Abstract
First identified as the master regulator of major histocompatibility complex II transcription, class II transactivator (CIITA) has since been implicated in a host of pathologies by modulating the transcription of multiple different genes. How CIITA caters to cell- and tissue-specific transcriptional needs is hotly debated and investigated. One of the possible mechanisms underlying spatiotemporal control of CIITA transcriptional activity is the posttranslational modification (PTM) machinery that refines certain amino acid residues of CIITA and hence alters its activity in response to specific cellular and environmental cues. This review discusses our current understanding of the PTM map of CIITA, how these modifications fine-tune its activity, and how the study of this area may lead to potential therapeutic strategies.
Class II transactivator (CIITA) was initially identified in patients with a rare hereditary disease called bare lymphocyte syndrome characterized by a complete loss of major histocompatibility complex class II (MHC II) expression and severe deficiency in CD4+ T-cell-dependent adaptive immunity (31). Complementation experiments with CIITA cDNA rescued the phenotype and established CIITA as the master regulator of MHC II activation (27). CIITA can be placed in the family of NLR/CATERPILLER proteins, the members of which share key structural modules and are believed to actively participate in the regulation of the immune response (29, 37). Investigations over the last few decades have greatly expanded the spectrum of target genes that are transcriptionally regulated by CIITA, which now include collagen type I (35, 40), interleukin-4 (IL-4) (24), IL-10 (39), E-cathepsin (38), MMP-9 (21), plexin (33), various thyroid-specific genes (18), FasL (6), and the scavenger receptor CD36 (X. Wu and Y. Xu, unpublished data). A comprehensive comparison of gene expression profiles in wild-type versus CIITA-deficient cells reveals more than 60 potential targets for CIITA (19), attesting to the importance of CIITA as a pluripotent modulator of transcription. The versatility of CIITA also presents a critical question: how is the transcriptional activity of CIITA tailored to meet the specific requirements of different cells in distinct physiological and pathological microenvironments? The answer lies in the fact that the ability of CIITA to modulate transcription is itself fine-tuned by a complex layer of regulatory pathways, ranging from promoter selectivity to mRNA stability and posttranslational modification (PTM).
PTM refers to the reversible reaction that covalently transfers to or removes from proteins certain chemical groups (e.g., phosphate) and small polypeptides (e.g., ubiquitin). PTM confers both specificity and plasticity on existing protein functions, thus greatly expanding the size of the functional genome (11). The importance of PTMs in modulating CIITA activity is increasingly being recognized, and this minireview summarizes recent advances in this area.
PTM: A NEW TERRITORY IN CIITA RESEARCH
Phosphorylation is probably the most common form of PTM, with more than 500 different kinases being identified (11). Accumulating data point to a critical role for phosphorylation in regulating the activity of CIITA (Fig. 1). Li et al. first reported that certain immune modulators (e.g., prostaglandins) trigger the protein kinase A (PKA)-dependent phosphorylation of CIITA at S834 and S1050 (within the leucine-rich repeat region [LRR]), leading to strong inhibition of MHC II synthesis in monocyte/macrophage cell lines (16). Multiple additional PKA sites in CIITA which could have other functions were detected (Table 1). A later paper, however, outlined a contradictory picture suggesting that phosphorylation of CIITA by PKA at distinct sites (S374 and S375) within the proline/serine/threonine (PST)-rich domain (N terminal to the LRR) is capable of stimulating the transcription of MHC II (25). These authors did not detect the S834 and S1050 phosphorylation, possibly because they used different cell types or different culture conditions (for instance, the former group chose to add ZnCl2 to the media to potentiate PKA). Yet another possibility is that PKA first activates CIITA at the MHC II promoter and later phosphorylates S834 and