Abstract
Notch signaling requires a series of proteolytic cleavage events to release the Notch intracellular domain (NICD) that functions directly in signal transduction. The Notch receptor is locked down in a protease-resistant state by a negative regulatory region (NRR) that protects an ADAM (a disintegrin and metalloprotease) cleavage site. Engagement with ligand-bearing cells induces global conformational movements in Notch that unfold the NRR structure to expose the ADAM cleavage site and initiate proteolytic activation. Although both ADAM10 and ADAM17 have been reported to cleave Notch to facilitate NICD release by γ-secretase, the relevant ADAM has remained controversial. Our study provides new insight into this conflict, as we find that although Notch1 (N1) is a substrate for both ADAM10 and ADAM17, the particular ADAM required for receptor activation is context dependent. Specifically, ADAM10 was absolutely required for N1 signaling induced by ligands, while signaling independent of ligands required ADAM17. In contrast to the strict and differential use of ADAM10 and ADAM17 in normal and dysregulated signaling, respectively, both proteases participated in signaling intrinsic to N1 mutations associated with leukemia. We propose that in addition to exposing the ADAM cleavage site, activating N1 conformational changes facilitate selective cleavage by specific proteases.
Tight control over Notch signaling is critical to normal development as both gains and losses in signaling produce developmental defects (1, 2). Signaling from the Notch receptor is dependent on, and highly regulated by, three distinct types of proteases that cleave Notch at defined sites (35). The first cleavage takes place at a site in the ectodomain (S1) by a furin-like convertase (28) to generate an intramolecular, heterodimeric cell surface receptor that exhibits a protease-resistant conformation in the absence of ligand (10). Interactions with Notch ligands override the autoinhibitory state to allow additional proteolysis at a second site (S2) within the Notch extracellular juxtamembrane region by ADAM proteases. S2 cleavage allows subsequent proteolysis within the membrane-spanning region at additional sites (S3 and S4) by the aspartyl protease γ-secretase. Completion of this proteolytic cascade allows Notch intracellular domain (NICD) membrane release and nuclear translocation resulting in transcriptional complex formation with the CSL [CBF-1/Su(H)/Lag-1] DNA binding protein to directly activate target gene expression (2, 35).
Numerous functional studies have identified a negative regulatory region (NRR) located within the ectodomain that prevents Notch proteolytic activation (13, 20, 25). Recent structural studies have provided mechanistic insight into how the Notch NRR conformation contributes to the protease-resistant state and, importantly, how ligands might override this to activate Notch signaling (10). Specifically, the ADAM cleavage site is buried deep within the NRR structure, and extensive noncovalent interactions within the NRR function to stabilize the heterodimer and occlude the S2 site in the absence of ligand. Moreover, based on ADAM17 structure data, the S2 site might need to be completely unstructured to gain access to the metalloprotease catalytic site (32). Thus, it has been suggested that ligand binding must produce substantial movement within the Notch heterodimer to destabilize the NRR conformation and allow full exposure of the S2 cleavage site (11, 12). In this regard, studies of flies and mammalian cells have identified a strict requirement for ligand endocytosis in Notch signaling (23, 39), and we have found that ligand endocytosis is required to both dissociate the ligand-bound heterodimer and promote Notch proteolysis for downstream signaling (38). Importantly, our studies indicate that ligand binding alone is not sufficient to dissociate the Notch1 (N1) heterodimer and activate signaling. Accordingly, we have proposed that ligand endocytosis generates a mechanical force to destabilize the NRR and completely dissociate the heterodimer, events that would clearly expose the S2 site and allow ADAM cleavage to initiate the activation of proteolysis.
Although it is clear that prior S2 cleavage is required for efficient γ-secretase cleavage to generate the active NICD, it is less clear which ADAM family members actually cleave Notch. Biochemical studies of flies have indicated that ADAM10 interacts with and cleaves Notch to activate signaling, and genetic manipulations that result in losses in protease activity lead to developmental defects similar to those described for defects in Notch signaling (26, 41, 48, 53). Together, these data provide strong support for the idea that ADAM10 directly participates in the proteolytic activation of Notch to regulate signaling. However, genetic studies of Caenorhabditis elegans have identified functional redundancy for the orthologs of ADAM10 and ADAM17, SUP-17 and ADM-4, respectively, for a subset of developmental decisions mediated by the Notch-related LIN-12 receptor (19). Further complicating these issues are the in vitro studies of mammalian cells that have both identified ADAM17 as the relevant metalloprotease in S2 cleavage and excluded a role for ADAM10 in this event (3, 36). However, a role for ADAM17 in the proteolytic activation of N1 is difficult to reconcile with genetic studies demonstrating that ADAM10 knockout mice display classic Notch loss-of-function phenotypes (4, 14, 56), while ADAM17 mutant mice clearly do not (42).
Despite a large body of evidence supporting a role for ADAM10 in Notch cleavage and activation of signaling, ADAM17 is often considered in the literature as the relevant ADAM responsible for activating Notch proteolysis. We reasoned that this discrepancy is due to the fact that the studies implicating ADAM17 in S2 cleavage employed forms of N1 that either lacked sequences required to form the NRR or encoded destabilizing NRR mutations (3, 36), which, based on structure data, likely expose the S2 site to proteolysis independent of ligand. Given the strong dependency on Notch ligands to expose the S2 site and release autoinhibition, we reevaluated the requirement of these ADAMs in Notch signaling by using cell coculture assays that rely on direct interactions between Notch ligands and receptor-expressing cells to activate N1 proteolysis and downstream signaling. Our findings indicate that N1 proteolytic processing by ADAM10 is strictly ligand dependent and, importantly, ADAM17 cannot functionally replace ADAM10 activity, suggesting an exclusive role for this protease in ligand-induced signaling. Interestingly, while ADAM17 cannot activate N1 in response to ligand, it can nonetheless activate signaling in a ligand-independent manner.
MATERIALS AND METHODS
Mammalian expression constructs.
The following cDNA constructs have been previously described: pBos-N1 (52), pBos-N1Δmyc (59), pBos-NICD (52), pBOS-N1CC→SS (34) and pBOS-Dll1HA (14). The following plasmids were generously provided: wild-type CSL reporter (pGL3P-JH26) from M. Hancock and A. Orth, pBABE-DNKUZ and pMSCV2.2-DNTACE from E. Robey (31, 41), pcDNA3-ADAM10HA from F. Fahrenholz (22), pcDNA3-ADAM17 from R. Black (42), and human N1 T-ALL mutants pcDNA3-N1-L1601P, -N1-L1679P, -N1-V1577E, -N1-L1594P, -N1-R1599P, and -N1-P12 in full-length and ΔEGF (where EGF is epidermal growth factor) forms from J. Aster and S. Blacklow (30).
Cell lines.
Parental C2C12 and NIH 3T3 cell lines were obtained from the American Type Culture Collection and propagated as directed. ADAM10+/+ and ADAM10−/− mouse embryonic fibroblasts (MEFs) were gifts from D. J. Pan, P. Saftig, and C. Blobel and were cultured in Dulbecco modified Eagle medium (Invitrogen) containing 10% fetal bovine serum (HyClone). ADAM17+/+ and ADAM17ΔZn/ΔZn MEFs were gifts from R. Black (Amgen) and cultured as described previously (47).
Stable L-cell lines expressing Dll1 or J1 have been described previously (15, 27, 38, 59). C2C12, ADAM10+/+, ADAM10−/−, ADAM17+/+, and ADAM17ΔZn/ΔZn (described below) MEFs stably expressing N1 in which C-terminal sequences were replaced with six myc epitope tags (N1Δmyc) were generated by cotransfection with pBos-N1Δmyc and an expression vector for either the puromycin or neomycin resistance gene by the use of the Lipofectamine reagent (Invitrogen) according to the manufacturer's specifications, followed by selection with either 1 μg/ml puromycin (Sigma) or 0.2 μg/ml G418 (Sigma).
Stable ADAM10+/+ and ADAM10−/− MEFs expressing the hemagglutinin (HA)-tagged form of the Notch ligand Delta-like 1 (Dll1HA) were generated by cotransfection with pBos-Dll1HA and an expression vector for the neomycin fresistance gene by the use of the Lipofectamine reagent (Invitrogen), followed by selection with 0.2 μg/ml G418 (Sigma).
CSL reporter assays.
NIH 3T3 cells and MEFs grown in 12-well tissue culture plates were transfected with a total of 1 μg DNA using 3 μl Lipofectamine reagent (Invitrogen) in serum-free media according to the manufacturer's protocol. The transfection mixture included 0.05 μg of full-length or mutated N1, 0.125 μg of the Notch-dependent CSL reporter containing eight CSL binding sites upstream of the luciferase gene (pGL3JH26), and also, to control for transfection efficiency, 0.0025 μg of Renilla luciferase reporter (pRLCMV; Promega). As indicated, pBABE-DNKUZ, pcDNA3-ADAM10HA, pcDNA3-ADAM17, and control vectors (0.1 μg) were also included in the transfection mixture. At 5 h posttransfection, cells were cocultured with either Dll1- or J1-expressing L-cell lines or the parental L cells as previously described (21, 38, 59). The metalloprotease inhibitor BB94 (10 μM; British Biotech), γ-secretase inhibitor DAPT (N-S-phenyl-glycine-t-butyl ester; 25 μM; Calbiochem) or vehicle dimethyl sulfoxide (DMSO; Sigma) was included at the time of coculture. Lysates were collected 16 to 24 h posttransfection and assayed for luciferase activity by using the dual-luciferase assay kit (Promega) using a Turner Designs luminometer (TD-20/20) following the manufacturer's instructions. Experiments were performed in triplicate, and luciferase values were normalized to transfection control Renilla values and expressed as relative luciferase units (RLU).
Proteolytic cleavage of Notch.
Detection of ligand-induced proteolytic cleavage of ectopic N1Δmyc or endogenous full-length N1 was carried out as previously described (59). Briefly, N1Δmyc-expressing cells were cocultured with Dll1-expressing or parental L cells for 5 h in the absence or presence of the proteasome inhibitor MG132 (N-CBZ-Leu-Leu-Leu-AL; 10 μM; Sigma Aldrich), the metalloprotease inhibitor BB94 (10 μM; British Biotech), the γ-secretase inhibitor DAPT (25 μM; Calbiochem), or the vehicle DMSO (Sigma). Cocultures were lysed in Triton X buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 10% glycerol, and 1% Triton X-100 supplemented with 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, and 10 mg/ml aprotinin and phosphatase inhibitors). Equal amounts of cellular protein were immunoprecipitated with anti-myc antibody (9E10, 1:100; Santa Cruz), and immune complexes were collected on protein A-agarose beads (20 μl/lysate; Roche) prebound with rabbit anti-mouse antiserum. Beads were washed three times in lysis buffer, and eluted protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti-myc antibody (9E10; 1:500; Santa Cruz) followed by horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:5,000; Amersham Biosciences) or rabbit anti-Val1744 antibody (1:1,000; Cell Signaling) followed by horseradish peroxidase-conjugated protein A (1:5,000; Amersham Biosciences). The ECL Plus Western blot detection system (Amersham Biosciences), Typhoon 9410 scanner (Amersham Biosciences), and ImageQuant software (Molecular Dynamics) were used for protein detection and quantification. Band intensity corresponding to the cleavage fragments in whole-cell lysates was normalized for protein loading by immunoblotting with anti-α-tubulin antibody (1:10,000; Sigma).
