Abstract
GMX1777 is a prodrug of the small molecule GMX1778, currently in phase I clinical trials for the treatment of cancer. We describe findings indicating that GMX1778 is a potent and specific inhibitor of the NAD+ biosynthesis enzyme nicotinamide phosphoribosyltransferase (NAMPT). Cancer cells have a very high rate of NAD+ turnover, which makes NAD+ modulation an attractive target for anticancer therapy. Selective inhibition by GMX1778 of NAMPT blocks the production of NAD+ and results in tumor cell death. Furthermore, GMX1778 is phosphoribosylated by NAMPT, which increases its cellular retention. The cytotoxicity of GMX1778 can be bypassed with exogenous nicotinic acid (NA), which permits NAD+ repletion via NA phosphoribosyltransferase 1 (NAPRT1). The cytotoxicity of GMX1778 in cells with NAPRT1 deficiency, however, cannot be rescued by NA. Analyses of NAPRT1 mRNA and protein levels in cell lines and primary tumor tissue indicate that high frequencies of glioblastomas, neuroblastomas, and sarcomas are deficient in NAPRT1 and not susceptible to rescue with NA. As a result, the therapeutic index of GMX1777 can be widended in the treatment animals bearing NAPRT1-deficient tumors by coadministration with NA. This provides the rationale for a novel therapeutic approach for the use of GMX1777 in the treatment of human cancers.
The cyanoguanidinopyridine GMX1778 (previously known as CHS828) is the active form of the prodrug GMX1777 and has potent antitumor activity in vitro and in vivo against cell lines derived from several different tumor origins (11). The antitumor activity of GMX1778 has been widely studied since its discovery (1, 11, 19-21, 24), but positive identification of the molecular target and the mechanism of action of GMX1778 has been elusive. Here, we demonstrate that GMX1778 exerts its antitumor activity via its potent and selective antagonism of NAD+ biosynthesis. GMX1777 is currently being assessed in phase I clinical trials for treatment of patients with refractory solid tumors.
The pyridine nucleotide NAD+ plays a major role in the regulation of several essential cellular processes (7, 22, 25, 38). In addition to being a biochemical cofactor for enzymatic redox reactions involved in cellular metabolism, including ATP production, NAD+ is important in diverse cellular pathways responsible for calcium homeostasis (17), gene regulation (5), longevity (18), genomic integrity (33), and apoptosis (36). Cancer cells exhibit a significant dependence on NAD+ for support of the high levels of ATP production necessary for rapid cell proliferation. They also consume large amounts of this cofactor via reactions that utilize poly(ADP) ribosylation, including DNA repair pathways (10, 37, 39).
In eukaryotes, the biosynthesis of NAD+ occurs via two biochemical pathways: the de novo pathway, in which NAD+ synthesis occurs through the metabolism of l-tryptophan via the kynurenine pathway, and the salvage pathway. The NAD+ salvage pathway can use either nicotinamide (niacinamide) (NM) or nicotinic acid (niacin) (NA) (via the Preiss-Handler pathway) as a substrate for NAD+ production. Saccharomyces cerevisiae species predominantly use NA as the substrate for NAD+ biosynthesis, through the deamidation of NM by the nicotinamidase PNC1 (25). However, mammalian cells do not express a nicotinamidase enzyme and use NM as the preferred substrate for the NAD+ salvage pathway. The mammalian NAD+ biosynthesis salvage pathway using NM is composed of NA phosphoribosyltransferase (NAMPT), which is the rate-limiting and penultimate enzyme that catalyzes the phosphoribosylation of NM to produce nicotinamide mononucleotide (NMN) (27, 29). NMN is subsequently converted to NAD+ by NMN adenyltransferases (NMNAT). The gene encoding NAMPT was originally identified as encoding a cytokine named pre-B-cell colony-enhancing factor (PBEF1) (30). NAMPT was also identified as a proposed circulating adipokine named visfatin (thought to be secreted by fat cells) and was suggested to function as an insulin mimetic; however, this role of NAMPT currently remains controversial (8). In mice, NAMPT has been shown to act as a systemic NAD+ biosynthetic enzyme that regulates insulin secretion from β cells (28). The molecular structure of NAMPT from human (15), rat (16) and mouse (35) tissue, containing either NMN or the inhibitor APO866, have been determined by X-ray crystallography. These structures revealed that NAMPT is a dimeric type II phosphoribosyltransferase.
Here, we report that the anticancer compound GMX1778 is a specific inhibitor of NAMPT in vivo and in vitro and is itself a substrate for the enzyme. Phosphoribosylated GMX1778 inhibits NAMPT as potently as GMX1778 but is preferentially retained within cells. Finally, we have identified a novel anticancer strategy utilizing NA rescue of GMX1778 cytotoxicity to increase the therapeutic index of GMX1777 activity in tumors that are deficient in NA phosphoribosyltransferase 1 (NAPRT1).
MATERIALS AND METHODS
Biochemical pathway profiling studies.
IM-9 cells were treated with 0.2% dimethyl sulfoxide (DMSO) or GMX1778 at 30 nM (six replicate experiments each). At 6 h after GMX1778 treatment, 2 × 106 cells were harvested from each sample, rinsed three times in cold phosphate-buffered saline (PBS), and snap-frozen in liquid nitrogen. Frozen cell pellets were thawed and extracted using an automated MicroLab STAR system (Hamilton Company). The resulting extracts were divided into two fractions, one for liquid chromatography (LC) and one for gas chromatography. Statistical analysis of the data was performed using JMP (SAS), a commercial software package, and R software (http://cran.r-project.org/). A log transform was applied to the data observed for the relative concentrations of each biochemical. Biochemicals with detectable levels in at least two-thirds of the samples in any group were included in the analyses. Biochemicals considered to be significantly changed relative to time-matched control samples had a q value ≤ 0.2 and a P value ≤ 0.1.
Soft-agar clonogenic assay.
IM-9 cells were treated with 25 nM GMX1778 for 72 h, and equal volumes were then plated in 0.35% agar. Colonies were counted after 21 days.
Caspase 3 activity and cleavage.
IM-9 cells exposed to 25 nM GMX1778 for 72 h were collected, washed with PBS, and resuspended in lysis buffer (50 mM HEPES [pH 7.4], 5 mM EDTA, 1% Triton X-100). Cell lysate (25 μg) was assayed for caspase 3 activity in buffer containing 2 mM dithiothreitol and 50 mM N-acetyl-DEVD-(7-amino-4-methylcoumarin) substrate. Cleaved caspase 3 was detected by separation of 25 μg of cell extract by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunodetection with a cleaved caspase 3-specific antibody (Cell Signaling).
Cell lysis and apoptosis assays.
IM-9 cells were removed at various times after GMX1778 treatment, diluted 1:3 in PBS containing propidium iodide (Molecular Probes) (50 ng/ml) and annexin V (Biovision), and analyzed by flow cytometry within 1 h using a FACSCalibur flow cytometer (BD Biosciences) and CellQuest Pro version 5.2.1 software. Data from 10,000 cells were collected for each time point.
Determination of intracellular NAD+ levels.
Cells were harvested by trypsinization and centrifugation, and cell pellets were snap-frozen in liquid nitrogen and stored at −80°C before extraction. Extraction and NAD+ levels were measured as described in reference 2.
Cell culture and test compounds.
Cell lines were obtained from the National Cancer Institute (NCI) and from the American Type Culture Collection. Cells were cultured in RPMI-1640 medium (HyClone) supplemented with 10% fetal bovine serum (HyClone), penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine (Invitrogen). GMX1777 and GMX1778 were synthesized by Propharma Ltd. (Glasgow, United Kingdom) and dissolved in DMSO (Sigma). NA, NM, and NMN (Sigma-Aldrich) were dissolved in RPMI-1640 medium and filter sterilized before use. Doxorubicin (Sigma-Aldrich) was dissolved in DMSO. APO866 was dissolved in DMSO. Bortezomib was from Millenium Pharmaceuticals Inc.
Viability assay.
Serial dilutions of GMX1778 in DMSO were performed to achieve a final concentration of 0.2% DMSO. Relative ATP levels after 72 h were determined using a ViaLight HS high-sensitivity cytotoxicity and cell proliferation BioAssay kit (Cambrex Bioproducts) per the manufacturer's instructions. For GMX1778 cytotoxicity rescue experiments, cells were treated with NA (10 μM) or NMN (100 μM) simultaneously with GMX1778. Sigmoidal dose-response curves were generated by nonlinear regression analysis of variable slope using GraphPad Prism version 4.00 (GraphPad Software) to calculate 50% inhibitory (IC50) values.
NF-κB-regulated gene expression.
HeLa cells were cotransfected with a plasmid harboring an NF-κB-regulated firefly luciferase gene (NFκB-luc; Panomics) and a plasmid containing a basal-promoter-regulated Renilla luciferase gene (phRL-TK) (Promega) to serve as an internal control. DNA transfection (15 μg; NF-κB-luc to TK-RLluc ratio of 9.5 to 1) was done using Lipofectamine 2000 (Invitrogen). At 24 h later, transfected cells were harvested, replated, and treated with DMSO or GMX1778 (100 nM) in the presence or absence of 10 μM NA for 24 h followed by 4 h of stimulation with tumor necrosis factor alpha (TNF-α) (50 ng/ml) to induce NF-κB transcription factors. Firefly and Renilla luciferase levels were quantified using a Dualglo luciferase assay (Promega) per manufacturer's instructions and a luminometer (EG&G Berthold). NF-κB-regulated firefly luciferase activity was normalized to the Renilla luciferase activity in each of the replicate experiments, and results were expressed as percentages of the means of untreated sample data.
NA rescue of IκBα phosphorylation.
HeLa cells were treated with 100 nM GMX1778 in the presence or absence of 10 μM NA for 30 h. Cells were then treated with 1 μM bortezomib for 1 h prior to treatment with TNF-α (Upstate Biotechnologies) (50 ng/ml) for 5 min. Cells were harvested, and 25 μg of protein was separated by denaturing gel electrophoresis. IκBα phosphorylation was detected using Western blot analysis and a rabbit IκBα antibody (SC-371; Santa Cruz).
Synthesis of NAD+ from NM or NA.
HeLa cells (1 × 106) were treated with DMSO or GMX1778 (20 nM) for 2 h and incubated with 0.5 μCi of [14C]NM (1 μM) or 0.05 μCi of [14C]NA (100 nM) (Moravek Biochemicals) for 6 h. Cell pellets were resuspended in 50 μl of 10 mM NaH2PO4 and subjected to six freeze-thaw cycles to liberate the radiolabeled nucleotide metabolites. A total of 10 μl was separated overnight using thin-layer chromatography (TLC) and silica gel 60 plates (Merck KGaA) in a solvent composed of isobutyric acid/ammonium hydroxide/water (66:1:33). Dried TLC plates were exposed by the use of BAS-mass spectroscopy (BAS-MS) screens (Fuji Photo Film Co.) for 2 to 3 days and scanned on a Typhoon imager (Amersham).
