Abstract
HOX DNA-binding proteins control patterning during development by regulating processes such as cell aggregation and proliferation. Recently, a possible involvement of HOX proteins in replication origin activity was suggested by results showing that a number of HOX proteins interact with the DNA replication licensing regulator geminin and bind a characterized human origin of replication. The functional significance of these observations, however, remained unclear. We show that HOXD13, HOXD11, and HOXA13 bind in vivo all characterized human replication origins tested. We furthermore show that HOXD13 interacts with the CDC6 loading factor, promotes pre-replication complex (pre-RC) proteins assembly at origins, and stimulates DNA synthesis in an in vivo replication assay. HOXD13 expression in cultured cells accelerates DNA synthesis initiation in correlation with the earlier pre-RC recruitment onto origins during G1 phase. Geminin, which interacts with HOXD13 as well, blocks HOXD13-mediated assembly of pre-RC proteins and inhibits HOXD13-induced DNA replication. Our results uncover a function for Hox proteins in the regulation of replication origin activity and reveal an unforeseen role for the inhibition of HOX protein activity by geminin in the context of replication origin licensing.
Hox proteins belong to the large family of homeodomain-containing DNA-binding proteins. During embryonic development they control cell fates, eventually leading to the generation of different regional identities along the primary body and limb axes (17, 35). Genetic analyses, including loss- and gain-of-function experiments, showed that vertebrate Hox genes modulate morphogenetic processes by controlling crucial aspects such as cell proliferation and aggregation (16). This is particularly true of 5′ HoxA and HoxD genes, which are involved in limb patterning (reviewed in references 49 and 65). The inactivation of the Hoxd13 gene in mice, for instance, causes a phenotype resulting from defects in the proliferation and/or condensation of limb mesenchymal cells (11, 14).
Hox proteins have been shown to act as transcription factors, which are thought to regulate sets of target genes by binding to specific DNA sequences within the transcriptional regulatory regions of these (7, 33, 38, 43, 50, 51, 61, 62). Recent findings, however, have hinted at a possible additional function for this family of DNA-binding proteins. Hox proteins have been found to associate with the DNA replication licensing regulator geminin (39), and a number of them have been shown to bind the human LaminB2 and other origins of replication, suggesting their possible role in origin definition and/or assembly of the replication machinery (9, 13, 20). The functional significance of these observations, however, remained elusive.
Origins of replication are poorly defined in metazoa. A consensus sequence appears not to be required for replication initiation in human cells, and only a few origins where replication initiates from a localized site in each cell cycle have been well characterized (reviewed in reference 10). The stepwise association of licensing factors leads to the formation of a replication origin bound multisubunit complex. Initially, origins are bound by the origin recognition complex (ORC), subsequently, two loading factors Cdc6 and Cdt1 mediate the association of the MCM helicase complex and endow origins the license to replicate (reviewed in reference 42). Several mechanisms are active within eukaryotic cells to ensure that DNA is replicated only once per cell cycle (42). One of them is based on the tight interaction between Cdt1 and the geminin protein. Geminin, which accumulates during S phase, by binding to Cdt1 prevents the loading of the MCM complex onto origins, thus inhibiting relicensing of replication during S phase (reviewed in reference 44).
In the present study we explored the possible role of HOX proteins in the control of replication origin function. We show that the HOXD13, HOXD11, and HOXA13 proteins bind in vivo all human DNA replication origins tested. HOXD13, which binds origins primarily during G1 phase of the cell cycle, interacts with the CDC6 loading factor, promotes the assembly of pre-replication complex (pre-RC) proteins at replication origins, and stimulates DNA synthesis in an in vivo transient DNA replication assay. Moreover, HOXD13 exogenous expression within cultured cells accelerates DNA synthesis initiation, in correlation with the earlier G1 phase recruitment of pre-RC proteins onto origins. We furthermore show that the licensing regulator geminin, which interacts with HOXD13, blocks the HOXD13-mediated assembly of pre-RC proteins and inhibits HOXD13-induced DNA replication. Conversely, HOXD13 expression relieves the G1 block induced by a nondegradable form of geminin. These results establish a role for Hox proteins in the context of the regulation of replication origin activity and assign a functional significance to the HOX-geminin interaction in this process.
MATERIALS AND METHODS
Plasmid constructs.
The LD13IΔN construct was described in reference 51). Constructs expressing geminin or gemininΔDB were generated by cloning murine geminin cDNA in the Migr1 (46) or in the pSport6 vector. GemininΔDB was generated by PCR mutagenesis and sequence verified. Origins within the LAMININB2 (LMNB2, 760 bp) and MCM4 (UPR, 579 bp) genes were PCR amplified using human genomic DNA, sequence verified, and cloned in pT81luc and pBluescript SK(+). The pT81-TTAC reporter contains a six-mer of the TTACGAG HOXD13 binding site. The pT81-Shh reporter was described previously (3). The pSG5-HA-HOXD13, pSG5-HA-HOXD13IQN, and pGEX4T-HOXD13HD expression constructs were as described previously (4). Expression vectors for Myc-tagged Orc2 and Cdc6 were as described previously (58).
Cell culture, retroviral transduction, and transfection.
SW1353 cells were cultured in Leibovitz l-15 medium (Invitrogen). HEK293, NIH 3T3, and Cos7 cells were cultured in Dulbecco modified Eagle medium. P19 cells were cultured in α-minimal essential medium. Media were supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U penicillin/ml, and 100 μg of streptomycin/ml. Viral stocks of LXIΔN and LD13IΔN were obtained as described elsewhere (32). SW1353 and HEK293 cells were transduced with viral supernatants and 0.8 μg of Polybrene/ml.
P19 and NIH 3T3 cells were transfected and used for luciferase and β-galactosidase assays or for formaldehyde cross-linking, as described previously (51). HA-HOXD13, geminin, Myc-Orc2, and Cdc6 expression in COS7 cells was achieved by transfecting 5 to 10 μg of expression vectors per 10-cm dish. GemininΔDB was expressed in HEK293, HEK-LXIΔN, or HEK-LD13IΔN cells by transfecting 20 μg of Migr1-gemininΔDB or 1, 5, and 10 μg of pSport-GMNΔDB per 10-cm dish.
siRNA depletion of HOXD13.
