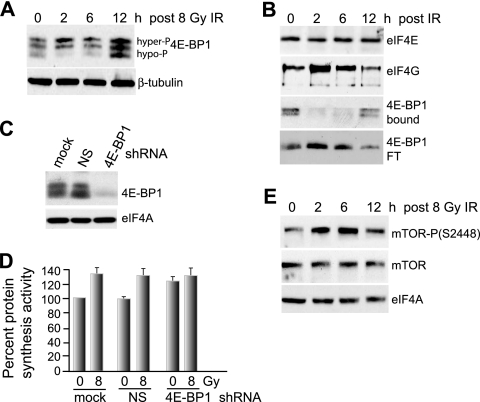
We investigated the mechanism by which IR stimulates protein synthesis shortly after irradiation. IR-sensitive MCF10A cells were treated with 8 Gy IR, and the effect on 4E-BP1 phosphorylation was investigated. IR mediated an increase in the hyperphosphorylation of 4E-BP1 within 2 h, which was sustained at 6 h and decreased to control levels by 12 h (Fig. 3A). The hyperphosphorylation of 4E-BP1 was associated with an increase in eIF4F/cap initiation complexes between 2 and 6 h, as shown by m7GTP chromatography recovery of increased levels of eIF4G bound to eIF4E and decreased eIF4E interaction with 4E-BP1 (Fig. 3B). Silencing of 4E-BP1 (Fig. 3C) stimulated protein synthesis in control cells and eliminated the IR-mediated increase observed 2 h after irradiation (Fig. 3D). These data suggest that IR inhibition of 4E-BP1 at 8 Gy is primarily responsible for the early transient increase in protein synthesis in MCF10A cells. Since mTOR is a major regulator of 4E-BP1 phosphorylation and activity, we assessed the effect of IR on mTOR by investigating its activating phosphorylation (Fig. 3E). IR at 8 Gy induced a significant but transient increase in mTOR phosphorylation at activating site S2448 in MCF10A cells without changing the total mTOR protein levels, consistent with IR-mediated hyperphosphorylation of 4E-BP1 and stimulation of protein synthesis.
FIG. 3.
Increased hyperphosphorylation of 4E-BP1 early after IR promotes eIF4F complex formation and protein synthesis. (A) 4E-BP1 was resolved by high-resolution SDS-15% PAGE using equal amounts of protein extracts obtained from MCF10A cells irradiated with a single fraction of 8 Gy IR and harvested at the indicated time points. Hyper- and hypophosphorylated forms of 4E-BP1 are indicated. (B) Cells irradiated at 8 Gy were harvested at the times shown, equal amounts of protein extracts were subjected to m7GTP-Sepharose cap chromatography, and retentates were recovered by elution with SDS, resolved by SDS-PAGE, and detected by immunoblot analysis with antibodies as indicated. The flowthrough fraction (FT) of 4E-BP1 that did not bind to m7GTP-Sepharose is shown. The data were quantified by densitometry of autoradiograms from at least three independent experiments. Representative results are shown. (C) MCF10A cells were stably transformed with lentivirus vectors expressing NS or 4E-BP1-silencing shRNAs, and equal amounts of protein lysates were subjected to immunoblot analysis with antibodies as shown. (D) MCF10A cells silenced as for panel C were subjected to 8 Gy irradiation, and protein synthesis rates were determined 2 h later by metabolic labeling with [35S]methionine/cysteine. (E) mTOR abundance and activating phosphorylation at Ser2448 were determined in MCF10A cells irradiated at 8 Gy at the times shown by immunoblot analysis using specific antibodies as shown. All results are representative of at least three independent experiments, with standard errors of the mean determined from all studies.
IR stimulates protein synthesis via ATM-dependent ERK phosphorylation.
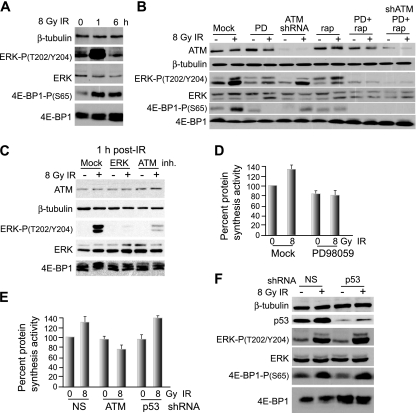
We investigated the signaling pathway by which IR promotes the rapid increase in protein synthesis. Immunoblot analysis following IR revealed strong but transient phosphorylation of 4E-BP1 at Ser65 (Fig. 4A), a key inactivating site, consistent with stimulation of protein synthesis. While the IR-activated protein kinase ATM has been identified as a direct kinase for 4E-BP1, it has been linked solely to phosphorylation of 4E-BP1 at Ser112, a nonconventional site, and only in response to insulin (77). It should be noted that a low level of ATM protein has been reported to be cytoplasmic and to have cytoplasmic targets in a variety of cell types (38, 41, 55, 74). The effect of Ser112 phosphorylation on 4E-BP1 function is also not clear (17, 29). The cdc2-cyclin B complex is also known to phosphorylate 4E-BP1 during mitosis (following the G2/M checkpoint) as part of cell cycle regulation of translational control. However, it was unlikely that cdc2 activity was responsible for the rapid phosphorylation of 4E-BP1, as most cells were not in G2/M and treatment of cells with the cdc2 inhibitor roscovitine did not block transient 4E-BP1 phosphorylation by IR (data not shown). Phosphorylation of 4E-BP1 has also been linked to MAPK signaling (42). Specifically, ERK1/2 has been shown to stimulate mTOR in a variety of systems through the canonical phosphatidylinositol 3-kinase/Akt pathway, possibly by acting on PDK1 and RSK (70), and independently of Akt, probably through RSK inhibition of the mTOR inhibitor TSC2 (45). ERK can also directly phosphorylate 4E-BP1 at Ser65, which, in cooperation with hierarchical phosphorylation at Thr37/46 by mTOR, reduces 4E-BP1 sequestration of eIF4E (14, 22, 28, 32). IR has been shown to rapidly activate ERK in an ATM-dependent and transient manner (23, 72). Consistent with these findings, IR at 8 Gy was found to activate ERK in MCF10A cells, shown by phosphorylation at T202/Y204, which was rapid but transient, in accord with increased phosphorylation of 4E-BP1 at activating site S65 (Fig. 4A). Analysis of several highly transformed breast cancer cell lines revealed constitutively activated ERK that was not further stimulated by IR, consistent with the established constitutive activation of ERK with transformation and the lack of effect of IR on 4E-BP1 phosphorylation and overall protein synthesis rates in highly transformed cells (data not shown).
FIG. 4.