To measure NAD+ production in a cell-free system, HeLa cell extracts were prepared as described above and centrifuged at 23,000 × g for 90 min at 4°C. A 20-μl volume of extract was diluted to 50 μl with a solution consisting of 5 mM MgCl2, 2 mM ATP, 0.5 mM phosphoribosyl pyrophosphate (PRPP), and 500 nM [14C]NM or 50 nM [14C]NA and incubated in the presence or absence of 20 nM GMX1778 at 37°C for 1 (NM) or 2 (NA) h in 50 μl of the solution. After incubation at 100°C for 2 min, radiolabeled nucleotides were separated by TLC.
Expression of yeast Pnc1 in HeLa cells.
The S. cerevisiae PNC1 gene was amplified from yeast genomic DNA and cloned into pcDNA3 vector (Invitrogen) to generate a plasmid (pcFLAGPNC1) encoding a FLAG epitope at the NH2 terminus of the protein. Plasmids were transfected into HeLa cells by the use of Lipofectamine 2000 (Invitrogen) and after 24 h were treated with GMX1778. Cell viability was assessed at 72 h as described above. The expression of FLAG-Pnc1 protein was monitored using Western blot analysis of cell extracts and monoclonal anti-FLAG antibody (Sigma). The Western blot signal was revealed with enhanced chemiluminescence (GE) per the manufacturer's instructions and detected with a Versadoc detection system (Bio-Rad).
Expression and purification of recombinant NAMPT.
Human NAMPT cDNA (Origene) was cloned into pET151 vector (Invitrogen) to produce a recombinant NAMPT protein with a hexahistidine and a FLAG epitope tag at the NH2 terminus expressed in BL21(DE3) pLysS Escherichia coli cells. Recombinant NAMPT from the bacteria was purified using nickel-nitrilotriacetic acid chromatography (Qiagen), concentrated using Amicon Ultra-15 centrifugal filter concentrators (Millipore), and stored at −80°C. Protein concentrations were determined by the use of a Bradford assay (Bio-Rad).
In vitro coupled-enzyme NAMPT assay.
Recombinant NAMPT activity was assessed using a coupled-enzyme assay based on the quantitation of NAD+ (4). Recombinant NMNAT type 1 (NMNAT1) was from Alexis Biochemicals. Reactions were performed at room temperature for 180 min using mixtures consisting of 50 mM HEPES (pH 7.4), 50 mM KCl, 5 mM MgCl2, 0.5 mM β-mercaptoethanol, 0.005% bovine serum albumin, 1% DMSO, 2.0 U/ml lactate dehydrogenase (Sigma), 4 mM sodium l-lactate (Sigma), 0.4 U/ml diaphorase (Sigma), 6 μΜ resazurin sodium salt, 0.4 mM PRPP, 3.0 nM NMNAT1, 125 μM ATP, 50 μM NM, and 2 to 5 μM recombinant NAMPT. Fluorescence was measured with a Tecan Safire plate reader (excitation wavelength, 560 nm; emission wavelength, 590 nm). Ki values were calculated using Graphpad Prism 4.0 software and the Cheng-Prusoff equation.
Fluorescence polarization.
GMX1778-Alexa Fluor (GMX2240) was synthesized at Gemin X. For competition experiments, 20 nM GMX2240 was incubated in a solution consisting of 50 mM HEPES (pH 7.4), 50 mM KCl, 5 mM MgCl2, 0.5 mM β-mercaptoethanol, 0.005% bovine serum albumin, 0.4 mM PRPP, and 125 μM ATP with 200 nM recombinant NAMPT and titrated with increasing amounts of GMX1778 or NM in a 100-μl volume in black 96-well plates (Corning). Fluorescence polarization was measured in a Tecan Ultra plate reader with an excitation wavelength of 485 nm and an emission wavelength of 535 nm.
siRNA-mediated knockdown of NAMPT in HeLa cells.
HeLa cells were transfected twice (24 h apart) using 50 nM ON-TARGETplus SMARTpool human NAMPT small interfering RNA (siRNA) (Dharmacon) or ON-TARGETplus siControl (nontargeting) siRNA (Dharmacon) or Oligofectamine (Invitrogen) per the manufacturer's instructions. At 24 h following the second transfection, cells were replated and treated with various concentrations of GMX1778 for 72 h. Cell viability was assessed as described above. NAMPT protein levels were detected using Western blotting of cell lysates and rabbit polyclonal NAMPT antibody (Abcam) at a 1:1,000 dilution.
Transient inducible overexpression of NAMPT.
Human NAMPT cDNA was cloned into pcDNA4-TO-Hygro vector and transfected into a HEK-293T-TR cell line that expresses the tet repressor from stable plasmid integration (31). Plasmid (1 μg) was transfected into HEK-293T-TR cells. Transfected cells were treated with GMX1778 (with or without 1 μg/ml doxycycline) to determine the IC50 of GMX1778. Cell viability was determined after 72 h as described above. NAMPT protein levels were detected by Western blotting of cell lysates with a monoclonal anti-FLAG antibody (Sigma) at a 1:1,000 dilution.
GMX1778-resistant cell line.
HCT-116 cells were grown in GMX1778. Surviving cells were exposed to increasing amounts of GMX1778. The resulting clone was called HCT-116R.
Pearson correlation analysis.
The mean GMX1778 IC50 values were determined by 72-h cell viability assays in two to four independent experiments using 25 cell lines from the NCI panel of 60 cell lines. Relative mRNA expression level values (microarray data determined using Affymetrix U95Av2 arrays) for the same 25 cell lines were obtained from the NCI DTP molecular target program (http://dtp.nci.nih.gov/mtargets/download.html), experiment 89629, pattern GC95997. Pearson correlation analysis of the data sets was performed using GraphPad Prism software, version 4.00.
NAPRT1 protein expression in lung tumor cell lines.
Western analysis of cell extracts (25 μg) was performed using a 1/500 dilution of a mouse polyclonal NAPRT1 antibody (a gift from Nobumasa Hara).
In vitro phosphoribosylation of [14C]GMX1778.
[14C]GMX1778 was synthesized by Vitrax Inc. [14C]GMX1778 was incubated with 2.5 μM recombinant NAMPT for 1 h at 37°C. Phosphoribosyl [14C]GMX1778 was partially purified on an Oasis SPE column (Waters) (3 c3). For assessment of inhibitory activity seen with NAMPT, the amount of phosphoribosyl GMX1778 was determined by scintillation counting, taking into consideration the specific activity of [14C]GMX1778. The purity of phosphoribosyl [14C]GMX1778 was assessed by TLC analysis as described above.
Dephosphorylation of phosphoribosyl [14C]GMX1778.
Phosphoribosyl [14C]GMX1778 (25 pmol), produced in vitro, was incubated with 2.5 U of calf intestinal alkaline phosphatase (Roche) for 1 h per the manufacturer's instructions. Products were separated by TLC and exposed for phosphorimaging as described above.
Cellular retention of phosphoribosyl GMX1778.
HeLa cells were treated with 100 nM [14C]GMX1778 for 1 h in the presence or absence of 500 nM APO866. Half the cells were harvested, and the other half were washed (three times using 1 ml each time) in culture media and reincubated (in the presence or absence of APO866) for 1 h at 37°C and then rinsed and harvested. Cell extracts were separated by silica TLC and exposed for autoradiography as described above.
RNA purification, cDNA preparation, and qRT-PCR.
Frozen tissue samples were from the Brain Tumor Tissue Bank (London, Canada) and ProteoGenex, Inc. Total RNA was isolated using an RNeasy Plus Mini kit (Qiagen) per the manufacturer's instructions and quantified by measurement at an optical density of 260 nm. First-strand cDNA synthesis was carried out using a Transcriptor first-strand cDNA synthesis kit (Roche). Total RNA (0.1 μg) was transcribed using 60 μM random hexamer primers, 2.5 μM anchored oligonucleotide (dT)18 primers, 1 μM deoxynucleotide mix, 1 U/μl protector RNase inhibitor, and 0.5 U of Transcriptor reverse transcriptase (RT) in 20 μl and incubated at 25°C for 10 min, 55°C for 30 min, and 85°C for 5 min in an Mx3005P quantitative RT-PCR (qRT-PCR) system (Stratagene). qRT-PCR primers and probe (labeled with 6-carboxyfluorescein dye and a minor groove binder) were from TaqMan gene expression assays (Applied Biosystems). The ribosomal protein-large P0 primers and probe were from a TaqMan endogenous control assay and were labeled with VIC dye and a minor groove binder. Reactions were prepared using a 20-μl volume with a final concentration consisting of 1× TaqMan gene expression master mix, the NAPRT1 primer pair (225 nM), the RPLP0 primer pair (450 nM), the NAPRT1 probe (62.5 nM), the RPLP0 probe (125 nM), and 2 ng of cDNA. Thermal cycling was performed at 50°C for 2 min and 95°C for 10 min, followed by 60 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. Threshold cycle values were determined using triplicate experiments. A standard curve obtained with 12 serial dilutions of HeLa cDNA was determined for each run. NAPRT1 mRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or RPLP0 mRNA levels were calculated using the standard curve. NAPRT1 mRNA levels were normalized and expressed as copies of NAPRT1 mRNA relative to HeLa levels (×1,000).
Immunohistochemistry.
Tumor array slides (US Biomax Inc.) (5-μm sections) were baked at 60°C for 2 h, deparaffinized in xylene, and then rehydrated. Antigen retrieval performed using antigen-unmasking solution (Vector Labs) (high pH) was followed by endogenous peroxidase inactivation with 3% hydrogen peroxide (Sigma). Slides were blocked for 1 h at room temperature with tyramide signal amplification blocking solution (Perkin Elmer) and then incubated with 1:5,000 dilution of rabbit polyclonal NAPRT1 antibody (Proteintech) in CanGet signal immunoreaction enhancer solution 1 (Toyobo) for 36 h at 4°C in a humidified chamber. Slides were incubated with a 1:1,000 dilution of goat anti-rabbit antibody-horseradish peroxidase (Jackson ImmunoResearch Labs) in PBS for 30 min at room temperature. Signal amplification using a tyramide signal amplification kit (Perkin Elmer) was performed per the manufacturer's instructions followed by incubation with Vectastain ABC reagent (Vector Labs) for 30 min at room temperature. 3-3′-Diaminobenzidine (DAB) was used for colorimetric detection (the peroxidase substrate DAB kit was from Vector Labs). Slides were mounted with Permount (Fisher Scientific) and scanned using a Nanozoomer instrument (Olympus).
Xenograft studies.
CB17 SCID/SCID female mice (6 to 8 weeks of age) were from Charles River Laboratories and were injected subcutaneously with 100 μl of an HT1080, HCT-116, or HCT-116R cell suspension (1 × 106 cells). Each treatment group included eight mice, and body weight and tumor size were measured three times per week. Relative tumor volumes were calculated using the following equation: length (in millimeters) × [width (in millimeters)]2/2. Animals were treated with a 24-h intravenous (iv) infusion of vehicle (0.9% NaCl) or GMX1777 (in 10 mM citrate buffer [pH 4.8]). For NA rescue experiments, these treatments were followed by a 4-h infusion of NA (Sigma Aldrich) prepared in 5% dextrose (USP) and filtered (using a 0.2-μm-pore-size filter) before use. Infusions were delivered from an external syringe pump (Lomir Biomedical Inc.), and mice were connected via a catheter inserted in the right jugular vein implanted under conditions of general anesthesia administered using isoflurane gas (Baxter Corporation). Significant differences in tumor growth in mice treated with GMX1777 from that seen in mice treated with vehicle only were determined using two-way analysis of variance and GraphPad Prism software version 4.00 for Windows (GraphPad Software, San Diego, CA). Mispro Biotechnology Services, Inc., is an organization that strictly complies with the requirements of the Canadian Council on Animal Care. External reviewers and the McGill University Animal Care Committee approved this study.