Three different small interfering RNA (siRNA) duplexes were designed targeting the HOXD13 mRNA, within regions spanning nucleotides 700 to 724 (siRNA1), 930 to 954 (siRNA2), and 960 to 984 (siRNA3) (Stealth siRNA; Invitrogen). siRNA transfection was performed with 100 or 200 pM RNAi duplexes using Lipofectamine 2000 (Invitrogen) according to the manufacturer. HOXD13 depletion was verified by reverse transcription-PCR (RT-PCR) and immunoblotting 48 h after siRNA transfection.
Cell synchronization and cytofluorimetric analysis.
HEK293 cells were synchronized in G1 phase by using 20 μM mevastatin (Sigma), 800 ng of rapamycin (Sigma)/ml, and 30 μM hydroxyurea (Sigma) for 16 h; in S phase by adding 5 μg of aphidicolin (Sigma)/ml for 48 h; and in G2/M phase by adding 0.2 μg of nocodazole (Sigma)/ml for 16 h. SW1353 synchronization was obtained by doubling the incubation times. Cell synchronization was verified by cytofluorimetry: cells were permeabilized with Triton X-100 0.1% and stained with 50 μg of propidium iodide/ml. The transduction efficiency of SW1353 and HEK293 with LXIΔN and LD13IΔN was monitored by ΔNGFR expression using an anti-NGFR antibody (Becton Dickinson) (23). Transduced cells were purified to near homogeneity by immunomagnetic purification using anti-hNGFR monoclonal antibody (Dynabeads; Invitrogen) according to the manufacturer's guidelines.
To monitor DNA synthesis, exponentially growing HEK-LD13ΔN and HEK-LXIΔN cells were nocodazole arrested and released in nocodazole-free medium. The cells were collected 0, 2, 4, 6, and 8 h after release; pulsed with 1 μM bromodeoxyuridine (BrdU; Sigma) for 30 min at 37°C, and then fixed with ethanol 70% overnight at −20°C. BrdU incorporation was detected by cytofluorimetry using an anti-BrdU antibody (Becton Dickinson) (29). Hoechst 33342 staining was performed as described earlier (19), and cells were analyzed using the CyFlow ML (Partec GmbH, Muenster, Germany) cytofluorimeter. Cells were stained with TO-PRO3 vital dye (Molecular Probes) before analysis as described previously (60). Immunofluorescence was carried out as described by Kilstrup-Nielsen et al. (30), and stained cells were observed with a Zeiss AxioSkop 40 fluorescence microscope (Carl Zeiss, Germany).
ChIP and PCR analysis.
Chromatin immunoprecipitation (ChIP) assays were performed on SW1353, HEK293, or NIH 3T3 cells as described earlier (51) using the antibodies anti-Flag (Sigma), anti-HOXD13 (Imgenex), anti-geminin (Santa Cruz), anti-Cdt1 (Santa Cruz), anti-CDC6 (Santa Cruz), anti-Orc1 (Abcam), anti-Mcm7 (Santa Cruz), anti-HOXD11 (Abcam), or anti-HOXA13 (Abcam) or with the anti-ProtA (Sigma) control antibody. Enrichments of DNA sequences in immunoprecipitates from at least three independent ChIP results were analyzed by quantitative real-time PCR (qPCR) using SYBR GreenER qPCR SuperMix (Invitrogen). The Bio-Rad iQSybr platform was used with the following cycling conditions: 2 min at 50°C, 8 min and 30 s at 95°C, and then 40 cycles of 15 s at 95°C and 1 min at 60°C. After 40 amplification cycles, threshold cycle values were automatically calculated, and femtograms of starting cDNA were calculated from a standard curve covering a range of 4 orders of magnitude. Total input and immunoprecipitation sample standard curves ranged from 10 to 10−3 pg per 25-μl reaction. Melting curves of the amplified products were used to determine the specificity of the reaction. Enrichment of amplified DNA sequences (primers in Table S1 in the supplemental material) in immunoprecipitates was calculated as the ratio between the DNA amount in immunoprecipitation samples and that in the total input chromatin. Enrichments in some ChIP experiments were analyzed by semiquantitative PCR in three independent ChIP assays. PCR analyses were performed using 1.5 μl of immunoprecipitated DNA with 0.3 mM deoxynucleoside triphosphates, 30 ng of both primers/μl, 10 μl of 5× Go-Taq buffer, and 0.3 μl of GoTaq (Promega). Primers are listed in Table S1 in the supplemental material.
Transient-transfection replication assays.
HEK293 cells were transfected with 50 ng of the pBS-LMNB2, pBS-MCM4(UPR), or empty pBluescript plasmids, together with 5 μg of expression vectors for HOXD13, HOXD13IQN, and/or gemininΔDB. At 96 h after transfection the cells were harvested, and extrachromosomal DNA was extracted according to the method of Hirt (26) and resuspended in 100 μl of H2O. The amount of replicated plasmid was determined essentially as described previously (59). Then, 5 μl of extrachromosomal DNA was digested with DpnI overnight; the samples were then treated with exonuclease III (Promega) for 30 min to reduce the background. Bacterially derived plasmids are dam methylated at specific sites on both strands. ExoIII was then heat inactivated at 70°C, and 0.1 μl of each sample was analyzed in triplicate by quantitative real-time PCR. The replication fraction for each sample was calculated as the ratio between the DNA content of samples digested with DpnI+ExoIII and digested with ExoIII alone.
RT-PCR analysis and Affymetrix array analysis.
RNA was extracted from SW1353 and HEK293 cells by using RNeasy (Qiagen). Synthesis of cDNA was done starting from 3 μg of RNA using SSII reverse transcriptase kit (Invitrogen). Semiquantitative PCR was performed with the oligonucleotides indicated in Table S1 in the supplemental material. A GAPDH (glyceraldehyde-3-phosphate dehydrogenase) control RT-PCR was done using standard oligonucleotides (see Table S1 in the supplemental material). Affymetrix U133 plus 2.0 GeneChip array hybridizations, stainings, and scannings, were performed, starting from 3 μg of total cellular RNA extracted from SW1353 LXIΔN or SW1353 LD13IΔN cells, using standard protocols (Affymetrix, Santa Clara, CA). The results were analyzed as described elsewhere (40) to compare the gene expression profiles of the two samples.