4E-BP1 is transiently phosphorylated at early times following IR treatment in a p53-independent, ATM-, ERK-, and mTOR-dependent manner. (A) Immunoblot analysis of total cell lysate from MCF10A cells treated with 8 Gy IR and harvested at the times shown. Equal amounts of total protein were resolved by SDS-12% PAGE and subjected to immunoblot analysis with protein- and phosphoprotein-specific antibodies as shown. (B) Immunoblot analysis of MCF10A cells at 1 h following 8 Gy IR, treated simultaneously with vehicle alone or the ERK inhibitor PD98059 (PD) at 20 μM or subjected to shRNA gene silencing for ATM alone or in combination with PD98059. Equal amounts of total protein (25 μg) were loaded for analysis on SDS-10% PAGE. Immunoblot analysis used protein- and phosphoprotein-specific antibodies as shown. rap, rapamycin. (C) Study conducted as for panel B but including high-resolution SDS-15% PAGE analysis of 4E-BP1 and use of the ATM inhibitor (inh.) KU55933 at 10 μM added 30 min prior to IR treatment and washed out 20 min following treatment. (D) Protein-synthetic rates in MCF10A cells treated with vehicle or the MEK inhibitor PD98059 at 20 μM for 2 h immediately following treatment with 8 Gy IR. Protein synthesis activity was determined by [35S]methionine/cysteine metabolic labeling of MCF10A cells, either untreated or treated with 8 Gy IR, with or without inhibition of ERK with PD98059. (E) Protein synthesis activity was determined as for panel D for MCF10A cells silenced for ATM or p53 or expressing NS shRNA from lentivirus vectors at 2 h following treatment with 8 Gy IR. (F) Immunoblot analysis of total cell lysate from MCF10A cells stably transformed with lentiviruses expressing NS or p53 shRNA. The lysates were reduced by SDS-10% PAGE and probed with protein- and phosphoprotein-specific antibodies as shown. Typical results of at least three independent experiments are shown for all studies. Standard errors of the mean (D and E) were calculated from the means of three independent experiments.
ERK phosphorylation of 4E-BP1 was also found to be important for the IR-induced transient increase in protein synthesis in MCF10A cells, as shown by treatment with the ERK inhibitor PD958059. ERK inhibition blocked the increase in 4E-BP1 Ser65 inactivating phosphorylation at 1 h following treatment with 8 Gy IR in MCF10A cells (Fig. 4B) and the slower electrophoretic migration of 4E-BP1, indicative of its hyperphosphorylation (Fig. 4C). Similar results were found with the ERK1/2 inhibitor U0126 (data not shown). Inhibition of 4E-BP1 hyperphosphorylation mediated by ERK also prevented the early increase in protein synthesis by IR (Fig. 4D). Moreover, prevention of ATM activity by either shRNA silencing or chemical inhibitor reduced IR-mediated 4E-BP1 phosphorylation at Ser65 (Fig. 4B) and its hyperphosphorylation (Fig. 4C) and eliminated the early transient increase in protein synthesis by IR (Fig. 4E). In contrast, silencing of p53 was not associated with any change in 4E-BP1 phosphorylation (Fig. 4F) or protein synthesis (Fig. 4E). ERK can potentially also block TSC2, leading to greater mTOR activity and increased phosphorylation of 4E-BP1. Inhibition of mTOR with rapamycin partially blocked IR-mediated phosphorylation of 4E-BP1 at Ser65 (Fig. 4B), but not to the extent of ERK inhibition. Inhibition of ERK and mTOR most strongly blocked 4E-BP1 phosphorylation equivalent to ERK inhibition or ATM silencing. Thus, it is likely that ERK acts directly on 4E-BP1 and indirectly via TSC2/mTOR following IR. These results demonstrate that nontransformed MCF10A breast epithelial cells respond to IR with an early transient, but significant, escalation of global protein synthesis, which is dependent upon signaling by the ATM and ERK pathways.
Inhibition of translation by IR involves stabilization of 4E-BP1.
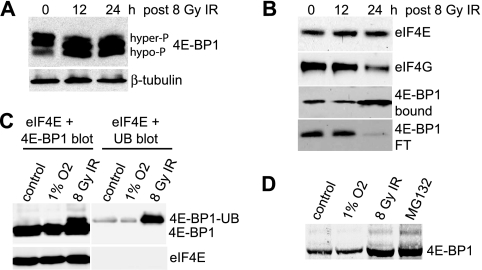
The inhibition of protein synthesis following IR has not been well studied, but it has been suggested to involve inhibition of mTOR activity (68). We therefore examined the abundance of cap initiation complexes (eIF4F) and 4E-BP1 activity during protein synthesis inhibition in control and 8-Gy-irradiated MCF10A cells. In addition to an expected shift to the hypophosphorylated form, there was an unexpected striking increase in the total abundance of 4E-BP1 in MCF10A cells at late times (between 12 and 24 h) following 8-Gy-IR treatment (Fig. 5A). Analysis of cap initiation complex stability by m7GTP-Sepharose chromatography of eIF4E and associated proteins revealed a significant decrease in the formation of eIF4F complexes at 24 h, which was associated with increased abundance of 4E-BP1, its increased association with eIF4E, and decreased presence of 4E-BP1 in the flowthrough fraction (Fig. 5B). While transcriptional induction of 4E-BP1 has been reported for some conditions of stress (60), measurement of 4E-BP1 mRNA levels by real-time qRT-PCR revealed no significant transcriptional induction of 4E-BP1 by IR (data not shown). 4E-BP1 has been previously reported to undergo rapid proteasome-mediated degradation in primary fibroblasts infected by human cytomegalovirus (73). MCF10A cells were therefore transfected with an expression vector for Myc-His-tagged ubiquitin, followed by IR treatment, and accumulation of ubiquitinated 4E-BP1 was examined 24 h following treatment (Fig. 5C). A low level of ubiquitinated 4E-BP1 was found in untreated MCF10A cells, which was strongly increased by 8 Gy IR but not another stress, such as hypoxia (1% O2; 24 h) (Fig. 5C). There was also no ubiquitin labeling of control eIF4E under hypoxia conditions. Addition of the proteasome inhibitor MG132 to untreated cells resulted in fourfold-increased accumulation of 4E-BP1, equal to that with IR (Fig. 5D), suggesting that IR inhibits rapid turnover of 4E-BP1 protein by the proteasome. While p53 activation has been implicated in proteolysis-mediated N-terminal truncation of 4E-BP1 to a hyperactive eIF4E-binding moiety (9, 67), we found no evidence for this truncated form at the IR doses employed in these studies (data not shown). Thus, IR uniquely stabilizes 4E-BP1 against proteasome-mediated degradation and promotes its dephosphorylation at late time points in IR-sensitive MCF10A cells.
FIG. 5.