For analysis of proteolytic cleavage fragments of endogenous N1, WCLs were immunoprecipitated using a rabbit polyclonal antibody raised against the intracellular domain of N1 (anti-NICD; PCR12, 1:100) and immune complexes collected on protein A-agarose beads (20 μl/lysate; Roche), followed by SDS-PAGE and immunoblotting with another rabbit polyclonal anti-NICD antibody (93-4, 1:5,000) or anti-Val1744 antibody (1:1,000; Cell Signaling).
EDTA-induced Notch activation.
C2C12 cells were washed twice with Hank's buffered saline solution (HBSS; Gibco) and then incubated in 1 mM EDTA or HBSS buffer alone for 10 min at 37°C as previously described (45). Following buffer removal, cultures were incubated in Dulbecco modified Eagle medium containing 10% fetal bovine serum at 37°C for 6 to 8 h and assayed for luciferase activity as described above.
ConA pulldown.
Cells washed twice with ice-cold phosphate-buffered saline were lysed in concanavalin A (ConA) lysis buffer (500 mM NaCl, 1 mM MnCl2, 1 mM CaCl2, 1 mM EDTA, 1% Nonidet P-40, and 1× Tris-buffered saline) supplemented with protease inhibitors (as described above) and 10 μM BB94 (British Biotech) to block metalloprotease autocatalytic activity during cell lysis. Cleared lysates were then incubated with 10 μl of ConA-Sepharose 4B beads (Amersham Biosciences) for 1 h at 4°C to enrich glycosylated proteins as previously described (29). Bound material was eluted from beads and analyzed by SDS-PAGE followed by immunoblotting with anti-ADAM10 (1:1,000; Cell Signaling) or anti-ADAM17 (1:1,000; Cell Signaling) antibodies.
Coimmunoprecipitation of N1 with ADAM10 or ADAM17.
C2C12 cells transfected with 1 μg N1 and/or 1 μg ADAM10HA or 1 μg ADAM17 constructs, using Lipofectamine reagent (Invitrogen), were lysed 48 h posttransfection in Triton X buffer supplemented with protease inhibitors and 10 μM BB94 (British Biotech). Lysates were then immunoprecipitated with anti-N1 antiserum (1:100; PCR12) and immunoblotted with anti-ADAM10 (1:1,000; Cell Signaling), anti-ADAM17 (1:1,000; Cell Signaling) or anti-N1 (1:5,000; 93-4) antibodies.
siRNA-mediated knockdown.
C2C12 cells or MEFs were reverse transfected with 20 nM small interfering RNA (siRNA) duplexes by the use of Lipofectamine RNA interference MAX reagent (Invitrogen) according to the manufacturer's instructions. The target sequences of the siRNAs specific for mouse ADAM10, ADAM17, or scrambled (SCR) control have been described previously (37, 60) and are as follows: ADAM10, 5′-GCAAAGATGATTGCTGCTTCG-3′; ADAM17-1, 5′-CAAAGAGACAGAGTGCTAGT-3′, ADAM17-2, 5′-GAGAAGCTTGATTCTTTGC-3′, and SCR, 5′-GGTATATGCGCCATACACTACCC-3′. At 24 h post-siRNA treatment, cells were transfected with the CSL-luciferase reporter construct and then either cocultured with ligand cells or EDTA-treated for reporter assays, or the cells were cocultured with ligand cells for analysis of N1 proteolytic cleavage fragments. In parallel, cell lysates were incubated with ConA beads and subjected to Western blot analysis to monitor the knockdown efficiency of ADAMs.
For ADAM10 rescue experiments, C2C12 cells treated with siRNA duplexes for 24 h were cotransfected with 0.1 μg pcDNA3 vector encoding either ADAM10HA or ADAM17 and the CSL reporter. Cell lysates were analyzed at 24 h posttransfection for reporter activity or ADAM protein expression.
RNA isolation and endogenous Herp2 expression analysis.
RNA isolated from C2C12 cultures with the mRNA isolation kit (Qiagen) was subjected to first-strand cDNA synthesis using oligo(dT) primers according to the manufacturer's protocol (cDNA synthesis kit; Invitrogen). Herp2 and β-tubulin transcript levels were determined in duplicate samples by using Sybr green (Roche)-based quantitative real-time PCR (qRT-PCR) analysis with the following primer sets: mouse Herp2 Forward, CAG CCC TAA GCA CTC TCA GTC; mouse Herp2 Reverse, GCA CCA AAA GGA AAA CAC AAC; mouse β-tubulin Forward, CAT CCA GGA GCT CTT CAA GC; and mouse β-tubulin Reverse, CAC CAT TTA CCC CCA ATG AG. The qRT-PCR analysis was performed with MX-3000 software (Stratagene), and Herp2 transcript levels were normalized to β-tubulin levels.
Statistical analysis.
Statistical significance was calculated by Student's t test for two-tailed distribution with equal variances, using Microsoft Excel software (Microsoft). Error bars indicate the mean ± standard deviation of the mean (SD).
RESULTS
ADAM10 activity is required by Notch signal-receiving cells but not ligand signal-sending cells to activate signaling.
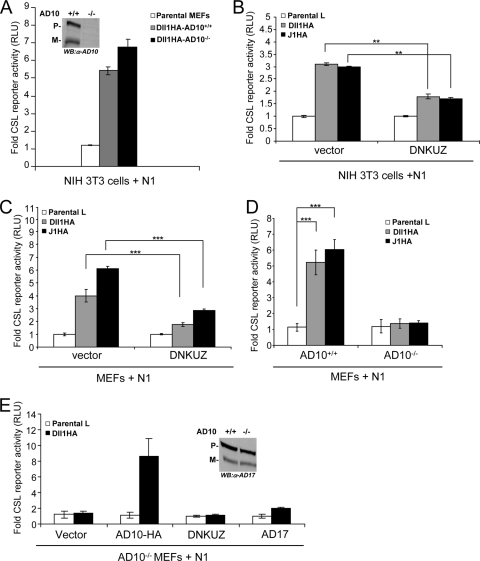
Our previous studies using broad-spectrum metalloprotease inhibitors to block ligand-induced Notch signaling are consistent with a requirement for ADAM10 (38, 59). However, these inhibitors are not specific for ADAM10 activity, and this approach could not determine which of the interacting cells required ADAM10, which is important since both cell-autonomous and non-cell-autonomous roles for ADAM10 in Notch signaling have been proposed (41, 43, 48, 53). To investigate a requirement for ADAM10 in the ligand signal-sending cell, MEFs isolated from ADAM10-targeted embryos (ADAM10−/− MEFs) (14), lacking both precursor and mature forms of ADAM10 (Fig. 1A, inset), were engineered along with wild-type MEFs (ADAM10+/+ MEFs) to stably express a C-terminal Dll1HA. These Dll1HA-expressing MEFs and control parental MEFs were cocultured with NIH 3T3 fibroblasts transiently expressing N1 and a CSL reporter, and the level of luciferase activity was used as a measure of Notch signaling as previously described (21, 38, 59). In this study, Dll1HA-expressing ADAM10−/− MEFs induced levels of luciferase activity similar to those measured for Dll1HA-expressing ADAM10+/+ MEFs (Fig. 1A), indicating that ligand cells do not require ADAM10 to activate signaling in cocultured N1 cells.
FIG. 1.
ADAM10 functions cell autonomously in Notch signaling. (A) ADAM10 is not required by Dll1 cells to activate signaling. NIH 3T3 cells coexpressing N1 and a CSL reporter were cocultured with stable Dll1HA-ADAM10+/+ (AD10+/+) or Dll1HA-ADAM10−/− (AD10−/−) MEFs and assayed for activation (activation over parental MEFs; n = 3). ADAM10 protein expression was determined for ADAM10+/+ and ADAM10−/− MEF ConA-enriched lysates by Western blotting (WB) (inset). (B and C) Ectopic expression of DNKUZ reduces ligand-induced Notch signaling. Reporter activity for NIH 3T3 cells (B) or wild-type MEFs (C) coexpressing DNKUZ with N1 induced by Dll1 or J1 cells and assayed for luciferase activity (activation over parental L cells; n = 4). (D) Ligand-induced Notch signaling is defective in ADAM10−/− MEFs. Dll1- or J1-induced reporter activity was measured in ADAM10−/− and ADAM10+/+ MEFs (activation over parental L cells; n = 6). (E) Ectopic ADAM10HA expression rescues ADAM10−/− MEF reporter activity. Reporter activity for ADAM10−/− MEFs cocultured with Dll1 cells following ectopic ADAM10HA (AD10-HA), DNKUZ, or ADAM17 (AD17) expression was measured as described for panel D (n = 4). ADAM17 protein expression was determined for ADAM10+/+ and ADAM10−/− MEF ConA-enriched lysates by Western blotting (inset). P, precursor form; M, mature; α, anti; **, P < 0.01; ***, P < 0.001.
These data specifically eliminated a requirement for ADAM10 in the ligand signal-sending cell, a finding more consistent with the proposed cell-autonomous role for ADAM10 in Notch signaling. To investigate the requirement for ADAM10 in the Notch cell during ligand-induced signaling, we used a mutant form of ADAM10 lacking the metalloprotease domain that functions as a dominant negative (DNKUZ) to block Notch signaling (31, 41). Expression of DNKUZ in either NIH 3T3 fibroblasts or MEFs programmed to express N1 and the CSL reporter significantly reduced luciferase activity induced by cocultured Dll1HA cells or cells expressing another Notch ligand, Jagged1 (J1HA), relative to that induced by vector control cells (Fig. 1B and C). These findings are consistent with a requirement for ADAM10 in the Notch cell; however, high expression of this mutant protein could directly or indirectly affect other metalloproteases implicated in Notch activation, in particular ADAM17. Therefore, to determine a specific requirement for ADAM10 in the Notch cell, we tested ADAM10−/− MEFs ectopically expressing N1 and the CSL reporter for signaling induced by cocultured ligand cells. Notch reporter activity induced in Dll1 or J1 cocultures was strongly reduced in ADAM10−/− MEFs relative to that detected with ADAM10+/+ MEFs (Fig. 1D). In fact, the low level of activation induced in ADAM10−/− MEFs cocultured with cells ectopically expressing Notch ligands was similar to that induced by control parental L cells. Moreover, ectopic expression of full-length ADAM10 rescued the losses in endogenous Notch signaling associated with ADAM10−/− MEFs (Fig. 1E), indicating a specific requirement for ADAM10 in the Notch cell for ligand-induced signaling. Importantly, DNKUZ, lacking the metalloprotease domain, was unable to rescue the ADAM10−/− MEFs signaling defect (Fig. 1E), indicating that the ADAM10 requirement relies on functional metalloprotease activity.