RESULTS
GMX1778 induces NAD+ depletion through inhibition of NAD+ biosynthesis.
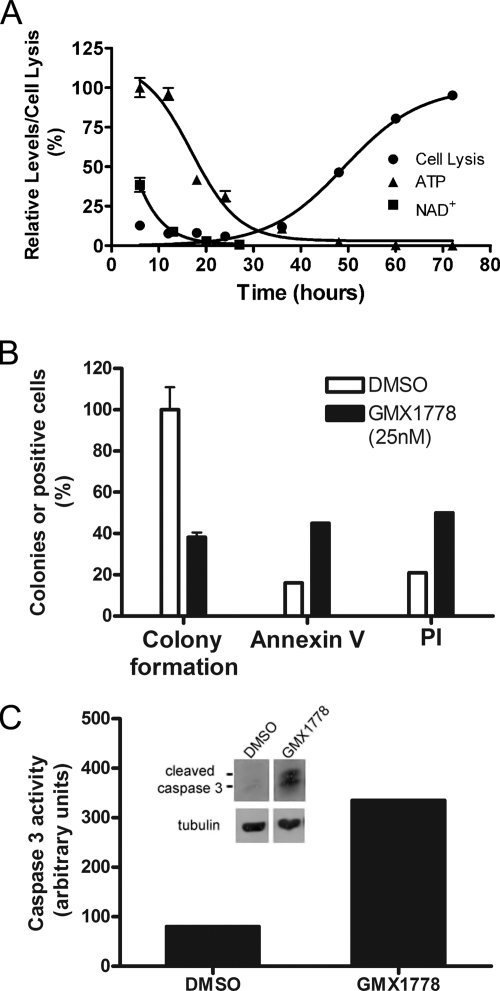
A global metabolomic analysis was conducted to characterize the metabolic changes occurring in IM-9 multiple myeloma cells upon exposure to GMX1778. A time course metabolism profile was determined by analysis using extracts of IM-9 cells treated with GMX1778. Following 6 h of treatment with 30 nM GMX1778, 88 cellular metabolites were profiled and the levels of 4 of these were found to have changed >1.5-fold. NAD+ and NM levels had decreased compared to those seen in cytosolic extracts of untreated cells (data not shown) and continued to decrease throughout the remainder of the time course experiment. The metabolite NAD+ level was found to be the most profoundly changed, and so we carried out a comparison of the kinetics of NAD+ depletion, ATP depletion, and cell lysis (Fig. 1A). The NAD+ decline occurred between the 6- and 20-h time points and preceded the loss of cellular ATP, which occurred between the 24- and 36-h time points of continuous exposure to GMX1778. Finally, cell lysis occurred between the 48- and 72-h time points of treatment with GMX1778. Since one of the characteristics of GMX1778 cytotoxicity is ATP depletion, we confirmed that the method used to determine cell viability (employing a ViaLight HS high-sensitivity cytotoxicity and cell proliferation BioAssay kit), which uses ATP levels as a measure of viability, gave results similar to those seen with soft-agar clonogenic assays and uptake of propidium iodide (Fig. 1B) as well as other cell viability assays, including WST-1 and real-time electronic impedance measurements (data not shown) of changes seen in response to GMX1778 treatment. We also demonstrated that cell death has some classic apoptosis features, such as annexin V staining and cleavage of caspase 3 concomitant with increased caspase 3 enzymatic activity in IM-9 cell extracts in response to GMX1778 treatment (Fig. 1B and C).
FIG. 1.
Characterization of GMX1778 cytotoxicity. (A) Kinetics of NAD+ depletion, ATP depletion, and cell lysis. Cellular NAD+ levels, measured by LC/MS, and ATP depletion data are from one experiment, while the cell lysis data are from an independent experiment. Data are presented as means ± standard deviations. (B) Clonogenicity, annexin V reactivity, and cell lysis (propidium iodide [PI] permeability) of IM-9 cells after 72 h of exposure to GMX1778 (25 nM). Clonogenicity was assessed in soft agar by counting colonies, and data are presented as percentages (± standard errors of the means) of colonies compared to levels of DMSO-treated cells. Annexin V reactivity and PI permeability were assessed by flow cytometric analyses of 10,000 events. (C) Caspase 3 activity and cleavage in IM-9 cells treated with GMX1778. Caspase 3 activity was measured in 25 μg of cell extract. Inset, Western blot of the accumulation of cleaved caspase 3 in 25 μg of extracts from cells treated with GMX1778. The panels are from a single Western blot, with intervening lanes removed.
Restoration of NAD+ levels rescues cells from GMX1778 cytotoxicity.
A schematic representation of the main pathways involved in eukaryotic NAD+ biosynthesis is presented (see Fig. 3A). To determine whether NAD+ depletion was the causal factor leading to GMX1778-induced ATP depletion and, ultimately, cell death in human tumor cell lines, we artificially increased the level of intracellular NAD+ in HeLa cells through supplementation with NA, a substrate of the NAD+ biosynthetic salvage pathway, and measured cell sensitivity to GMX1778 exposure. Figure 2A shows that the presence of NA (10 μM) completely rescued HeLa cells from GMX1778 cytotoxicity. NA increased intracellular NAD+ levels about 2.5-fold in DMSO-treated cells and protected cells from GMX1778-induced NAD+ reduction (inset, Fig. 2A). By contrast, cell death induced by doxorubicin, a DNA-intercalating agent exerting no direct effect on cellular NAD+ levels, was not rescued by increasing intracellular NAD+ levels through the addition of exogenous NA (data not shown).
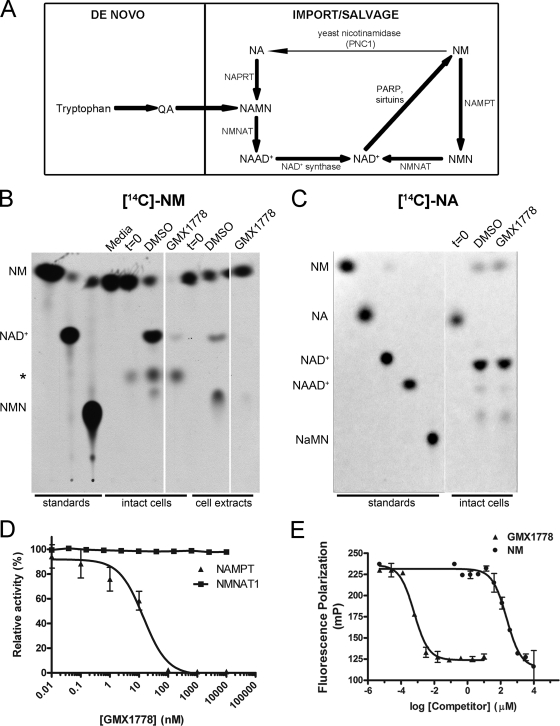
FIG. 3.
The salvage NAD+ biosynthetic pathway from nicotinamide is inhibited by GMX1778. (A) NAD+ biosynthetic pathways in mammalian cells are the de novo pathways which synthesize NAD+ from tryptophan and quinolinic acid (QA) and the salvage pathways where NAD+ is generated from either NA or NM that is taken up by cells. NA is converted to NAD+ through a three-step enzymatic process involving NAPRT1, NMNAT, and NAD+ synthetase in sequence. NAD+ synthesis from NM is a two-step process involving NAMPT and NMNAT. The ribose portion of NAD+ is utilized or broken down by multiple enzymes, including poly(ADP-ribose) polymerase (PARP) and sirtuin proteins, to regenerate NM, which is in turn recycled back to NAD+. (B and C) Intact HeLa cells were treated with GMX1778 (20 nM) for 2 h and then with [14C]NM (1 μM) (B) or [14C]NA (100 nM) (C) for an additional 6 h. Cell extracts were as indicated. Metabolic products were separated by TLC and visualized by autoradiography or phosphorimaging. Media, [14C]NM added to cell culture media; t = 0, cultured cells harvested immediately following the addition of [14C]NM or [14C]NA; asterisk, unidentified NM metabolite; NAAD+, NA adenine dinucleotide. The three columns in panel B are from one autoradiogram with intervening lanes removed. The two columns in panel C are from one phosphorimager image with an intervening lane removed. (D) Effect of GMX1778 on recombinant NAMPT and NMNAT1. Fluorescence coupled-enzyme assay for the measurement of NAD+ production after 180 min. For NMNAT1 activity, NMNAT1 and NMN concentrations were 3 nM and 125 μM, respectively. For NAMPT activity, NAMPT and NM concentrations were 2 μM and 50 μM, respectively. Data are presented as means ± standard errors of the means. (E) Fluorescence polarization competition assay. Disruption of GMX1778-Alexa Fluor (20 nM) and NAMPT (200 nM) complex by GMX1778 or NM. Data are presented as means ± standard errors of the means.
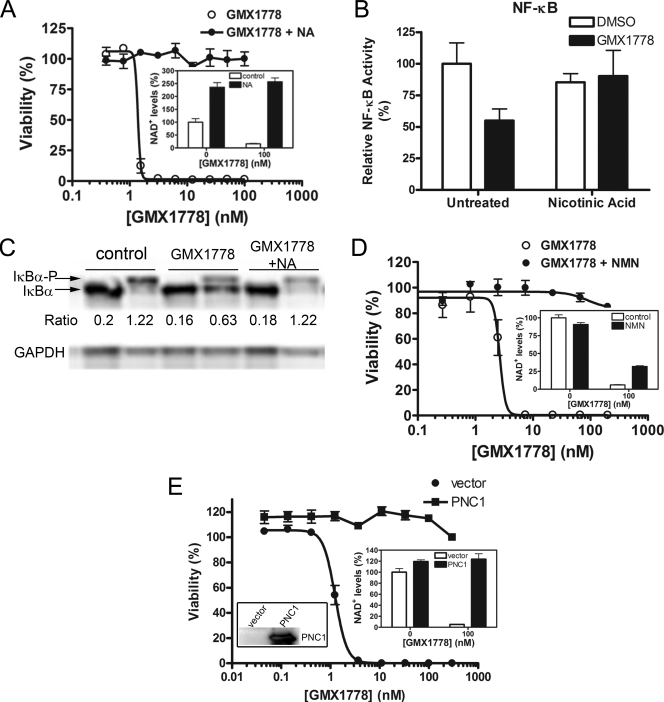
FIG. 2.