S1050 to inactivate CIITA and remove it from the MHC II promoter to prevent prolonged (and uncontrolled) activation of MHC II transcription. Intriguingly, in our hands, H89 (a PKA-specific inhibitor) failed to affect CIITA-mediated repression of collagen transcription in lung fibroblast cells (Y. Xu and B. D. Smith, unpublished data), indicating that transcriptional outcome followed by PKA-induced phosphorylation of CIITA may also be reliant on different extracellular, intracellular, or even local chromatin environments. Our recent results demonstrate that CIITA is phosphorylated at S373 within the PST region by glycogen synthase kinase 3 (GSK3) once S377 is primed by casein kinase (CKI) (34). Interestingly, combinatorial phosphorylation of CIITA by GSK3 and CKI enhances the repression of collagen transcription but minimally affects the activation of MHC II transcription. Possibly when the GSK3/CKI sites are phosphorylated, the neighboring PKA sites cannot be modified in these cells. Adding to the complexity of this scheme is the finding that the PST region of CIITA alone contains more than 20 putative phosphorylation sites as predicted by software simulation. Protein kinase C (PKC)-catalyzed phosphorylation of CIITA within the PST region promotes its oligomerization and recruitment to the MHC II promoter, suggesting a critical role for PKC in sustaining the activity of CIITA (30). On the other hand, Greer et al. demonstrated that the nucleus-localized form of CIITA is heavily phosphorylated within the PST region whereas the cytoplasmic CIITA is predominantly unphosphorylated (7). A more detailed analysis reveals that phosphorylation of the PST region is confined to three serine residues, 286, 288, and 293. Surprisingly, replacement of these key serines by alanines has little impact on the self-association of CIITA (as would have been predicted by the report from Tosi et al. [30]), indicating that an alternative kinase may be involved in phosphorylation-dependent subcellular shuttling of CIITA. More recently, it has been demonstrated by Voong et al. (32) that extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylates these sites (286, 288, and 293) in CIITA and inhibition of ERK1/2 activity causes retention of CIITA in the nucleus by blocking the interaction of CIITA with the nuclear export factor CRM1. Lending further support to the notion that phosphorylation modulates CIITA activity in a promoter- and cell-type-specific manner is the observation that mutation of serine 293 prevents the repression of CD36 transcription by CIITA in macrophages but leaves the MHC II activation in macrophages and collagen repression in smooth muscle cells intact (Wu and Xu, unpublished). In aggregate, phosphorylation (by different kinases) clearly affords much elasticity to CIITA in regulating transcription.
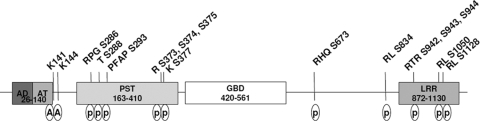
FIG. 1.
A schematic map of known modification sites in CIITA. AD, activation domain; AT, acetyltransferase domain; PST, PST domain; GBD, GTP binding domain.
TABLE 1.
Possible PTMs of CIITA
| Modification | Site(s) detected | Enzyme | Consequence(s) | Reference or source |
|---|---|---|---|---|
| Phosphorylation | S834, S1050 | PKA | Inhibition of MHC II transcription | 16 |
| S286, S374, S375, S673, S942, S943, S944, S1128 | PKA | NDa (S374 and S375 called S375 and S376, respectively, in reference 16) | 16 | |
| S374, S375 | PKA | Increased MHC II transcription, interaction with coactivator p300, and oligomerization | 25 | |
| S373, S377 | GSK3, CKI | Inhibition of collagen transcription by interaction with corepressor Sin3B | 34 | |
| S286, S288, S293 | ND | Inhibition of MHC II, nuclear import of phosphorylated CIITA | 7 | |
| S286, S288, S293 | ERK1/2, cdc2 | Nuclear export | 32 | |
| S293 | CKII | Repression of CD36 | Wu and Xu, unpublished | |
| 253-321 | PKC | Increased phosphorylation and oligomerization of CIITA PST domain | 30 | |
| Ubiquitination | ND | ND | Increased MHC II activation | 8 |
| Acetylation | K141, K144 | CBP/p300, pCAF | Enhanced nuclear localization of CIITA | 26 |
| Deacetylation | ND | HDAC2 | Decreased stability of CIITA, diminished interaction with RFX5 | 14 |
| ND | SIRT1 | Increased MHC II activation, prolonged CIITA stability | Kong and Xu, unpublished |
ND, not determined.
