Abstract
Transcription factor USF is a ubiquitously expressed member of the helix-loop-helix family of proteins. It binds with high affinity to E-box elements and, through interaction with coactivators, aids in the formation of transcription complexes. Previous work demonstrated that USF regulates genes during erythroid differentiation, including HoxB4 and β-globin. Here, we show that the erythroid cell-specific expression of a dominant-negative mutant of USF, A-USF, in transgenic mice reduces the expression of all β-type globin genes and leads to the diminished association of RNA polymerase II with locus control region element HS2 and with the β-globin gene promoter. We further show that the expression of A-USF reduces the expression of several key erythroid cell-specific transcription factors, including EKLF and Tal-1. We provide evidence demonstrating that USF interacts with known regulatory DNA elements in the EKLF and Tal-1 gene loci in erythroid cells. Furthermore, A-USF-expressing transgenic mice exhibit a defect in the formation of CD71+ progenitor and Ter-119+ mature erythroid cells. In summary, the data demonstrate that USF regulates globin gene expression indirectly by enhancing the expression of erythroid transcription factors and directly by mediating the recruitment of transcription complexes to the globin gene locus.
The human β-globin gene locus consists of five genes that are expressed in a developmental stage- and tissue-specific manner in erythroid cells (37, 52). The high-level expression of the β-like globin genes requires a locus control region (LCR), which is located upstream of the globin genes (24). The LCR is composed of many erythroid cell-specific DNase I-hypersensitive (HS) sites (20, 55). These HS sites harbor clusters of DNA binding motifs for ubiquitously expressed or tissue-restricted transcription factors (25, 44). Among these factors are erythroid krüppel-like factor (EKLF), GATA-1, NF-E2 (p45), Tal-1, and USF. These proteins bind to specific sequences in the LCR HS sites and recruit chromatin-modifying activities, coactivators, or components of the basal transcription apparatus, including RNA polymerase II (RNA Pol II) (30). Most of the proteins that interact with the LCR also associate with the globin gene promoters in a developmental stage-specific manner (39). Recent data suggest that the LCR-mediated activation of globin gene expression is associated with physical proximity between the LCR and globin gene promoters, and that highly expressed β-globin genes associate with transcription factories in an LCR-dependent manner (7, 47). To gain better insight into regulatory mechanisms, it is important to identify proteins that recruit transcription complexes to the globin gene locus or that mediate the association of the LCR and the globin genes with RNA Pol II transcription factories.
USF is a ubiquitously expressed transcription factor that binds to DNA E-box motifs and has been associated with the transcription of many cellular and viral genes (12). It belongs to a family of transcription factors characterized by their basic helix-loop-helix leucine zipper (bHLH-LZ) DNA binding domains (50). Currently, there are two known members of this family: USF1 (44 kDa) and USF2 (43 kDa). The predominant form of USF is a USF1/USF2 heterodimer, although homodimers are known to exist in various degrees across cell types (51). Interestingly, most genes activated by USF are expressed at high levels in differentiated cells, including the β-globin gene (12, 13). Previous studies have shown that USF interacts with conserved E-box elements located in LCR element HS2 as well as in the adult β-globin downstream promoter region (5, 13, 17, 35). The expression of a dominant-negative mutant form of USF, A-USF, in mouse erythroleukemia cells leads to the inhibition of βmaj-globin gene expression and a reduction in the recruitment of RNA Pol II to LCR element HS2 and to the βmaj-globin gene promoter (13). A-USF contains the USF heterodimerization domain but lacks the USF-specific region, which is required for transcriptional activation (41, 45). Additionally, the basic DNA binding region has been mutated to contain an acidic extension, which renders A-USF-containing heterodimers unable to bind DNA (45). USF interacts with coactivators and histone modifiers in erythroid cells, suggesting that it functions through chromatin remodeling and RNA Pol II recruitment (14, 26). The genome-wide mapping of USF interaction sites in hepatocytes revealed that it preferentially binds DNA in close proximity to transcription start sites, supporting the hypothesis that USF is involved in transcription complex recruitment (46). However, USF also is known to function at chromatin barrier elements (21, 58).
In the present study, we demonstrate that the erythroid cell-specific expression of A-USF in transgenic mice leads to a decrease in both adult βmaj-globin gene expression and the recruitment of RNA Pol II to the adult βmaj-globin gene promoter. The erythroid cell-specific expression of A-USF also reduces the expression of other erythroid cell-specific genes in the embryonic yolk sac, including embryonic globin genes, Band3, EKLF, Tal-1, and NF-E2 (p45). Furthermore, we show that USF associates with E-box-containing regulatory elements in the EKLF and Tal-1 gene loci.
MATERIALS AND METHODS
Plasmid construction and generation of transgenic mice.
The plasmid expressing dominant-negative USF under the control of erythroid cell-specific regulatory elements was created by digesting pITRp543f2beta4 with NcoI and PmeI, which removed the β-globin gene, the 3′ enhancer, and the 3′ chicken HS4 insulator (29). The A-USF coding region was isolated from a pCMV/A-USF plasmid provided by Charles Vinson (NIH) by digestion with NcoI and PmeI (13, 45). The two restriction fragments then were ligated, creating pITRp543f2A-USF. The 3′ enhancer and 3′ chicken HS4 elements were amplified using pITRp543f2beta4 as the template with primers containing PmeI sites and were cloned into the pTOPO vector (Invitrogen). The pTOPO/3′ enhancer, 3′ chicken HS4, and pITRp543f2A-USF subsequently were digested with PmeI, and the appropriate fragments were isolated and ligated to form the vector pITRp543f2A-USF4. The inserts of the plasmid were sequenced to verify that no mutations were introduced. The plasmid pITRp543f2A-USF4 was linearized and purified through agarose gel electrophoresis followed by DNA extraction from gel slices using a QIAquick gel extraction kit (Qiagen) per the manufacturer's protocol. The linearized plasmid DNA was resuspended in injection buffer at a concentration of 2 ng/μl and injected into fertilized murine oocytes as described by Bungert et al. (6). DNA was prepared from tail clips of the offspring by overnight digestion in DNA lysis buffer containing proteinase K and was purified by a series of one phenol, one phenol-chloroform-isoamyl alcohol, and one chloroform-isoamyl alcohol extraction. The presence of the A-USF expression construct in transgenic mice was determined by PCR.
Cell culture.
Murine erythroleukemia (MEL) cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 5% penicillin-streptomycin. Cells were grown in 5% CO2 at 37°C and maintained at a density between 1 × 105 and 2 × 106 cells/ml. The induction of the erythroid differentiation of MEL cells was achieved by incubating cells in medium containing 1.5% dimethylsulfoxide (DMSO) for 72 h.
RNA isolation and analysis.