IR increases the stability of 4E-BP1 by preventing its ubiquitination and proteasome-mediated rapid decay. (A) 4E-BP1 was resolved by high-resolution SDS-15% PAGE using equal amounts of protein extract from MCF10A cells irradiated at 8 Gy and harvested at the times shown post-IR treatment. Proteins were detected by immunoblot analysis. hyper-P, hyperphosphorylated; hypo-P, hypophosphorylated. (B) MCF10A cells subjected to 8 Gy IR were harvested at the times shown, equal amounts of protein extract were subjected to m7GTP-Sepharose cap chromatography, and retentates were recovered by elution with SDS, resolved by SDS-10% PAGE, and detected by immunoblot analysis with antibodies as indicated. The flowthrough fraction (FT) of 4E-BP1 that did not bind to m7GTP-Sepharose is shown. Representative results are shown for three independent experiments. (C) MCF10A cells were transfected with a His-Myc-tagged pentameric ubiquitin (UB) expression vector. The cells were either untreated (control), hypoxia treated (24 h; 1% O2), or treated with 8 Gy IR, and equal amounts of protein extracts were resolved by SDS-PAGE and detected by immunoblotting for 4E-BP1 or ubiquitin and eIF4E. 4E-BP1 blots were overexposed to reveal the ubiquitinated 4E-BP1 protein. (D) MCF10A cells were treated as described for panel C, resolved by low-resolution SDS-8% PAGE, and detected by immunoblot analysis within the linear range of radiological imaging.
Depletion of 4E-BP1 blocks IR inhibition of protein synthesis.
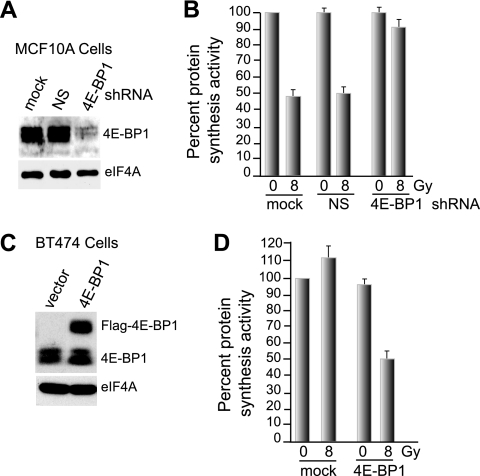
Given the increased abundance of 4E-BP1, greater association with eIF4E, and disruption of eIF4F complexes at late times following IR, we sought to determine whether 4E-BP1 is the primary factor involved in translational inhibition by IR. MCF10A cells were stably silenced for 4E-BP1 or nonsilencing (NS) control using stable lentivirus shRNAs (Fig. 6A) and then subjected to 8 Gy IR, followed by the determination of protein-synthetic rates at 24 h post-IR treatment (Fig. 6B). Depletion of 4E-BP1 significantly protected against IR-induced translational inhibition at 24 h post-IR treatment (Fig. 6B). 4E-BP1 silencing also prevented the disruption of eIF4F complexes, as expected (data not shown).
FIG. 6.
Increased abundance of 4E-BP1 is the primary means of protein synthesis inhibition in IR-treated cells. (A) MCF10A cells were stably transformed with lentivirus vectors expressing NS or 4E-BP1-silencing shRNAs, and equal amounts of protein lysates were subjected to immunoblot analysis with antibodies as shown. (B) MCF10A cells stably transformed with NS and 4E-BP1-silencing lentivirus vectors were subjected to 8 Gy IR, and protein synthesis activity in the cells was determined by [35S]methionine/cysteine labeling. Cells were harvested 24 h post-IR treatment. The results are the average of three independent experiments with standard errors of the mean shown. (C) A Flag-tagged 4E-BP1 protein was overexpressed in radioresistant BT474 cells transformed with a cDNA-expressing lentvirus vector. Proteins were detected by immunoblotting with antibodies to 4E-BP1. The decreased electrophoretic mobility of Flag-4E-BP1 is due to the additional Flag motif. (D) Vector control and 4E-BP1-overexpressing BT474 cells were subjected to 8 Gy IR, and protein synthesis rates were determined 24 h later as described for panel B. The data are the averages of three independent experiments, with standard errors of the mean shown.
To independently confirm the importance of 4E-BP1 in mediating IR-induced inhibition of protein synthesis, a Flag-tagged form of the protein was overexpressed in highly transformed BT474 cells, which are resistant to IR inhibition (Fig. 6C) and in which there is no change in 4E-BP1 or other translation factor abundance with IR (see Fig. S1A in the supplemental material). Whereas 4E-BP1 overexpression slightly reduced protein synthesis in untreated control cells, it conferred a significant (>50%) reduction in protein synthesis at 24 h, mediated by IR (Fig. 6D). These data further support the primary role of 4E-BP1 abundance in IR inhibition of protein synthesis.
Under conditions of severe stress, such as exposure to anoxia (<0.02% O2) combined with serum starvation, the initiation factor eIF2 undergoes inactivation by phosphorylation of its α subunit, thereby inhibiting global protein synthesis (1). Since we observed an increase in the overall levels of eIF2α phosphorylation at late time points following high doses of IR in MCF10A cells (>8 Gy) (see Fig. S1B in the supplemental material), a retrovirus vector was used to express the GADD34 C-terminal fragment to constitutively dephosphorylate eIF2α at the inactivating site Ser51 (49). Stable expression of the Flag-GADD34 C-terminal fragment fully blocked eIF2α phosphorylation in response to IR compared to vector-expressing cells (see Fig. S1C in the supplemental material). However, despite the inhibition of eIF2α phosphorylation by the GADD34 protein, protein synthesis was inhibited to the same extent as in control cells after treatment with 8 Gy IR (see Fig. S1D in the supplemental material). There was no change in the inactivating phosphorylation state of elongation factor kinase EF2K (see Fig. S1B in the supplemental material). These data demonstrate that 4E-BP1 is the major effector of translation inhibition following IR.
4E-BP1 inhibition of protein synthesis at late times following IR requires ATM and involves p53.
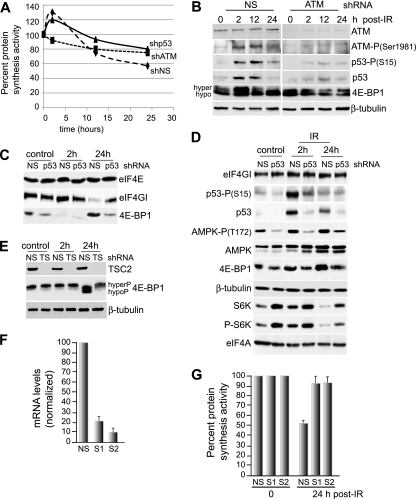
Stimulation (hypophosphorylation) of 4E-BP1 involves inhibition of mTOR activity. The inhibition of mTOR activity is mediated by multiple upstream components that act on the TSC2 protein, particularly AMPK and AKT (53). The LKB1 protein has been associated with p53 effector functions through direct interaction (33, 50). Moreover, LKB1 was shown to directly activate AMPK in response to energy stress (56, 57, 78), which linked LKB1 to inhibition of mTOR through AMPK and TSC2 (40, 56). It was also recently shown that p53 target genes encoding Sestrin 1 and 2 are activators of AMPK and inhibitors of mTOR activity through AMPK phosphorylation of TSC2 (4). Thus, p53 activation by IR may inhibit mTOR via a Sestrin-LKB1-AMPK-TSC2 pathway. We therefore examined the roles of ATM, p53, and other key upstream mediators of IR responses in translational control pathways by silencing ATM and p53 in MCF10A cells (Fig. 7A). Silencing of ATM (Fig. 7B) eliminated the initial translational increase following IR and partially blocked the inhibition of protein synthesis at later times (Fig. 7A). Silencing of p53 (Fig. 7D) did not impair the early increase in protein synthesis but did decrease the extent of inhibition at late times, similar to ATM silencing (Fig. 7A). This was further reflected in a greater fraction of hyperphosphorylated 4E-BP1 in ATM-silenced cells at 24 h following treatment with 8 Gy IR (Fig. 7B). Also apparent was greater preservation of eIF4F complexes at 24 h after treatment with 8 Gy IR, recoverable by cap chromatography in p53-silenced (Fig. 7C) and ATM-silenced MCF10A cells (Fig. 8C), again consistent with roles for ATM and p53 in IR-mediated activation of 4E-BP1. It was also apparent that silencing of ATM or p53 eliminated the increased levels of 4E-BP1 in cells 24 h following treatment with 8 Gy IR (Fig. 7B and D). These data suggest that hyperphosphorylation of 4E-BP1 is associated with its decreased stability and proteasome-mediated turnover.