Acetylation is yet another dynamic process that regulates the activity of CIITA. Two ubiquitous histone acetyltransferases (HATs), p300 (and the closely related protein CBP) and pCAF, catalyze the acetylation of CIITA at two N-terminal lysine residues (K141 and K144) and, in so doing, promote the transactivation of the MHC II gene by CIITA (26). Evidently, acetylation of CIITA by p300/CBP and pCAF may be dispensable because HAT-defective forms of these cofactors synergize with CIITA just as well as do their wild-type counterparts in stimulating MHC II transcription (9). Alternatively, acetylation of CIITA may be required for target genes other than MHC II. Acetyl groups can be removed by a large family of proteins collectively called histone deacetylases (HDACs) that can be further categorized, based on sequence homology, into three subtypes. More recently, Kong et al. identified CIITA as a target for deacetylation by HDAC2, a type I HDAC (14). HDAC2-induced deacetylation commits CIITA to proteasomal degradation, thus dampening its activity. In contrast, SIRT1, a type III HDAC, deacetylates CIITA (likely at sites different from those of HDAC2) and upregulates its activity (X. Kong and Y. Xu, unpublished data). Interestingly, an intrinsic HAT activity has been mapped to the most N-terminal area of CIITA between amino acids 94 and 132, which renders TAF(II)250 dispensable for activation of MHC I transcription (22), and yet the in vivo target for CIITA-mediated acetylation has yet to be elucidated.
Ubiquitination of CIITA was initially implicated because an N-terminally truncated form of CIITA exhibited enhanced stability, suggesting a possible role for this modification in controlling the half-life of CIITA via proteasome-mediated degradation (23). In contrast, the research group led by Ting reported that both mono- and polyubiquitination are required for CIITA to form an enhanceosome with other transcription factors on the MHC II promoter, indicating that it potentiates, rather than inhibits, CIITA activity (8). Of note, both studies failed to map the exact site for ubiquitination in CIITA, and it is quite likely that multiple ubiquitination sites determine the ultimate fate of CIITA.
Available evidence indicates that the different PTM machineries are likely to form cross talk to coordinate their activities such that a given transcriptional factor can be “processed” down the PTM assembly line with distinct characteristics in response to specific cues. For instance, a growing body of evidence indicates that there is an antagonism between phosphorylation and ubiquitination-dependent protein degradation (1, 15), a pathway that likely contributes to the regulation of CIITA activity. As reported by Tosi et al. (30), treatment of the monocytic cell line U937 stably expressing CIITA with a PKC activator or a phosphatase inhibitor results in a rapid accumulation of CIITA proteins. Since this form of CIITA is transcribed from a cytomegalovirus promoter (which usually does not respond to these aforementioned chemicals), this piece of data strongly argues for a role of negative cross talk between phosphorylation and ubiquitination in regulating the stabilization of CIITA protein. Similarly, it is well known that acetylation/deacetylation is often coupled to ubiquitination (2, 12), often resulting in altered protein stability. As indicated in our recent paper (14), HDAC2-induced deacetylation of CIITA promotes its degradation possibly via a ubiquitination-proteasomal pathway. However, it has also been shown that augmenting CIITA acetylation and inhibiting CIITA deacetylation both contribute to its ubiquitination and enhanced activity irrelevant to proteasomal degradation (8).
MECHANISMS UNDERLYING PTM-DEPENDENT MODULATION OF CIITA ACTIVITY
PTM-specific modulation of CIITA activity, which involves multiple forms of modifications usually in different combinations, is a complex matter. Mechanistic exploration is further confounded by the fact that any given modification can be catalyzed by more than one group of enzymes. Several theories have been put forward to explain how PTMs affect CIITA activity.
Altered subcellular compartmentation of CIITA.