Even though the ADAM10−/− MEFs are defective in ligand-induced signaling, they express wild-type levels of precursor and mature forms of ADAM17 protein (Fig. 1E, inset), suggesting that ADAM17 cannot functionally replace the loss of ADAM10 activity. However, this idea is inconsistent with reports that ADAM17 can cleave N1 to activate signaling. To address this conflict, we ectopically expressed ADAM17 in ADAM10−/− MEFs and found that even though ADAM17 is active in shedding tumor necrosis factor alpha (TNF-α) when ectopically expressed in cells (49, 61) (data not shown), it could not rescue the defect in ligand-induced signaling as was found for ectopic ADAM10 expression (Fig. 1E). Together, our data suggest that ADAM10 is the relevant ADAM that functions in the Notch cell during signal reception and that the related ADAM17 cannot functionally substitute for ADAM10 in ligand-induced activation of Notch signaling.
ADAM17 does not function in ligand-induced N1 signaling.
Based on the data described above, we wanted to reevaluate the requirement for ADAM17 (also known as TACE for TNF-α), specifically in the receiving cell during ligand-induced activation of Notch signaling. To this end, we took advantage of the MEF line derived from mouse embryos carrying a targeted deletion of the TACE zinc-binding domain (TACEΔZn/ΔZn), which inactivates ADAM17 metalloprotease activity (42, 47). This TACEΔZn/ΔZn MEF line has been used extensively to assess ADAM17 substrate specificity and biological function. Indeed, these cells (referred to here as ADAM17ΔZn/ΔZn) are the same cells previously used to implicate a role for ADAM17 in the cleavage of N1 (3). Thus, to assess the requirement for ADAM17 specifically in ligand-induced Notch signaling, ADAM17ΔZn/ΔZn MEFs were assayed as described above for ADAM10−/− MEFs.
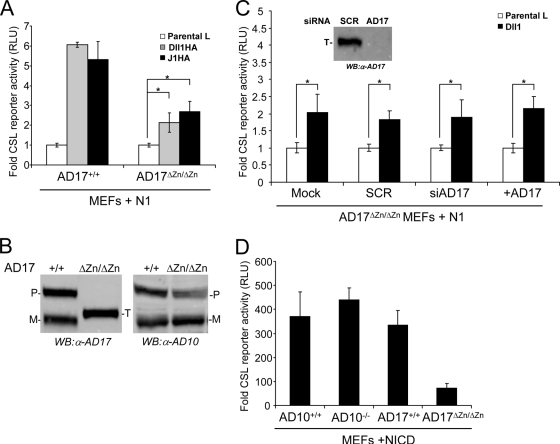
CSL reporter activity detected with ADAM17ΔZn/ΔZn cells cocultured with Dll1 or J1 was approximately two- to threefold less than that obtained with wild-type ADAM17+/+ MEFs (Fig. 2A). Although the lower level of reporter activity detected with ADAM17ΔZn/ΔZn cells could indicate a requirement for ADAM17, these cells reproducibly displayed a statistically significant two- to threefold increase in reporter activity induced by Dll1 or J1 cells, relative to that induced by control parental L cells (Fig. 2A), indicative of a ligand-specific activation of N1. Given this, we asked if the low level of activation detected with ADAM17ΔZn/ΔZn cells was due to the expression of a truncated ADAM17 protein from the targeted taceΔZn/ΔZn gene, which migrates between the full-length precursor and mature forms on immunoblots (Fig. 2B). Even though this truncated protein is enzymatically inactive (42, 47), we considered that it might function as a dominant negative and inhibit ADAM10. Antagonism of ADAM10 activity could account for the significant, yet low, level of signaling detected with ADAM17ΔZn/ΔZn MEFs. To address this idea, we used siRNA duplexes designed to target ADAM17 transcripts to specifically deplete the ADAM17ΔZn/ΔZn protein. ADAM17ΔZn/ΔZn MEFs treated with either ADAM17-specific siRNA duplexes to deplete ADAM17ΔZn/ΔZn protein (Fig. 2C, inset) or SCR were cotransfected with N1 and the CSL reporter followed by coculture with Dll1 or parental L cells. Depletion of the ADAM17ΔZn/ΔZn protein did not alter reporter activity compared to that detected for either mock- or SCR-treated MEFs (Fig. 2C), ruling out interference by the mutant ADAM17 protein. Moreover, ectopic expression of ADAM17 did not rescue the signaling defect of ADAM17ΔZn/ΔZn MEFs (Fig. 2C).
FIG. 2.
ADAM17 (AD17) is not required for ligand-induced Notch signaling (A) ADAM17ΔZn/ΔZn MEFs (AD17ΔZn/ΔZn) displayed a significant, but low level of reporter activity induced by ligand cells. Reporter activities for N1-expressing ADAM17ΔZn/ΔZn and ADAM17+/+ MEFs cocultured with Dll1 or J1 cells were assayed for luciferase activity (induction over parental L cells; n = 4). (B) ADAM17ΔZn/ΔZn MEFs express a targeted (T) ADAM17 protein and display ADAM10 (AD10) protein levels similar to those displayed by ADAM17+/+ MEFs. ADAM10 and ADAM17 protein expression was determined for ConA-enriched lysates isolated from ADAM17+/+ and ADAM17ΔZn/ΔZn MEFs by Western blotting (WB). (C) ADAM17ΔZn/ΔZn protein does not alter Notch signaling. ADAM17ΔZn/ΔZn MEFs treated with SCR or ADAM17 siRNA (siAD17) were cotransfected with ADAM17 and the CSL reporter constructs and then cocultured with Dll1 cells and assayed for luciferase activity (induction over parental L cells; n = 3). Specific siRNA depletion of ADAM17ΔZn/ΔZn protein was assayed by Western blotting (inset). (D) ADAM17ΔZn/ΔZn MEFs have intrinsically low Notch signaling activity. Reporter activity induced by constitutively active NICD in ADAM17ΔZn/ΔZn MEFs was assayed for luciferase activity (activation over vector transfected cells; n = 3). P, precursor form; M, mature; α, anti; *, P < 0.05.
Alternatively, the lower level of Notch signaling associated with ADAM17ΔZn/ΔZn MEFs could reflect clonal variability as reported for different ADAM10−/− MEFs in amyloid precursor protein α-cleavage (14), rather than defects in ADAM17 enzymatic activity. Therefore, to assess the variability in Notch signaling among ADAM10, ADAM17, wild-type, and targeted MEFs, reporter activity induced by a constitutively active N1 (NICD) was determined. NICD signaling was lowest in ADAM17ΔZn/ΔZn MEFs (Fig. 2D), and given that this form of N1 does not require ADAM or γ-secretase proteolysis to signal (40, 52), we favor the idea that clonal variation in Notch signaling, rather than loss in ADAM17 activity, accounts for the reduced activity detected (Fig. 2A). Together, our findings suggest that defects in ADAM17 enzymatic activity do not perturb signaling induced by Dll1 or J1, consistent with a role for ADAM10, rather than ADAM17, in ligand-induced Notch signaling.
RNA interference studies confirm a requirement for ADAM10 exclusive of ADAM17 in ligand-induced N1 signaling.
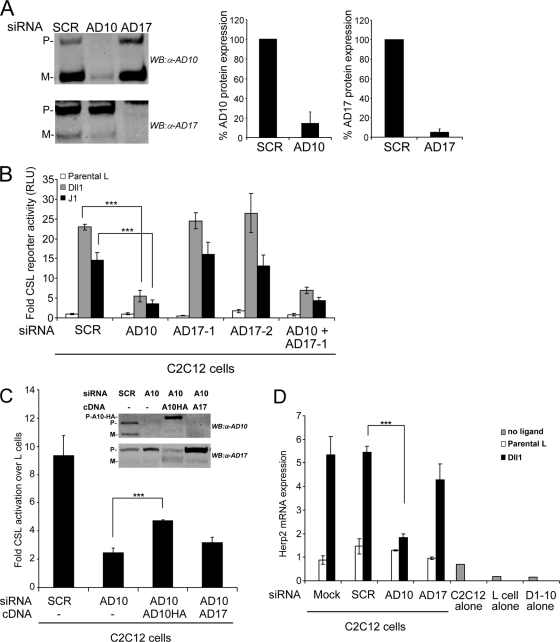
Given the clonal variability associated with genetically targeted MEF lines, we used siRNA duplexes designed to deplete ADAM10 and ADAM17 in the same cell line. Treatment of C2C12 cells with either ADAM10 or ADAM17 siRNA duplexes specifically depleted ADAM10 and ADAM17 proteins by more than 90% relative to the levels in SCR siRNA-treated cells (Fig. 3A). When C2C12 cells specifically depleted of either ADAM10 or ADAM17 were cotransfected with a CSL reporter and cocultured with ligand cells, losses in ADAM10 resulted in a dramatic decrease in reporter activity (Fig. 3B). In contrast, neither of the two ADAM17 siRNAs produced losses in reporter activity following coculture with ligand cells, and cotreatment with both ADAM10 and ADAM17 siRNAs yielded losses in reporter activity similar to those detected for ADAM10 siRNA treatment alone (Fig. 3B). Importantly, losses in ligand-induced reporter activity in ADAM10-depleted cells could be rescued by ectopic expression of an ADAM10 cDNA (Fig. 3C, inset), indicating that losses in activity mediated by the ADAM10 siRNAs are specifically linked to ADAM10 protein depletion. In contrast, ectopic ADAM17 expression did not significantly rescue the signaling defect associated with ADAM10 depletion (Fig. 3C), indicating that ADAM17 cannot functionally replace ADAM10. Moreover, losses in ADAM10 but not ADAM17 protein following siRNA treatments led to significant losses in mRNA expression of the endogenous Notch target gene, the Herp2 gene (17), induced by Dll1 cells (Fig. 3D). Together, our data indicate that ADAM10 is absolutely required for both Dll1 and J1 to activate signaling and drive target gene expression.
FIG. 3.
ADAM10, but not ADAM17, is required for ligand-induced Notch-dependent reporter activity and endogenous target gene (Herp2 gene) expression. (A) siRNA depletion of ADAM10 (AD10) and ADAM17 (AD17) proteins is specific and efficient. Western blot (WB) analysis and quantification of ADAM10 and ADAM17 proteins following siRNA depletion were performed (percentage relative to SCR-treated cells arbitrarily set to 100; representative of 10 experiments). (B) Depletion of ADAM10, but not ADAM17, decreased CSL reporter activity. C2C12 cells depleted for ADAM10 and/or ADAM17 protein by siRNA were transfected with the CSL reporter followed by coculture with Dll1 or J1 cells and assayed for luciferase activity (induction over parental L cells; n = 4). (C) Ectopic ADAM10HA (AD10HA or A10HA) expression rescued reporter activity in ADAM10 (AD10 or A10)-depleted C2C12 cells. Dll1-induced reporter activity was measured in C2C12 cells treated with SCR or ADAM10 siRNAs and then transfected with ADAM10 or ADAM17 (AD17 or A17) cDNAs (induction over parental L cells; n = 4). siRNA depletion and ectopic expression of ADAM10 and ADAM17 in ConA-enriched lysates were assayed by Western blotting (inset). (D) Depletion of ADAM10, but not ADAM17, decreased Dll1-induced endogenous Herp2 expression. C2C12 cells treated with indicated siRNAs were cocultured with Dll1 cells, and Herp2 expression was detected by qRT-PCR analysis (see Materials and Methods for details) (n = 4). P, precursor form; M, mature; α, anti; ***, P < 0.001.