Characterization of GMX1778 cytotoxicity. (A) HeLa cells are protected from GMX1778 cytotoxicity by NA. HeLa cells were exposed to GMX1778 in the presence or absence of 10 μM NA. Viability was measured after 72 h and is presented as means ± standard deviations. The inset shows cellular NAD+ levels from HeLa cells treated with 100 nM GMX1778. Cellular NAD+ levels were measured by LC/MS and are presented as means ± standard deviations. (B) NA (10 μM) rescue of GMX1778 cytotoxicity also rescues TNF-α-induced NF-κB activity. HeLa cells were exposed to GMX1778 in the presence or absence of NA. NFκB transcriptional activity was measured by luminescence generated from expression of a reporter gene and is presented as means ± standard deviations. (C) NA rescues GMX1778-mediated inhibition of IκBα phosphorylation. IκBα phosphorylation was induced by a 5-min stimulation with TNF-α following treatment of cells with bortezomib (1 h) and 100 nM GMX1778 (30 h) with or without 10 μM NA. (D) GMX1778 cytotoxicity is rescued by NMN. HeLa cells were exposed to GMX1778 in the presence or absence of 100 μM NMN. Viability was measured after 72 h and is presented as means ± standard deviations. The inset shows cellular NAD+ levels from HeLa cells treated with 100 nM GMX1778. Cellular NAD+ levels were measured by LC/MS and are presented as means ± standard deviations. (E) Yeast nicotinamidase (PNC1) expression rescues mammalian cells from GMX1778 cytotoxicity. HeLa cells were transiently transfected with vector carrying FLAG-PNC1 or empty vector and challenged by exposure to GMX1778. Viability after 72 h is presented as means ± standard deviations. Left inset, Western blot of 25 μg of cell extract from cells transfected with either vector or PNC1 as indicated and probed with anti-FLAG. Right inset, cellular NAD+ levels from transfected cells treated with 100 nM GMX1778. Cellular NAD+ levels were measured by LC/MS and are presented as means ± standard deviations.
Previous studies suggested that the primary mechanism of action of GMX1778 was direct inhibition of IκB kinase (IKK) activity, resulting in the inhibition of NF-κB transcriptional activity (24). To address whether this effect was mediated indirectly by altered NAD+ levels, we measured the NF-κB transcriptional activity when NAD+ levels were maintained concomitant with GMX1778 treatment. Figure 2B shows that NF-κB transcriptional activity decreased upon treatment with GMX1778 but was restored in the presence of 10 μM NA. To directly address whether NA could rescue the inhibition of IKK, we monitored the TNF-α-induced phosphorylation of IκBα in response to GMX1778 treatment in the presence or absence of NA. In the absence of GMX1778, IκBα was phosphorylated by IKK in response to TNF-α treatment and the ratio of phosphorylated IκBα to unphosphorylated IκBα increased by over sixfold to 1.22 compared to the results seen with untreated cells (Fig. 2C). After a 30-h treatment with GMX1778, TNF-α stimulation of cells resulted in an IκBα phosphorylation ratio of 0.63, reflecting an almost 50% reduction in the amount of IκBα phosphorylation. Treatment of the cells with GMX1778 and NA for 30 h restored the ratio of IκBα phosphorylation to that seen with untreated cells, indicating that NA can block the inhibitory effects of GMX1778 on this pathway. The degree of inhibition of IκBα phosphorylation is consistent with the approximately 50% reduction in TNF-α-induced, NF-κB-mediated transcriptional activity. Taken together, these results indicate that the inhibition of IKK and the downstream NF-κB activity observed upon exposure of HeLa cells are secondary to the NAD+ depletion.
Figure 2D shows that exogenous addition of NMN, the product of the NAMPT enzyme in the NAD+ biosynthesis pathway that uses NM as a substrate, can also rescue the viability of HeLa cells treated with GMX1778. The inset in Fig. 2D shows that in the presence of exogenous NMN (100 μM), the reduction in NAD+ levels in response to GMX1778 (100 nM) treatment of HeLa cells is only partially abrogated. Figure 2E shows that the protection of HeLa cells from GMX1778 cytotoxicity could also be recapitulated, in the absence of exogenous NA, by expression of the yeast PNC1 protein. PNC1 is a yeast-specific nicotinamidase that catalyzes the deamidation of NM into NA, which is the preferred substrate for the biosynthesis of NAD+ through the salvage pathway in yeast (Fig. 3A). PNC1 overexpression by transient transfection in HeLa cells (left inset, Fig. 2E) conferred complete resistance to GMX1778 cytotoxicity by allowing the conversion of NM to NA, which is converted to NAD+ through the NA pathway, bypassing the GMX1778-induced block in NAD+ biosynthesis and maintaining NAD+ levels (right inset, Fig. 2E). Therefore, using two independent methods to increase NA levels in cultured tumor cell lines, we completely rescued cells from cell death induced by GMX1778 treatment, indicating that the NAD+ salvage pathway from NA was not inhibited. Taken together, these results suggest that GMX1778 cytotoxicity is caused by NAD+ depletion resulting from a block in the NAD+ biosynthesis salvage pathway by the use of NM as a substrate.
GMX1778 inhibits NAD+ biosynthesis from NM.
To determine which branch of the NAD+ biosynthetic pathway (Fig. 3A) was inhibited by GMX1778, we traced the metabolism of the radiolabeled substrates [14C]NM and [14C]NA in HeLa cells treated with GMX1778. Figure 3B (lanes marked “intact cells”) shows that GMX1778 treatment of intact HeLa cells blocked the production of [14C]NAD+ from [14C]NM. However, in this case, there was no detectable accumulation of NM in cells treated with GMX1778, as would have been predicted to have occurred if an enzymatic step in the NAD+ biosynthesis pathway from NM were blocked. There are at least two possible explanations for this. (i) Accumulated NM is used up in other pathways to produce unidentified NM metabolites. One such unidentified metabolite appears in intact cells only and yet is not affected by GMX1778 treatment (Fig. 3B). (ii) The lack of NM accumulation in intact cells treated with GMX1778 arises from inhibition of [14C]NM uptake. In order to examine this possibility, we monitored conversion by the use of a cell-free cytosolic extract system. GMX1778 blocked the production of [14C]NMN and [14C]NAD+ from the substrate [14C]NM in the cell-free cytosolic extracts (Fig. 3B, lanes marked “cell extracts”). The production of [14C]NAD+ from [14C]NM was reduced by more than 95% when intact cells or cell extracts were treated with GMX1778. Although it appears that the formation of [14C]NMN is also inhibited by GMX1778 treatment in intact cells and cell extracts, we cannot exclude the possibility that this slowest-migrating spot represents an unidentified product of [14C]NM metabolism. Figure 3C shows that GMX1778 treatment of HeLa cells had no effect on the conversion of [14C]NA to [14C]NAD+ in intact cells. Taken together, these results indicate that GMX1778 is an inhibitor of the branch of the NAD+ biosynthetic salvage pathway which uses NM as a substrate.
GMX1778 is a specific and potent inhibitor of NAMPT.
The biosynthesis of NAD+ from NM requires two enzymatic steps. NAMPT converts NM to NMN, which is used as a substrate by NMNAT to produce NAD+. To identify which enzyme was the target of GMX1778 inhibition, we used an in vitro coupled-enzyme assay to measure NAD+ production (4). Figure 3D shows that the phosphoribosyltransferase activity of recombinant NAMPT was sensitive to inhibition by GMX1778 (IC50 < 25 nM) whereas the adenyltransferase activity of recombinant NMNAT1 was not. This result indicated that GMX1778 is a potent inhibitor of NAMPT.
The binding affinity (Kd) for the interaction of NM with rat NAMPT, as determined using isothermal titration calorimetry, is 6 mM (16). During the characterization of the inhibition of NAMPT activity by GMX1778, we discovered that high concentrations (>10 mM) of NM inhibited NAMPT activity. This most likely resulted from a mechanism involving substrate inhibition (unpublished results) and precluded the demonstration of competitive derepression of GMX1778 inhibition of NAMPT activity by NM. Therefore, we used an alternative method, fluorescence polarization, to demonstrate that NM could compete for the interaction of GMX1778 with NAMPT. We determined that the Kd of recombinant NAMPT for GMX1778 labeled with a fluorescent tag (GMX1778-Alexa Fluor) was 120 nM (data not shown). Subsequent competitive binding analyses indicated that GMX1778 was very efficient at competing with the GMX1778-Alexa Fluor/NAMPT interaction and that NM was a significantly (>1,000-fold) less effective competitor (Fig. 3E). Taken together, these results indicate that GMX1778 interacts very tightly with NAMPT and that it is a competitive inhibitor of NAMPT activity.
Modulation of NAMPT levels alters the sensitivity of tumor cell lines to GMX1778.
To confirm that NAMPT is the physiological target of GMX1778, we manipulated the expression levels of NAMPT in cells through siRNA-mediated knockdown and regulated overexpression and measured the effect of GMX1778 on cell viability. First, NAMPT levels were reduced in HeLa cells by double transfection with a pool of siRNAs specific for human NAMPT (inset, Fig. 4A) and the effect on GMX1778 cytotoxicity was compared to that seen with cells transfected with a control siRNA. Figure 4A shows that siRNA knockdown of NAMPT in HeLa cells resulted in a 10-fold decrease in GMX1778 IC50 values compared to control siRNA-treated cell results. Figure 4B shows that reduction of NAMPT expression resulted in a concomitant decrease in the level of cellular NAD+ in both the absence and the presence of GMX1778. Thus, decreased NAMPT levels result in reduced NAD+ levels and in increased sensitivity of HeLa cells to GMX1778 cytotoxicity.
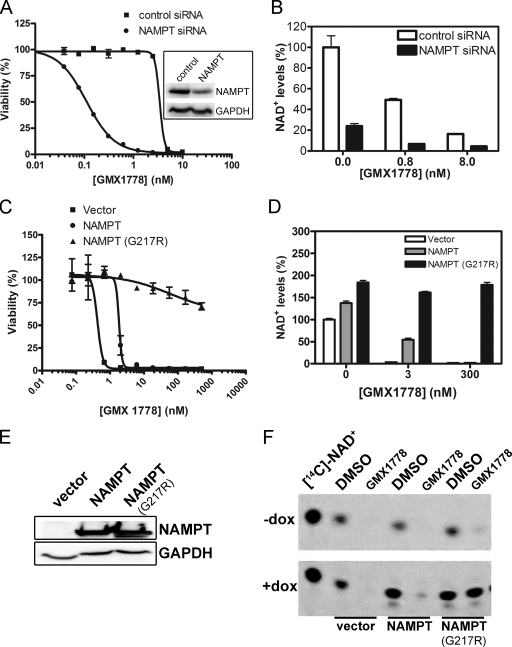
FIG. 4.
Modulation of NAMPT expression alters the sensitivity of tumor cell lines to GMX1778. (A) Cytotoxicity of GMX1778 in HeLa cells transfected with either control or NAMPT siRNA. Cell viability was measured after 72 h of GMX1778 treatment. The inset shows the results of Western blot analysis of 25 μg of cell extract from HeLa cells transfected with either control or NAMPT siRNA. The blot was probed with anti-NAMPT and anti-GAPDH as indicated. (B) Cellular NAD+ levels from HeLa cells transfected with NAMPT siRNA. Cellular NAD+ levels were measured by LC/MS and are presented as means ± standard deviations. (C) GMX1778 sensitivity of HEK-293-TR cells transfected with vector, NAMPT, or NAMPT(G217R). Cells were grown in the presence of doxycycline (to induce transfected NAMPT protein expression). Viability after 72 h is presented as means ± standard deviations. (D) Cellular NAD+ levels from HeLa cells transfected with expression constructs for NAMPT and NAMPT(G217R) and treated with indicated concentrations of GMX1778. Cellular NAD+ levels were measured by LC/MS and are presented as means ± standard deviations. (E) Western blot analysis of FLAG-NAMPT protein expression in 25 μg of doxycycline-induced cell extract of cells from the experiment whose results are presented in panel C. (F) TLC analysis of the [14C]NM metabolic process resulting in [14C]NAD+ in extracts from HeLa cells transfected with vector, NAMPT, or NAMPT(G217R) and treated with 1 μM [14C]NM in the presence or absence of 50 nM GMX1778. Data from an experiment examining the [14C]NM metabolic process resulting in [14C]NAD in extracts from cells grown in the absence of doxycycline to repress protein expression (−dox) or in the presence of doxycycline to induce protein expression (+dox) are shown. These data are from an experiment that was conducted independently of the experiment whose results are presented in panel C.