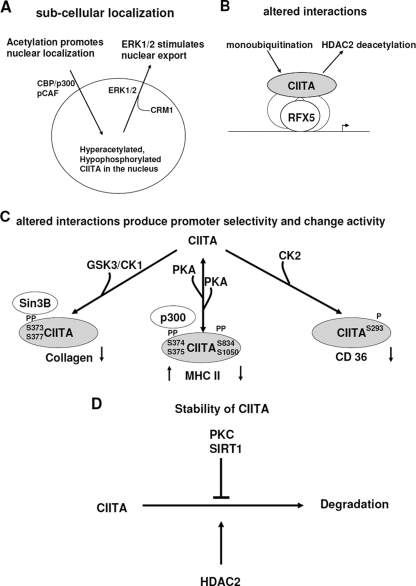
Under normal conditions, CIITA is distributed in both the cytoplasm and the nucleus (4). Since CIITA needs to be present in the nucleus to be a potent transcriptional modulator, several PTMs operate to control the subcellular shuttling of CIITA. CBP/pCAF-dependent acetylation of CIITA, for instance, stimulates the nuclear accumulation of CIITA by activating the N-terminal nuclear localization signal next to the acetylation site, leading to elevated transactivation of the MHC II gene (26). In contrast, ERK-dependent phosphorylation of CIITA within the PST region promotes its interaction with the nuclear export factor CAM1 (32). The ensuing nuclear exclusion of CIITA leads to diminished transcription of MHC II (Fig. 2A).
FIG. 2.
PTMs manipulate CIITA activity by different mechanisms. Specific covalent modifications can alter the subcellular localization of CIITA (A), interactions of CIITA with other factors and promoter selectivity (B and C), or stability of the CIITA protein (D).
Altered interactions of CIITA with other transcription factors.
Being unable to bind to DNA by itself, CIITA forms extensive protein-protein interactions, which in turn affect its intracellular localization, promoter recruitment, and/or transactivation. It has been proposed elsewhere that increased overall phosphorylation of CIITA by PKA promotes its interaction with the coactivator p300 through a region residing between the LRR and the PST domains (25). Ubiquitinated CIITA preferentially interacts with RFX5 and NF-Y, which are responsible for enhanced association with chromatin and activated MHC II transcription (8). Our recent data suggest that deacetylation mediated by HDAC2 disrupts the interaction of CIITA with RFX5, reduces the enlistment of CIITA to target promoters, and consequently suppresses its transcriptional activity (14) (Fig. 2B). The different PTMs can also alter the ability of CIITA to regulate transcription on different promoters by recruitment of either corepressors or coactivators, ultimately determining the promoter selectivity and/or transactivation of CIITA. For example, GSK3/CKI phosphorylation of CIITA induces its interaction with the corepressor Sin3B, which accumulates on the collagen promoter and represses collagen transcription (34). On the other hand, recruitment of the coactivator p300 to the MHC II promoter requires PKA phosphorylation on S374 and S375 (25). When PKA activity is sustained, it continues to phosphorylate CIITA at S834 and S1050, which may lead to the release of coactivators and diminished activity of CIITA. Finally, phosphorylation of S293 is necessary for repression of CD36 transcription (Wu and Xu, unpublished) (Fig. 2C).
Altered protein stability of CIITA.
The CIITA protein, once synthesized, survives for only a short period of time (23). Thus, certain PTMs also target the half-life of CIITA as a means of manipulating its activity. Phosphorylation by PKC greatly induces the accumulation of CIITA protein, probably through an increased stability. Kong et al. recently defined a deacetylation-dependent pathway mediated by HDAC2 for the degradation of CIITA in macrophages and smooth muscle cells (14) (Fig. 2D). However, deacetylation-associated modulation of CIITA stability seems to rely on specific deacetylases (probably through acting on specific sites within CIITA), since SIRT1-induced deacetylation of CIITA prolongs, rather than shortens, its half-life (Kong and Xu, unpublished).