Electrophoretic mobility shift assays, Western blotting, immunoprecipitations, and GST-pulldown assays.
The HOXD13 and geminin proteins were produced in reticulocyte lysates (Promega). Then, 5 μl of HOXD13 and 5 to 10 μl of geminin were used, and binding reactions and electrophoresis were performed as described earlier (51). A 32P-labeled oligonucleotide containing the TTACGAG HOXD13 binding site was used as a probe. Flag-HOXD13 expression in SW1353 and HEK293 cells, and its depletion after siRNA transfection in HEK-LD13IΔN cells, was verified by immunoblotting using an anti-Flag antibody (Sigma). NFY-B or β-actin was used as a loading control. In vitro-transcribed or -translated 35S-labeled HA-HOXD13, geminin, CDC6, and Myc-Orc2 proteins were produced in reticulocyte lysates (Promega). Next, 5-μl portions of labeled proteins were used in coimmunoprecipitation assays using anti-HA (Santa Cruz) or anti-CDC6 antibodies. Total (Cos7) or nuclear (HEK and HEK-LD13IΔN) extracts from cells expressing HA-HOXD13 and geminin, HA-HOXD13 alone, or HA-HOXD13 and Myc-Orc2 were used in coimmunoprecipitations. Six hundred micrograms of cell extract was immunoprecipitated with anti-HA antibody, anti-CDC6 antibody, or anti-HOXD13 antibody and detected by immunoblotting using anti-HA, anti-geminin, anti-CDC6, or anti-Myc antibodies. Glutathione S-transferase (GST) pulldown assays were performed as described by Zappavigna et al. (66). Bacterially produced GST-HOXD13HD fusion protein and GST were immobilized on a glutathione-Sepharose 4B resin (Amersham). Equal amounts of resin-bound GST and GST-HOXD13HD were challenged with 20 μl of in vitro-transcribed or -translated 35S-labeled CDC6 or Orc2.
RESULTS
HOXD13 binds in vivo to characterized human DNA replication origins.
We tested the binding in vivo of HOXD13 to replication origins by chromatin cross-linking and immunoprecipitation (ChIP). As cellular models we used HEK293 human embryonic kidney cells (56) and SW1353 human humeral bone chondroblast cells (45). SW1353 cells do not express paralogy group 13 and the majority of other Hox genes (V. Salsi and V. Zappavigna, unpublished results), and conserve many of the signaling pathways of primary chondrocytes (53). Conversely, HEK293 cells express HOXD13 endogenously (see Fig. S1A in the supplemental material). The enforced exogenous expression of Flag-tagged HOXD13 (Flag-HOXD13) was achieved by using a retroviral vector. Both SW1353 and HEK293 cells were transduced with the LD13IΔN, or with the empty LXIΔN, retroviruses and sorted to near homogeneity using the truncated NGF receptor (ΔN) as a selection marker (51). The expression of HOXD13 in transduced SW1353 and HEK293 cells was verified both by semiquantitative RT-PCR and immunoblotting (see Fig. 1A and B in the supplemental material).
FIG. 1.
HOXD13 binds human DNA replication origins. (A) ChIP analysis of HOXD13 binding to human origins in the following asynchronous cells: SW1353 and HEK293 exogenously expressing Flag-HOXD13 (SW-LD13IΔN, HEK-LD13IΔN), HEK293 expressing only endogenous HOXD13 (HEK), and LXIΔN-transduced SW1353 (SW-LXIΔN). Graphs represent the average enrichment of HOXD13-bound origin regions (ori), as assessed by semiquantitative PCR (a representative set of results is shown in Fig. S2A in the supplemental material). Values indicate the ratio (as a percentage) between the amount of DNA in immunoprecipitates (IP) and that in input chromatin (INPUT). Error bars represent the mean percent IP/INPUT ratio ± the standard errors of the mean (SEM) of at least three independent ChIPs for each sample. Two genomic regions, located 5′ (5′c) and 3′ (3′c) with respect to each origin were chosen as controls (see the text). (B) Quantitative real-time PCR analysis of HOXD13 binding in ChIP experiments to human origins using chromatin from cells arrested in G1, S, or G2/M phase. A genomic region, located ∼2 kb upstream to the EPHA7 gene (primers are listed in Table S1 in the supplemental material), which is not bound by HOXD13 (51), was chosen as a negative control. Values represent the ratio (as a percentage) between the amount of DNA in immunoprecipitates and that in input chromatin, normalized by subtracting the negative control region-binding ratio. Binding to origins was more abundant in the G1 phase in all analyzed cell types. Conversely, HOXD13 binds the EPHA7 promoter at comparable levels in the G1 and G2/M phases in LD13IΔN-transduced and in nontransduced HEK293 cells and at similar levels in all phases of the cell cycle in LD13IΔN-transduced SW1353 cells.
Formaldehyde-cross-linked chromatin was extracted from HEK293 cells and from HEK293 and SW1353 cells exogenously expressing Flag-HOXD13. Chromatins were immunoprecipitated with a control antibody and with a specific anti-HOXD13 or with an anti-Flag antibody. We analyzed the binding of HOXD13 to a set of well-characterized human replication origins, located within the LaminB2 (21), MCM4 (36), c-MYC (31), TOP1 (28), and FMR1 (2) loci. The enrichment for HOXD13-bound sequences in ChIPs was verified by PCR, using primers that amplify genomic regions spanning these origins, and two control regions for each origin, located on average ∼2Kb 5′ and 3′ to the tested origin region (see Table S1 in the supplemental material). The percentage of enrichment was determined from the ratio between the amount of PCR-amplified origin DNA in ChIP samples and in input chromatin. All tested origins were bound by endogenous HOXD13 in HEK293 cells (Fig. 1A and see Fig. S2A in the supplemental material). Binding of HOXD13 was detected at all origins tested also in SW1353 and HEK293 cells exogenously expressing HOXD13 (Fig. 1A and see Fig. S2A in the supplemental material). No binding was detected at the 5′ and 3′ control regions in HEK293 cells and in both HEK293 and SW1353 cells exogenously expressing HOXD13. Binding was also not detected in control LXIΔN-transduced SW1353 cells (Fig. 1A and see Fig. S2A in the supplemental material).