FIG. 7.
Late-phase inhibition of protein synthesis by IR is dependent upon p53 and p53-induced Sestrin 1 and 2 proteins. (A) MCF10A cells were transformed with shRNA lentiviruses to ATM, p53, or NS as described in Materials and Methods and treated with 8 Gy IR, and protein-synthetic rates were measured over a 24-h period. (B) MCF10A cells were transformed with lentivirus vectors expressing shRNAs against ATM or NS, treated with 8 Gy IR, and harvested at 2 h, 12 h, or 24 h post-IR treatment. Equal amounts of protein from total cell lysates were analyzed by SDS-10% PAGE and subjected to immunoblotting with protein- and phosphoprotein-specific antibodies as shown. hyper, hyperphosphorylated; hypo, hypophosphorylated. (C) m7GTP cap chromatography and immunoblot analysis of NS and p53-silenced MCF10A cells treated with 8 Gy IR and harvested at the indicated times. (D) MCF10A cells were transformed with lentivirus vectors, silenced for p53, treated with 8 Gy IR, and analyzed by immunoblot analysis at 2 h and 24 h post-IR treatment. (E) NS or TSC2-silenced (TS) MCF10A cells were treated with 8 Gy IR, harvested at the indicated times, and analyzed by immunoblot analysis using equal amounts of protein from total cell lysates. (F) Sestrin 1 (S1) and 2 (S2) were silenced in MCF10A cells using shRNA-expressing lentivirus vectors. mRNA levels were determined by real-time qRT-PCR. (G) Sestrin-silenced MCF10A cells and control NS cells were subjected to 8 Gy IR, and protein synthesis rates were determined by metabolic labeling with [35S]methionine/cysteine. The results represent the average of three independent experiments, with standard errors of the mean shown.
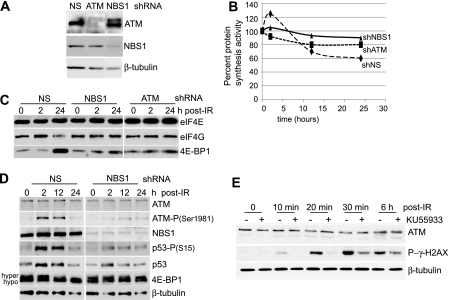
FIG. 8.
Late-phase inhibition of protein synthesis is signaled by the MRN DNA DSB complex. (A) MCF10A cells were transduced with lentiviruses expressing shRNAs to ATM, NBS1, or NS. Immunoblot analysis was conducted with antibodies as shown. (B) MCF10A cells were transduced with shRNA lentiviruses to ATM, NBS1, or NS and treated with 8 Gy IR, and protein-synthetic rates were measured over a 24-h period by [35S]methionine/cysteine incorporation. (C) MCF10A cells silenced with shRNA-expressing lentiviruses as shown were subjected to 8 Gy IR and harvested at the times shown; equal amounts of protein extract were subjected to m7GTP-Sepharose cap chromatography, and retentates were recovered by elution with loading dye at elevated temperature, resolved by SDS-10% PAGE, and detected by immunoblot analysis with antibodies as indicated. (D) MCF10A cells were silenced with control NS or NBS1 shRNA lentivirus vectors, treated with 8 Gy IR, and analyzed by immunoblot analysis at the times shown. hyper, hyperphosphorylated; hypo, hypophosphorylated. (E) MCF10A cells were treated with the ATM inhibitor KU55933 at 10 μM added 30 min prior to treatment with 8 Gy IR and washed out 20 min following treatment and were analyzed by immunoblot analysis at the times shown post-IR treatment.
IR promoted increased levels of p53 and activation (S15 phosphorylation) at early times and some continued activation at late times after 8-Gy irradiation of MCF10A cells (Fig. 7B), which was associated with increased phosphorylation (activation) of AMPK at T172 and decreased phosphorylation of S6K, as expected (Fig. 7D). Silencing of ATM increased the hyperphosphorylation of 4E-BP1 at late times following IR treatment (Fig. 7B). The role of TSC2 control of mTOR in mediating 4E-BP1 phosphorylation and stability following IR treatment was characterized using TSC2 silencing in MCF10A cells. TSC2 silencing prevented increased abundance of 4E-BP1 and blocked its dephosphorylation at 24 h following treatment with 8 Gy IR (Fig. 7E). The requirement for ATM, p53, AMPK, and TSC2 in the increased abundance and hypophosphorylation (activation) of 4E-BP1 at late times post-IR treatment implies the involvement of p53-induced Sestrin 1 and 2. These Sestrins were shown recently to promote AMPK activation and its inhibition of mTOR through phosphorylation of TSC2 (4). MCF10A cells were therefore silenced for Sestrin 1 and 2 (Fig. 7F) and irradiated at 8 Gy, and protein-synthetic rates were determined (Fig. 7G). Silencing of Sestrin 1 and 2 largely eliminated the inhibition of protein synthesis mediated by 8 Gy IR at 24 h. Sestrin 1 and 2 therefore provide a critical link between IR activation of ATM and p53 and the inhibition of protein synthesis by 4E-BP1 in MCF10A cells.
The MRN DNA DSB complex initiates inhibition of protein synthesis by IR.
The ATM pathway regulates the cellular response to DNA double-strand breaks (DSBs) that are induced by IR and is potentially disrupted during tumorigenesis as part of the promoted increased genomic instability (34). The initial steps of ATM recruitment to sites of IR-induced DNA damage indicate that the MRN complex initiates localization of ATM and other signaling factors to the sites of DSBs (71) with precise choreography of protein assembly at foci of DNA damage (24, 43). ATM phosphorylates a number of proteins involved in cell cycle checkpoint control, apoptotic responses, and DNA repair, including p53, Chk2, BRCA1, H2AX, SMC1, Rad17, Artemis, and NBS1 (35). We therefore silenced key components of the MRN complex to determine whether the DSB machinery is the IR sensor that signals to inhibit protein synthesis in response to DNA damage.