PERSPECTIVES
New potential PTMs for CIITA are emerging at a rapid pace. SUMO2, but not SUMO1 or SUMO3, appears to be specifically attached to CIITA and suppresses its activity (Wu and Xu, unpublished). Both type I (41) and type II (L. Luchsinger and B. D. Smith, unpublished data) protein arginine methyltransferases (PRMTs) have recently been demonstrated to interact with CIITA, although it has not been evaluated whether PRMTs directly methylate CIITA and, if so, how arginine methylation modulates CIITA activity. Despite these uncertainties, it is evident that there indeed exist comprehensive PTM machineries that program the spatiotemporal modulation of CIITA activity. This giant jigsaw puzzle, in which different PTMs fine-tune CIITA activity and steer CIITA-dependent transcriptional programs, is largely unfilled. First, all of the PTMs identified for CIITA are reversible processes that are catalyzed by a pair of enzymes with opposing activities. Yet in most cases, the chemical reactions are poorly defined. For instance, the E3 ligase(s) responsible for targeting CIITA to degradation is still elusive. Even for processes such as acetylation and deacetylation that are relatively well understood, compelling evidence is lacking to draw an unequivocal conclusion. Both HATs (p300/CBP and pCAF) and HDACs (HDAC2 and SIRT1) catalyze the addition to or removal from CIITA of the same chemical group (acetyl group). Current knowledge, however, is unable to place them on either end of an equation simultaneously in a site-specific manner. Second, some of these enzymes (HATs, HDACs, PRMTs, etc.) are capable of, in addition to modifying CIITA, altering the chromatin structure by manipulating specific histone residues. Previous studies have not been particularly well designed to differentiate between the possibilities as to whether the enzymes associating with CIITA modify the chromatin or CIITA directly. Third, the effect of PTM on CIITA activity has not been monitored on a genome-wide scale, as nearly all the investigations have used the transcription of MHC and (to a lesser extent) collagen genes as readouts. Currently, it is unknown whether other CIITA target genes are subject to the fine-tuning of PTM. Finally, the role of PTM in modulating CIITA activity has yet to be verified in vivo in animal models. Without verification in specific animal models, the pathophysiological relevance of CIITA modifications remains questionable. The knock-in mice harboring specific phosphorylation mutations within p53 have greatly facilitated the investigations of p53-dependent pathologies (13). The study of CIITA modifications would benefit from similar strategies.
Given the diverse range of genes regulated by CIITA, it is perceivable that dysregulated CIITA activity has been implicated in multiple pathophysiological processes, which include cancer (17, 36), atherosclerosis (3, 14), encephalitis (28), encephalomyelitis (20), carditis (5), and pulmonary fibrosis (Xu and Smith, unpublished). Because CIITA activity is modulated by the PTM machinery, it is now possible to target a host of PTM components in the development and/or optimization of therapeutic interventions against these pathologies. Atherosclerosis, for instance, is invariably initiated by the inflammatory injury, to a large extent dictated by CIITA-dependent MHC II expression that primes the activation of T cells, to the vasculature. Later development during atherogenesis is characterized by the formation in the artery of a mature atheroma with a fibrous cap (constituted mostly by type I collagen), whose acute rupture is responsible for many of the complications associated with atherosclerosis. Thus, it is desirable to curtail CIITA activity so that both the chronic inflammation and thinning of the fibrous cap can be attenuated. It should also be noted that since CIITA is integral to the homeostasis of the immune system, it is essential that CIITA activity be contained rather than eliminated. Therefore, reagents targeting the PTM machineries that function through suppressing CIITA activity should be chosen over those that operate by promoting CIITA degradation.
In summary, a number of mechanisms have been proposed to account for the modulation of CIITA activity and to dictate the verdict of CIITA-mediated transcriptional events, among which PTM plays a prominent role. Our understanding of the overall PTM program that regulates CIITA activity is, however, still in its infancy, as many important issues remain enigmatic. Future explorations should take advantage of recent advances in proteomic research (10) and focus on establishing reliable animal models. This will likely provide valuable insight into the development of therapeutic strategies.
Acknowledgments
We are mindful of the many excellent papers that could not be cited due to space constraints.