To analyze HOXD13 binding to origins during the cell cycle, ChIPs were performed on SW1353 and HEK293 cells expressing exogenous HOXD13, as well as on LXIΔN-transduced HEK293 cells expressing only endogenous HOXD13, arrested using specific cell cycle inhibitors in the G1, S, or G2/M phase. The enrichment for HOXD13-bound sequences in ChIPs was verified by quantitative real-time PCR analysis. G1-arrested LD13IΔN-transduced SW1353 and HEK293 cells, as well as LXIΔN-transduced HEK293 cells, showed the highest enrichments in HOXD13-bound origins (Fig. 1B). Detectable, albeit consistently lower, levels of HOXD13 binding were observed in some origins in G2/M phase in LD13IΔN-transduced or LXIΔN-transduced HEK293 cells, whereas S-phase cells showed only negligible levels of HOXD13 binding (Fig. 1B). Conversely, HOXD13 in vivo binding to the promoter of EPHA7, a transcriptionally regulated target of HOXD13 (51), was detected at similar levels in all phases of the cell cycle in LD13IΔN-transduced SW1353 cells (Fig. 1B). In LD13IΔN-transduced HEK293 and in LXIΔN-transduced HEK293 cells, HOXD13 binding to the EPHA7 promoter was found to be reduced only during S phase and was detected at comparable levels in both G1 and G2/M phases, with a visible predominance in the G2/M phase (Fig. 1B). HOXD13 transcription was constant throughout the cell cycle, as confirmed by RT-PCR on total RNA, extracted from G1-, S-, and G2/M-arrested HOXD13-expressing SW1353 and HEK293 cells, and from HEK293 cells (see Fig. S1A and B in the supplemental material). Similarly, exogenous HOXD13 protein levels throughout the SW1353 or HEK293 cell cycle were found to be constant in all phases (see Fig. S1B in the supplemental material).
Since the analyzed replication origins are located in the vicinity of transcribed genes, we tested whether the expression of these genes would be perturbed by HOXD13 expression and whether origin sequences would mediate transcriptional activation by HOXD13. The data were mined from Affymetrix U133 plus 2.0 GeneChip array hybridizations using total cellular RNA extracted from SW1353-LXIΔN or SW1353-LD13IΔN transduced cells, and the expression profiles of genes located nearby replication origins were compared. No significant perturbations in the expression of these genes were observed (see Fig. S3A in the supplemental material). We also generated two luciferase reporter constructs, one containing 760 bp spanning the LaminB2 origin (pT81-LMNB2) and the other containing 579 bp spanning the UPR region of the MCM4 origin (pT81-UPR). Both reporters were transiently transfected together with increasing amounts of the pSG5-HOXD13 expression construct. No significant changes in the activities of the reporters were observed (see Fig. 3B in the supplemental material).
FIG. 3.
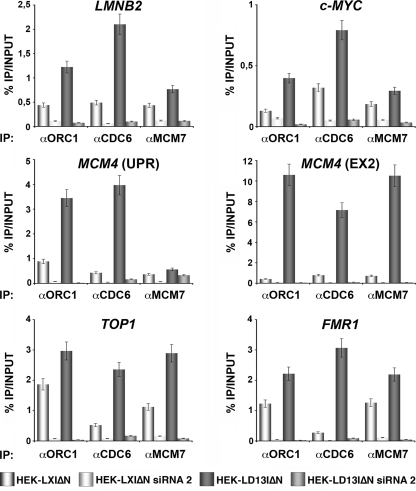
HOXD13 stimulates pre-RC assembly at origins. HEK293 cells transduced with LD13IΔN (HEK-LD13IΔN) or with the LXIΔN empty vector (HEK-LXIΔN) and LD13IΔN- or LXIΔN-transduced HEK293 cells treated with siRNA2 (see Fig. S5 in the supplemental material) to knock down HOXD13 expression were arrested in G2/M phase with nocodazole and then released from mitotic block. Six hours after release the cells were collected, and binding of the indicated pre-RC proteins at origins was analyzed by ChIP. Enrichment in origin regions was determined by quantitative real-time PCR. Values indicate the ratio (percent) between the amount of DNA in immunoprecipitate (IP) and that in input chromatin (INPUT).
These data show that HOXD13 binds in vivo to all tested human DNA replication origins, displaying a binding profile during the cell cycle that is different from that of the EPHA7 transcriptional target gene promoter. The result showing that origin regions do not mediate transcriptional activation by HOXD13 further suggested that the function of HOXD13 binding at replication origins is probably not that of the transcriptional regulation of nearby promoters.
HOXD13 binding to origins promotes the recruitment of pre-RC proteins.
To verify whether HOXD13 binding to origins would affect the assembly of licensing factors onto origins, we compared the recruitment of pre-RC proteins to episomal human origin sequences, transfected into murine NIH 3T3 cells, in the presence or absence of HOXD13. Plasmids containing the human LaminB2 and MCM4 UPR region origins (pBS-LMNB2 and pBS-UPR) were transiently transfected in NIH 3T3 cells, together with the pSG5-HOXD13 expression construct (Fig. 2A and B). Binding of HOXD13 and of Orc1, Cdc6, and Mcm7 to exogenous origin DNA was analyzed by ChIP and semiquantitative PCR. Control cells transfected with the pBS-LMNB2 and pBS-UPR plasmids alone (Fig. 2, CTRL) showed only a low-level binding of the pre-RC proteins to the LaminB2 and MCM4 origins. Conversely, both origins were found to bind more efficiently Orc1, Cdc6, and Mcm7 in the presence of HOXD13 (Fig. 2A and B). The expression of a mutant derivative of HOXD13 (D13IQN) that was shown to be unable to bind DNA (4) did not significantly stimulate Orc1, Cdc6, and Mcm7 binding. The coexpression of geminin (Fig. 2, GMN) led to a substantial reduction of pre-RC proteins binding to both replication origins (Fig. 2A and B). Interestingly, the expression of a nondegradable geminin mutant derivative (gemininΔDB) (see below) caused a substantial reduction of endogenous HOXD13 binding to replication origins in HEK293 cells (Fig. 2C).
FIG. 2.