RNA was isolated using the guanidinium-thiocyanate method as described previously and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad) (6, 11). RNA was subjected to analysis by quantitative real-time reverse transcription PCR (qRT-PCR) or reverse transcription PCR (RT-PCR) using the MyiQ (Bio-Rad) system, and reactions were carried out using the iQ SYBR green super mix (Bio-Rad). qRT-PCR conditions were the following: 95°C for 5 min, followed by 40 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 1 min (readings were taken after every cycle). A melting curve was performed from 60 to 95°C (readings every 0.5°C). Standard curves were generated using 10-fold serial dilutions of wild-type cDNA from the appropriate source. Final quantification analysis was performed using the relative standard curve method. Gene expression analysis was as described previously, with results reported as expression relative to wild-type levels after the normalization of the transcript data to those of a control gene, GAPDH (13, 14). Primers for amplifying murine βmaj-globin, murine β-actin, and murine GAPDH have been described previously (13). RNA from wild-type and transgenic mice was isolated from embryonic yolk sac, fetal liver, or the spleen of phenylhydrazine-treated anemic mice as described in Bungert et al. (6). Primers for the murine βmaj-globin gene and murine LCR element HS2 used to amplify cDNA in qRT-PCR analyses were as described in Crusselle-Davis et al. (13). In addition, the following primers were used to amplify cDNA: mouse Hba α1, 5′-CCTGGGGGAAGATTGGTG-3′ (upstream [US]) and 5′-GCCGTGGCTTACATCAAAGT-3′ (downstream [DS]); mouse βmin, 5′-TGAGCTCCACTGTGACAAGC-3′ (US) and 5′-TACTTGTGAGCCAGGGCAGT-3′ (DS); mouse HoxB4, 5′-TGGATGCGCAAAGTTCACG-3′ (US) and 5′-GGTCTTTTTTCCACTTCATGCG-3′ (DS); mouse USF1, 5′-GATGAGAAACGGAGGGCTCAACATA-3′ (US) and 5′-TTAGTTGCTGTCATTCTTGATGACG-3′ (DS); mouse p45, 5′-TCAGCAGAACAGGAACAGGT-3′ (US) and 5′-GCTTTGACACTGGTATAGCT-3′ (DS); and mouse Tal-1, 5′-TAGCCTTAGCCAGCCGCTCG-3′ (US) and 5′-GCGGAGGATCTCATTCTTGC-3′ (DS). Primers for mouse GATA-1 and EKLF analysis were described by Tanabe et al., primers for mouse βH1 were described by Basu et al., and primers for mouse Band3 were described by Nilson et al. (4, 42, 54).
ChIP and μChIP.
Conventional chromatin immunoprecipitation (ChIP) assays were performed as described previously (13, 35). Spleens from anemic mice were homogenized, and cell suspensions were subjected to ChIP analysis. The following antibodies were used in this study: USF1, USF2 (H-86 and N-18, respectively; Santa Cruz Biotechnology), and RNA Pol II (CTD45H8; Upstate Biotechnology, Inc.). In addition, for the yolk sac samples, MicroChIP (μChIP) was used according to a previous protocol with minor modifications (15, 16). Antibody-bead complexes were prepared using Dynabeads Protein A beads (Invitrogen). Embryonic yolk sacs were cross-linked with 1% formaldehyde in 500 μl phosphate-buffered saline (PBS) and quenched with 125 mM glycine. Prior to sonication, yolk sacs were homogenized using a glass tissue grinder (Radnoti) and washed with PBS. Sonication conditions were optimized to yield fragments of ∼500 bp, and sonication products were diluted 10-fold. Sonicated chromatin was incubated with various antibody-bead complexes, and after a series of washes, DNA was purified using a QIAprep Spin Miniprep kit (Qiagen). Quantitative real-time PCR (q-PCR) conditions were the following: 95°C for 5 min, followed by 40 cycles of 94°C for 30 s, 59°C for 20 s, and 72°C for 30 s. A melting curve was performed from 60 to 95°C (reading every 0.5°C). Negative control experiments were performed using (i) immunoglobulin G (IgG) antibodies and (ii) primers amplifying a region between LCR elements HS2 and HS3 that does not interact with USF and Pol II (14, 57 and data not shown). Standard curves were generated using 10-fold serial dilutions of the input DNA. Final quantification analysis was performed using the relative standard curve method. ChIP primers used for amplifying murine LCR element HS2 and the murine βmaj-globin promoter have been described previously (13). In addition, the following primers were used: mouse GATA-1 promoter, 5′-AGCCTCTGCTTGAAATGCTC-3′ (US) and 5′-CCTTTGGCTTCTGTGGAGTC-3′ (DS); mouse Tal-1 promoter, 5′-CAGATCCGTTAGAGGGTTCG-3′ (US) and 5′-CTGGGAATTACCTCGTGTGC-3′ (DS); mouse NF-E2 (p45) promoter, 5′-GCAGACACAGTGAGCACTCC-3′ (US) and 5′-GAGGGTCCTTAGGTGGGAGA-3′ (DS); and mouse Necdin promoter, 5′-TTTACATAAGCCTAGTGGTACCCTCC-3′ (US) and 5′-ATCGCTGTCCTGCATCTCACAGTCG-3′ (DS). Primers for the mouse EKLF gene promoter were described by Vakoc et al. (56).
Protein isolation and Western blotting.
Proteins were isolated and analyzed by Western blotting as described by Leach et al. (35). A total of 20 μg was loaded onto 7.5% Ready Gel (Bio-Rad). After transfer, proteins were detected using the ECL plus system (Amersham Pharmacia). Primary antibodies for USF1 and USF2 are the same as those used in the ChIP assays.
FACS analysis.
Cells obtained from yolk sac of transgenic and wild-type murine male embryos at 10.5 days postcoitum (dpc), as determined by Y chromosome-specific PCR, were subjected to fluorescence-activated cell sorting (FACS) analysis using antibodies against fluorescein isothiocyanate CD71 (BD Biosciences) and phycoeryrthrin-Cy7 Ter-119 (eBioscience). Cells were homogenized in PBS containing 2% FBS using a glass tissue grinder (Radnoti), passed through a 70-μm cell strainer, and incubated on ice with antibodies for 30 min. After a series of washes to remove unbound antibodies, cells were subjected to FACS using a BD LSRII system. Ter-119+ yolk sac cells collected for RNA extraction were homogenized with collagenase in PBS containing 20% FBS. Y chromosome primers used for identifying male embryos were described by Kunieda et al. (33). CD71+ cells represent early erythroid progenitor cells, while Ter-119+ cells represent more mature erythroid cells (40).