This work was funded in part by grants from the National Natural Science Foundation of China 30800436, the Natural Science Foundation of Jiangsu Province BK2008443, and the NJMU Scientific Development Fund 07NMUZ004 (all to Y.X.) and National Institutes of Health HL068094-9 and HL013262-31 (to B.D.S.). X.W. was supported by a grant from Nanjing Medical University (08NMUM004).
Footnotes
Published ahead of print on 31 August 2009.
REFERENCES
- 1.Abbas, T., D. White, L. Hui, K. Yoshida, D. A. Foster, and J. Bargonetti. 2004. Inhibition of human p53 basal transcription by down-regulation of protein kinase Cdelta. J. Biol. Chem. 279:9970-9977. [DOI] [PubMed] [Google Scholar]
- 2.Asher, G., D. Gatfield, M. Stratmann, H. Reinke, C. Dibner, F. Kreppel, R. Mostoslavsky, F. W. Alt, and U. Schibler. 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134:317-328. [DOI] [PubMed] [Google Scholar]
- 3.Buttice, G., J. Miller, L. Wang, and B. D. Smith. 2006. Interferon-gamma induces major histocompatibility class II transactivator (CIITA), which mediates collagen repression and major histocompatibility class II activation by human aortic smooth muscle cells. Circ. Res. 98:472-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cressman, D. E., K. C. Chin, D. J. Taxman, and J. P. Ting. 1999. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity 10:163-171. [DOI] [PubMed] [Google Scholar]
- 5.Fikrig, E., S. W. Barthold, M. Chen, C. H. Chang, and R. A. Flavell. 1997. Protective antibodies develop, and murine Lyme arthritis regresses, in the absence of MHC class II and CD4+ T cells. J. Immunol. 159:5682-5686. [PubMed] [Google Scholar]
- 6.Gourley, T. S., and C. H. Chang. 2001. Cutting edge: the class II transactivator prevents activation-induced cell death by inhibiting Fas ligand gene expression. J. Immunol. 166:2917-2921. [DOI] [PubMed] [Google Scholar]
- 7.Greer, S. F., J. A. Harton, M. W. Linhoff, C. A. Janczak, J. P. Ting, and D. E. Cressman. 2004. Serine residues 286, 288, and 293 within the CIITA: a mechanism for down-regulating CIITA activity through phosphorylation. J. Immunol. 173:376-383. [DOI] [PubMed] [Google Scholar]
- 8.Greer, S. F., E. Zika, B. Conti, X. S. Zhu, and J. P. Ting. 2003. Enhancement of CIITA transcriptional function by ubiquitin. Nat. Immunol. 4:1074-1082. [DOI] [PubMed] [Google Scholar]
- 9.Harton, J. A., E. Zika, and J. P. Ting. 2001. The histone acetyltransferase domains of CREB-binding protein (CBP) and p300/CBP-associated factor are not necessary for cooperativity with the class II transactivator. J. Biol. Chem. 276:38715-38720. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman, M. D., M. J. Sniatynski, and J. Kast. 2008. Current approaches for global post-translational modification discovery and mass spectrometric analysis. Anal. Chim. Acta 627:50-61. [DOI] [PubMed] [Google Scholar]
- 11.Hunter, T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28:730-738. [DOI] [PubMed] [Google Scholar]
- 12.Insinga, A., S. Monestiroli, S. Ronzoni, R. Carbone, M. Pearson, G. Pruneri, G. Viale, E. Appella, P. Pelicci, and S. Minucci. 2004. Impairment of p53 acetylation, stability and function by an oncogenic transcription factor. EMBO J. 23:1144-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwakuma, T., and G. Lozano. 2007. Crippling p53 activities via knock-in mutations in mouse models. Oncogene 26:2177-2184. [DOI] [PubMed] [Google Scholar]