MCF10A cells were stably transformed and silenced for ATM, p53, Rad50, or NBS1 using lentivirus vectors (Fig. 7D and 8A). The cells were irradiated at 8 Gy, and the effect on protein synthesis was determined by metabolic labeling with [35S]methionine/cysteine at the indicated times post-IR treatment. As shown above, silencing of ATM impaired the early IR-mediated increase in protein synthesis and significantly blocked protein synthesis inhibition at late times following IR treatment (Fig. 8B). Similarly, silencing NBS1 largely blocked both the early increase and the late decrease in protein synthesis by 8 Gy IR (Fig. 8B), as did silencing of Rad50 (data not shown). Thus, the DSB machinery itself is required for IR sensing and transduction of translation-regulatory signals in MCF10A cells.
The effect of NBS1 and ATM silencing on the integrity of the eIF4F/cap initiation complex during irradiation was examined by cap affinity chromatography and immunoblot analysis (Fig. 8C). Consistent with the effect on protein synthesis, silencing of NBS1 or ATM in MCF10A cells largely eliminated the increased interaction of eIF4G with eIF4E early after IR treatment, their decreased interaction at late times, and the increased binding of 4E-BP1 to eIF4E at late times. Silencing of NBS1 also blocked signaling to the key transducers of the 8-Gy-IR response, shown earlier to regulate mTOR/4E-BP1 abundance and availability (Fig. 8D). Silencing of NBS1 blocked IR activation of ATM, shown by reduced Ser1981 phosphorylation; activation of p53, shown by reduced Ser15 phosphorylation; and increased accumulation of 4E-BP1 protein at late times post-IR treatment. That functional assembly of the MRN complex is required for the response of protein synthesis to IR was shown by control studies. Inhibition of ATM with the potent inhibitor KU55933 blocked accumulation and activating phosphorylation of γ-H2AX in response to IR (Fig. 8E).
DISCUSSION
There have been surprisingly few studies directed to understanding the regulation of protein synthesis by IR, a ubiquitous and highly relevant genotoxic stress. Studies have previously explored a connection between p53, mTOR signaling, and protein synthesis. Using a temperature-sensitive p53 mutant, dephosphorylation of 4E-BP1 (activation) and p70S6K (inactivation) were observed as functions of p53 activity at permissive temperature (31). No contribution by eIF2α phosphorylation to the inhibition of protein synthesis was found, consistent with our findings. In addition, p53-dependent increased expression of PTEN was observed under protracted conditions of stress (61) that involved transcriptional upregulation of AMPK and TSC2 activity but required prolonged p53 activation (>18 h) in engineered cell lines (19, 20). Importantly, induction of p53 following IR is relatively transient and typically much lower (18).
A transient, rapid increase in protein synthesis observed as part of a biphasic response to IR (Fig. 1B) has been noted previously in other studies, as well. With IR treatment of <2 Gy, the EGF and ErbB2 receptors were found to mediate a rapid increase in p70S6K activity and increased protein synthesis activity (10). IR-induced phosphorylation of p70S6K was inhibited by MAPK blockade or rapamycin treatment, consistent with our results for transient increased 4E-BP1 phosphorylation (inactivation) following IR treatment, which we showed was mediated by increased ERK and mTOR activity (Fig. 4). It is well established that ERK is involved in survival signaling, is activated in response to a variety of genotoxic stresses, and may function in response to DNA damage (66). Moreover ERK has been recognized as an activator of mTOR and p70S6K and as an inhibitor of 4E-BP1 (30, 58).
The mechanism for ATM in the stimulation of ERK activity remains unclear. Several reports demonstrated a requirement for ATM that is independent of p53 and for subsequent ERK activation following activation of the DNA damage response (23, 66). These data are also consistent with our results, particularly the requirement for ATM in both early and late phases of IR protein synthesis regulation (Fig. 7) and the ability to significantly prevent translation inhibition by silencing MRN complex proteins (Fig. 8). Furthermore, mTOR activity was found to be required for maximal ERK activation following IR treatment (48), reported to be potentially mediated by mTOR regulation of protein phosphatase 2A activity on ERK (26). The lack of a pronounced biphasic translation response in highly transformed cells is likely the result of a largely constitutively active mTOR signaling pathway and increased basal ERK activity.
The inhibition of protein synthesis at later times following IR treatment involves ATM signaling and activation of AMPK, leading to inhibition of mTOR. Nuclear accumulation of mTOR following IR treatment was previously reported (47, 51), but it was not observed in our studies (data not shown). A shift in mTOR compartmentalization as a means for IR inhibition can therefore be excluded.
Our results indicate that inhibition of mTOR is largely p53 dependent, acting by Sestrin 1 and 2 protein activation of AMPK and inhibition of mTOR (Fig. 7). The failure to fully recover protein synthesis at later time points with p53 silencing may be related to residual p53 protein, similar to the inability to fully recover protein synthesis with ATM or NBS1 silencing. Low levels of ATM may be sufficient to activate downstream effectors (37), and there are several recognized redundant signaling interactions within the DNA damage response, including the phosphorylation of p53 by ATR (69) and the phosphorylation of LKB1 by DNA-PK (54). Our analysis of the assembly of the γH2AX/DNA damage response MRN complex (Fig. 8E) is consistent with this interpretation.
Several reports have demonstrated that ATM directly phosphorylates and activates LKB1 (21, 54). LKB1 is a recognized activator of AMPK, which then activates TSC2, resulting in mTOR inhibition (11). Moreover, a recent study has indicated that ATM can directly phosphorylate and activate AMPK in an LKB1-independent fashion (62, 64). Alternatively, p53-induced Sestrin 1 and 2 proteins also activate AMPK (4), which was substantiated for IR action by our work. Thus, in combination with Sestrin inhibition of mTOR through AMPK activation, there are multiple mechanisms for inhibition of mTOR activity. A p53-mediated TSC2 activation that requires AMPK was also shown by inhibition of AMPK with compound “C” (data not shown), a selective AMPK small-molecule inhibitor.
The late IR inhibition of protein synthesis was directly linked to activation (hypophosphorylation) of 4E-BP1 and inhibition of cap-dependent protein synthesis. Surprisingly, we found that the abundance of 4E-BP1 was upregulated >3-fold by IR at late times and was a result of enhanced protein stability due to decreased proteasome-mediated decay. Stabilization of 4E-BP1 may be associated with a general inhibition of proteasome activity associated with IR in a dose-dependent manner (52). In addition, it has been shown that hypophosphorylated 4E-BP1, as observed at late times following IR treatment, is also associated with increased 4E-BP1 stability against proteasome-mediated protein degradation (15).