RESULTS
We previously demonstrated that USF activates the expression of the adult β-globin gene in erythroid cell lines, while a dominant-negative mutant of USF (A-USF) inhibits expression (13). Targeted deletions of either the USF1 or USF2 coding region in mice do not result in obvious hematopoietic defects, suggesting that USF1 and USF2 are able to partially compensate for each other during mouse embryonic development (8). This is supported by the observation that the compound homozygous mutations lead to early embryonic lethality (8). We thus hypothesized that the expression of A-USF exclusively or preferentially in erythroid cells interferes with the function of both USF1 and USF2 without affecting vital functions of these proteins in other tissues and organs. A-USF contains the USF-specific dimerization domain and sufficiently inhibits the function of both USF1 and USF2 (45). As shown in Fig. 1A, the A-USF coding region is under the control of human β-globin LCR elements HS2 and HS3, the human β-globin promoter, and the human β-globin downstream enhancer element. The expression cassette also is flanked by two copies of the chicken HS4 insulator sequence to protect the transgene from position effects (Fig. 1A). We previously demonstrated that this construct is able to express a β-globin gene at high levels at various integration sites in transgenic mice, including a region close to the centromere (29). However, it should be noted that the construct may be subject to autoregulation by A-USF itself because it contains two USF recognition sites: one in HS2 and one in the β-globin promoter. This also may limit the potential deleterious effect of expressing the dominant-negative mutant.
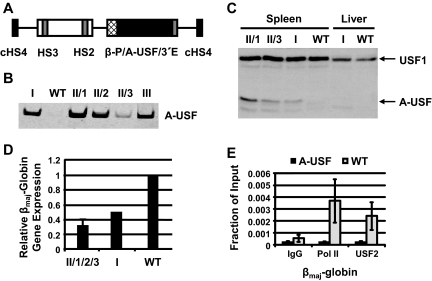
FIG. 1.
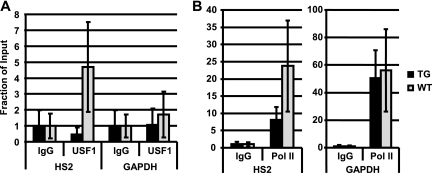
Generation and analysis of mice expressing A-USF. (A) DNA construct pITRp543f2A-USF4 used to generate transgenic mice expressing dominant-negative USF (A-USF). The A-USF coding region is under the control of the human β-globin gene promoter (β-P) and 3′ enhancer (3′E) as well as human LCR elements HS2 and HS3 plus flanking DNA. The DNA construct is flanked on either site by insulator elements derived from the chicken β-globin gene locus (cHS4). (B) SYBR green stain of the RT-PCR analysis of A-USF expression in transgenic (founders I and III and line II F1 littermates 1 to 3 [II/1 to II/3]) and wild-type (WT) mice. RNA was isolated from the spleens of phenylhydrazine-treated mice, reverse transcribed, subjected to PCR analysis with primers specific to the A-USF coding region, and electrophoresed in 5% Tris-borate-EDTA polyacrylamide gels. (C) Western blot analysis of A-USF expression in transgenic or wild-type mice. Protein was isolated from the spleen or liver of phenylhydrazine-treated mice and subjected to Western blot analysis using an antibody against USF1, which also detects A-USF. (D) qRT-PCR analysis of βmaj-globin gene expression in spleens of A-USF transgenic line II F1 littermates, A-USF founder mouse I, and wild-type mice. Data from the three line II F1 littermates were combined and are designated II/1/2/3. GAPDH was used as a loading control, and results from samples were normalized to those of the wild type. (E) ChIP analysis of RNA Pol II and USF2 interactions with the βmaj-globin gene promoter control mice (WT) and transgenic mice (A-USF). Spleens taken from two phenylhydrazine-treated F1 females (derived from line II) or wild-type mice were homogenized and subjected to ChIP analysis using antibodies against IgG, RNA Pol II, or USF2. Error bars reflect standard deviations from two independent experiments.
Three transgenic founders were generated with this construct (founders I, II, and III), although two of these founders did not transmit the transgene (founders I and III). A-USF was expressed in the spleen of all transgenic mice but not in the liver (Fig. 1B and C and data not shown). The analysis of A-USF in the spleen of three F1 females from founder II revealed that the expression of A-USF varied between littermates (Fig. 1B and C). The founder of line II is male, and we failed to obtain transgenic male offspring; we thus reason that the transgene integrated into the X chromosome and that the expression of A-USF in all erythroid cells is not compatible with survival. Females showed a somewhat variegated phenotype, likely because of differences in the silencing of the transgene on the X chromosome.
To analyze the effect of expressing A-USF on βmaj-globin gene expression, we treated transgenic (founder I and three F1 females from founder II) and four control wild-type (WT) littermates with phenylhydrazine, which induces hemolytic anemia and increases the number of nucleated red blood cells in the spleen. We found that βmaj-globin gene expression was reduced by 50% in the nontransmitting transgenic mouse (founder I) compared to expression in a wild-type control mouse (Fig. 1D). The expression of A-USF in the spleen of phenylhydrazine-treated F1 animals from the transmitting line varied (Fig. 1B), and it resulted in a two- to fivefold decrease in βmaj-globin gene expression in three transgenic littermates (line II samples 1 to 3) (Fig. 1D) compared to that of three wild-type littermates. Five transgenic female mice but none of the wild-type mice died as a result of the phenylhydrazine treatment, indicating a possible defect in erythropoiesis. Consistently with this observation is the fact that 4-week-old transgenic female mice weighed 2 to 3 g less than wild-type littermates (23 ± 1 and 26 ± 1 g, respectively).
The results shown in Fig. 1 demonstrate that USF is required for the high-level expression of the adult βmaj-globin gene. We also observed a reduction in α-globin gene expression that was comparable to a reduction in βmaj-globin (data not shown). The expression of A-USF does not globally affect gene expression; we did not observe a change in β-actin or GAPDH gene expression (data not shown). We next analyzed the recruitment of RNA Pol II at the murine βmaj-globin gene in mice expressing or not expressing A-USF (Fig. 1E). The expression of A-USF in erythroid cells led to a reduction in USF2 and Pol II binding to the βmaj-globin promoter. The recruitment of RNA Pol II to the control β-actin gene was not affected in A-USF-expressing mice (data not shown).