We increased NAMPT levels, in an inducible manner, in HEK-293T-TR cells which had been engineered for stable expression of the tetracycline repressor (31). Following transient transfection with an NAMPT expression vector under the control of a tetracycline-inducible promoter, NAMPT expression was induced by the addition of the tetracycline analogue doxycyline. Figure 4C shows that the overexpression of NAMPT decreased the sensitivity of HEK-293T-TR cells to GMX1778 by 4.3-fold. The cellular level of NAD+ was increased by about 40% upon overexpression of NAMPT. Overexpression of wild-type NAMPT was able to maintain a certain level of NAD+ under conditions of challenge with 3 nM GMX1778, but this effect was lost when cells were exposed to 300 nM GMX1778 (Fig. 4D). Figure 4E shows the results of Western blot analysis of cell extracts, confirming that the NAMPT protein is overexpressed in the presence of doxycyline.
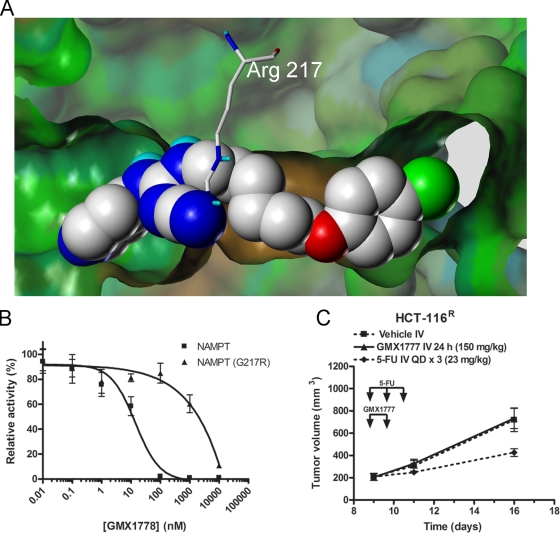
During our investigation into the mechanism of GMX1778 cytotoxicity, we derived a GMX1778-resistant clone of the HCT-116 colon cancer cell line (HCT-116R) through serial selection with increasing sublethal concentrations of GMX1778. The IC50 value for GMX1778-induced cell killing of the resistant cell line was 2,500-fold greater than that determined for the parental cell line, whereas there was no change in the level of expression of the NAMPT enzyme (data not shown). Sequencing of the genomic copy of the NAMPT gene in the GMX1778-resistant cell line revealed a single mutation of amino acid residue 217 from glycine to arginine [NAMPT(G217R)] that differed from the equivalent residue in the enzyme in the GMX1778-sensitive cell line. Glycine 217 is located near the active site of NAMPT, as determined by studies of the crystal structure of the enzyme (15, 16, 35). Figure 5A is a space-filling model of the crystal structure of NAMPT with GMX1778 in the active site. It is evident from the model that mutant NAMPT with an arginine at residue 217 exhibits a steric clash of the arginine side chains with GMX1778, which may explain the in vitro resistance of the enzyme to inhibition by GMX1778. Overexpression of NAMPT(G217R) increased basal levels of NAD+ by about 80% and completely protected HEK-293T-TR cells from GMX1778 cytotoxicity (Fig. 4C), as high NAD+ levels were maintained when HeLa cells were exposed to all concentrations of GMX1778 (Fig. 4D). Figure 4F shows that the NM salvage pathway was blocked in GMX1778-treated cells overexpressing the vector or NAMPT, whereas in the cells overexpressing NAMPT(G217R), the production of [14C]NAD+ from [14C]NM was intact. In vitro studies using recombinant NAMPT(G217R) indicated that the mutation had no effect on the catalytic activity involved in the conversion of NM to NMN. However, the mutation reduced the sensitivity of the recombinant protein to GMX1778 inhibition by about 40-fold (Fig. 5B). This suggests that the observed resistance to GMX1778 upon overexpression of NAMPT(G217R) results from the ability of the mutant enzyme (which is resistant to GMX1778 inhibition) to provide sufficient NAD+ for cell survival. In addition, xenograft tumors derived from the GMX1778-resistant cell line (HCT-116R) were resistant to the antitumor activity of GMX1777 in a mouse model (Fig. 5C). Taken together, these experiments indicate that the sensitivity of cultured cells to killing by GMX1778 treatment can be modulated by changes in the level of expression or in the genotype of NAMPT, confirming that GMX1778 induces cell death in tumor cell lines exclusively by inhibiting this enzyme.
FIG. 5.
The G217R mutation in NAMPT results in resistance to GMX1778 inhibition. (A) Docked structure of GMX17778 (space-filling model) in the active site of NAMPT (surface) superimposed with arginine (cylinders) at position 217. GMX1778 and the side chain of arginine 217 are colored according to atom type. The NAMPT structure is based on the X-ray crystal structure 2GVJ coordinates in the Protein Data Bank. (B) Inhibition of phosphoribosyltransferase activity of recombinant NAMPT and NAMPT(G217R) in vitro. The data are presented as means ± standard errors of the means. (C) SCID mouse xenograft assays using HCT-116R tumor cells. Mice carrying tumors were treated with a 24-h iv infusion of 150 mg/kg GMX1777 or vehicle. As a positive control for antitumor activity, mice carrying tumors were treated with iv infusions of 5-FU (5-fluororacil).
Cellular expression of NAMPT inversely correlates with GMX1778 cytotoxicity.
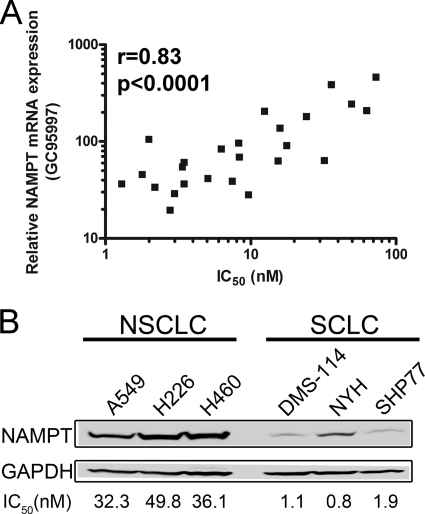
The observation that modulation of NAMPT expression levels altered sensitivity to GMX1778, coupled with the fact that NAMPT levels can be upregulated in some cancer cases (12, 34, 26), led us to examine a broad range of tumor cell lines to determine whether this relationship held for them. We compared the mRNA expression levels of NAMPT (32) with IC50 values determined by gene chip analysis for 25 tumor cell lines from the NCI panel of 60 cell lines. Figure 6A shows that there is a significant inverse correlation (Pearson correlation value of 0.83) between NAMPT mRNA expression levels and GMX1778 cytotoxicity across several tumor cell lines. To verify prospectively this inverse correlation using cell lines that were not included in the NCI panel, we measured the expression of NAMPT protein by Western blot analysis using three small-cell lung carcinoma (SCLC) cell lines and three non-SCLC (NSCLC) cell lines. We selected NSCLC and SCLC cell lines because they have GMX1778 IC50 values that differ by an order of magnitude. As predicted, NAMPT protein levels were significantly higher in the NSCLC cell lines that are less sensitive to GMX1778 than in the SCLC cells (Fig. 6B), confirming the correlation we identified in the comparisons of the levels of mRNA expression of NAMPT. Despite the fact that elevated NAMPT levels require higher GMX1778 IC50s, those higher concentrations are well within the achievable therapeutic concentrations of the drug (<100 nM).
FIG. 6.
Inverse correlation of NAMPT expression with GMX1778 cytotoxicity in tumor cell lines. (A) Relative expression of NAMPT mRNA with 72-h GMX1778 cytotoxicity IC50 values for 25 tumor cell lines. The Pearson correlation coefficient (r) and the P value are indicated. (B) Western blot analysis of 25 μg of cell extracts from SCLC and NSCLC cell lines for NAMPT and GAPDH protein expression. The corresponding 72-h GMX1778 IC50 values are indicated below the lanes for the respective cell lines.
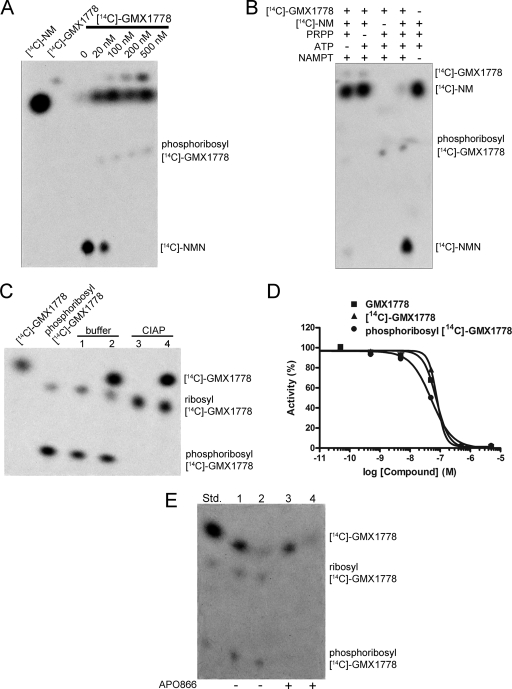
GMX1778 is a substrate for phosphoribosylation by NAMPT.
Figure 7A shows that [14C]GMX1778 is modified by recombinant NAMPT, as seen by a change in the migration of the reaction products analyzed by TLC. The modification of [14C]GMX1778 also correlated with the inhibition of NAMPT-mediated conversion of [14C]NM into [14C]NMN (Fig. 7A). We suspected that, because of its similarity with respect to the pyridinyl ring of NM, GMX1778 might be a substrate for NAMPT and might be phosphoribosylated on the pyridinyl nitrogen. Several lines of evidence confirmed that the modification did indeed represent phosphoribosylation. First, a quadrupole time-of-flight MS determination of the exact mass of modified GMX1778, purified either by the use of a TLC plate or by high-performance LC, resulted in a mass of 584.17, which is consistent with the molecular formula of phosphoribosyl-GMX1778 (C24H32N5O8PCl). Second, the modification of GMX1778 was dependent upon the presence of PRPP and ATP, which are essential cofactors for the activity of NAMPT (Fig. 7B). Third, incubation of modified [14C]GMX1778, generated in vitro with recombinant NAMPT, by the use of alkaline phosphatase shifted the migration of the product to that of a less polar molecule, as is consistent with a loss of the phosphate residue, resulting in ribosyl-[14C]GMX1778 (Fig. 7C). Fourth, fragmentation of the modified GMX1778 by triple-quadrupole MS identified the parental GMX1778 fragment and, as expected, did not identify the phosphoribosyl moiety due to lack of ionization of the sugar. Taken together, these data confirmed that the modification of GMX1778 by NAMPT did indeed represent phosphoribosylation.