It is well recognized that exposure of tumor cells to IR leads to activation of multiple survival signaling pathways that maintain cell viability (13, 25, 75, 76). Our results demonstrated an initial increase in protein synthesis associated with production of components of the DNA damage response (53BP1 and p21), as well as prosurvival proteins (XIAP and survivin), and abrogation of this translational response by treatment with cycloheximide or ERK inhibition, resulting in greater radiotoxicity. Several reports have substantiated the role of early activation of growth factor signaling in radioprotective effects following IR treatment (65). Activation of mTOR, in particular, following IR treatment has been demonstrated to be vital in yeast for radioresistance, and it has been shown to suppress mutagenesis and increase survival via induction of ribonucleotide reductase activity that is critical for DNA repair during S phase (58).
Cellular IRES-dependent translation has emerged as particularly significant under conditions of severe stress and apoptosis, which are also observed with high-dose IR treatment. As we have shown here, profound repression of cap-dependent protein synthesis appears to be protracted, with recovery of protein synthesis delayed until >48 h following high-dose IR treatment, and thus noncanonical modes of gene expression may be particularly vital for cellular survival and repair functions.
Supplementary Material
Acknowledgments
This work was supported by grants from the Breast Cancer Research Foundation, the Department of Defense Breast Cancer Research Program, and the National Institutes of Health to R.J.S. and S.C.F.
S.B., M.L.B., and Q.X. performed research and designed studies; R.J.S. and S.C.F. designed studies and wrote the paper.
We declare no conflict of interest.
Footnotes
Published ahead of print on 24 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abedin, M. J., D. Wang, M. A. McDonnell, U. Lehmann, and A. Kelekar. 2007. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 14:500-510. [DOI] [PubMed] [Google Scholar]
- 2.Albert, J. M., K. W. Kim, C. Cao, and B. Lu. 2006. Targeting the Akt/mammalian target of rapamycin pathway for radiosensitization of breast cancer. Mol. Cancer Ther. 5:1183-1189. [DOI] [PubMed] [Google Scholar]
- 3.Bhandari, B. K., D. Feliers, S. Duraisamy, J. L. Stewart, A. C. Gingras, H. E. Abboud, G. G. Choudhury, N. Sonenberg, and B. S. Kasinath. 2001. Insulin regulation of protein translation repressor 4E-BP1, an eIF4E-binding protein, in renal epithelial cells. Kidney Int. 59:866-875. [DOI] [PubMed] [Google Scholar]
- 4.Budanov, A. V., and M. Karin. 2008. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buttgereit, F., and M. D. Brand. 1995. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 312:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, C., T. Subhawong, J. M. Albert, K. W. Kim, L. Geng, K. R. Sekhar, Y. J. Gi, and B. Lu. 2006. Inhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cells. Cancer Res. 66:10040-10407. [DOI] [PubMed] [Google Scholar]
- 7.Clemens, M. J. 2001. Translational regulation in cell stress and apoptosis. Roles of the eIF4E binding proteins. J. Cell. Mol. Med. 5:221-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly, E., S. Braunstein, S. Formenti, and R. J. Schneider. 2006. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell. Biol. 26:3955-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constantinou, C., and M. J. Clemens. 2005. Regulation of the phosphorylation and integrity of protein synthesis initiation factor eIF4GI and the translational repressor 4E-BP1 by p53. Oncogene 24:4839-4850. [DOI] [PubMed] [Google Scholar]
- 10.Contessa, J. N., J. Hampton, G. Lammering, R. B. Mikkelsen, P. Dent, K. Valerie, and R. K. Schmidt-Ullrich. 2002. Ionizing radiation activates Erb-B receptor dependent Akt and p70 S6 kinase signaling in carcinoma cells. Oncogene 21:4032-4041. [DOI] [PubMed] [Google Scholar]
- 11.Corradetti, M. N., K. Inoki, N. Bardeesy, R. A. DePinho, and K. L. Guan. 2004. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18:1533-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuesta, R., G. Laroia, and R. J. Schneider. 2000. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 14:1460-1470. [PMC free article] [PubMed] [Google Scholar]
- 13.Dent, P., D. B. Reardon, J. S. Park, G. Bowers, C. Logsdon, K. Valerie, and R. Schmidt-Ullrich. 1999. Radiation-induced release of transforming growth factor alpha activates the epidermal growth factor receptor and mitogen-activated protein kinase pathway in carcinoma cells, leading to increased proliferation and protection from radiation-induced cell death. Mol. Biol. Cell 10:2493-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diggle, T. A., S. K. Moule, M. B. Avison, A. Flynn, E. J. Foulstone, C. G. Proud, and R. M. Denton. 1996. Both rapamycin-sensitive and -insensitive pathways are involved in the phosphorylation of the initiation factor-4E-binding protein (4E-BP1) in response to insulin in rat epididymal fat cells. Biochem. J. 316:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elia, A., C. Constantinou, and M. J. Clemens. 2008. Effects of protein phosphorylation on ubiquitination and stability of the translational inhibitor protein 4E-BP1. Oncogene 27:811-822. [DOI] [PubMed] [Google Scholar]
- 16.Eshleman, J. S., B. L. Carlson, A. C. Mladek, B. D. Kastner, K. L. Shide, and J. N. Sarkaria. 2002. Inhibition of the mammalian target of rapamycin sensitizes U87 xenografts to fractionated radiation therapy. Cancer Res. 62:7291-7297. [PubMed] [Google Scholar]
- 17.Fadden, P., T. A. Haystead, and J. C. Lawrence, Jr. 1998. Phosphorylation of the translational regulator, PHAS-I, by protein kinase CK2. FEBS Lett. 435:105-109. [DOI] [PubMed] [Google Scholar]
- 18.Fei, P., and W. S. El-Deiry. 2003. P53 and radiation responses. Oncogene 22:5774-5783. [DOI] [PubMed] [Google Scholar]