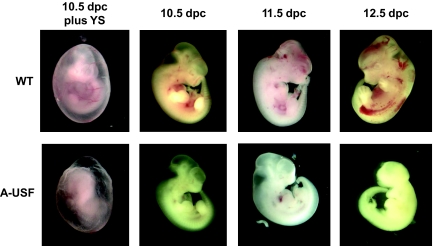
Because we were unable to obtain transgenic males, we examined embryos at different stages of development. At 14.5 dpc, the fetal liver is the major site of erythropoiesis. In two different litters at 14.5 dpc, which were obtained by mating a transgenic female (line II) with a wild-type male, we detected several reabsorbed and pale embryos. Genotyping with primers specific for the A-USF transgenic construct and for the Y chromosome revealed that the reabsorbed embryos were transgenic males (data not shown). We next examined embryos at earlier stages: 10.5, 11.5, and 12.5 dpc. At 10.5 and 11.5 dpc, all embryos appeared to be alive and normally developed; however, several of the embryos were pale, and these were identified by PCR as transgenic males (Fig. 2). At day 12.5, the male transgenic embryos ceased to develop further, demonstrating that the male transgenic embryos did not survive beyond 11.5 dpc.
FIG. 2.
Analysis of transgenic mouse embryos at different stages of development. Male embryos were isolated at the indicated time points of development from A-USF transgenic females (F1 females from line II) mated with wild-type (WT) males. Embryos were placed in a culture dish with PBS either in the presence or absence of the yolk sac (YS) and photographed using a Leica MZ16F4 instrument and the Qcapture program. Embryos were genotyped for sex and determined to be wild-type or transgenic (A-USF).
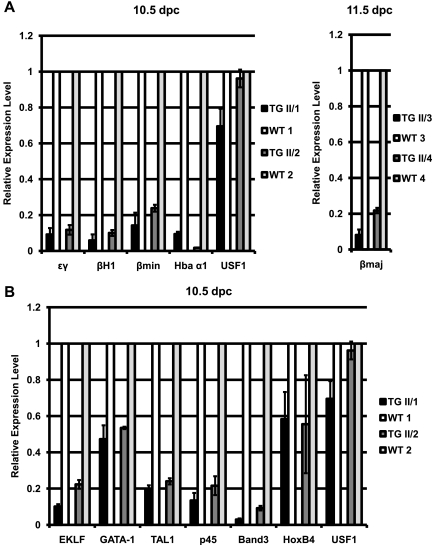
We next examined the expression of globin genes in 10.5- and 11.5-dpc embryos. The expression of A-USF caused a reduction in the expression of all globin genes compared to that of wild-type littermates (Fig. 3A). In 10.5-dpc yolk sac samples, the expression of the embryonic ɛγ and βH1 genes as well as that of the Hba α1- and βmin-globin genes was reduced 5- to 10-fold, and in 11.5-dpc fetal liver samples there was a 5- to 10-fold reduction in the expression of the adult βmaj-globin gene. We also examined the effect of A-USF on the expression of other erythroid cell-specific genes, including those encoding transcription factors regulating erythropoiesis, like GATA-1, EKLF, Tal-1, NF-E2 (p45), and HoxB4 (Fig. 3B). HoxB4, a homeobox transcription factor expressed in primitive hematopoietic stem cells, previously has been shown to be regulated by USF in K562 cells (22). We found that the expression of HoxB4 and GATA-1 is reduced by only about twofold in the yolk sac of transgenic males. In contrast, the expression of transcription factors EKLF, Tal-1, and NF-E2 (p45) was reduced by 5- to 10-fold, suggesting that USF is required for the expression of these genes during primitive erythropoiesis. The expression of another well-characterized erythroid cell-specific gene, Band3, also was reduced by more than fivefold in the transgenic yolk sac samples. The expression of A-USF did not affect the transcription of USF1 or USF2 (Fig. 3A and B and data not shown) or that of the housekeeping genes GAPDH and β-actin (data not shown).
FIG. 3.
Effects of A-USF expression on the expression of erythroid genes and erythroid cell-specific transcription factors. RNA was extracted from 10.5- or 11.5-dpc embryos, reverse transcribed, and subjected to qRT-PCR performed in triplicate. (A) qRT-PCR analysis of ɛγ-globin, βH1-globin, βmin-globin, Hba α1, USF1 (left; 10.5 dpc), and βmaj-globin (right; 11.5 dpc) gene expression in A-USF transgenic (TG II/1 to II/4) and wild-type (WT 1 to 4) mouse embryos. Two sets of four embryos, each containing two TG and two WT animals, were examined. GAPDH was used as an internal control, and sample data were normalized to those for a respective WT littermate. Data are represented as means ± standard errors of the means of at least three PCRs on each sample. (B) qRT-PCR analysis of EKLF, GATA-1, Tal-1, p45, Band3, HoxB4, and USF1 gene expression in the yolk sac of the transgenic (TG II/1 and II/2) and wild-type (WT 1 and 2) 10.5-dpc embryos examined in panel A (left). Data are presented as described for panel A.
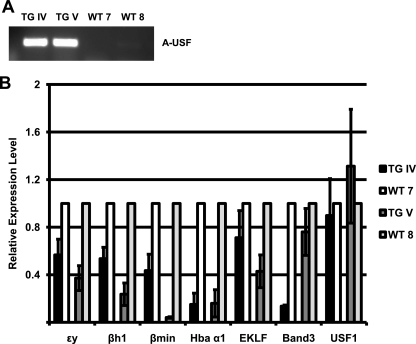
To exclude the possibility that the phenotype we observed in the male transgenic embryos is due to the integration of the transgene and the consequent disruption of a specific cellular function, we generated and analyzed two transient transgenic embryos at 10.5 dpc. Both embryos appeared pale, expressed A-USF as measured by RT-PCR (Fig. 4A), and revealed reductions in the expression of α- and β-globin genes as well as that of EKLF and Band3 (Fig. 4B). The expression of USF1 was not affected in these mice. The data demonstrate that the expression of A-USF in erythroid cells of transgenic mice leads to consistent defects in erythropoiesis in multiple independent transgenic embryos. Therefore, the erythroid phenotype observed in the transmitting line (line II) is unlikely to be due to the disruption of gene expression patterns at the site of transgene integration.
FIG. 4.
Generation and analysis of transient transgenic mouse embryos expressing A-USF. Fertilized oocytes were injected with the A-USF expression construct and implanted into the uterus of a pseudopregnant foster mother. Embryos (11.5 dpc) were isolated and subjected to DNA (embryo) and RNA (yolk sac) extraction. (A) cDNA from the embryos was analyzed by RT-PCR using primers specific for the A-USF transgene to verify A-USF expression. All four embryos, two transgenic (TG IV and TG V) and two wild-type (WT 7 and WT 8) embryos, were taken from the same litter. (B) RNA was subjected to qRT-PCR performed in triplicate for the analysis of ɛγ-globin, βH1-globin, βmin-globin, Hba α1, EKLF, Band3, and USF1 gene expression. Data were analyzed and are represented as described in the legend to Fig. 3A.