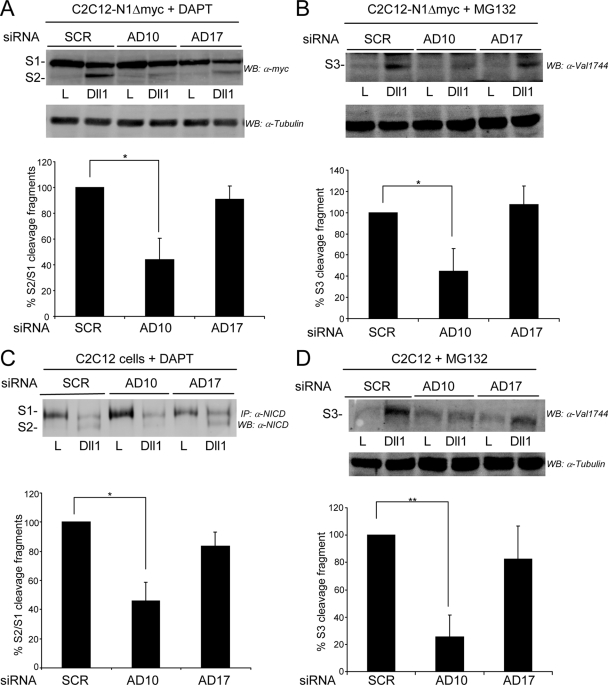
ADAM10 but not ADAM17 is required for S2 cleavage induced by Dll1 cells.
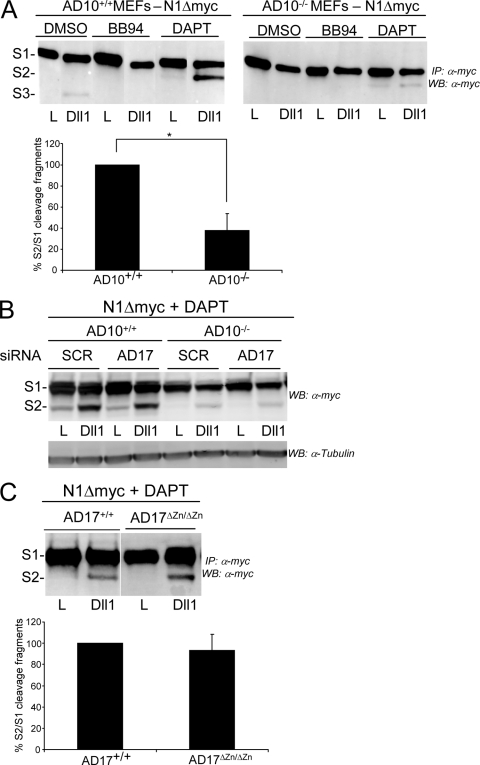
Having established that ADAM10 but not ADAM17 is required in the Notch cell for activation of signaling by ligand, we tested the requirement for these related ADAMs in the ligand-induced cleavage of N1. For these experiments, we established ADAM10+/+, ADAM10−/−, ADAM17+/+, and ADAM17ΔZn/ΔZn MEF lines stably expressing N1Δmyc to facilitate detection of the S1, S2, and S3 N1 cleavage products by immunoblotting (59). Important to our studies, S2 fragment detection can be enhanced by the γ-secretase inhibitor DAPT (6), which blocks the rapid conversion of S2 to S3 (Fig. 4A, compare the S2 bands for DMSO and DAPT treatments), allowing identification and quantification of the S2 fragment produced in N1Δmyc cells following coculture with Dll1 or J1 cells (59). Specifically, when ADAM10−/− N1Δmyc MEFs were cocultured with Dll1 cells in the presence of DAPT, a slight increase in S2 was detected compared to that induced by parental L cells (Fig. 4A). Quantification of the results for four independent experiments revealed that the amount of S2 produced relative to the S1 precursor was decreased >60% for ADAM10−/− N1Δmyc MEFs compared to that detected for ADAM10+/+ N1Δmyc MEFs (Fig. 4A).
FIG. 4.
ADAM10 expression is required for ligand-induced S2 cleavage of N1 in MEFs. (A) ADAM10−/− (AD10−/−) MEFs are defective in S2 cleavage. Dll1-induced N1Δmyc proteolytic fragments produced in stable ADAM10+/+ and ADAM10−/− MEFs treated with metalloprotease (BB94) or γ-secretase (DAPT) inhibitors (DMSO vehicle) were analyzed following immunoprecipitation (IP) and Western blotting (WB). The percent quantification of S1 fragment converted to S2 fragment from DAPT-treated Dll1 cocultures with ADAM10+/+ was arbitrarily set to 100% (lower panel; n = 4). (B) The residual S2 cleavage fragment detected with ADAM10−/− MEFs is not ADAM17 derived. The ligand-induced S2 cleavage fragment was analyzed by Western blotting in N1Δmyc ADAM10+/+ or ADAM10−/− MEFs following siRNA depletion of ADAM17 and coculture with Dll1 cells. The results shown are representative of one of two experiments. (C) Dll1-induced S2 cleavage is unaltered in ADAM17ΔZn/ΔZn MEFs. N1Δmyc-expressing ADAM17+/+ and ADAM17ΔZn/ΔZn MEFs were assayed and quantified as described for panel A (n = 4). *, P < 0.05.
Even though ligand cells are unable to activate the Notch reporter in ADAM10−/− cells (Fig. 1D and E), a low level of S2 is detected in ADAM10−/− cells cocultured with Dll1 cells (Fig. 4A). Detection of this fragment is prevented by BB94, indicating that it is metalloprotease dependent, and accumulation in the presence of DAPT indicates that it can be further cleaved by γ-secretase (Fig. 4A). Since the metalloprotease ADAM17 has also been implicated in S2 cleavage, we asked if the residual S2 detected with ADAM10−/− N1Δmyc cells in response to Dll1 was a product of endogenous ADAM17 (Fig. 1E, inset). Treatment of ADAM10−/− N1Δmyc MEFs with either SCR or ADAM17 siRNAs produced similarly low levels of S2 following coculture with Dll1 cells (Fig. 4B), suggesting that ADAM17 is not responsible for the residual cleavage detected with ADAM10−/− N1Δmyc cells. In fact, depletion of ADAM17 in ADAM10+/+ MEFs did not reduce the amount of S2 detected compared to control SCR treatments (Fig. 4B), indicating that ADAM17 is not involved in ligand-induced S2 cleavage. Consistent with this idea, the level of S2 detected for ADAM17ΔZn/ΔZn N1Δmyc MEFs cocultured with Dll1 cells was not significantly different from that induced in ADAM17+/+ N1Δmyc cocultures (Fig. 4C). Although ADAM17 is not required for S2 generation, our data suggest that in the absence of ADAM10, an alternative metalloprotease cleaves N1 to generate a similarly sized fragment that does not detectably activate signaling (Fig. 1D and E). Together, our data suggest that ADAM10 is required for efficient S2 cleavage and signaling induced by Dll1 and that ADAM17 does not participate in these events.
To confirm our findings with ADAM10 and ADAM17 targeted MEFs, we treated stable N1Δmyc-expressing C2C12 cells with specific siRNAs to deplete ADAM10 or ADAM17 prior to coculture with either Dll1 or parental L cells. In the presence of DAPT, the level of accumulated S2 was significantly decreased in cells treated with ADAM10 siRNA compared to that in cells treated with SCR siRNA; however, ADAM17 depletion did not significantly alter S2 generation induced by Dll1 (Fig. 5A). Since γ-secretase cleavage of S2 produces the biologically active S3 form, we also monitored this product by using an antibody that specifically detects S3. Consistent with the losses in ligand-induced reporter activity (Fig. 1C to E and 3B to C) and Herp2 expression (Fig. 3D) detected for cells deficient in ADAM10, S3/NICD generation was also significantly reduced in ADAM10-depleted cells compared to that detected in SCR or ADAM17 siRNA-treated cells (Fig. 5B). Although these data monitoring ligand-induced proteolysis support a role for ADAM10, rather than ADAM17, in S2 cleavage, endogenous N1 in C2C12 cells was also monitored to rule out any potential artifacts that may result from ectopic expression of N1Δmyc. This analysis revealed that Dll1-induced cleavage at both S2 and S3 sites in endogenous N1 was also significantly decreased by siRNA-mediated depletion of ADAM10, but not ADAM17 (Fig. 5C and D). Together, our data implicate ADAM10 and exclude ADAM17 as functioning in S2 cleavage of N1 in response to ligand.
FIG. 5.
ADAM10, but not ADAM17, expression is required for ligand-induced S2 and S3 cleavage of N1Δmyc and endogenous N1 in C2C12 cells. (A and B) Depletion of ADAM10, but not ADAM17, decreased ligand-induced S2 and S3 cleavage of ectopic N1Δmyc. Dll1-induced S2 (A) and S3 (B) N1Δmyc cleavage fragments produced in ADAM10 (AD10) or ADAM17 (AD17) siRNA-depleted C2C12 cells in the presence of the γ-secretase (DAPT) (A) or proteasome (MG132) inhibitors (B) were detected by Western blotting (WB). The percent quantification of S1 fragment converted to S2 fragment from DAPT-treated Dll1 cocultures (A) and the percent quantification of S3 cleavage fragment in MG132-treated Dll1 cocultures (B) with SCR siRNA were arbitrarily set to 100% (lower panels; n = 4). (C and D) Depletion of ADAM10, but not ADAM17, decreased ligand-induced S2 and S3 cleavage of endogenous N1. Dll1-induced S2 (C) and S3 (D) endogenous N1 cleavage fragments produced in parental C2C12 cells were assayed and quantified as described for panels A and B, respectively. Lysates were immunoprecipitated (IP), and endogenous N1 proteolytic cleavage fragments were detected by Western blotting (n = 3). α, anti; *, P < 0.05; **, P < 0.01.
Overexpression of ADAM17 and N1 leads to ligand-independent signaling.