- 14.Kong, X., M. Fang, P. Li, F. Fang, and Y. Xu. 2009. HDAC2 deacetylates class II transactivator and suppresses its activity in macrophages and smooth muscle cells. J. Mol. Cell. Cardiol. 46:292-299. [DOI] [PubMed] [Google Scholar]
- 15.Leung, C. H., W. Lam, W. J. Zhuang, N. S. Wong, M. S. Yang, and W. F. Fong. 2001. PKCdelta-dependent deubiquitination and stabilization of Gadd45 in A431 cells overexposed to EGF. Biochem. Biophys. Res. Commun. 285:283-288. [DOI] [PubMed] [Google Scholar]
- 16.Li, G., J. A. Harton, X. Zhu, and J. P.-Y. Ting. 2001. Downregulation of CIITA function by protein kinase A (PKA)-mediated phosphorylation: mechanism of prostaglandin E, cyclic AMP, and PKA inhibition of class II major histocompatibility complex expression in monocytic lines. Mol. Cell. Biol. 21:4626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meissner, M., T. L. Whiteside, R. Kaufmann, and B. Seliger. 2009. CIITA versus IFN-gamma induced MHC class II expression in head and neck cancer cells. Arch. Dermatol. Res. 301:189-193. [DOI] [PubMed] [Google Scholar]
- 18.Mori-Aoki, A., M. Pietrarelli, M. Nakazato, P. Caturegli, L. D. Kohn, and K. Suzuki. 2000. Class II transactivator suppresses transcription of thyroid-specific genes. Biochem. Biophys. Res. Commun. 278:58-62. [DOI] [PubMed] [Google Scholar]
- 19.Nagarajan, U. M., A. Bushey, and J. M. Boss. 2002. Modulation of gene expression by the MHC class II transactivator. J. Immunol. 169:5078-5088. [DOI] [PubMed] [Google Scholar]
- 20.Nikodemova, M., J. J. Watters, S. J. Jackson, S. K. Yang, and I. D. Duncan. 2007. Minocycline down-regulates MHC II expression in microglia and macrophages through inhibition of IRF-1 and protein kinase C (PKC)alpha/betaII. J. Biol. Chem. 282:15208-15216. [DOI] [PubMed] [Google Scholar]
- 21.Nozell, S., Z. Ma, C. Wilson, R. Shah, and E. N. Benveniste. 2004. Class II major histocompatibility complex transactivator (CIITA) inhibits matrix metalloproteinase-9 gene expression. J. Biol. Chem. 279:38577-38589. [DOI] [PubMed] [Google Scholar]
- 22.Raval, A., T. K. Howcroft, J. D. Weissman, S. Kirshner, X. S. Zhu, K. Yokoyama, J. Ting, and D. S. Singer. 2001. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol. Cell 7:105-115. [DOI] [PubMed] [Google Scholar]
- 23.Schnappauf, F., S. B. Hake, M. M. Camacho Carvajal, S. Bontron, B. Lisowska-Grospierre, and V. Steimle. 2003. N-terminal destruction signals lead to rapid degradation of the major histocompatibility complex class II transactivator CIITA. Eur. J. Immunol. 33:2337-2347. [DOI] [PubMed] [Google Scholar]
- 24.Sisk, T. J., T. Gourley, S. Roys, and C. H. Chang. 2000. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF-AT to bind the coactivator CREB binding protein (CBP)/p300. J. Immunol. 165:2511-2517. [DOI] [PubMed] [Google Scholar]
- 25.Sisk, T. J., K. Nickerson, R. P. Kwok, and C. H. Chang. 2003. Phosphorylation of class II transactivator regulates its interaction ability and transactivation function. Int. Immunol. 15:1195-1205. [DOI] [PubMed] [Google Scholar]
- 26.Spilianakis, C., J. Papamatheakis, and A. Kretsovali. 2000. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol. Cell. Biol. 20:8489-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steimle, V., L. A. Otten, M. Zufferey, and B. Mach. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 75:135-146. [PubMed] [Google Scholar]