NIH 3T3 cells were transfected with plasmids containing the human LMNB2 (A) or the human MCM4(UPR) (B) replication origins, together with expression constructs for the HOXD13 (D13), HOXD13IQN (D13IQN), and gemininΔDB (GMN) proteins. CTRL, transfection of the LMNB2 (A) or the MCM4(UPR) (B) plasmids alone. Binding of HOXD13, Cdc6, Orc1, and Mcm7 pre-RC proteins to the exogenous human LMNB2 and MCM4(UPR) origins was analyzed by ChIP using specific antibodies and primers selectively amplifying exogenous human LMNB2 (A) or MCM4(UPR) (B) origin sequences. Enrichment of origin sequences was analyzed by semiquantitative PCR analysis. Values in the graphs represent the percent ratio between the amount of DNA in immunoprecipitated samples and that in input chromatin. Transfections were performed in triplicate, three ChIPs were carried out for each experiment, and error bars represent the mean percent IP/INPUT ratio ± the SEM of all experiments. Pre-RC proteins are recruited on exogenous human origins more efficiently only in cells expressing HOXD13. No recruitment was observed in cells expressing the HOXD13IQN mutant. Pre-RC assembly was inhibited by the coexpression of geminin. C, geminin exogenous expression inhibits, in a dose-dependent manner, endogenous HOXD13 in vivo binding to origins. ChIP experiments, using anti-HOXD13 antibodies, on HEK293 cells transfected with increasing amounts of the pSport-GMNΔDB expression vector are shown.
We then tested the effect of HOXD13 expression on pre-RC formation on endogenous origins in HEK293 cells. LD13IΔN-transduced and control LXIΔN-transduced HEK293 cells were nocodazole-arrested in G2/M and subsequently released in G1. All tested origins showed, 6 h after nocodazole release, a significantly greater proportion of ORC1, CDC6, and MCM7 binding in HEK293 cells exogenously expressing HOXD13 (HEK-LD13IΔN) in comparison with control LXIΔN-transduced HEK293 cells (Fig. 3), indicating an earlier recruitment of pre-RC proteins to origin DNA in the presence of HOXD13. No differences in ORC1, CDC6, and MCM7 protein levels were observed in HOXD13-expressing versus control transduced cells or in HOXD13-depleted cells, indicating that HOXD13 exogenous expression or depletion does not alter the expression of these factors (see Fig. S4A in the supplemental material). We then used siRNAs to knock down both endogenous and exogenous HOXD13 in HEK293 cells and analyzed the binding of ORC1, CDC6, and MCM7 to origins. Of the three siRNAs tested, siRNA2 and siRNA3 efficiently knocked down both endogenous and exogenously expressed HOXD13 in HEK293 cells (see Fig. S5A and B in the supplemental material), causing a reduction of HOXD13 binding at the origins (see Fig. S5C in the supplemental material). The knockdown of exogenous plus endogenous HOXD13 with siRNA2 caused a significant decrease in the binding of ORC1, CDC6, and MCM7 to the origins at 6 h after release from G2/M arrest. Similarly, the knockdown of only endogenous HOXD13 in LXIΔN-transduced HEK293 cells led to a reduction of ORC1, CDC6, and MCM7 binding to the origins (Fig. 3).
Taken together, these data show that HOXD13 promotes the assembly of pre-RC proteins to origins. This activity requires DNA binding by HOXD13 and is antagonized by geminin.
HOXD13 interacts with the replication-licensing factor geminin.
We next verified whether HOXD13, as reported for several AbdB-like Hox proteins (39), would interact with geminin. HA-tagged HOXD13 (HA-HOXD13) was produced together with geminin by in vitro transcription and translation and immunoprecipitated using an anti-HA (α-HA) antibody. Geminin was coimmunoprecipitated by anti-HA in the presence of HA-HOXD13 and was not detected in a control immunoprecipitation experiment lacking HA-HOXD13 (see Fig. S6A, top, in the supplemental material). Geminin also coimmunoprecipitated with HA-HOXD13 in extracts from COS cells coexpressing HA-HOXD13 and geminin but was not immunoprecipitated in the absence of HA-HOXD13 (see Fig. S6A, bottom, in the supplemental material). The addition of increasing amounts of geminin led to a significant reduction of HOXD13 binding in electrophoretic mobility shift assays (see Fig. 6B in the supplemental material). The capability of geminin to block HOXD13 DNA binding in vivo and hence to inhibit its transcriptional activation was tested in transfection assays on two different luciferase reporter constructs, pT81-TTAC (4) and pT81-Shh (3). Both reporters were previously shown to be activated by HOXD13. Geminin efficiently antagonized HOXD13 activation on both reporters (see Fig. S6C and D in the supplemental material).
FIG. 6.
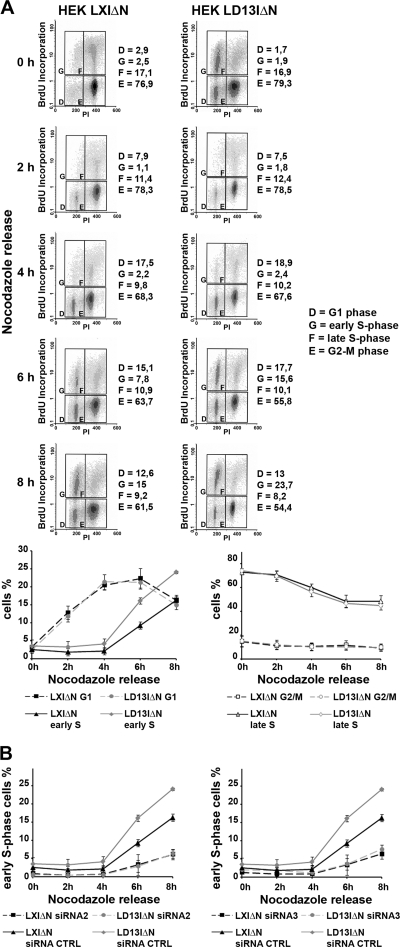
HOXD13 promotes DNA synthesis initiation in HEK293 cells. (A) HEK293 cells transduced with LD13IΔN or the control LXIΔN were arrested in G2/M phase by nocodazole treatment, released for the indicated times, pulse-labeled with BrdU, and analyzed by flow cytometry after propidium iodide staining. Gated cell populations represent G1-phase cells (D), early S-phase cells (G), late S-phase cells (F), and G2/M-phase cells (E). The progression of the cells in the different cell cycle phases is plotted in the charts at the bottom. Progressions through the G1 and early S phases are shown on the left; late S and G2/M progressions are shown on the right. Error bars represent the means ± the SEM of three independent experiments. (B) HEK-LD13IΔN and HEKLXIΔN cells were nocodazole arrested in G2/M phase after transfection with D13-siRNA2 (left graph), D13-siRNA3 (right graph), or with the control siRNA (siRNA CTRL). Cells were released from G2/M block for the indicated times, pulsed with BrdU, and analyzed by flow cytometry after propidium iodide staining. Graphs show changes in the proportion of cells in early S-phase over time.