FIG. 7.
GMX1778 is a substrate for phosphoribosylation by NAMPT. (A) TLC of the in vitro phosphoribosylation of [14C]GMX1778 by recombinant NAMPT. The concentrations of NAMPT and [14C]NM were 1 μM and 50 μM, respectively. (B) TLC of products from the reactions performed with recombinant NAMPT (2.5 μM), [14C]NM (500 nM), and [14C]GMX1778 (100 nM) in the presence or absence of ATP (2 mM) and PRPP (100 μM). (C) Phosphoribosyl-[14C]GMX1778 was incubated with calf intestinal alkaline phosphatase (CIAP) or buffer as indicated, and the products were separated by TLC and exposed to a phosphorimaging plate. Lanes 2 and 4 represent the same material as lanes 1 and 3, respectively, but were spiked with phosphoribosyl-[14C]GMX1778 to identify the migration pattern. (D) Phosphoribosyl-[14C]GMX1778 is a potent inhibitor of recombinant NAMPT. Phosphoribosyl-[14C]GMX1778 was produced by using recombinant NAMPT to convert [14C]GMX1778. Data represent the results of a fluorescence coupled-enzyme assay of NAD+ production after 180 min. The concentrations of NAMPT and NM were 1 μM and 50 μM, respectively. (E) Cellular retention of phosphoribosyl-[14C]GMX1778 in HeLa cells exposed to [14C]GMX1778. HeLa cells were exposed to 100 nM [14C]GMX1778 for 1 h (−APO866) and were then either washed (lane 1) or washed and incubated at 37°C for 1 h in media (lane 2) before extracts were prepared and resolved using TLC. HeLa cells were exposed to 100 nM [14C]GMX1778 and 500 nM APO866 for 1 h (+ APO866) and were then either washed (lane 3) or washed and incubated at 37°C for 1 h in medium (lane 4) before extracts were prepared and resolved by TLC. Std., phosphoribosyl-[14C]GMX1778 standard.
Phosphoribosylated GMX1778 has NAMPT inhibitory activity and exhibits preferential cell retention.
We tested the ability of purified phosphoribosyl-[14C]GMX1778 (produced in vitro) to inhibit recombinant NAMPT. As shown in Fig. 7D, phosphoribosyl-[14C]GMX1778 inhibits NAMPT to an extent similar to that seen with the parental [14C]GMX1778. Furthermore, phosphoribosyl-[14C]GMX1778 did not inhibit NMNAT1 (data not shown). Thus, phosphoribosyl-GMX1778 is as potent an inhibitor of recombinant NAMPT as GMX1778. Synthetic analogues of GMX1778 with different modifications of the pyridinyl nitrogen also showed NAMPT inhibitory activity similar to that of GMX1778 (data not shown), confirming that this activity does not require an unsubstituted pyridinyl nitrogen and consistent with the ability of phosphoribosyl-[14C]GMX1778 to inhibit NAMPT.
We predicted that the addition of a phosphoribosyl moiety to a lipophilic small molecule such as GMX1778 would increase its polarity and reduce its propensity to cross the plasma membrane, thereby trapping it within the cell. To test this hypothesis, we probed for the difference in the cellular retention of phosphoribosyl-[14C]GMX1778 generated by NAMPT compared to the cellular retention of parental [14C]GMX1778, whose conversion to phosphoribosyl-[14C]GMX1778 was prevented by the NAMPT inhibitor APO866. Figure 7E shows that intact HeLa cells were competent for the conversion of [14C]GMX1778 to phosphoribosyl-[14C]GMX1778 (lane 1), which was retained in the cells upon washing (lane 2), while parental [14C]GMX1778 was lost from the cells. By contrast, intact HeLa cells exposed to APO866 showed no conversion of [14C]GMX1778 to phosphoribosyl-[14C]GMX1778 (lane 3) and the parental [14C]GMX1778 was also lost from cells upon washing (lane 4). Therefore, phosphoribosylation of GMX1778 leads to increased cellular retention of phosphoribosyl-GMX1778.
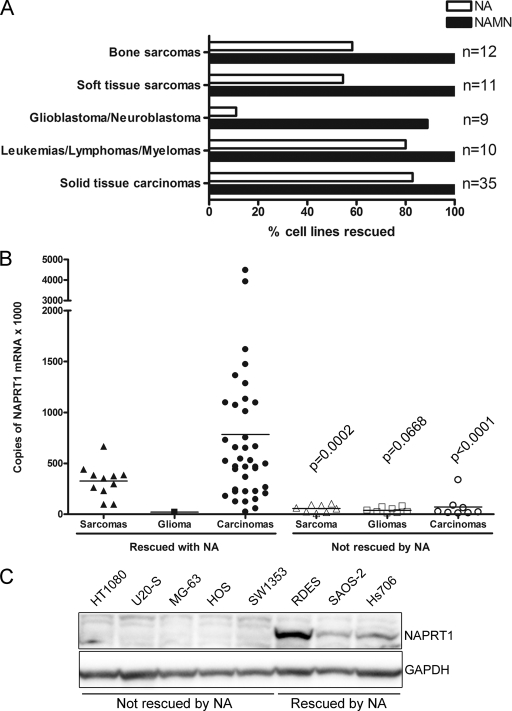
Certain tumor cell lines are functionally deficient in NAPRT1 activity.
During our analysis of the NA rescue of GMX1778 cytotoxicity in the tumor cell line panel, we observed that certain cell lines were not protected by NA. In addition, our in vitro NA rescue data determined using this panel of cell lines, which exhibit various levels of NAMPT expression, showed no indication that NAMPT expression levels influence the protection exerted by NA in excess quantities, indicating that NAPRT1 is universally able to protect against GMX1778 at the concentrations of NA that are used for rescue. Cytotoxicity of GMX1778 in cell lines derived from brain tumors (glioblastomas and neuroblastomas) and sarcomas was prevented by the presence of NA at a much lower frequency than in cell lines derived from carcinomas (Fig. 8A). To investigate whether the lack of protection by NA was due to a deficiency in NAPRT1 activity, the ability of these cell lines to be rescued by NA mononucleotide (NAMN), the product of NAPRT1 in the Preiss-Handler pathway, was tested. Figure 8A shows that all of the cell lines, with the exception of one neuroblastoma cell line, were rescued by the presence of NAMN, suggesting that the defect in cell lines not rescued by NA was in the conversion of NA to NAMN induced by the presence of the NAPRT1 enzyme.
FIG. 8.
NAPRT1 status of human tumor cell lines. (A) Frequency of rescue from GMX1778 cytotoxicity (30 nM) by 10 μM NA or 10 μM NAMN in a panel of human tumor cell lines. Cytotoxicity was measured at 72 h. (B) NAPRT1 mRNA levels measured by qRT-PCR and normalized to GAPDH mRNA expression. Statistical significance was determined by Student's t test with Welch's correction; P values are indicated. (C) Western blot of NAPRT1 protein expression in cytosolic extracts (30 μg) from human tumor cell lines that were rescued or not rescued from GMX1778 cytotoxicity by NA as indicated.
The existence of functional NAPRT1 deficiency in certain cancer cell lines suggests an opportunity for a novel therapeutic strategy whereby NA coadministration could increase the therapeutic index and improve the tolerability of GMX1777 without affecting antitumor activity. However, this combination therapy would be specifically beneficial for the treatment of NAPRT1-deficient tumors. To establish the frequency of NAPRT1 deficiency in the tumor cell line panel, we used qRT-PCR analysis to characterize the level of NAPRT1 mRNA expression. Cell lines that were not rescued from GMX1778 cytotoxicity by NA were also low in NAPRT1 mRNA expression (Fig. 8B). The level of NAPRT1 protein expression in selected cell lines was tested, and the results showed that it was significantly lower in the nonrescued group of cell lines (Fig. 8C).
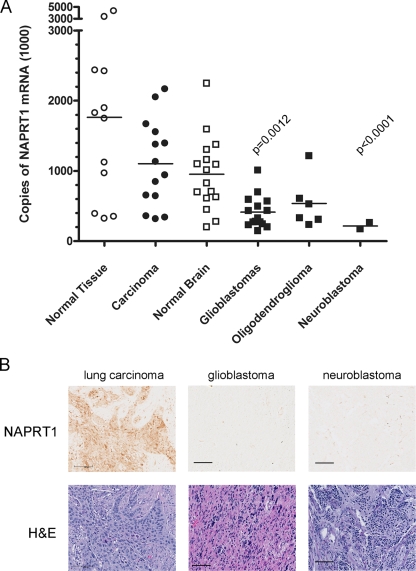
To extend this observation from tumor cell lines to primary human cancer tissue, the level of NAPRT1 mRNA expression was measured using qRT-PCR and a collection of frozen tumor tissue samples obtained from commercial sources and tumor banks. We found that glioblastoma and neuroblastoma tumor tissue did indeed exhibit lower levels of NAPRT1 mRNA than did a variety of carcinoma tumor tissues (Fig. 9A). An immunohistochemical assay was developed to detect expression of NAPRT1 protein in sections of paraffin-embedded tumor tissues. Figure 9B shows representative examples of NAPRT1 expression in lung carcinoma, glioblastoma, and neuroblastoma tumor tissue sections. As predicted from the mRNA results, glioblastomas and neuroblastomas expressed little NAPRT1 compared to carcinoma tissues.
FIG. 9.
NAPRT1 mRNA and protein expression in primary human tumor tissues. (A) Comparison of NAPRT1 mRNA levels in normal tissue to levels in malignant tissue of the same origin. Levels of NAPRT1 mRNA from normal, carcinoma, brain, normal brain, glioblastoma, oligodendroglioma, and neuroblastoma frozen tissue samples were measured by qRT-PCR. Values were normalized to ribosomal protein-large P0 mRNA levels; solid bars indicate the means. Statistical significance was determined by Student's t test with Welch's correction; P values are indicated. (B) Immunohistochemical detection of NAPRT1 protein in lung, glioblastoma, and neuroblastoma tumor tissue sections. Upper panel, sections (5 μm) were stained with a rabbit polyclonal NAPRT1 antibody. Lower panel, sequential sections were stained with hematoxylin and eosin (H&E) to reveal cellular structures. Scale bars, 100 μm.
NA coadministration widens the therapeutic index of GMX1777 treatment of NAPRT1-deficient human tumor xenografts in mice.