- 19.Feng, Z., W. Hu, E. de Stanchina, A. K. Teresky, S. Jin, S. Lowe, and A. J. Levine. 2007. The regulation of AMPK β1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 67:3043-3053. [DOI] [PubMed] [Google Scholar]
- 20.Feng, Z., H. Zhang, A. J. Levine, and S. Jin. 2005. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. USA 102:8204-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandes, N., Y. Sun, S. Chen, P. Paul, R. J. Shaw, L. C. Cantley, and B. D. Price. 2005. DNA damage-induced association of ATM with its target proteins requires a protein interaction domain in the N terminus of ATM. J. Biol. Chem. 280:15158-15164. [DOI] [PubMed] [Google Scholar]
- 22.Gingras, A. C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golding, S. E., E. Rosenberg, S. Neill, P. Dent, L. F. Povirk, and K. Valerie. 2007. Extracellular signal-related kinase positively regulates ataxia telangiectasia mutated, homologous recombination repair, and the DNA damage response. Cancer Res. 67:1046-1053. [DOI] [PubMed] [Google Scholar]
- 24.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 25.Hagan, M., L. Wang, J. R. Hanley, J. S. Park, and P. Dent. 2000. Ionizing radiation-induced mitogen-activated protein (MAP) kinase activation in DU145 prostate carcinoma cells: MAP kinase inhibition enhances radiation-induced cell killing and G2/M-phase arrest. Radiat. Res. 153:371-383. [DOI] [PubMed] [Google Scholar]
- 26.Harwood, F. C., L. Shu, and P. J. Houghton. 2008. mTORC1 signaling can regulate growth factor activation of p44/42 mitogen. J. Biol. Chem. 282:2575-2585. [DOI] [PubMed] [Google Scholar]
- 27.Hay, N., and N. Sonenberg. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926-1945. [DOI] [PubMed] [Google Scholar]
- 28.Haystead, T. A., C. M. Haystead, C. Hu, T. A. Lin, and J. C. Lawrence, Jr. 1994. Phosphorylation of PHAS-I by mitogen-activated protein (MAP) kinase. Identification of a site phosphorylated by MAP kinase in vitro and in response to insulin in rat adipocytes. J. Biol. Chem. 269:23185-23191. [PubMed] [Google Scholar]
- 29.Heesom, K. J., M. B. Avison, T. A. Diggle, and R. M. Denton. 1998. Insulin-stimulated kinase from rat fat cells that phosphorylates initiation factor 4E-binding protein 1 on the rapamycin-insensitive site (serine-111). Biochem. J. 336:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbert, T. P., A. R. Tee, and C. G. Proud. 2002. The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J. Biol. Chem. 277:11591-11596. [DOI] [PubMed] [Google Scholar]
- 31.Horton, L. E., M. Bushell, D. Barth-Baus, V. J. Tilleray, M. J. Clemens, and J. O. Hensold. 2002. p53 activation results in rapid dephosphorylation of the eIF4E-binding protein 4E-BP1, inhibition of ribosomal protein S6 kinase and inhibition of translation initiation. Oncogene 21:5325-5334. [DOI] [PubMed] [Google Scholar]
- 32.Karim, M. M., J. M. Hughes, J. Warwicker, G. C. Scheper, C. G. Proud, and J. E. McCarthy. 2001. A quantitative molecular model for modulation of mammalian translation by the eIF4E-binding protein 1. J. Biol. Chem. 276:20750-20757. [DOI] [PubMed] [Google Scholar]
- 33.Karuman, P., O. Gozani, R. D. Odze, X. C. Zhou, H. Zhu, R. Shaw, T. P. Brien, C. D. Bozzuto, D. Ooi, L. C. Cantley, and J. Yuan. 2001. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol. Cell 7:1307-1319. [DOI] [PubMed] [Google Scholar]
- 34.Kastan, M. B., and J. Bartek. 2004. Cell-cycle checkpoints and cancer. Nature 432:316-323. [DOI] [PubMed] [Google Scholar]
- 35.Kastan, M. B., and D. S. Lim. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell Biol. 1:179-186. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, V., D. Sabatini, P. Pandey, A. C. Gingras, P. K. Majumder, M. Kumar, Z. M. Yuan, G. Carmichael, R. Weichselbaum, N. Sonenberg, D. Kufe, and S. Kharbanda. 2000. Regulation of the rapamycin and FKBP-target 1/mammalian target of rapamycin and cap-dependent initiation of translation by the c-Abl protein-tyrosine kinase. J. Biol. Chem. 275:10779-10787. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J. H., and T. T. Paull. 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26:7741-7748. [DOI] [PubMed] [Google Scholar]
- 38.Leemput, J., C. Masson, K. Bigot, A. Errachid, A. Dansault, A. Provost, S. Gadin, S. Aoufouchi, M. Menasche, and M. Abitbol. 2009. ATM localization and gene expression in the adult mouse eye. Mol. Vis. 15:393-416. [PMC free article] [PubMed] [Google Scholar]
- 39.Lelouard, H., E. K. Schmidt, V. Camosseto, G. Clavarino, M. Ceppi, H. T. Hsu, and P. Pierre. 2007. Regulation of translation is required for dendritic cell function and survival during activation. J. Cell Biol. 179:1427-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine, A. J., Z. Feng, T. W. Mak, H. You, and S. Jin. 2006. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 20:267-275. [DOI] [PubMed] [Google Scholar]
- 41.Lim, D. S., D. G. Kirsch, C. E. Canman, J. H. Ahn, Y. Ziv, L. S. Newman, R. B. Darnell, Y. Shiloh, and M. B. Kastan. 1998. ATM binds to beta-adaptin in cytoplasmic vesicles. Proc. Natl. Acad. Sci. USA 95:10146-10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, T. A., X. Kong, T. A. Haystead, A. Pause, G. Belsham, N. Sonenberg, and J. C. Lawrence, Jr. 1994. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266:653-656. [DOI] [PubMed] [Google Scholar]
- 43.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 44.Lu, X., L. de la Pena, C. Barker, K. Camphausen, and P. J. Tofilon. 2006. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 66:1052-1061. [DOI] [PubMed] [Google Scholar]
- 45.Ma, L., Z. Chen, H. Erdjument-Bromage, P. Tempst, and P. P. Pandolfi. 2005. Phosphorylation and functional inactivation of TSC2 by Erk: implications for tuberous sclerosis and cancer pathogenesis. Cell 121:179-193. [DOI] [PubMed] [Google Scholar]
- 46.Meisenberg, G., and W. H. Simmons. 1998. Principles of medical biochemistry. Mosby, St. Louis, MO.