HOXD13 interacts with the loading factor CDC6.
We next tested whether HOXD13 would directly interact with components of the pre-RC. We performed coimmunoprecipitation experiments using CDC6, Orc2, and HA-tagged HOXD13 produced in vitro in rabbit reticulocyte lysates. HOXD13 efficiently coimmunoprecipitated with CDC6 and, reciprocally, CDC6 coimmunoprecipitated with HA-tagged HOXD13 (Fig. 4A). We also tested the interaction between HOXD13 and CDC6 by coimmunoprecipitation using extracts from transfected COS cells and from HEK293 cells. Both exogenously expressed (COS and HEK293 cells) and endogenous (HEK293 cells) HOXD13 efficiently coimmunoprecipitated CDC6 (Fig. 4B). Conversely, Orc2 did not coimmunoprecipitate with HOXD13 both in rabbit reticulocyte lysates and in cotransfected COS cells (Fig. 4C). We also performed GST-pulldown assays using a bacterially produced GST fusion containing the homeodomain and C-terminal portion of HOXD13 (GST-D13HD). The CDC6 protein synthesized by in vitro transcription/translation was specifically retained by resin-bound GST-D13HD, whereas the Orc2 protein was not (Fig. 4D). Significantly, the exogenous expression of gemininΔDB in HEK293 cells led to a visible dose-dependent reduction of the amount of CDC6 coimmunoprecipitated with HOXD13 (Fig. 4E).
FIG. 4.
HOXD13 interacts with the pre-RC protein CDC6. (A) Coimmunoprecipitation using in vitro-transcribed/translated 35S-labeled HA-HOXD13 and CDC6. In the left panel, HA-HOXD13 alone or HA-HOXD13 plus CDC6 was immunoprecipitated with the anti-CDC6 antibody. In the right panel, CDC6 alone or CDC6 plus HA-HOXD13 was immunoprecipitated with the anti-HA antibody. (B) For the upper panel, coimmunoprecipitations were performed using Cos7 whole-cell extracts expressing HA-HOXD13 and using the anti-CDC6 (α-CDC6) or control immunoglobulin G (IgG) antibodies. The immunoprecipitated CDC6 and HA-HOXD13 proteins were revealed by Western blotting (WB) using anti-CDC6 (α-CDC6) and anti-HA (α-HA) antibodies, respectively. An asterisk indicates a nonspecific band revealed by the anti-CDC6 antibody. For the middle panel, coimmunoprecipitations were performed using LD13IΔN-transduced HEK nuclear extracts and anti-HOXD13 (α-D13) or IgG antibodies. For the lower panel, coimmunoprecipitations were performed using HEK293 nuclear extracts and anti-HOXD13 or IgG antibodies. (C) Coimmunoprecipitation using in vitro-transcribed/translated 35S-labeled HA-HOXD13 and Orc2. For the upper panel, Orc2 alone or Orc2 plus HA-HOXD13 was immunoprecipitated with the anti-HA antibody. For the lower panel, coimmunoprecipitations were performed using Cos7 whole-cell extracts expressing Myc-Orc2 alone, or Myc-Orc2, and HA-HOXD13 and using the anti-HA (α-HA) antibody. HOXD13 and Orc2 were revealed by immunoblotting using anti-HA and anti-Myc antibodies, respectively. (D) GST-pulldown assay using bacterially produced GST and a GST fusion with the HOXD13 homeodomain/C-terminus region (GST-D13HD) (lower panel). Resins were challenged with in vitro-transcribed/translated 35S-labeled CDC6 (upper panel) or Orc2 (central panel). Retained material was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and revealed by autoradiography. (E) For the upper panel, coimmunoprecipitations were performed using nuclear extracts from HEK293 cells transfected with increasing amounts of the gemininΔDB expression vector (pSport-GMNΔDB). Immunoprecipitations were performed using anti-HOXD13 (α-D13) or control IgG antibodies. For the lower panel, control Western blots (WB) using nuclear extracts from transfected HEK293. L, load; B, bound; Ig, immunoglobulin heavy chains. An asterisk indicates nonspecific bands.
These data show that HOXD13 interacts with CDC6 via its homeodomain region and that geminin inhibits this interaction. Moreover, these results suggest that HOXD13 promotes pre-RC formation via the direct recruitment and/or stabilization of one of its components onto origin DNA.
HOXD13 stimulates DNA replication.
We then analyzed whether HOXD13 expression would stimulate DNA replication of plasmids containing the LaminB2 (pBS-LMNB2) and MCM4 UPR (pBS-UPR) origins. The pBS-LMNB2, the pBS-UPR, or the control pBluescript plasmids were cotransfected in HEK293 cells together with expression constructs for HOXD13, HOXD13IQN, or geminin. Since bacterially derived plasmids are subjected to dam methylation at specific sites on both strands and mammalian cells are unable to dam methylate DNA, replication of plasmids in these cells produces hemimethylated or unmethylated plasmid DNA that is resistant to digestion with the DpnI restriction enzyme. The quantitative recovery of DpnI-resistant plasmid DNA after transfection is therefore a measure of plasmid DNA replication within mammalian cells (5). The replication rate of pBS-LMNB2, pBS-UPR, and pBluescript was thus determined by quantitative real-time PCR after DpnI digestion. The expression of HOXD13 was found to significantly stimulate replication of both pBS-LMNB2 and pBS-UPR, whereas no increase in replication of the empty pBluescript (pBlue) plasmid was observed (Fig. 5). The coexpression of geminin caused a considerable reduction of the HOXD13-stimulated replication. Moreover, the expression of the HOXD13IQN mutant protein did not appreciably stimulate pBS-LMNB2 or pBS-UPR replication (Fig. 5). These results show that HOXD13 expression stimulates DNA synthesis and that this activity, which requires HOXD13 binding to DNA, is antagonized by geminin.