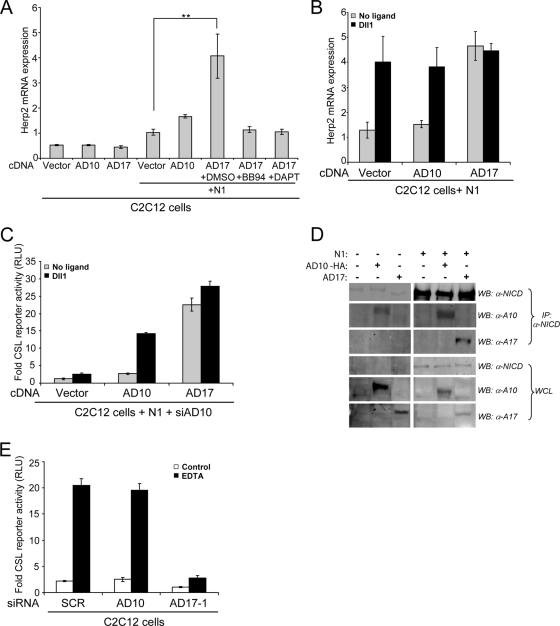
Previous studies implicating ADAM17 in S2 cleavage and Notch signaling used N1 mutant proteins that are cleaved independent of ligand (3, 36). Moreover, recent studies have reported that high levels of matrix metalloproteinase 7 (MMP7) in mammalian cells (51) or TACE in Drosophila cells (5) activate Notch signaling independent of ligand. Therefore, to determine if ADAM10 or ADAM17 could activate Notch in a ligand-independent manner, C2C12 cells were transiently transfected with ADAM10 or ADAM17 cDNAs, and Herp2 mRNA expression was used as a readout of Notch activation. Although, ectopic expression of either ADAM10 or ADAM17 in C2C12 cells did not increase Herp2 expression over the vector control, coexpression of N1 and ADAM17 significantly increased Herp2 expression (Fig. 6A), suggestive of ligand-independent activation of N1 by ADAM17. Both BB94 and DAPT suppressed Herp2 gene expression (Fig. 6A), indicating that transcriptional activation of this Notch target gene by ADAM17 involves proteolytic activation by both metalloprotease and γ-secretase. Moreover, Herp2 expression induced by ADAM17 ectopically expressed with N1 was not further enhanced by ligand (Fig. 6B), consistent with ADAM17-mediated, ligand-independent Notch signaling. In contrast to these findings for ADAM17, cells coexpressing both ADAM10 and N1 significantly induced Herp2 expression in response to only ligand (Fig. 6A and B), providing further support that ADAM10 is required for ligand-induced proteolytic activation of N1. Furthermore, siRNA depletion of ADAM10 did not diminish the level of reporter activity detected with C2C12 cells overexpressing ADAM17 and N1, either in the presence or absence of Dll1 cells (Fig. 6C), suggesting that ADAM10 is not required for the ADAM17-mediated, ligand-independent activation of N1. Since metalloprotease blockade suppressed ADAM17-induced Herp2 expression (Fig. 6A), it seems likely that ADAM17 functions directly in the proteolytic activation of N1 that occurs independent of ligand.
FIG. 6.
ADAM17 activates Notch signaling independent of ligand. (A) Overexpression of ADAM17 (AD17) with N1 induced ligand-independent Notch signaling that requires both ADAM and γ-secretase activity. C2C12 cells cotransfected with N1 and either ADAM10 (AD10) or ADAM17 (AD17) were incubated with metalloprotease (BB94) and γ-secretase (DAPT) inhibitors (DMSO vehicle), and Herp2 expression was detected by qRT-PCR analysis (see Materials and Methods for details) (n = 3). (B) Herp2 expression induced by ADAM17 is not sensitive to ligand. Herp2 mRNA was detected by qRT-PCR analysis in C2C12 cells, as described for panel A, and cocultured with Dll1 cells (n = 2). (C) ADAM10 is not required for reporter activity induced by ADAM17 independent of ligand. CSL reporter activity in ADAM10 siRNA depleted C2C12 cells overexpressing N1 with either ADAM10 or ADAM17 cDNAs was measured, following coculture with Dll1 cells (induction over parental L cells; n = 2). (D) ADAM17 interacts with ectopic but not endogenous N1. C2C12 cells expressing ADAM10 or ADAM17 either alone or together with N1 were either immunoblotted (WB) with anti-NICD (93-4), anti-AD10, or anti-AD17 antibodies or immunoprecipitated (IP) with anti-NICD (PCR 12), followed by WB with the indicated antibodies. The p120 furin-cleaved NICD fragment is shown for both NICD blots. (E) EDTA-induced reporter activity is ADAM17 dependent. C2C12 cells treated with the indicated siRNAs and expressing a CSL reporter were treated with either 1 mM EDTA or HBSS buffer for 10 min and assayed for luciferase activity 6 to 8 h later (n = 2). α, anti; **, P < 0.01.
ADAM17 but not ADAM10 is required for ligand-independent activation of N1.
Our findings indicate that ADAM10 cleaves either endogenous or ectopically expressed N1 (Fig. 4 and 5), but cleavage at the S2 site is highly dependent on ligand. While ADAM17 was not required for ligand-induced S2 cleavage of N1, it could nonetheless activate signaling independent of ligand when ectopically coexpressed with N1 (Fig. 6A and B). The fact that ADAM17 coimmunoprecipitated with ectopic N1, but not endogenous N1 (Fig. 6D), offers a plausible explanation as to why ligand-independent signaling requires coexpression of both ADAM17 and N1 (Fig. 6A to C). In contrast, ADAM10 coimmunoprecipitated with both ectopic and endogenous N1, indicating that ADAM10 constitutively interacts with N1; however, ADAM10 can activate N1 only in the presence of ligand (Fig. 6A to C), suggesting that ligand binding to N1 facilitates ADAM10 cleavage.
Although coexpression of N1 with ADAM17 identified a cellular context in which ADAM17 could activate N1, the mechanism of this ligand-independent activation is unclear. Therefore, we used EDTA to activate signaling independent of ligand since EDTA-mediated sequestration of calcium is thought to destabilize the NRR structure and expose the S2 cleavage site similar to that proposed for ligand (11, 45). Cells treated with EDTA strongly induced reporter activity; however, cells depleted of ADAM17 were defective in EDTA-induced reporter activity (Fig. 6E), suggesting that ADAM17 is required for N1 activation induced by EDTA, consistent with a role for this protease in the ligand-independent signaling. Importantly, the loss of ADAM10 did not diminish EDTA-induced reporter activity, underscoring the importance of ligand in ADAM10-mediated activation of N1.
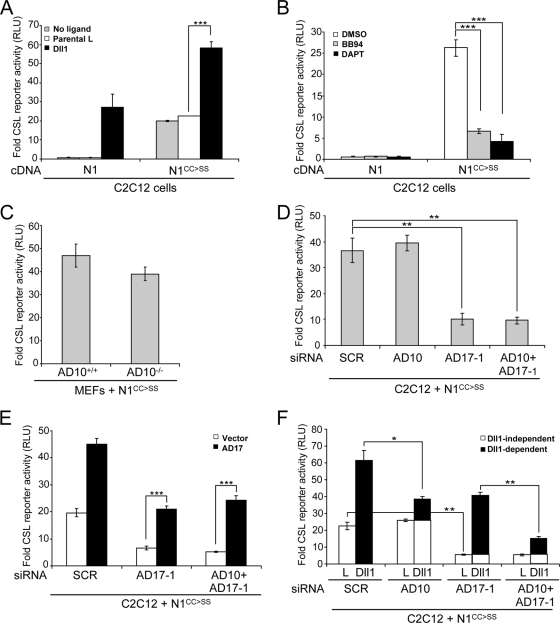
To further explore the protease dependency in ligand-independent Notch signaling, we examined an N1 mutant in which two highly conserved cysteine residues within the NRR (C1675 and C1682) were converted to serine (N1CC→SS). These mutations in Drosophila Notch produce a hyperactive ligand-independent form (25), and N1CC→SS expressed in C2C12 cells strongly activated a CSL reporter in the absence of ligand (Fig. 7A). Although the activity detected with N1CC→SS in the absence of ligand was similar to that induced by wild-type N1 in response to Dll1 cells, signaling was further enhanced by ligand (Fig. 7A), suggesting that although this mutant is constitutively active, it can respond to ligand. Furthermore, the ligand-independent activity intrinsic to N1CC→SS was sensitive to BB94, indicating a strict dependence on metalloprotease activity (Fig. 7B). Based on the strong requirement for ligand in ADAM10 activation of N1, it seemed likely that this protease would not be required for N1CC→SS to signal in the absence of ligand. Indeed, N1CC→SS activated a Notch reporter to similar levels when expressed by wild-type or ADAM10−/− MEFs (Fig. 7C). It is important to note that this result is reminiscent of a previous report for normal proteolytic processing of a truncated N1CC→SS in ADAM10−/− MEFs, a finding that was used to exclude a role for ADAM10 in the activation of N1 (36).
FIG. 7.
ADAM17 is required for constitutive N1CC→SS signaling activity. (A) N1CC→SS reporter activity is enhanced by ligand. C2C12 cells coexpressing either N1 or N1CC→SS and a CSL reporter were assayed for activation in the presence and absence of Dll1 cells (activation over N1 cells in the absence of ligand; n = 3). (B) Constitutive N1CC→SS activity requires ADAM and γ-secretase cleavage. Reporter activity was measured for N1- or N1CC→SS-expressing C2C12 cells treated with metalloprotease (BB94) or γ-secretase (DAPT) inhibitors (activation over DMSO-treated N1 cells; n = 3). (C) ADAM10 (AD10) is not required for constitutive signaling by N1CC→SS. ADAM10+/+ or ADAM10−/− MEFs were cotransfected with either N1 or N1CC→SS and CSL reporter constructs and assayed for luciferase activity (activation over N1 cells; n = 3). (D) N1CC→SS constitutive activity requires ADAM17 (AD17). CSL reporter activity for N1CC→SS-expressing C2C12 cells depleted for ADAM10, ADAM17, or both by siRNAs and assayed for luciferase activity (activation over SCR siRNA-treated N1 cells; n = 3). (E) Losses in N1CC→SS reporter activity following ADAM17 or both ADAM10 and ADAM17 siRNA depletion can be rescued by ectopic expression of ADAM17 cDNA (activation over SCR-treated, vector-transfected N1 cells; n = 2). (F) N1CC→SS signaling induced by Dll1 is a composite of ligand-independent (open bars) and -dependent (filled bars) activation that requires both ADAM10 and ADAM17. Reporter activity of N1CC→SS-expressing C2C12 cells treated with ADAM10, ADAM17, or both siRNAs were cocultured with either L or Dll1 cells (activation over SCR-treated N1 cells cocultured with parental L cells; n = 3).
To determine the ADAM requirement in ligand-independent signaling, we treated C2C12 cells ectopically expressing N1CC→SS with siRNAs specific for ADAM10 or ADAM17. Efficient ADAM10 knockdown in N1CC→SS cells induced signaling to levels similar to those measured for cells treated with SCR siRNA (Fig. 7D), corroborating findings with ADAM10−/− and wild-type MEFs. That cells treated with ADAM17 siRNA either alone or in combination with ADAM10 siRNA displayed a dramatic loss in activity (Fig. 7D) suggests that ADAM17 is required for signaling by the constitutively active N1CC→SS mutant protein. In support of this, ectopic expression of ADAM17 rescued the loss in signaling associated with the siRNA-mediated depletion (Fig. 7E), ruling out nonspecific siRNA effects. Moreover, ectopic expression of ADAM17 enhanced transcription by N1CC→SS (Fig. 7E, SCR), as found for wild-type N1 (Fig. 6A to C).
Further evidence for selective use of ADAM10 and ADAM17 in ligand-dependent and -independent activation of N1, respectively, was obtained following specific ADAM depletion in N1CC→SS-expressing cells cocultured with either L or Dll1 cells. This analysis allowed identification of signaling induced in Dll1-independent and -dependent contexts (Fig. 7F), following coculture with either L or Dll1 cells. From this analysis it can be deduced that reporter activity detected for Dll1 cocultures represents the product of signaling intrinsic to N1CC→SS and that induced by ligand. ADAM10 depletion did not diminish signaling intrinsic to N1CC→SS (L cells) but did decrease signaling induced by Dll1 cells, consistent with the requirement for ligand in the ADAM10 activation of N1. In contrast, depletion of ADAM17 strongly suppressed the N1CC→SS activity measured for L cocultures, supporting the requirement for this protease in ligand-independent constitutive signaling. Moreover, depletion of both ADAMs suppressed signaling induced in either the absence (L coculture) or presence (Dll1 coculture) of ligand. Together, these N1CC→SS data provide support for the intriguing idea that ligand-dependent and -independent activation of N1 requires distinct ADAMs.