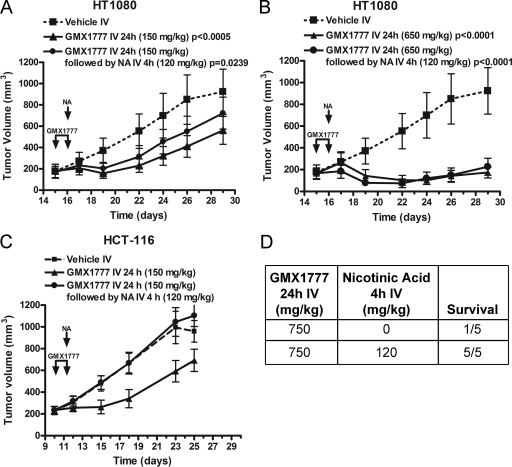
To determine whether the combination of GMX1777 and NA could improve GMX1777 tolerability without affecting the antitumor activity in vivo, we compared the effect of NA coadministration on xenograft tumors derived from NAPRT1-deficient (HT1080) and NAPRT1-proficient (HCT-116) cell lines in mice. A 4-h iv infusion of NA (120 mg/kg of body weight) did not adversely affect the antitumor activity of a 24-h iv infusion of GMX1777 at a dose of 150 mg/kg (Fig. 10A) or 650 mg/kg (Fig. 10B) in the NAPRT1-deficient xenograft experiments. However, antitumor activity against the NAPRT1-proficient HCT-116 cell line was abolished by coadministration with NA (Fig. 10C). We have previously shown that GMX1777 at 650 mg/kg is above the maximum tolerated dose (2). In addition, Fig. 10D shows that the administration of NA as a 4-h iv infusion immediately following treatment with 750 mg/kg GMX1777 reduced the mortality associated with toxic doses of GMX1777. Taken together, these results suggest that NA coadministration can improve the therapeutic index of GMX1777 for the treatment of NAPRT1-deficient tumors.
FIG. 10.
Antitumor activity of combination treatment with GMX1777 and NA in NAPRT1-deficient tumors. (A) SCID mice carrying NAPRT1-deficient HT1080 tumors were treated with a 24-h iv infusion of GMX1777 (150 mg/kg of body weight) followed by a 4-h iv infusion of NA (120 mg/kg) or left without further treatment (Vehicle IV). (B) SCID mice carrying HT1080 tumors were treated with a 24-h iv infusion of GMX1777 (650 mg/kg) followed by a 4-h iv infusion of NA (120 mg/kg) or left without further treatment (Vehicle IV). One mouse out of eight died from the GMX1777 treatment in the absence of NA. (C) SCID mice carrying NAPRT1-proficient HCT-116 tumors were treated with a 24-h iv infusion of GMX1777 (150 mg/kg) followed by a 4-h iv infusion of NA (120 mg/kg) or left without further treatment (Vehicle IV). (D) NA protects SCID mice from a toxic dose of GMX1777. Two groups (five mice each) were treated with a 24-h iv infusion of GMX1777 (750 mg/kg). One group was subsequently treated with a 4-h iv infusion of NA (120 mg/kg), and survival rates were assessed.
DISCUSSION
In the present study we established that NAMPT is the molecular target of the GMX1778 small-molecule cancer therapeutic candidate. A global metabolic profiling study was instrumental in the discovery that NAD+ levels were rapidly depleted in GMX1778-treated cells. NAD+ depletion was followed by ATP depletion and ultimately resulted in cell death. Previous results suggested that the mechanism of action of GMX1778 involved the inhibition of IKK leading to decreased NF-κB activity (24). However, our results show that the inhibition of IKK and the subsequent inhibition of NF-κB activity in cells following GMX1778 treatment are secondary to the initial NAD+ depletion, since it can be recovered by restoration of NAD+ levels. Furthermore, NAD+ repletion completely rescues cells from the cytotoxicity of GMX1778. There appears to be a threshold level of NAD+ required to protect cells from GMX1778 cytotoxicity, given that NMN supplementation only partially restores the cellular NAD+ levels while completely rescuing cell viability (ATP levels) whereas NA supplementation completely restores cellular NAD+ levels and rescues viability (ATP levels). The differences in the levels of NAD+ repletion in cells treated with these compounds may reflect differences in cellular permeability and availability as well as the robustness of the respective metabolic pathways.
The metabolism of [14C]NM in intact cells exposed to GMX1778 did not result in the accumulation of [14C]NM, as would be predicted upon NAMPT inhibition (Fig. 3B). One possible explanation is that NM uptake was coupled to conversion of NM by NAMPT and that unused [14C]NM was rinsed away from the intact cells in the presence of GMX1778 in our experiment. Our results cannot rule out the possibility that accumulating NM, in intact cells, is converted to metabolites other than NMN which contribute to the cellular cytotoxicity of GMX1778. While it is possible that factors other than NAD+ depletion might play a role in causing cell death induced by GMX1778, our interpretation is that the primary event leading to cell death induced by GMX1778 exposure is most likely the depletion of NAD+. NA can effectively rescue cells from GMX1778 cytotoxicity by bypassing NAMPT inhibition to produce NAD+ through the Preiss-Handler pathway. The APO866 (FK866) cytotoxic compound has previously been identified as a potent noncompetitive inhibitor of NAMPT (9) whose activity leads to NAD+ depletion and delayed cell death. Olesen et al. (23) reported that GMX1778 (CHS828) treatment led to decreased NAD+ levels in NYH cells and suggested that it may inhibit the NAD+ biosynthesis enzyme NAMPT. Furthermore, they reported that the biologic effects of APO866 are very similar to those of GMX1778. Our report confirms the results showing in vivo NAD+ depletion in response to GMX1778 treatment and extends them to show that GMX1778 is a direct inhibitor of recombinant NAMPT in vitro.
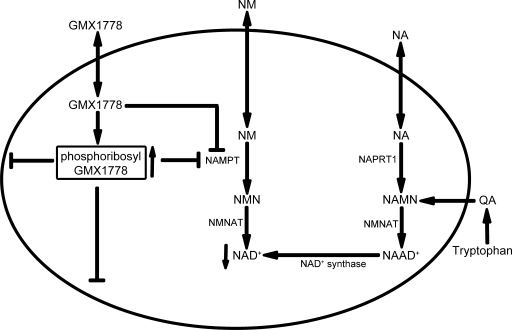
The potent inhibition of recombinant NAMPT by GMX1778 is consistent with the low-nanomolar IC50 values observed for cytotoxicity toward several human tumor cell lines (11). GMX1778 exhibits a strong affinity for NAMPT (Kd of 120 nM), leading to potent inhibition of phosphoribosylation activity through competition with NM for occupancy of the active site of NAMPT. A surprising finding of this study was that GMX1778 is a substrate for phosphoribosylation by NAMPT and that the resulting product retains NAMPT inhibitory activity. To our knowledge, this is a novel mechanism that could lead to increased levels of the inhibitor within the cell. We propose a model (Fig. 11) in which phosphoribosylation imparts polar characteristics to the lipophilic parental GMX1778, leading to preferential accumulation within cells without affecting its inhibitory capacity toward NAMPT. Inhibition of NAMPT leads to a decrease in the cellular levels of NAD+ and ultimately to cell death. However, NAD+ depletion by GMX1778 exposure can be overcome by replenishment of NAD+ levels through supplementation with NA, which is converted to NAD+. Taken together, these data suggest a novel mechanism through which NAMPT activity can be effectively inhibited in tumor cells, leading to NAD+ depletion and ultimately to cell death.
FIG. 11.
Model for the inhibition of NAD+ biosynthesis by GMX1778 in tumor cells. See text for details. NAAD+, NA adenine dinucleotide; QA, quinolinic acid.
Increased trapping of purine bases within cells by phosphoribosylation in response to mitogenic stimulation in Swiss-3T3 cells has been described previously (3). Furthermore, enzymatic polyglutamylation of methotrexate (MTX) results in MTX polyglutamates that inhibit dihydrofolate reductase as well as MTX. MTX polyglutamates are preferentially retained in cells following the removal of extracellular drug (6). By comparison, GMX1778 acts as both a substrate and an inhibitor of its cellular target, providing a mechanism by which intracellular levels of inhibitor could accumulate. One implication of the cellular retention of phosphoribosyl GMX1778 could be an increase in the effective concentration of the inhibitor, making it more available to bind and inhibit cellular NAMPT.
There are several characteristics of tumor cells that make NAMPT an attractive target for chemotherapeutic intervention in the treatment of cancer. First, tumor cells exhibit a high rate of NAD+ turnover due to the high levels of ADP-ribosylation activity required for DNA repair, genome stability, and telomere maintenance (10, 37, 39). For example, the base excision-repair pathway is dependent on poly(ADP-ribose) polymerase 1 for efficient DNA repair and poly(ADP-ribose) polymerase 1 is known to be one of the major cellular NAD+-consuming enzymes. This dependence on NAD+ levels makes tumor cells more susceptible to NAMPT inhibition than normal cells. Second, NAMPT is the rate-limiting enzyme in the salvage NAD+ biosynthesis pathway from NM and the expression of NAMPT is upregulated in several cancers (12, 34, 26). Third, NAD+ biosynthesis is increased in response to DNA damage in primary human breast cancer cells (13). These characteristics indicate that NAMPT is the main enzyme responsible for the maintenance of NAD+ levels in tumors (14) and may provide an “Achilles heel” that can be exploited for chemotherapeutic intervention with NAMPT inhibitors.
We have previously shown that NA can be used as an antidote to rescue the mortality of mice treated with a lethal dose of GMX1777 (2). This observation, coupled with the current result indicating that some tumors are deficient in NAPRT1, allows an opportunity to extend the therapeutic index of GMX1777 by coadministration of NA. The combination of NA with GMX1777 exhibits antitumor activity only in cancers that are deficient in NAPRT1. NAD+ is produced through the intact NA pathway, bypassing the inhibition of NAMPT and serving to protect NAPRT1-proficient normal cells from GMX1777 cytotoxicity while not compromising the antitumor effect of GMX1777 on NAPRT1-deficient tumor cells. The present study revealed that glioblastomas and neuroblastomas exhibit a high frequency of NAPRT1 deficiency, at the level of both mRNA expression and protein expression, and are therefore promising candidates for treatment of cancer with this combination therapy. We are currently testing several cancer tissue microarrays to identify other candidates for treatment with the GMX1777/NA combination. In addition, and particularly for patients suffering from cancers such as carcinomas or sarcomas that are generally not deficient in NAPRT1, immunohistochemical analysis for determinations of NAPRT1 deficiency could be applied to the screening of sections of tumor biopsy samples in order to identify cancer patients who would benefit from this novel therapy using the combination of GMX1777 and NA.
In conclusion, we have shown that the GMX1778 anticancer compound is a potent and specific inhibitor of NAMPT, causing tumor cell death through a mechanism involving NAD+ depletion. Furthermore, we identified a novel mechanism by which phosphoribosylation of GMX1778 occurs while binding and inhibiting its NAMPT cellular target. Phosphoribosylation does not affect the inhibitory potency of GMX1778 but rather leads to its accumulation within tumor cells. Finally, we have established that NA can be used in a novel combination treatment to increase the therapeutic index of GMX1777 in the treatment of NAPRT1-deficient tumors. These data place GMX1777 in a class of NAMPT inhibitors with exciting novel possibilities for the treatment of cancer through the modulation of NAD+ levels.
Acknowledgments
We thank colleagues at Gemin X Pharmaceuticals for valuable scientific discussions. We thank Marcela White (Brain Tumor Bank) for tissue samples, Louis Gaboury (Universite de Montréal) for help with immunohistochemical analyses, and Nobumasa Hara for the generous gift of the NAPRT1 antibody.