FIG. 5.
HOXD13 promotes DNA replication at the MCM4(UPR) and LAMB2 origins. HEK293 cells were transfected with plasmids containing the human LAMB2 or the human MCM4(UPR) origins or with the empty pBluescript plasmid, together with expression constructs for HOXD13 (D13), HOXD13IQN (D13IQN), and gemininΔDB (GMN). At 96 h after transfection extrachromosomal DNA was extracted and digested with DpnI+ExoIII or ExoIII alone, and the replication rate was measured by quantitative real-time PCR (qPCR) using primers amplifying the respective origin sequences or a region within pBluescript. The indicated replication fraction represents the amount of DpnI-resistant DNA as calculated from the ratio between qPCR values of DpnI+ExoIII-digested DNA and ExoIII-digested DNA.
HOXD13 expression accelerates DNA synthesis initiation in cultured cells.
We next sought to verify whether HOXD13 exogenous expression would promote the initiation of DNA synthesis in cultured cells. HEK293 cells exogenously expressing HOXD13 (HEKLD13IΔN) and LXIΔN-transduced control HEK293 cells were arrested in G2/M by nocodazole treatment and subsequently released in nocodazole-free medium. DNA synthesis was measured by BrdU incorporation at 2, 4, 6, and 8 h after release. HOXD13-expressing HEK293 cells showed at 6 to 8 h from nocodazole release a significantly higher proportion of BrdU-incorporating early S-phase cells (Fig. 6A). The same proportion of BrdU-incorporating cells was reached in the LXIΔN-transduced HEK293 control only starting from 8 h after nocodazole release. No increase was observed for late S phase and for the other phases of the cell cycle (Fig. 6A). We subsequently analyzed whether a knockdown of both endogenous and exogenous HOXD13 using siRNAs would conversely delay the initiation of DNA synthesis in HEK293 cells. HEK293 cells exogenously expressing HOXD13 and LXIΔN-transduced HEK293 cells expressing only endogenous HOXD13 were transfected with siRNA2 and siRNA3, arrested in G2/M by nocodazole treatment, and subsequently released in nocodazole-free medium. DNA synthesis was measured at 2, 4, 6, and 8 h after release. Depletion of both endogenous and exogenous HOXD13 in HEK293 cells (LD13IΔN siRNA2 or siRNA3) caused a significant reduction in the fraction of early S phase cells at 6 and at 8 h after nocodazole release (Fig. 6B). Notably, also the depletion of endogenous HOXD13 in LXIΔN-transduced HEK293 cells (30LXIΔN siRNA2 or siRNA3) delayed their entry into S phase (Fig. 6B), uncovering the role of endogenous HOXD13 in DNA synthesis initiation. Taken together, these results show that HOXD13 exogenous expression promotes the initiation of DNA synthesis in cultured cells. Consistently, depletion of both exogenous and endogenous HOXD13, as well as of endogenous HOXD13 alone, leads to a delay in DNA synthesis initiation.
HOXD13 relieves the geminin-mediated accumulation of cells in G1 phase of the cell cycle.
Geminin has been shown to be degraded as the cells exit from mitosis, to be absent during G1 phase, and to accumulate again during the S, G2, and M phases (41). We explored the possible effect of HOXD13 expression on geminin-induced G1 phase prolongation (55, 64). HEK293 cells, transduced with LD13IΔN or with LXIΔN, were transiently transfected with an expression construct for the nondegradable gemininΔDB mutant derivative (41) (Migr-gemininΔDB) or with the empty Migr1 expression vectors (see Fig. S7A in the supplemental material). The cell cycle of transfected cells was analyzed by flow cytometric analysis of DNA content. GemininΔDB expression in control LXIΔN-transduced HEK293 cells caused an increase in the proportion of G1-phase cells (Fig. 7) in accordance with previously reported results (55, 64). The coexpression of HOXD13 together with gemininΔDB essentially restored the proportion of G1-phase cells observed in control cells transduced with the LXIΔN and transfected with Migr1 empty expression vectors (Fig. 7 and see Fig. S7B in the supplemental material), whereas the expression of HOXD13 alone caused a decrease in the proportion of G1-phase cells (Fig. 7).
FIG. 7.

HOXD13 relieves the geminin-induced G1-phase accumulation of HEK293 cells. Flow cytometry of the DNA content (Hoechst 33342) of HEK293 cells transduced with LD13IΔN (HOXD13) or with the LXIΔN control, sorted to homogeneity, and transfected with Migr1-gemininΔDB (gemininΔDB) or with the Migr1 control was performed (see also Fig. S7 in the supplemental material). A chart representing the percent variations in cell numbers in the different cell cycle phases as observed comparing the cell subpopulations (indicated below) is shown. Error bars represent the mean percent variations ± the SEM of at least three independent experiments.
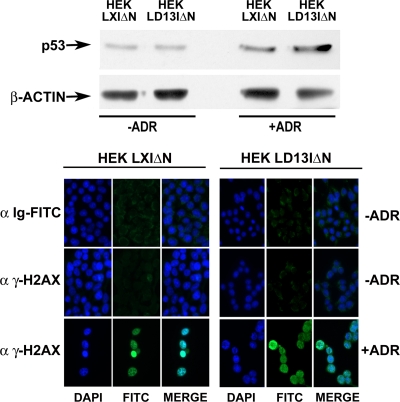
HOXD13 enforced exogenous expression did not cause replication stress and/or DNA damage leading to checkpoint activation, since no increase in p53 levels, as well as no accumulation of γH2AX, was observed in LD13IΔN-transduced HEK293 cells (Fig. 8). Moreover, HOXD13 expression did not significantly induce rereplication in HEK293 cells, as revealed by the accumulation of only a small fraction (15% ± 0.9%) of cells with >4 N DNA content in flow cytometric analysis with respect to control LXIΔN HEK293 cells (6.9% ± 1.4%) (data not shown). Thus, in accordance with the capability of HOXD13 to interact with geminin and to stimulate pre-RC recruitment to origins, HOXD13 expression antagonized the geminin-induced accumulation of cells in the G1 phase.