To verify that the expression of A-USF affects the binding of USF in transgenic embryos, we examined the binding of USF1 to LCR element HS2 in yolk sac samples taken from 10.5-dpc transgenic embryos (line II) and wild-type litter mates using the μChIP assay, which allows the detection of protein-chromatin interactions with a small number of cells. The binding of USF1 to the LCR was reduced in transgenic embryos compared to that of wild-type littermates (Fig. 5A). The interaction of RNA Pol II with LCR element HS2 also was reduced in 10.5-dpc yolk sac samples from transgenic mice compared to that of littermates (Fig. 5B), whereas there was no change in the association of RNA Pol II with the GAPDH gene between wild-type and transgenic embryos (Fig. 5B).
FIG. 5.
μChIP analysis of RNA Pol II and USF1 association with LCR element HS2 and the GAPDH gene in the yolk sac of wild-type and A-USF transgenic embryos. Embryos (10.5 dpc) were taken from an A-USF transgenic female (mated to a wild-type male). Yolk sacs were isolated and subjected to μChIP analysis. (A) μChIP was performed with antibodies against negative control IgG and USF1. DNA was analyzed by qPCR using primers specific for LCR element HS2 as well as for the control GAPDH gene, as indicated. (B) μChIP was performed with antibodies against the negative control IgG and RNA Pol II. The DNA was analyzed by qPCR using primers specific for LCR element HS2 as well as for the control GAPDH gene, as indicated. Data were normalized to those for IgG and are represented as means ± standard errors of the means of three independent μChIP experiments with qPCRs performed in triplicate.
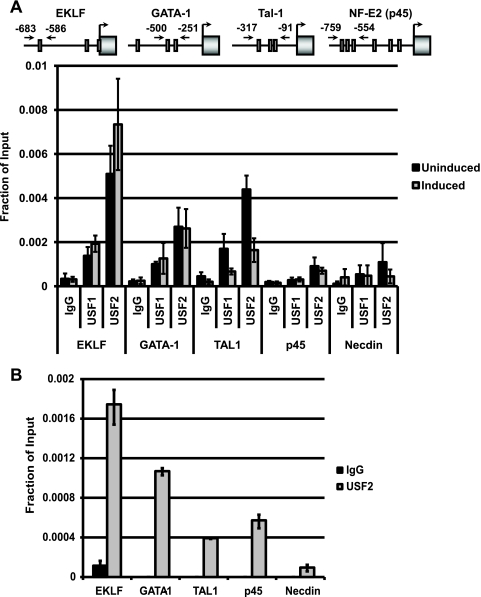
Because EKLF, Tal-1, and NF-E2 (p45) failed to be expressed at high levels in the hematopoietic tissue of transgenic mice, we examined the possibility that USF directly regulates these genes. We performed ChIP to examine the interaction of USF with the gene loci encoding these transcription factors during the differentiation of murine erythroleukemia (MEL) cells. One of the multiple DNA regulatory elements in the EKLF gene locus contains an E-box, which previously has been shown to interact with Tal-1 (1, 2). Both subunits of USF associated with the E-box-containing regulatory region of the EKLF gene in MEL cells (Fig. 6A). We also observed interactions of USF with the Tal-1 gene locus, which also contains an E-box motif in a regulatory element. Interestingly, the interaction of USF with the Tal-1 gene decreased during DMSO-induced MEL cell differentiation. USF binding also was detectable at the GATA-1 gene locus (Fig. 6A). There are no previous data concerning E-box elements regulating the GATA-1 gene. We failed to detect significant interactions of USF1 with the NF-E2 (p45) gene; however, the recovery of USF2-precipitated p45 gene fragments was higher than that of the IgG control. The data suggest that the Tal-1 and EKLF genes are direct targets of both USF1 and USF2 in differentiating erythroid cells. There was no significant binding of USF to the control Necdin gene, which is not expressed in erythroid cells (Fig. 6A). We confirmed the interactions of USF2 with the erythroid cell-specific gene loci in primary erythroid cells taken from 16.5-dpc mouse fetal liver samples (Fig. 6B). The ChIP results demonstrated that USF2 interacts with the EKLF, GATA-1, and Tal-1 gene loci but not with the Necdin gene locus. We observed a reproducible interaction of USF2 with the NF-E2 (p45) gene locus in fetal liver cells.
FIG. 6.
Interaction of USF with regulatory elements of genes encoding hematopoietic-specific transcription factors. ChIP analysis of the interaction of USF1 and USF2 with regulatory elements of the EKLF, GATA-1, Tal-1, and NF-E2 (p45) genes as well as with the Necdin promoter serving as a negative control. (A) The diagrams at the top indicate the position of E-boxes with respect to the transcription start site of each individual gene, with arrows indicating the location of primers used to amplify each region. ChIP was performed on uninduced or induced MEL cells. Cells were induced to differentiate for 3 days in the presence of 1.5% DMSO. Uninduced and induced cells were incubated with 1% formaldehyde. After being quenched with 125 mM glycine, the cells were lysed and chromatin was fragmented by sonication prior to precipitation with antibodies specific for IgG, USF1, or USF2. The isolated DNA was analyzed by qPCR with primers specific for the EKLF, GATA-1, Tal-1, NF-E2 (p45), and Necdin gene promoters, as indicated. Results are represented as means ± standard errors of the means of three independent experiments, with each PCR performed in duplicate. (B) ChIP performed on 16.5-dpc liver cells, examining the interaction of USF2 with the regions examined in panel A. Results are represented as means ± standard errors of the means of two independent experiments with PCRs performed in duplicate.
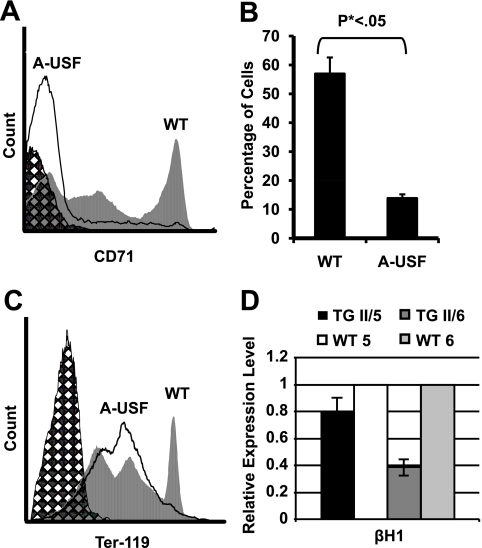
We next examined the possibility that the expression of A-USF in erythroid cells impairs their differentiation potential. We began these studies by examining 10.5-dpc yolk sac cells from transgenic embryos and wild-type littermates for the expression of the transferrin receptor CD71, which is expressed at high levels in developing erythroid cells and serves as a marker for erythroid progenitors (40). The CD71-mediated sorting of yolk sac cells revealed that the number of CD71+ cells was about threefold lower in the transgenic embryos than that of the wild-type embryos (Fig. 7A and B). Furthermore, we observed a decrease in the number of cells that express high levels of Ter-119 (Fig. 7C), which is a marker for more differentiated erythroid cells (32). The number of benzidine-positive cells also was reduced by three to fourfold in the yolk sac cells from transgenic embryos compared to those taken from wild-type littermates (data not shown). Taken together, these results demonstrate that USF is an important contributor to erythroid cell differentiation and mediates the high-level expression of erythroid transcription factors and the expression of the globin genes.