T-ALL activating Notch1 mutations require both ADAM10 and ADAM17.
The N1CC→SS mutations map to the heterodimerization (HD) domain within the NRR, a region that contains the majority of activating human N1 mutations isolated from T-ALL patients (12, 30, 58). Although a mutation equivalent to C1675 has been detected in T-ALL (11), the additional C1682S change in N1CC→SS has so far not been reported either alone or in combination with C1675. To determine whether our findings with N1CC→SS could be extended to the aberrant signaling associated with T-ALL, we examined the requirement for ADAM10 and -17 in signaling by several well-characterized activating T-ALL mutants.
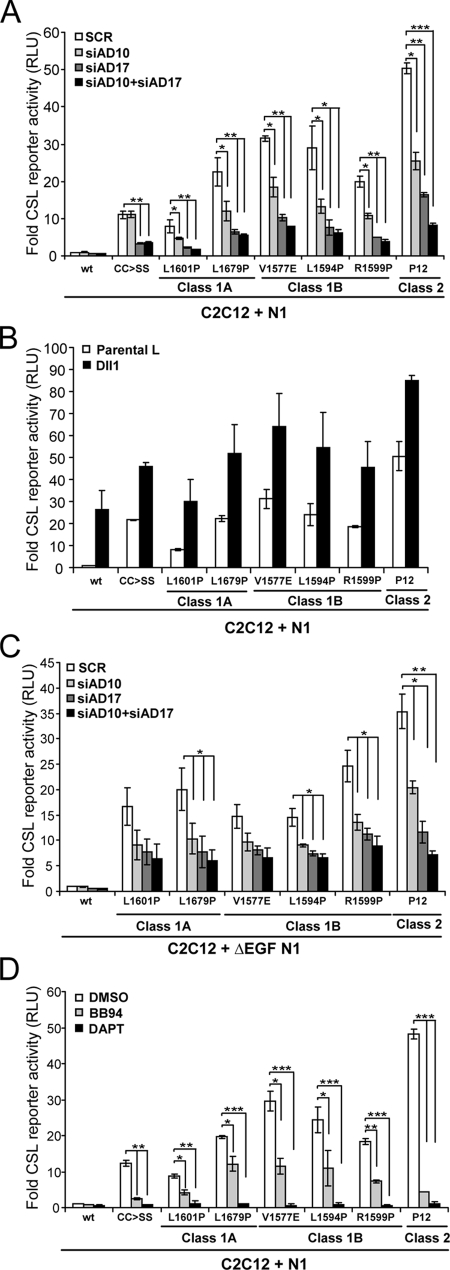
Approximately 40% of the isolated T-ALL mutations in human N1 map to the NRR and have been classified according to the destabilizing effects on heterodimer structure and level of activation (30, 58). Briefly, class 1A are weakly activating and associated with strong and spontaneous heterodimer dissociation, while mutations that weakly destabilize but still produce ligand-independent activity are classified as class 1B. On the other hand, class 2 mutations produce high levels of Notch signaling activity without affecting the stability of the heterodimer. Significantly, all NRR mutations are thought to allow constitutive cleavage through S2 exposure or access; however, the relevant ADAMs have not been identified. To this end, we tested full-length forms of N1 mutants from class 1A (L1601P, L1679P), class 1B (V1577E, L1594P, R1599P), and class 2 (P12) for reporter activity following siRNA knockdown of ADAM10 and ADAM17, either alone or in combination. Although equivalent levels of expression of the T-ALL mutants (data not shown) produced various levels of reporter activity, the activities were nonetheless significantly higher than those for wild-type N1 in the absence of ligand (Fig. 8A). As found with N1CC→SS, depletion of ADAM17 alone or together with ADAM10 resulted in a similar and strong reduction in reporter activity relative to that for SCR siRNA-treated cells; however, unlike N1CC→SS that did not display a requirement for ADAM10, all T-ALL mutations showed some level of dependence on ADAM10. Nonetheless, this analysis suggested a stronger requirement for ADAM17 over ADAM10 for constitutive signaling intrinsic to T-ALL mutations.
FIG. 8.
Signaling activity intrinsic to N1 T-ALL mutants have enhanced protease sensitivity. (A and C) T-ALL activating mutants require both ADAM10 and ADAM17 for constitutive signaling activity. C2C12 cells treated with ADAM10 (siAD10) and/or ADAM17 (siAD17) siRNAs were transfected with the indicated full-length (A) or EGF-truncated (ΔEGF) (C) class1 and 2 T-ALL N1 mutants and assayed for luciferase activity (activation over SCR-treated wild-type N1 cells; n = 2). (B) Signaling by T-ALL mutants can be enhanced by ligand. C2C12 cells cotransfected with the CSL reporter and the full-length N1 cDNAs with indicated mutations were cocultured with Dll1 cells (activation over wild-type N1 cells cocultured with L cells; n = 2). (D) Inhibition of γ-secretase but not metalloproteases efficiently suppressed signaling by class 1 T-ALL mutants. Reporter activity was detected for C2C12 cells expressing the indicated full-length N1 T-ALL mutants following treatment with metalloprotease (BB94) or γ-secretase (DAPT) inhibitors (DMSO vehicle) (activation over wild-type N1 cells treated with DMSO; n = 2).
Given the strong requirement for ligand in ADAM10 activation of N1, we reasoned that the requirement for ADAM10 in activation of the T-ALL mutants might reflect input by ligand. To address this possibility, we determined whether the signaling activity intrinsic to these T-ALL mutants could be further enhanced by ligand as found for N1CC→SS. In every case, reporter activity was increased following coculture with Dll1 cells (Fig. 8B), indicating that the mutant proteins were expressed at the cell surface and that signaling could be enhanced by ligand. Since the level of enhancement was similar to that induced by either N1CC→SS or wild-type N1 in response to ligand, the ADAM10 requirement could reflect ligand-dependent activation. To directly address this, we tested forms lacking the EGF-like repeats (ΔEGF-N1) that facilitate ligand binding (30), following siRNA depletion of ADAM10, ADAM17, or both. Losses in either ADAM10 or ADAM17 reduced reporter activity induced by the ΔEGF-N1 mutants (Fig. 8C), indicating a requirement for both proteases in ligand-independent signaling. In contrast to the full-length forms that showed a 30 to 50% and 60 to 80% reduction in reporter activity following ADAM10 and -17 depletion, respectively (Fig. 8A), the ΔEGF-N1 mutants did not display an obvious preference for ADAM17 over ADAM10 in this assay (Fig. 8C). The exception to this was the class 2 mutant P12 that had a stronger dependence on ADAM17 compared to ADAM10 for both full-length and truncated forms (Fig. 8A and C). Even more surprising were the findings that in contrast to γ-secretase blockade, not all the class 1 mutants were strongly inhibited by BB94 (Fig. 8D), suggesting that the signaling activity intrinsic to these mutations is not completely dependent on constitutive metalloprotease cleavage. Interestingly, the activity associated with the class 2 mutation that does not involve NRR destabilization for S2 cleavage was strongly inhibited by both BB94 and DAPT (Fig. 8D).
DISCUSSION
Metalloproteases can both positively and negatively regulate Notch signaling; yet the exact roles that these proteases perform have remained controversial. Further complicating this issue is the proteolytic processing of both Notch ligands and receptors by a number of different metalloproteases (7, 62). Moreover, ADAM-mediated shedding of ligand from the signal-sending cell can downregulate signaling (33, 37, 55), while proteolytic removal of ligand from the Notch cell enhances signal reception (50). We have focused our studies on cell-autonomous roles for ADAM10 and ADAM17 in proteolytic activation of N1, which have identified selective use of ADAM10 in ligand-induced activation. The strict dependence on ADAM10 in the ligand-induced cleavage of N1 is consistent with the overlap in developmental defects displayed by mice lacking either N1 or ADAM10 (4, 14, 31, 41, 44, 56, 57). Even though our findings indicate that ADAM17 cannot cleave Notch in response to ligand, we have uncovered a specific role for this metalloprotease in the activation of Notch independent of ligand. Although Notch is a substrate for both ADAM10 and ADAM17, our studies suggest that context-dependent conformational changes in N1 may dictate which protease cleaves to facilitate receptor activation.
We find that ADAM10 is not required in the ligand cell to send a signal but, rather, is required by the Notch cell to receive the signal, and this is more consistent with a role for ADAM10 in regulating the rate-limiting S2 cleavage of N1 in response to ligand. Specifically, defects in ADAM10 activity meditated by expression of a dominant negative form, or depletion of ADAM10 protein through gene deletion or RNA interference all lead to losses in ligand-induced signaling. Moreover, ADAM10 losses resulting in signaling defects correlate with decreased S2 generation, which is necessary for γ-secretase cleavage to generate the active NICD signaling fragment. Despite previous reports identifying N1 as an ADAM17 substrate (3, 36), as well as high ADAM17 expression in cells lacking ADAM10, ADAM17 could not cleave or activate N1 in response to ligand. Consistent with this finding, losses in ADAM17 activity or protein did not significantly diminish S2 cleavage, reporter activity, or Herp2 expression in response to Notch ligands. It is unclear why ADAM17, a principal sheddase for a large number of cell surface proteins (8), cannot cleave cell surface N1 in response to ligand. Perhaps ADAM17 and N1 occupy distinct cell surface microdomains, thereby precluding interactions, consistent with our inability to coimmunoprecipitate ADAM17 with endogenous N1. Nonetheless, our findings indicate that ADAM17 does not compete with ADAM10 in ligand-induced cleavage of N1 and that these closely related ADAMs are not functionally redundant in signaling regulated by ligand.
The exclusive use of ADAM10 over ADAM17 in ligand-induced signaling is in keeping with our findings that ADAM10, but not ADAM17, interacts with endogenous N1. However, ADAM17 could interact with N1 when ectopically coexpressed, and this resulted in dysregulated receptor activation in the absence of ligand. The constitutive signaling associated with ADAM17 did not require ADAM10 but was absolutely dependent on ADAM17 activity, which could reflect differential localization of these proteases to specific subcellular compartments. In fact, ADAM17 activation of N1 did not involve ectodomain shedding (data not shown), excluding the possibility that ADAM17 cleaves N1 at the cell surface. Although it is unclear where in the cell ADAM17 cleaves and activates N1, we speculate that this ligand-independent activation may involve an accumulation of high levels of ADAM17 and N1 proteins intracellularly, which could facilitate interactions between these proteins and induce conformational changes in N1 that would expose the S2 site and allow cleavage. In fact, the intracellular accumulation of Notch in trafficking mutants in flies is associated with constitutive activation of signaling that appears to occur independent of ligand (9).