- 28.Suter, T., U. Malipiero, L. Otten, B. Ludewig, A. Muelethaler-Mottet, B. Mach, W. Reith, and A. Fontana. 2000. Dendritic cells and differential usage of the MHC class II transactivator promoters in the central nervous system in experimental autoimmune encephalitis. Eur. J. Immunol. 30:794-802. [DOI] [PubMed] [Google Scholar]
- 29.Ting, J. P., and K. L. Williams. 2005. The CATERPILLER family: an ancient family of immune/apoptotic proteins. Clin. Immunol. 115:33-37. [DOI] [PubMed] [Google Scholar]
- 30.Tosi, G., N. Jabrane-Ferrat, and B. M. Peterlin. 2002. Phosphorylation of CIITA directs its oligomerization, accumulation and increased activity on MHCII promoters. EMBO J. 21:5467-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touraine, J. L., and H. Betuel. 1983. The bare lymphocyte syndrome: immunodeficiency resulting from the lack of expression of HLA antigens. Birth Defects Orig. Artic. Ser. 19:83-85. [PubMed] [Google Scholar]
- 32.Voong, L. N., A. R. Slater, S. Kratovac, and D. E. Cressman. 2008. Mitogen-activated protein kinase ERK1/2 regulates the class II transactivator. J. Biol. Chem. 283:9031-9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong, A. W., W. J. Brickey, D. J. Taxman, H. W. van Deventer, W. Reed, J. X. Gao, P. Zheng, Y. Liu, P. Li, J. S. Blum, K. P. McKinnon, and J. P. Ting. 2003. CIITA-regulated plexin-A1 affects T-cell-dendritic cell interactions. Nat. Immunol. 4:891-898. [DOI] [PubMed] [Google Scholar]
- 34.Xu, Y., J. A. Harton, and B. D. Smith. 2008. CIITA mediates interferon-gamma repression of collagen transcription through phosphorylation-dependent interactions with co-repressor molecules. J. Biol. Chem. 283:1243-1256. [DOI] [PubMed] [Google Scholar]
- 35.Xu, Y., L. Wang, G. Buttice, P. K. Sengupta, and B. D. Smith. 2004. Major histocompatibility class II transactivator (CIITA) mediates repression of collagen (COL1A2) transcription by interferon gamma (IFN-gamma). J. Biol. Chem. 279:41319-41332. [DOI] [PubMed] [Google Scholar]
- 36.Yazawa, T., H. Kamma, M. Fujiwara, M. Matsui, H. Horiguchi, H. Satoh, M. Fujimoto, K. Yokoyama, and T. Ogata. 1999. Lack of class II transactivator causes severe deficiency of HLA-DR expression in small cell lung cancer. J. Pathol. 187:191-199. [DOI] [PubMed] [Google Scholar]
- 37.Ye, Z., and J. P. Ting. 2008. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr. Opin. Immunol. 20:3-9. [DOI] [PubMed] [Google Scholar]
- 38.Yee, C. S., Y. Yao, P. Li, M. J. Klemsz, J. S. Blum, and C. H. Chang. 2004. Cathepsin E: a novel target for regulation by class II transactivator. J. Immunol. 172:5528-5534. [DOI] [PubMed] [Google Scholar]
- 39.Yee, C. S., Y. Yao, Q. Xu, B. McCarthy, D. Sun-Lin, M. Tone, H. Waldmann, and C. H. Chang. 2005. Enhanced production of IL-10 by dendritic cells deficient in CIITA. J. Immunol. 174:1222-1229. [DOI] [PubMed] [Google Scholar]
- 40.Zhu, X.-S., and J. P.-Y. Ting. 2001. A 36-amino-acid region of CIITA is an effective inhibitor of CBP: novel mechanism of gamma interferon-mediated suppression of collagen α2(I) and other promoters. Mol. Cell. Biol. 21:7078-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zika, E., L. Fauquier, L. Vandel, and J. P. Ting. 2005. Interplay among coactivator-associated arginine methyltransferase 1, CBP, and CIITA in IFN-gamma-inducible MHC-II gene expression. Proc. Natl. Acad. Sci. USA 102:16321-16326. [DOI] [PMC free article] [PubMed] [Google Scholar]