FIG. 7.
Transgenic (TG) A-USF embryos reveal a reduction in the number of CD71-positive and Ter-119-positive erythroid cells. Yolk sac cells from 10.5-dpc male embryos were isolated and subjected to flow cytometry against CD71 or Ter-119. Hatched areas indicate unstained yolk sac cells analyzed separately. Solid lines represent the analysis of cells from A-USF-expressing transgenic embryos, while gray-shaded areas represent cells from wild-type embryos. (A) FACS analysis using antibodies against CD71. (B) Number of CD71-positive cells in the 10.5-dpc yolk sac of wild-type (WT) and A-USF-expressing (A-USF) embryos. (C) FACS analysis using antibodies against Ter-119. (D) βH1-globin gene expression in Ter-119+ embryonic yolk sac cells. Yolk sac cells from 10.5-dpc embryos were sorted using Ter-119 antibodies, and a subset of Ter-119+ cells was collected and subjected to RNA extraction. Data were analyzed as described in the legend to Fig. 3A and are represented as the means ± standard errors of the means of two qRT-PCRs performed in duplicate.
To examine whether USF not only regulates the differentiation of erythroid cells but also functions within the context of differentiating cells, we analyzed the expression of the βH1-globin gene in Ter-119-sorted cells obtained from transgenic or wild-type embryos (Fig. 7D). The expression of the βH1-globin gene was reduced in Ter-119+ cells isolated from two transgenic embryos compared to that of their wild-type littermates.
DISCUSSION
It currently is unknown how ubiquitously expressed and tissue-specific transcription factors coordinate the activation of highly expressed genes during differentiation. Perhaps tissue-specific factors mediate the accessibility of regulatory sites, whereas ubiquitously expressed proteins perform basic functions involved in the local remodeling of nucleosomes and the recruitment of transcription complexes.
The β-globin genes are competitively regulated by the LCR (10, 19). During the activation of the β-like globin genes, the LCR comes in close proximity to the genes. Furthermore, it appears that during the differentiation of erythroid cells, certain factors and protein complexes, including RNA Pol II, first associate with the LCR before they interact with the globin gene promoters (31, 38, 57). The LCR could serve as the primary site of recruitment for activities involved in globin gene regulation (34, 37). These activities could be transferred to the globin genes by looping mechanisms (18). If mechanisms are known that mediate the stage-specific association of the globin genes with the LCR, strategies could be developed to change these association patterns. This could lead to novel therapies for the treatment of sickle cell anemia or other hemoglobinopathies, e.g., by favoring interactions of the LCR with therapeutic γ-globin genes over those with the mutant β-globin genes.
USF was one of the first transcription factors shown to activate transcription mediated by RNA Pol II, and it plays a role in the high-level expression of many genes in differentiated cells (12, 48). Accumulating evidence points to the possibility that highly expressed genes are transcribed in specialized nuclear domains enriched for splicing factors and RNA Pol II, often referred to as transcription factories or transcription domains (9, 27, 53). It is possible that the LCR nucleates such a transcription domain in erythroid cells (39). USF is a likely candidate protein that could mediate the association of genes or regulatory elements with transcription domains in the nucleus. USF mediates the high-level expression of genes during cellular differentiation, and the global analysis of the interaction of USF with chromatin revealed that USF mostly binds to regions close to transcription start sites (46).
The data presented here suggest that USF regulates many genes involved in erythropoiesis, including genes encoding key erythroid transcription factors. Inactivating USF thus causes a defect in the differentiation of erythroid cells. This is supported by our observation that the expression of A-USF in transgenic mice causes reductions in the number of CD71+ and Ter-119+ cells in the yolk sac (Fig. 7). In addition to regulating erythropoiesis, several lines of evidence suggest that USF also directly regulates the recruitment of transcription complexes to the β-globin gene locus. First, both USF1 and USF2 interact with LCR element HS2 and with the adult β-globin gene promoter in vitro and in the context of intact erythroid cells (5, 13, 17, 35). Electrophoretic mobility shift assays using protein extracts from erythroid cells demonstrated that a single complex is formed on the E-box derived from the adult β-globin downstream promoter region (35). This complex is supershifted with USF antibodies. This result suggests that no other helix-loop-helix protein present in the protein extract, e.g., Tal-1, is capable of interacting with this site in vitro. The expression of a dominant-negative mutant of USF in MEL cells (13) or transgenic mice (shown here) reduces the recruitment of USF and Pol II to the β-globin gene locus. Furthermore, we demonstrated that A-USF reduces the recruitment of RNA Pol II to immobilized LCR templates in vitro (Z. Zhou and J. Bungert, unpublished data). Finally, we observed a reduction in globin gene expression in Ter-119+ cells isolated from A-USF-expressing transgenic embryos (Fig. 7D). All of these data are consistent with the hypothesis that USF plays a direct role in the regulation of erythroid-specific genes, including the globin genes. However, USF does not appear to act globally on transcription, as we did not observe changes in the expression of housekeeping genes in the A-USF-transgenic mice.
The reduction of embryonic globin gene expression in A-USF-expressing transgenic mice could be due to reduced interactions of USF and transcription complexes with the LCR, which may impair its activity. In addition, we also found a reduction in the expression of EKLF, Tal-1, and NF-E2 (p45), whereas the expression of GATA-1 and HoxB4 was only mildly reduced. Therefore, the reduced expression of these key erythroid transcription factors likely also contributes to decreased globin gene expression in the yolk sac of A-USF transgenic mice. We provide evidence that USF interacts with E-box-containing regulatory elements in the EKLF and Tal-1 gene loci. We propose that USF functions within the context of erythroid-specific transcription domains in the nucleus, and that genes expressed during erythropoiesis associate with these domains, which is consistent with data from Osborne et al. (43).