In contrast to ADAM17, interactions detected between ectopically expressed ADAM10 and N1 did not produce constitutive signaling, providing additional support for the importance of ligand in ADAM10 cleavage of N1. It is important to note that N1 was first identified as an ADAM17 substrate through its ability to cleave a short N1 fragment containing the S2 site but lacking the protective NRR sequences and ligand binding sites (3). This S2-containing N1 fragment was not cleaved by ADAM10, which the authors interpreted to mean that ADAM10 is not the relevant protease in Notch signaling. However, we would argue that the inability of ADAM10 to cleave this short N1 fragment more reflects our findings that interactions between ADAM10 and N1 do not activate signaling in the absence of ligand.
To further explore differences between ADAM10 and ADAM17 activation of N1, we determined the protease requirement for signaling induced by treatment of cells with EDTA, which is proposed to mimic that induced by ligand (45). Since the NRR structure is dependent on calcium and zinc binding (11) and EDTA induces shedding of the Notch ectodomain (45), one might predict that structural changes induced by EDTA expose the S2 site, as do those induced by ligand. However, ADAM17 was absolutely required for EDTA-induced activation of N1, while losses in ADAM10 did not affect signaling, findings that are in complete opposition to those identified for ligand activation of N1. Although the molecular basis for this selectivity is unknown, it is possible that the EDTA-induced conformational changes that lead to heterodimer dissociation provide a more suitable ADAM17 substrate, while ligand-induced dissociation of N1 produces a conformation conducive to ADAM10 cleavage. Moreover, since ADAM17 is unable to cleave N1 in response to ligand, even in the absence of ADAM10, it seems that ligand-induced conformational changes in N1 are not sufficient for ADAM17 proteolysis. Importantly, our findings have identified mechanistic differences between ligand- and EDTA-induced Notch signaling that indicate that these two modes of Notch activation cannot be considered equivalent.
Similarly, the constitutive signaling capacity associated with N1CC→SS presumably reflects changes in NRR conformation that promote ADAM17, yet disfavor ADAM10 proteolytic activation. Based on structure data, mutating these two conserved cysteine residues within the NRR would prevent the formation of a disulfide bond, which is predicted to contribute to the tight packing interactions between the Lin12/Notch repeats (LNR) and HD domains required to stabilize the heterodimer and prevent S2 exposure and cleavage (11). Interestingly, these changes did not accommodate S2 cleavage by ADAM10 in the absence of ligand. Findings from a previous study also indicated that ADAM10 is not required for N1CC→SS cleavage; however, this was used to exclude ADAM10 as a relevant protease in Notch signaling (36). In contrast, our findings have identified an obligatory role for ADAM10 in signaling induced by ligand and indicate that ADAM17 expressed by ADAM10 deficient cells is capable of activating N1CC→SS independent of ligand, further establishing differential requirements for these proteases in ligand-dependent and -independent modes of Notch activation.
The differential and strict requirements for ADAM10 and ADAM17 in regulated and dysregulated Notch signaling, respectively, suggested that NRR-activating T-ALL mutations might also display similar ADAM preferences. If this were the case, ADAM17-specific inhibitors could be considered an alternative therapeutic rationale to target pathological signaling induced by ADAM17, while excluding normal Notch signaling mediated by ADAM10. Indeed, the current stumbling block to effective treatment of T-ALL using γ-secretase inhibitors is the severe toxicity resulting from disruption of normal Notch signaling (46). However, examination of a number of well-characterized T-ALL mutations revealed a requirement for ADAM10, suggesting that this protease might contribute to the constitutive signaling intrinsic to these activating mutations. Although the T-ALL mutations displayed an anticipated preference for ADAM17 in constitutive signaling, not all were as sensitive to metalloprotease blockade as they were to inhibition of γ-secretase. The increased resistance to metalloprotease inhibition suggests that either additional proteases activate these T-ALL mutants or perhaps they signal independent of S2 cleavage and/or rely on S1 cleavage by furin proteases for intrinsic activity. Even though the signaling activity of the T-ALL mutants could be increased by ligand, suggesting cleavage at the cell surface, the specific cellular compartment that facilitates ligand-independent signaling by these mutants is unknown. Nonetheless, the observed differences in requirements for activating proteases suggest that the conformational changes mediated by these T-ALL mutations must be distinct from those induced by either ligand, EDTA, or the specific mutations encoded by the N1CC→SS construct. Together, our studies suggest that the N1 mutations associated with leukemia have undergone selection to expand the repertoire of proteases competent to activate N1. Enhanced protease sensitivity would ensure efficient signaling by γ-secretase, underscoring the importance of developing γ-secretase-based therapies that selectively inhibit pathological signaling with limited effects on normal Notch signaling.
ADAM10 and ADAM17 have overlapping and distinct substrates, and ectodomain shedding by these proteases can be induced by a number of stimuli (8); however, the molecular basis of ADAM substrate specificity is not well understood. Interestingly, under certain conditions, ADAM10 can be induced to process substrates normally cleaved only by ADAM17 (24), and the alteration in substrate specificity is linked to changes in ADAM activity (16). In contrast, our studies suggest that while N1 is a substrate for both ADAM10 and ADAM17, the particular activating conformation induced in either a ligand-dependent or -independent context determines which protease cleaves N1. In this regard, unlike many proteases that recognize and cleave consensus primary amino acid sequences, ADAMs rely on structural motifs that harbor the cleavage site close to the membrane (8). In fact, T-ALL mutations that displace the S2 site in N1 away from the membrane facilitate constitutive metalloprotease processing in the absence of NRR destabilization (30, 54), suggesting that positioning the S2 site near the cell surface prevents unregulated proteolysis and activation of signaling. In fact, placement of the S2 site close to the membrane offers an additional level of regulation, since ADAM membrane tethering would further restrict substrate access and cleavage. In support of this idea, large amounts of soluble MMP7, which is unrestricted in membrane orientation, are able to cleave Notch independent of ligand (51).
The molecular basis for the ADAM10 selectivity in ligand-induced activation of Notch signaling is unclear; however, it does not reflect the need for ADAM10 to bind EGF-like repeats that contain the ligand binding domain, since T-ALL mutants lacking these sequences required ADAM10 for full constitutive signaling capacity. The strong dependence on ligand for ADAM10 in normal Notch signaling is reminiscent of the regulated proteolytic cleavage of ephrin-A2 by ADAM10, which requires the formation of ligand-receptor signaling complexes for efficient proteolysis (18). In this regard, ligand could position ADAM10 for efficient Notch cleavage, or ADAM10 interactions with ligand-Notch complexes could enhance protease activity. However, given the strong role for ligand in overriding the Notch protease-resistant state, we favor the idea that ligand induces global conformational changes in the NRR that are critical for ADAM10 to recognize, access, and/or cleave N1 at the S2 site. Our findings suggest that structural changes in NRR conformation, induced by ligand and required to override the autoinhibitory state, must not allow ADAM17 recognition and/or cleavage. Our data identifying ADAM10 as the relevant S2 protease may appear inconsistent with previous reports; however, it is important to note that the previous studies (3, 36) were actually measuring ligand-independent Notch activation that we show requires ADAM17, yet extrapolated their findings to signaling induced by ligand that is ADAM10 dependent.
Based on our finding with three different mammalian cell types, we conclude that cleavage of N1 by either ADAM10 or ADAM17 may require distinct N1 conformations. Therefore, in addition to exposing the S2 site, we propose that ligand-dependent and -independent global conformational changes in N1 might also direct selection of the activating protease. Together, our studies have identified mechanistic differences between ligand-induced and ligand-independent Notch signaling not previously appreciated. Future structural studies are required to understand the molecular basis of these differences.
Acknowledgments
We thank Luisa Iruela-Arispe, Alison Miyamoto, Carl Blobel, Steve Blacklow, and Jon Aster for helpful discussions on this work, and Brendan D'Souza and Abdi Musse for useful comments on the manuscript. We also thank Christine Yao for the N1Δmyc cell line; Carl Blobel, Paul Saftig, Roy Black (Amgen), and D. J. Pan for ADAM10 and ADAM17 targeted MEF lines; Roy Black (Amgen) for ADAM17; Falk Fahrenholz for ADAM10HA; Carl Blobel for TNF-α-AP; Jon Aster and Steven Blacklow for T-ALL mutant N1 cDNA clones; and British Biotechnology for BB94.
This work was supported by funding from the NIH NS31885 (NINDS Javits Award to G.W.) and the JCCF (predoctoral fellowship to E.C.B.).
Footnotes
Published ahead of print on 24 August 2009.