- 47.Moeller, B. J., and M. W. Dewhirst. 2006. HIF-1 and tumour radiosensitivity. Br. J. Cancer 95:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molhoek, K. R., D. L. Brautigan, and C. L. Slingluff, Jr. 2005. Synergistic inhibition of human melanoma proliferation by combination treatment with B-Raf inhibitor BAY43-9006 and mTOR inhibitor rapamycin. J. Transl. Med. 3:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novoa, I., H. Zeng, H. P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okoshi, R., T. Ozaki, H. Yamamoto, K. Ando, N. Koida, S. Ono, T. Koda, T. Kamijo, A. Nakagawara, and H. Kizaki. 2008. Activation of AMP-activated protein kinase (AMPK) induces p53-dependent apoptotic cell death in response to energetic stress. J. Biol. Chem. 283:3979-3987. [DOI] [PubMed] [Google Scholar]
- 51.Paglin, S., N. Y. Lee, C. Nakar, M. Fitzgerald, J. Plotkin, B. Deuel, N. Hackett, M. McMahill, E. Sphicas, N. Lampen, and J. Yahalom. 2005. Rapamycin-sensitive pathway regulates mitochondrial membrane potential, autophagy, and survival in irradiated MCF-7 cells. Cancer Res. 65:11061-11070. [DOI] [PubMed] [Google Scholar]
- 52.Pervan, M., K. S. Iwamoto, and W. H. McBride. 2005. Proteasome structures affected by ionizing radiation. Mol. Cancer Res. 3:381-390. [DOI] [PubMed] [Google Scholar]
- 53.Reiling, J. H., and D. M. Sabatini. 2006. Stress and mTORture signaling. Oncogene 25:6373-6383. [DOI] [PubMed] [Google Scholar]
- 54.Sapkota, G. P., M. Deak, A. Kieloch, N. Morrice, A. A. Goodarzi, C. Smythe, Y. Shiloh, S. P. Lees-Miller, and D. R. Alessi. 2002. Ionizing radiation induces ataxia telangiectasia mutated kinase (ATM)-mediated phosphorylation of LKB1/STK11 at Thr-366. Biochem. J. 368:507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scata, K. A., and W. S. El-Deiry. 2006. Taming NEMO to slay cancer cells. Cancer Biol. Ther. 5:1096-1097. [DOI] [PubMed] [Google Scholar]
- 56.Shaw, R. J., N. Bardeesy, B. D. Manning, L. Lopez, M. Kosmatka, R. A. DePinho, and L. C. Cantley. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6:91-99. [DOI] [PubMed] [Google Scholar]
- 57.Shaw, R. J., M. Kosmatka, N. Bardeesy, R. L. Hurley, L. A. Witters, R. A. DePinho, and L. C. Cantley. 2004. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 101:3329-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen, C., C. S. Lancaster, B. Shi, H. Guo, P. Thimmaiah, and M. A. Bjornsti. 2007. TOR signaling is a determinant of cell survival in response to DNA damage. Mol. Cell. Biol. 27:7007-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shinohara, E. T., C. Cao, K. Niermann, Y. Mu, F. Zeng, D. E. Hallahan, and B. Lu. 2005. Enhanced radiation damage of tumor vasculature by mTOR inhibitors. Oncogene 24:5414-5422. [DOI] [PubMed] [Google Scholar]
- 60.Southgate, R. J., B. Neill, O. Prelovsek, A. El-Osta, Y. Kamei, S. Miura, O. Ezaki, T. J. McLoughlin, W. Zhang, T. G. Unterman, and M. A. Febbraio. 2007. FOXO1 regulates the expression of 4E-BP1 and inhibits mTOR signaling in mammalian skeletal muscle. J. Biol. Chem. 282:21176-21186. [DOI] [PubMed] [Google Scholar]
- 61.Stambolic, V., D. MacPherson, D. Sas, Y. Lin, B. Snow, Y. Jang, S. Benchimol, and T. W. Mak. 2001. Regulation of PTEN transcription by p53. Mol. Cell 8:317-325. [DOI] [PubMed] [Google Scholar]
- 62.Sun, Y., K. E. Connors, and D. Q. Yang. 2007. AICAR induces phosphorylation of AMPK in an ATM-dependent, LKB1-independent manner. Mol. Cell. Biochem. 306:239-245. [DOI] [PubMed] [Google Scholar]
- 63.Sunavala-Dossabhoy, G., S. K. Balakrishnan, S. Sen, S. Nuthalapaty, and A. De Benedetti. 2005. The radioresistance kinase TLK1B protects the cells by promoting repair of double strand breaks. BMC Mol. Biol. 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki, A., G. Kusakai, A. Kishimoto, Y. Shimojo, T. Ogura, M. F. Lavin, and H. Esumi. 2004. IGF-1 phosphorylates AMPK-alpha subunit in ATM-dependent and LKB1-independent manner. Biochem. Biophys. Res. Commun. 324:986-992. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki, K., S. Kodama, and M. Watanabe. 2001. Extremely low-dose ionizing radiation causes activation of mitogen-activated protein kinase pathway and enhances proliferation of normal human diploid cells. Cancer Res. 61:5396-5401. [PubMed] [Google Scholar]
- 66.Tang, D., D. Wu, A. Hirao, J. M. Lahti, L. Liu, B. Mazza, V. J. Kidd, T. W. Mak, and A. J. Ingram. 2002. ERK activation mediates cell cycle arrest and apoptosis after DNA damage independently of p53. J. Biol. Chem. 277:12710-12717. [DOI] [PubMed] [Google Scholar]
- 67.Tee, A. R., and C. G. Proud. 2002. Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol. Cell. Biol. 22:1674-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tee, A. R., and C. G. Proud. 2000. DNA-damaging agents cause inactivation of translational regulators linked to mTOR signalling. Oncogene 19:3021-3031. [DOI] [PubMed] [Google Scholar]
- 69.Tibbetts, R. S., K. M. Brumbaugh, J. M. Williams, J. N. Sarkaria, W. A. Cliby, S. Y. Shieh, Y. Taya, C. Prives, and R. T. Abraham. 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsokas, P., T. Ma, R. Iyengar, E. M. Landau, and R. D. Blitzer. 2007. Mitogen-activated protein kinase upregulates the dendritic translation machinery in long-term potentiation by controlling the mammalian target of rapamycin pathway. J. Neurosci. 27:5885-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valerie, K., A. Yacoub, M. P. Hagan, D. T. Curiel, P. B. Fisher, S. Grant, and P. Dent. 2007. Radiation-induced cell signaling: inside-out and outside-in. Mol. Cancer Ther. 6:789-801. [DOI] [PubMed] [Google Scholar]
- 73.Walsh, D., and I. Mohr. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 20:461-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watters, D., P. Kedar, K. Spring, J. Bjorkman, P. Chen, M. Gatei, G. Birrell, B. Garrone, P. Srinivasa, D. I. Crane, and M. F. Lavin. 1999. Localization of a portion of extranuclear ATM to peroxisomes. J. Biol. Chem. 274:34277-34282. [DOI] [PubMed] [Google Scholar]
- 75.Yacoub, A., R. McKinstry, D. Hinman, T. Chung, P. Dent, and M. P. Hagan. 2003. Epidermal growth factor and ionizing radiation up-regulate the DNA repair genes XRCC1 and ERCC1 in DU145 and LNCaP prostate carcinoma through MAPK signaling. Radiat. Res. 159:439-452. [DOI] [PubMed] [Google Scholar]
- 76.Yacoub, A., A. Miller, R. W. Caron, L. Qiao, D. A. Curiel, P. B. Fisher, M. P. Hagan, S. Grant, and P. Dent. 2006. Radiotherapy-induced signal transduction. Endocr. Relat. Cancer 13(Suppl. 1):S99-114. [DOI] [PubMed] [Google Scholar]
- 77.Yang, D. Q., and M. B. Kastan. 2000. Participation of ATM in insulin signalling through phosphorylation of eIF-4E-binding protein 1. Nat. Cell Biol. 2:893-898. [DOI] [PubMed] [Google Scholar]
- 78.Yoo, L. I., D. C. Chung, and J. Yuan. 2002. LKB1—a master tumour suppressor of the small intestine and beyond. Nat. Rev. Cancer 2:529-535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.