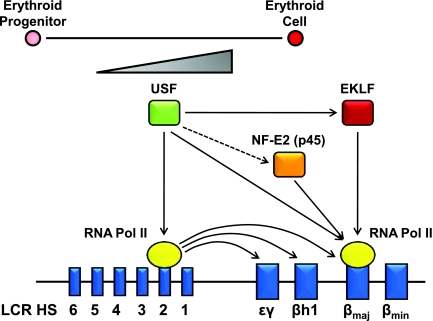
Previous studies have shown that EKLF is required for the recruitment of chromatin remodeling complexes to the LCR and to the adult β-globin gene promoter (3, 23, 36). Interestingly, a recent report from Sengupta et al. revealed a requirement for EKLF in the recruitment of TAF9, which interacts with a basal promoter element in the β-globin gene, suggesting that EKLF is directly involved in recruiting transcription complexes to the β-globin gene promoter (49). NF-E2 (p45) also is required for the efficient recruitment of RNA Pol II to the adult β-globin gene promoter, but it is dispensable for its initial recruitment to the LCR (28). Since both proteins appear to be regulated by USF, our data strongly suggest that USF regulates β-globin gene expression indirectly by enhancing the expression of erythroid-specific transcription factors and directly by cooperating with these factors in the recruitment of transcription complexes to the globin gene locus (Fig. 8).
FIG. 8.
Model depicting USF-mediated regulation of β-globin gene expression. The expression of USF increases during the differentiation of erythroid cells. USF regulates the recruitment of transcription complexes to the β-globin gene locus by interacting with E-boxes located in LCR element HS2 and in the adult β-globin gene promoter. Through the LCR, USF regulates the expression of the embryonic genes. USF further regulates the expression of the globin genes indirectly by enhancing the expression of erythroid cell-specific transcription factors with which it cooperates in mediating the recruitment of transcription complexes to the globin gene locus.
Acknowledgments
We thank Mike Rule, Dan Tuttle, Gary Brown, and Ed Scott from the UF transgenic core facility for the generation of A-USF transgenic mice and Cortney M. Bouldin (UF) for help with microscopy. We thank Joeva Barrow as well as Neal Benson and Steve McClellan from the UF ICBR for assistance with flow cytometry. We are grateful to Justin S. Bickford (UF) for assistance with copyediting and Thomas P. Yang (UF) and Zhuo Zhou (UP) for helpful discussions. We thank Keiji Tanimoto (Tsukuba, Japan) for reading the manuscript and helpful advice.
This project was supported by grants from the NIH (DK 52356 to J.B. and HL 090589 to S.H.).
Footnotes
Published ahead of print on 24 August 2009.
REFERENCES
- 1.Anderson, K. P., S. C. Crable, and J. B. Lingrel. 2000. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood 95:1652-1655. [PubMed] [Google Scholar]
- 2.Anderson, K. P., S. C. Crable, and J. B. Lingrel. 1998. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J. Biol. Chem. 273:14347-14354. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 4.Basu, P., P. E. Morris, J. L. Haar, M. A. Wani, J. B. Lingrel, K. M. Gaensler, and J. A. Lloyd. 2005. KLF2 is essential for primitive erythropoiesis and regulates the human and murine embryonic beta-like globin genes in vivo. Blood 106:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnick, E. H., and G. Felsenfeld. 1993. Evidence that the transcription factor USF is a component of the human beta-globin locus control region heteromeric protein complex. J. Biol. Chem. 268:18824-18834. [PubMed] [Google Scholar]
- 6.Bungert, J., U. Dave, K. C. Lim, K. H. Lieuw, J. A. Shavit, Q. Liu, and J. D. Engel. 1995. Synergistic regulation of human beta-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 9:3083-3096. [DOI] [PubMed] [Google Scholar]
- 7.Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 8.Casado, M., V. S. Vallet, A. Kahn, and S. Vaulont. 1999. Essential role in vivo of upstream stimulatory factors for a normal dietary response of the fatty acid synthase gene in the liver. J. Biol. Chem. 274:2009-2013. [DOI] [PubMed] [Google Scholar]
- 9.Chakalova, L., E. Debrand, J. A. Mitchell, C. S. Osborne, and P. Fraser. 2005. Replication and transcription: shaping the landscape of the genome. Nat. Rev. Genet. 6:669-677. [DOI] [PubMed] [Google Scholar]
- 10.Choi, O. R., and J. D. Engel. 1988. Developmental regulation of beta-globin gene switching. Cell 55:17-26. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 12.Corre, S., and M. D. Galibert. 2005. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 18:337-348. [DOI] [PubMed] [Google Scholar]
- 13.Crusselle-Davis, V. J., K. F. Vieira, Z. Zhou, A. Anantharaman, and J. Bungert. 2006. Antagonistic regulation of beta-globin gene expression by helix-loop-helix proteins USF and TFII-I. Mol. Cell. Biol. 26:6832-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crusselle-Davis, V. J., Z. Zhou, A. Anantharaman, B. Moghimi, T. Dodev, S. Huang, and J. Bungert. 2007. Recruitment of coregulator complexes to the beta-globin gene locus by TFII-I and upstream stimulatory factor. FEBS J. 274:6065-6073. [DOI] [PubMed] [Google Scholar]
- 15.Dahl, J. A., and P. Collas. 2008. MicroChIP—a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 36:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl, J. A., and P. Collas. 2007. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells 25:1037-1046. [DOI] [PubMed] [Google Scholar]
- 17.Elnitski, L., W. Miller, and R. Hardison. 1997. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region. Role of basic helix-loop-helix proteins. J. Biol. Chem. 272:369-378. [DOI] [PubMed] [Google Scholar]
- 18.Engel, J. D., and K. Tanimoto. 2000. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell 100:499-502. [DOI] [PubMed] [Google Scholar]
- 19.Enver, T., N. Raich, A. J. Ebens, T. Papayannopoulou, F. Costantini, and G. Stamatoyannopoulos. 1990. Developmental regulation of human fetal-to-adult globin gene switching in transgenic mice. Nature 344:309-313. [DOI] [PubMed] [Google Scholar]
- 20.Forrester, W. C., S. Takegawa, T. Papayannopoulou, G. Stamatoyannopoulos, and M. Groudine. 1987. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 15:10159-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher, P. G., D. G. Nilson, L. A. Steiner, Y. D. Maksimova, J. Y. Lin, and D. M. Bodine. 2009. An insulator with barrier-element activity promotes alpha-spectrin gene expression in erythroid cells. Blood 113:1547-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giannola, D. M., W. D. Shlomchik, M. Jegathesan, D. Liebowitz, C. S. Abrams, T. Kadesch, A. Dancis, and S. G. Emerson. 2000. Hematopoietic expression of HOXB4 is regulated in normal and leukemic stem cells through transcriptional activation of the HOXB4 promoter by upstream stimulating factor (USF)-1 and USF-2. J. Exp. Med. 192:1479-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillemans, N., R. Tewari, F. Lindeboom, R. Rottier, T. de Wit, M. Wijgerde, F. Grosveld, and S. Philipsen. 1998. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the beta-globin locus control region in vivo. Genes Dev. 12:2863-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosveld, F., G. B. van Assendelft, D. R. Greaves, and G. Kollias. 1987. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51:975-985. [DOI] [PubMed] [Google Scholar]