Disclosure of financial conflicts of interest: Mark Watson, Anne Roulston, Laurent Bélec, Xavier Billot, Richard Marcellus, Cynthia Bernier, Stephane Branchaud, Helen Chan, Kenza Dairi, Karine Gilbert, Henady Isakau, Daniel Goulet, Michel-Olivier Gratton, Anne Jang, Abdelkrim Khadir, Elizabeth Koch, Manon Lavoie, Denis Paquette, and Pierre Beauparlant are or were employees of Gemin X Pharmaceuticals Inc. Gordon C. Shore is a cofounder and shareholder of Gemin X Pharmaceuticals Inc.
Footnotes
Published ahead of print on 24 August 2009.
REFERENCES
- 1.Aleskog, A., S. Bashir-Hassan, P. Hovstadius, J. Kristensen, M. Höglund, B. Tholander, L. Binderup, R. Larsson, and E. Jonsson. 2001. Activity of CHS 828 in primary cultures of human hematological and solid tumors in vitro. Anticancer Drugs 12:821-827. [DOI] [PubMed] [Google Scholar]
- 2.Beauparlant, P., D. Bedard, C. Bernier, H. Chan, K. Gilbert, D. Goulet, M.-O. Gratton, M. Lavoie, A. Roulston, E. Turcotte, and M. Watson. 2009. Preclinical development of the nicotinamide phosphoribosyl transferase inhibitor prodrug GMX1777. Anticancer Drugs 20:346-354. [DOI] [PubMed] [Google Scholar]
- 3.Becker, M. A., P. Dicker, and E. Rozengurt. 1983. Mitogenic enhancement of purine base phosphoribosylation in Swiss mouse 3T3 cells. Am. J. Physiol. 244:C288-C296. [DOI] [PubMed] [Google Scholar]
- 4.Bembenek, M. E., E. Kuhn, W. D. Mallender, L. Pullen, P. Li, and T. Parsons. 2005. A fluorescence-based coupling reaction for monitoring the activity of recombinant human NAD synthetase. Assay Drug Dev. Technol. 3:533-541. [DOI] [PubMed] [Google Scholar]
- 5.Blander, G., and L. Guarente. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73:417-435. [DOI] [PubMed] [Google Scholar]
- 6.Chabner, B. A., C. J. Allegra, G. A. Curt, N. J. Clendeninn, J. Baram, S. Koizumi, J. C. Drake, and J. Jolivet. 1985. Polyglutamation of methotrexate. Is methotrexate a prodrug? J. Clin. Investig. 76:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., R. Petrelli, K. Felczak, G. Gao, L. Bonnac, J. S. Yu, E. M. Bennett, and K. W. Pankiewicz. 2008. Nicotinamide adenine dinucleotide based therapeutics. Curr. Med. Chem. 15:650-670. [DOI] [PubMed] [Google Scholar]
- 8.Fukuhara, A., M. Matsuda, M. Nishizawa, K. Segawa, M. Tanaka, K. Kishimoto, Y. Matsuki, M. Murakami, T. Ichisaka, H. Murakami, E. Watanabe, T. Takagi, M. Akiyoshi, T. Ohtsubo, S. Kihara, S. Yamashita, M. Makishima, T. Funahashi, S. Yamanaka, R. Hiramatsu, Y. Matsuzawa, and I. Shimomura. 2007. Retraction. Science 318:565. [DOI] [PubMed] [Google Scholar]
- 9.Hasmann, M., and I. Schemainda. 2003. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 63:7436-7442. [PubMed] [Google Scholar]
- 10.Hassa, P. O., S. S. Haenni, M. Elser, and M. O. Hottiger. 2006. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 70:789-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hjarnaa, P. J., E. Jonsson, S. Latini, S. Dhar, R. Larsson, E. Bramm, T. Skov, and L. Binderup. 1999. CHS 828, a novel pyridyl cyanoguanidine with potent antitumor activity in vitro and in vivo. Cancer Res. 59:5751-5757. [PubMed] [Google Scholar]
- 12.Hufton, S. E., P. T. Moerkerk, R. Brandwijk, A. P. de Bruïne, J. W. Arends, and H. R. Hoogenboom. 1999. A profile of differentially expressed genes in primary colorectal cancer using suppression subtractive hybridization. FEBS Lett. 463:77-82. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson, E. L., W. M. Shieh, and A. C. Huang. 1999. Mapping the role of NAD metabolism in prevention and treatment of carcinogenesis. Mol. Cell. Biochem. 193:69-74. [PubMed] [Google Scholar]
- 14.Khan, J. A., F. Forouhar, X. Tao, and L. Tong. 2007. Nicotinamide adenine dinucleotide metabolism as an attractive target for drug discovery. Expert Opin. Ther. Targets 11:695-705. [DOI] [PubMed] [Google Scholar]
- 15.Khan, J. A., X. Tao, and L. Tong. 2006. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat. Struct. Mol. Biol. 13:582-588. [DOI] [PubMed] [Google Scholar]
- 16.Kim, M., J. H. Lee, H. Kim, S. J. Park, S. H. Kim, G. B. Kang, Y. S. Lee, J. B. Kim, K. K. Kim, S. W. Suh, and S. H. Eom. 2006. Crystal structure of visfatin/pre-B cell colony-enhancing factor 1/nicotinamide phosphoribosyltransferase, free and in complex with the anti-cancer agent FK-866. J. Mol. Biol. 362:66-77. [DOI] [PubMed] [Google Scholar]
- 17.Lee, H. C. 2001. Physiological functions of cyclic ADP-ribose and NAADP as calcium messengers. Annu. Rev. Pharmacol. Toxicol. 41:317-345. [DOI] [PubMed] [Google Scholar]
- 18.Lin, S. J., P. A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126-2128. [DOI] [PubMed] [Google Scholar]
- 19.Lövborg, H., P. Martinsson, J. Gullbo, S. Ekelund, P. Nygren, and R. Larsson. 2002. Modulation of pyridyl cyanoguanidine (CHS 828) induced cytotoxicity by 3-aminobenzamide in U-937 GTB cells. Biochem. Pharmacol. 63:1491-1498. [DOI] [PubMed] [Google Scholar]
- 20.Lövborg, H., P. Nygren, and R. Larsson. 2004. Multiparametric evaluation of apoptosis: effects of standard cytotoxic agents and the cyanoguanidine CHS 828. Mol. Cancer Ther. 3:521-526. [PubMed] [Google Scholar]
- 21.Lövborg, H., J. Wojciechowski, R. Larsson, and J. Wesierska-Gadek. 2002. Action of a novel anticancer agent, CHS 828, on mouse fibroblasts: increased sensitivity of cells lacking poly (ADP-Ribose) polymerase-1. Cancer Res. 62:4206-4211. [PubMed] [Google Scholar]
- 22.Magni, G., A. Amici, M. Emanuelli, G. Orsomando, N. Raffaelli, and S. Ruggieri. 2004. Enzymology of NAD+ homeostasis in man. Cell Mol. Life Sci. 61:19-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olesen, U. H., M. K. Christensen, F. Björkling, M. J.äättelä, P. B. Jensen, M. Sehested, and S. J. Nielsen. 2008. Anticancer agent CHS-828 inhibits cellular synthesis of NAD. Biochem. Biophys. Res. Commun. 367:799-804. [DOI] [PubMed] [Google Scholar]
- 24.Olsen, L. S., P. V. Hjarnaa, S. Latini, P. K. Holm, R. Larsson, E. Bramm, L. Binderup, and M. W. Madsen. 2004. Anticancer agent CHS 828 suppresses nuclear factor-kappa B activity in cancer cells through downregulation of IKK activity. Int. J. Cancer 111:198-205. [DOI] [PubMed] [Google Scholar]
- 25.Pollak, N., C. Dölle, and M. Ziegler. 2007. The power to reduce: pyridine nucleotides—small molecules with a multitude of functions. Biochem. J. 402:205-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy, P. S., S. Umesh, B. Thota, A. Tandon, P. Pandey, A. S. Hegde, A. Balasubramaniam, B. A. Chandramouli, V. Santosh, M. R. S. Rao, P. Kondaiah, and K. Somasundaram. 2008. PBEF1/NAmPRTase/Visfatin: a potential malignant astrocytoma/glioblastoma serum marker with prognostic value. Cancer Biol. Ther. 7:663-668. [DOI] [PubMed] [Google Scholar]
- 27.Revollo, J. R., A. A. Grimm, and S. Imai. 2004. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 279:50754-50763. [DOI] [PubMed] [Google Scholar]
- 28.Revollo, J. R., A. Körner, K. F. Mills, A. Satoh, T. Wang, A. Garten, B. Dasgupta, Y. Sasaki, C. Wolberger, R. R. Townsend, J. Milbrandt, W. Kiess, and S. Imai. 2007. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 6:363-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rongvaux, A., R. J. Shea, M. H. Mulks, D. Gigot, J. Urbain, O. Leo, and F. Andris. 2002. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 32:3225-3234. [DOI] [PubMed] [Google Scholar]
- 30.Samal, B., Y. Sun, G. Stearns, C. Xie, S. Suggs, and I. McNiece. 1994. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol. Cell. Biol. 14:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarig, R., Y. Zaltsman, R. C. Marcellus, R. Flavell, T. W. Mak, and A. Gross. 2003. BID-D59A is a potent inducer of apoptosis in primary embryonic fibroblasts. J. Biol. Chem. 278:10707-10715. [DOI] [PubMed] [Google Scholar]
- 32.Scherf, U., D. T. Ross, M. Waltham, L. H. Smith, J. K. Lee, L. Tanabe, K. W. Kohn, W. C. Reinhold, T. G. Myers, D. T. Andrews, D. A. Scudiero, M. B. Eisen, E. A. Sausville, Y. Pommier, D. Botstein, P. O. Brown, and J. N. Weinstein. 2000. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 24:236-244. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber, V., F. Dantzer, J. Ame, and G. de Murcia. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7:517-528. [DOI] [PubMed] [Google Scholar]
- 34.Van Beijnum, J. R., P. T. M. Moerkerk, A. J. Gerbers, A. P. De Bruïne, J. Arends, H. R. Hoogenboom, and S. E. Hufton. 2002. Target validation for genomics using peptide-specific phage antibodies: a study of five gene products overexpressed in colorectal cancer. Int. J. Cancer 101:118-127. [DOI] [PubMed] [Google Scholar]
- 35.Wang, T., X. Zhang, P. Bheda, J. R. Revollo, S. Imai, and C. Wolberger. 2006. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat. Struct. Mol. Biol. 13:661-662. [DOI] [PubMed] [Google Scholar]
- 36.Wright, S. C., Q. S. Wei, D. H. Kinder, and J. W. Larrick. 1996. Biochemical pathways of apoptosis: nicotinamide adenine dinucleotide-deficient cells are resistant to tumor necrosis factor or ultraviolet light activation of the 24-kD apoptotic protease and DNA fragmentation. J. Exp. Med. 183:463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yalcintepe, L., L. Turker-Sener, A. Sener, G. Yetkin, D. Tiryaki, and E. Bermek. 2005. Changes in NAD/ADP-ribose metabolism in rectal cancer. Braz. J. Med. Biol. Res. 38:361-365. [DOI] [PubMed] [Google Scholar]
- 38.Ying, W. 2006. NAD+ and NADH in cellular functions and cell death. Front. Biosci. 11:3129-3148. [DOI] [PubMed] [Google Scholar]
- 39.Zong, W., D. Ditsworth, D. E. Bauer, Z. Wang, and C. B. Thompson. 2004. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 18:1272-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]