REFERENCES
- 1.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 2.Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678-689. [DOI] [PubMed] [Google Scholar]
- 3.Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail, J. R. Doedens, A. Cumano, P. Roux, R. A. Black, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5:207-216. [DOI] [PubMed] [Google Scholar]
- 4.Conlon, R. A., A. G. Reaume, and J. Rossant. 1995. Notch1 is required for the coordinate segmentation of somites. Development 121:1533-1545. [DOI] [PubMed] [Google Scholar]
- 5.Delwig, A., and M. D. Rand. 2008. Kuz and TACE can activate Notch independent of ligand. Cell. Mol. Life Sci. 65:2232-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dovey, H. F., V. John, J. P. Anderson, L. Z. Chen, P. de Saint Andrieu, L. Y. Fang, S. B. Freedman, B. Folmer, E. Goldbach, E. J. Holsztynska, K. L. Hu, K. L. Johnson-Wood, S. L. Kennedy, D. Kholodenko, J. E. Knops, L. H. Latimer, M. Lee, Z. Liao, I. M. Lieberburg, R. N. Motter, L. C. Mutter, J. Nietz, K. P. Quinn, K. L. Sacchi, P. A. Seubert, G. M. Shopp, E. D. Thorsett, J. S. Tung, J. Wu, S. Yang, C. T. Yin, D. B. Schenk, P. C. May, L. D. Altstiel, M. H. Bender, L. N. Boggs, T. C. Britton, J. C. Clemens, D. L. Czilli, D. K. Dieckman-McGinty, J. J. Droste, K. S. Fuson, B. D. Gitter, P. A. Hyslop, E. M. Johnstone, W. Y. Li, S. P. Little, T. E. Mabry, F. D. Miller, and J. E. Audia. 2001. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J. Neurochem. 76:173-181. [DOI] [PubMed] [Google Scholar]
- 7.D'Souza, B., A. Miyamoto, and G. Weinmaster. 2008. The many facets of Notch ligands. Oncogene 27:5148-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, D. R., M. M. Handsley, and C. J. Pennington. 2008. The ADAM metalloproteinases. Mol. Aspects Med. 29:258-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortini, M. E., and D. Bilder. 2009. Endocytic regulation of Notch signaling. Curr. Opin. Genet. Dev. 19:323-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon, W. R., K. L. Arnett, and S. C. Blacklow. 2008. The molecular logic of Notch signaling—a structural and biochemical perspective. J. Cell Sci. 121:3109-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, W. R., M. Roy, D. Vardar-Ulu, M. Garfinkel, M. R. Mansour, J. C. Aster, and S. C. Blacklow. 2009. Structure of the Notch1 negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood 113:4381-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon, W. R., D. Vardar-Ulu, G. Histen, C. Sanchez-Irizarry, J. C. Aster, and S. C. Blacklow. 2007. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 14:295-300. [DOI] [PubMed] [Google Scholar]
- 13.Greenwald, I. 1994. Structure/function studies of lin-12/Notch proteins. Curr. Opin. Genet. Dev. 4:556-562. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann, D., B. de Strooper, L. Serneels, K. Craessaerts, A. Herreman, W. Annaert, L. Umans, T. Lubke, A. Lena Illert, K. von Figura, and P. Saftig. 2002. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol. Genet. 11:2615-2624. [DOI] [PubMed] [Google Scholar]
- 15.Hicks, C., S. H. Johnston, G. diSibio, A. Collazo, T. F. Vogt, and G. Weinmaster. 2000. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat. Cell Biol. 2:515-520. [DOI] [PubMed] [Google Scholar]
- 16.Horiuchi, K., S. Le Gall, M. Schulte, T. Yamaguchi, K. Reiss, G. Murphy, Y. Toyama, D. Hartmann, P. Saftig, and C. P. Blobel. 2007. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol. Biol. Cell 18:176-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237-255. [DOI] [PubMed] [Google Scholar]
- 18.Janes, P. W., N. Saha, W. A. Barton, M. V. Kolev, S. H. Wimmer-Kleikamp, E. Nievergall, C. P. Blobel, J. P. Himanen, M. Lackmann, and D. B. Nikolov. 2005. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 123:291-304. [DOI] [PubMed] [Google Scholar]
- 19.Jarriault, S., and I. Greenwald. 2005. Evidence for functional redundancy between C. elegans ADAM proteins SUP-17/Kuzbanian and ADM-4/TACE. Dev. Biol. 287:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopan, R., E. H. Schroeter, H. Weintraub, and J. S. Nye. 1996. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 93:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladi, E., J. T. Nichols, W. Ge, A. Miyamoto, C. Yao, L. T. Yang, J. Boulter, Y. E. Sun, C. Kintner, and G. Weinmaster. 2005. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol. 170:983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lammich, S., E. Kojro, R. Postina, S. Gilbert, R. Pfeiffer, M. Jasionowski, C. Haass, and F. Fahrenholz. 1999. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. USA 96:3922-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Borgne, R. 2006. Regulation of Notch signalling by endocytosis and endosomal sorting. Curr. Opin. Cell Biol. 18:213-222. [DOI] [PubMed] [Google Scholar]
- 24.Le Gall, S. M., P. Bobe, K. Reiss, K. Horiuchi, X. D. Niu, D. Lundell, D. R. Gibb, D. Conrad, P. Saftig, and C. P. Blobel. 2009. ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as transforming growth factor alpha, l-selectin, and tumor necrosis factor alpha. Mol. Biol. Cell 20:1785-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieber, T., S. Kidd, E. Alcamo, V. Corbin, and M. W. Young. 1993. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 7:1949-1965. [DOI] [PubMed] [Google Scholar]
- 26.Lieber, T., S. Kidd, and M. W. Young. 2002. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 16:209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsell, C. E., C. J. Shawber, J. Boulter, and G. Weinmaster. 1995. Jagged: a mammalian ligand that activates Notch1. Cell 80:909-917. [DOI] [PubMed] [Google Scholar]
- 28.Logeat, F., C. Bessia, C. Brou, O. LeBail, S. Jarriault, N. Seiday, and A. Israel. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95:8108-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lum, L., M. S. Reid, and C. P. Blobel. 1998. Intracellular maturation of the mouse metalloprotease disintegrin MDC15. J. Biol. Chem. 273:26236-26247. [DOI] [PubMed] [Google Scholar]
- 30.Malecki, M. J., C. Sanchez-Irizarry, J. L. Mitchell, G. Histen, M. L. Xu, J. C. Aster, and S. C. Blacklow. 2006. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 26:4642-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manilay, J. O., A. C. Anderson, C. Kang, and E. A. Robey. 2005. Impairment of thymocyte development by dominant-negative Kuzbanian (ADAM-10) is rescued by the Notch ligand, delta-1. J. Immunol. 174:6732-6741. [DOI] [PubMed] [Google Scholar]
- 32.Maskos, K., C. Fernandez-Catalan, R. Huber, G. P. Bourenkov, H. Bartunik, G. A. Ellestad, P. Reddy, M. F. Wolfson, C. T. Rauch, B. J. Castner, R. Davis, H. R. Clarke, M. Petersen, J. N. Fitzner, D. P. Cerretti, C. J. March, R. J. Paxton, R. A. Black, and W. Bode. 1998. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc. Natl. Acad. Sci. USA 95:3408-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra-Gorur, K., M. D. Rand, B. Perez-Villamil, and S. Artavanis-Tsakonas. 2002. Down-regulation of Delta by proteolytic processing. J. Cell Biol. 159:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyamoto, A., R. Lau, P. W. Hein, J. M. Shipley, and G. Weinmaster. 2006. Microfibrillar proteins MAGP-1 and MAGP-2 induce Notch1 extracellular domain dissociation and receptor activation. J. Biol. Chem. 281:10089-10097. [DOI] [PubMed] [Google Scholar]
- 35.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151-165. [DOI] [PubMed] [Google Scholar]
- 36.Mumm, J. S., E. H. Schroeter, M. T. Saxena, A. Griesemer, X. Tian, D. J. Pan, W. J. Ray, and R. Kopan. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5:197-206. [DOI] [PubMed] [Google Scholar]
- 37.Muraguchi, T., Y. Takegami, T. Ohtsuka, S. Kitajima, E. P. Chandana, A. Omura, T. Miki, R. Takahashi, N. Matsumoto, A. Ludwig, M. Noda, and C. Takahashi. 2007. RECK modulates Notch signaling during cortical neurogenesis by regulating ADAM10 activity. Nat. Neurosci. 10:838-845. [DOI] [PubMed] [Google Scholar]
- 38.Nichols, J. T., A. Miyamoto, S. L. Olsen, B. D'Souza, C. Yao, and G. Weinmaster. 2007. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 176:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols, J. T., A. Miyamoto, and G. Weinmaster. 2007. Notch signaling—constantly on the move. Traffic 8:959-969. [DOI] [PubMed] [Google Scholar]
- 40.Nofziger, D., A. Miyamoto, K. M. Lyons, and G. Weinmaster. 1999. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126:1689-1702. [DOI] [PubMed] [Google Scholar]
- 41.Pan, D., and G. M. Rubin. 1997. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90:271-280. [DOI] [PubMed] [Google Scholar]
- 42.Peschon, J. J., J. L. Slack, P. Reddy, K. L. Stocking, S. W. Sunnarborg, D. C. Lee, W. E. Russell, B. J. Castner, R. S. Johnson, J. N. Fitzner, R. W. Boyce, N. Nelson, C. J. Kozlosky, M. F. Wolfson, C. T. Rauch, D. P. Cerretti, R. J. Paxton, C. J. March, and R. A. Black. 1998. An essential role for ectodomain shedding in mammalian development. Science 282:1281-1284. [DOI] [PubMed] [Google Scholar]
- 43.Qi, H., M. D. Rand, X. Wu, N. Sestan, W. Wang, P. Rakic, T. Xu, and S. Artavanis-Tsakonas. 1999. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science 283:91-94. [DOI] [PubMed] [Google Scholar]
- 44.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H. R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10:547-558. [DOI] [PubMed] [Google Scholar]
- 45.Rand, M. D., L. M. Grimm, S. Artavanis-Tsakonas, V. Patriub, S. C. Blacklow, J. Sklar, and J. C. Aster. 2000. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell. Biol. 20:1825-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Real, P. J., V. Tosello, T. Palomero, M. Castillo, E. Hernando, E. de Stanchina, M. L. Sulis, K. Barnes, C. Sawai, I. Homminga, J. Meijerink, I. Aifantis, G. Basso, C. Cordon-Cardo, W. Ai, and A. Ferrando. 2009. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat. Med. 15:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy, P., J. L. Slack, R. Davis, D. P. Cerretti, C. J. Kozlosky, R. A. Blanton, D. Shows, J. J. Peschon, and R. A. Black. 2000. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. J. Biol. Chem. 275:14608-14614. [DOI] [PubMed] [Google Scholar]
- 48.Rooke, J., D. Pan, T. Xu, and G. M. Rubin. 1996. KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science 273:1227-1231. [DOI] [PubMed] [Google Scholar]
- 49.Sahin, U., G. Weskamp, K. Kelly, H. M. Zhou, S. Higashiyama, J. Peschon, D. Hartmann, P. Saftig, and C. P. Blobel. 2004. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 164:769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sapir, A., E. Assa-Kunik, R. Tsruya, E. Schejter, and B. Z. Shilo. 2005. Unidirectional Notch signaling depends on continuous cleavage of Delta. Development 132:123-132. [DOI] [PubMed] [Google Scholar]
- 51.Sawey, E. T., J. A. Johnson, and H. C. Crawford. 2007. Matrix metalloproteinase 7 controls pancreatic acinar cell transdifferentiation by activating the Notch signaling pathway. Proc. Natl. Acad. Sci. USA 104:19327-19332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shawber, C., D. Nofziger, J. J.-D. Hsieh, C. Lindsell, O. Bogler, D. Hayward, and G. Weinmaster. 1996. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development 122:3765-3773. [DOI] [PubMed] [Google Scholar]
- 53.Sotillos, S., F. Roch, and S. Campuzano. 1997. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124:4769-4779. [DOI] [PubMed] [Google Scholar]
- 54.Sulis, M. L., O. Williams, T. Palomero, V. Tosello, S. Pallikuppam, P. J. Real, K. Barnes, L. Zuurbier, J. P. Meijerink, and A. A. Ferrando. 2008. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood 112:733-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, D., H. Li, and A. Zolkiewska. 2008. The role of Delta-like 1 shedding in muscle cell self-renewal and differentiation. J. Cell Sci. 121:3815-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swiatek, P. J., C. E. Lindsell, F. F. del Amo, G. Weinmaster, and T. Gridley. 1994. Notch1 is essential for postimplantation development in mice. Genes Dev. 8:707-719. [DOI] [PubMed] [Google Scholar]
- 57.Tian, L., X. Wu, C. Chi, M. Han, T. Xu, and Y. Zhuang. 2008. ADAM10 is essential for proteolytic activation of Notch during thymocyte development. Int. Immunol. 20:1181-1187. [DOI] [PubMed] [Google Scholar]
- 58.Weng, A. P., A. A. Ferrando, W. Lee, J. P. Morris IV, L. B. Silverman, C. Sanchez-Irizarry, S. C. Blacklow, A. T. Look, and J. C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306:269-271. [DOI] [PubMed] [Google Scholar]
- 59.Yang, L. T., J. T. Nichols, C. Yao, J. O. Manilay, E. A. Robey, and G. Weinmaster. 2005. Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Mol. Biol. Cell 16:927-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhan, M., B. Jin, S. E. Chen, J. M. Reecy, and Y. P. Li. 2007. TACE release of TNF-alpha mediates mechanotransduction-induced activation of p38 MAPK and myogenesis. J. Cell Sci. 120:692-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng, Y., J. Schlondorff, and C. P. Blobel. 2002. Evidence for regulation of the tumor necrosis factor alpha-convertase (TACE) by protein-tyrosine phosphatase PTPH1. J. Biol. Chem. 277:42463-42470. [DOI] [PubMed] [Google Scholar]
- 62.Zolkiewska, A. 2008. ADAM proteases: ligand processing and modulation of the Notch pathway. Cell. Mol. Life Sci. 65:2056-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]