- 25.Hardison, R., J. L. Slightom, D. L. Gumucio, M. Goodman, N. Stojanovic, and W. Miller. 1997. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene 205:73-94. [DOI] [PubMed] [Google Scholar]
- 26.Huang, S., X. Li, T. M. Yusufzai, Y. Qiu, and G. Felsenfeld. 2007. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol. Cell. Biol. 27:7991-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iborra, F. J., A. Pombo, J. McManus, D. A. Jackson, and P. R. Cook. 1996. The topology of transcription by immobilized polymerases. Exp. Cell Res. 229:167-173. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 29.Kang, S. H., P. P. Levings, F. Andersen, P. J. Laipis, K. I. Berns, R. T. Zori, and J. Bungert. 2004. Locus control region elements HS2 and HS3 in combination with chromatin boundaries confer high-level expression of a human beta-globin transgene in a centromeric region. Genes Cells 9:1043-1053. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S. I., and E. H. Bresnick. 2007. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene 26:6777-6794. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S. I., S. J. Bultman, C. M. Kiefer, A. Dean, and E. H. Bresnick. 2009. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl. Acad. Sci. USA 106:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kina, T., K. Ikuta, E. Takayama, K. Wada, A. S. Majumdar, I. L. Weissman, and Y. Katsura. 2000. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br. J. Haematol. 109:280-287. [DOI] [PubMed] [Google Scholar]
- 33.Kunieda, T., M. Xian, E. Kobayashi, T. Imamichi, K. Moriwaki, and Y. Toyoda. 1992. Sexing of mouse preimplantation embryos by detection of Y chromosome-specific sequences using polymerase chain reaction. Biol. Reprod. 46:692-697. [DOI] [PubMed] [Google Scholar]
- 34.Leach, K. M., K. Nightingale, K. Igarashi, P. P. Levings, J. D. Engel, P. B. Becker, and J. Bungert. 2001. Reconstitution of human beta-globin locus control region hypersensitive sites in the absence of chromatin assembly. Mol. Cell. Biol. 21:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leach, K. M., K. F. Vieira, S. H. Kang, A. Aslanian, M. Teichmann, R. G. Roeder, and J. Bungert. 2003. Characterization of the human beta-globin downstream promoter region. Nucleic Acids Res. 31:1292-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, C. H., M. R. Murphy, J. S. Lee, and J. H. Chung. 1999. Targeting a SWI/SNF-related chromatin remodeling complex to the beta-globin promoter in erythroid cells. Proc. Natl. Acad. Sci. USA 96:12311-12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levings, P. P., and J. Bungert. 2002. The human beta-globin locus control region. Eur. J. Biochem. 269:1589-1599. [DOI] [PubMed] [Google Scholar]
- 38.Levings, P. P., Z. Zhou, K. F. Vieira, V. J. Crusselle-Davis, and J. Bungert. 2006. Recruitment of transcription complexes to the beta-globin locus control region and transcription of hypersensitive site 3 prior to erythroid differentiation of murine embryonic stem cells. FEBS J. 273:746-755. [DOI] [PubMed] [Google Scholar]
- 39.Liang, S., B. Moghimi, T. P. Yang, J. Strouboulis, and J. Bungert. 2008. Locus control region mediated regulation of adult beta-globin gene expression. J. Cell Biochem. 105:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loken, M. R., V. O. Shah, K. L. Dattilio, and C. I. Civin. 1987. Flow cytometric analysis of human bone marrow: I. Normal erythroid development. Blood 69:255-263. [PubMed] [Google Scholar]
- 41.Luo, X., and M. Sawadogo. 1996. Functional domains of the transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol. Cell. Biol. 16:1367-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilson, D. G., D. E. Sabatino, D. M. Bodine, and P. G. Gallagher. 2006. Major erythrocyte membrane protein genes in EKLF-deficient mice. Exp. Hematol. 34:705-712. [DOI] [PubMed] [Google Scholar]
- 43.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 44.Peterson, K. R. 2003. Hemoglobin switching: new insights. Curr. Opin. Hematol. 10:123-129. [DOI] [PubMed] [Google Scholar]
- 45.Qyang, Y., X. Luo, T. Lu, P. M. Ismail, D. Krylov, C. Vinson, and M. Sawadogo. 1999. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol. Cell. Biol. 19:1508-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rada-Iglesias, A., A. Ameur, P. Kapranov, S. Enroth, J. Komorowski, T. R. Gingeras, and C. Wadelius. 2008. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 18:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ragoczy, T., M. A. Bender, A. Telling, R. Byron, and M. Groudine. 2006. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 20:1447-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawadogo, M., and R. G. Roeder. 1985. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell 43:165-175. [DOI] [PubMed] [Google Scholar]
- 49.Sengupta, T., N. Cohet, F. Morle, and J. J. Bieker. 2009. Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc. Natl. Acad. Sci. USA 106:4213-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sirito, M., Q. Lin, T. Maity, and M. Sawadogo. 1994. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 22:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sirito, M., S. Walker, Q. Lin, M. T. Kozlowski, W. H. Klein, and M. Sawadogo. 1992. Members of the USF family of helix-loop-helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 2:231-240. [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatoyannopoulos, G. 2005. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 33:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sutherland, H., and W. A. Bickmore. 2009. Transcription factories: gene expression in unions? Nat. Rev. Genet. 10:457-466. [DOI] [PubMed] [Google Scholar]
- 54.Tanabe, O., Y. Shen, Q. Liu, A. D. Campbell, T. Kuroha, M. Yamamoto, and J. D. Engel. 2007. The TR2 and TR4 orphan nuclear receptors repress Gata1 transcription. Genes Dev. 21:2832-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuan, D., W. Solomon, Q. Li, and I. M. London. 1985. The “beta-like-globin” gene domain in human erythroid cells. Proc. Natl. Acad. Sci. USA 82:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vakoc, C. R., D. L. Letting, N. Gheldof, T. Sawado, M. A. Bender, M. Groudine, M. J. Weiss, J. Dekker, and G. A. Blobel. 2005. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17:453-462. [DOI] [PubMed] [Google Scholar]
- 57.Vieira, K. F., P. P. Levings, M. A. Hill, V. J. Crusselle, S. H. Kang, J. D. Engel, and J. Bungert. 2004. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J. Biol. Chem. 279:50350-50357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.West, A. G., S. Huang, M. Gaszner, M. D. Litt, and G. Felsenfeld. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453-463. [DOI] [PubMed] [Google Scholar]