FIG. 8.
HOXD13 expression does not increase p53 or γ-H2AX levels. The top shows an immunoblot analysis of the expression of p53 in HEK293 cells transduced with LD13IΔN or LXIΔN. The bottom panel shows immunofluorescence staining of LXIΔN- or LD13IΔN-transduced HEK293 cells using the indicated antibodies. As a control, cells were treated with adriamycin to induce DNA damage and to promote the accumulation of p53- and γ-H2AX-positive foci.
Human DNA replication origins are bound in vivo by multiple HOX proteins.
Finally, we addressed whether HOX proteins other than HOXD13 would be capable of binding to the same set of characterized human replication origins bound by HOXD13. Formaldehyde-cross-linked chromatin was extracted from HEK293 cells, since these cells were found to endogenously express several HOX proteins, including HOXD11 and HOXA13 (data not shown), for which specific antibodies were available. Chromatin was immunoprecipitated with a control antibody and with anti-HOXD11 or anti-HOXA13 antibodies, and the enrichment for HOXD11 or HOXA13-bound sequences was verified by PCR analysis, using primers that amplify genomic regions spanning the analyzed human origins and two control regions for each origin (Fig. 1A and see Table S1 in the supplemental material). Binding of endogenous HOXD11 or HOXA13 was detected at all origins tested in HEK293 cells (Fig. 9). These results indicate that other HOX proteins besides HOXD13 bind in vivo to human replication origins, supporting the conclusion that the modulation of replication origin function is a general property of HOX homeodomain-containing proteins.
FIG. 9.
HOXD11 and HOXA13 bind human DNA replication origins. ChIP of endogenous HOXD11 and HOXA13 binding to human origins in HEK293 cells. Graphs represent the average enrichment of HOXD11- or HOXA13-bound origin regions (ori), as assessed by semiquantitative PCR. Values indicate the ratio (in percent) between the amount of DNA in immunoprecipitates (IP) and that in input chromatin (INPUT). Error bars represent the mean percent IP/INPUT ratio ± S.E.M. of at least three independent ChIPs for each sample. Two genomic regions, located respectively 5′ (5′c) and 3′ (3′c) to each origin were chosen as controls (see the text).
DISCUSSION
Hox proteins and DNA replication.
In this study we explored the possibility that Hox proteins might play a role in replication origin activity and that the HOX-geminin interaction would reveal its functional significance in this context. Only a defined set of human replication origins that show initiation of DNA synthesis from a localized site have been characterized (reviewed in reference 10), we hence focused our analysis on these. Our results show that both exogenously expressed and endogenous HOXD13 specifically bind in vivo all tested human replication origins. Despite the constant levels of exogenous HOXD13 protein expression throughout the cell cycle, the highest degree of HOXD13 binding to origin sequences was observed during the G1 phase of the cell cycle. This binding behavior differed from that detected at the promoter of EPHA7, a HOXD13 transcriptional target (51). This finding, together with the observations that the transcription of loci adjacent to origins was not perturbed by HOXD13 expression and that bound origin regions did not mediate transcriptional activation, suggested that the function of HOXD13 at replication origins is unlikely that of the transcriptional regulation of nearby located promoters. HOXD13 binding at replication origins instead promoted the assembly of pre-RC proteins onto origin DNA and stimulated, in a DNA-binding-dependent manner, the replication in vivo of plasmids containing human origins. Other HOX proteins (HOXD11 and HOXA13) were furthermore found to bind in vivo human replication origins, suggesting that the modulation of replication origin activity is a function potentially shared by multiple HOX proteins. Our results thus point to a direct role of HOXD13, and possibly of other HOX proteins, in promoting DNA replication origin licensing and replication initiation.
The mechanism by which HOXD13 promotes pre-RC assembly is probably based on the direct recruitment and/or stabilization onto DNA of one or more of its key components, since HOXD13 was found to interact with the pre-RC factor CDC6 via its homeodomain region. CDC6 is a critical factor for origin licensing, which functions as an ATP-dependent DNA-helicase loader. In yeast its conditional knockout causes the lack of pre-RC formation (8), and the knockdown by RNAi of human CDC6 prevents cell proliferation and induces apoptosis (18, 37). Moreover, the targeting of CDC6 to a plasmid has been shown to be sufficient to generate an artificial replication origin (58). The expression of HOXD13 within cells has a critical influence on the timing of origin licensing and of the initiation of DNA replication. HOXD13 promotes the earlier recruitment of pre-RC proteins onto origin during G1 phase and accelerates DNA synthesis initiation. In accordance, HOXD13 depletion was found to delay the assembly of the pre-RC and consequently to delay DNA synthesis initiation. These results correlate with previous reports showing that the enforced exogenous expression of CDC6 causes a premature S-phase entry in mammalian cells (24, 47, 57) and that injection of anti-CDC6 antibodies or dominant-negative CDC6 proteins delays entry into S phase (24, 25).
Increasing evidence has been gained for a tight link between transcription factor DNA binding and replication. In fission yeast, replication origins are located in close proximity to promoter regions (22). Moreover, in both Xenopus laevis and Drosophila melanogaster during development, replication origins are localized initially randomly and, at later stages, preferentially within promoters (27, 52). The choice and timing of origin usage in metazoa appear to be closely regulated, despite the fact that ORC proteins lack DNA-binding specificity and that metazoan origins of replication display low sequence specificity. Sequence-specific transcription factors in metazoa are held to play an important role in the selection of replication origins (reviewed in references 10 and 34). The molecular mechanism(s), however, by which transcription factors increase origin selectivity are still largely unknown (34). Transcription factors can also play crucial roles in origin activation, such as the c-Myc proto-oncogene, whose overexpression causes an increase in the number of active replicons (15). Since the origins examined in our study were all bound by pre-RC factors in the cells analyzed, independently of exogenous HOXD13 gene expression (see Fig. S2B in the supplemental material), we expect Hox proteins to have no, or at best a minor, role in origin specification. Moreover, as HOXD13, HOXD11, and HOXA13 were found to be associated in vivo with all human replication origins tested, we hypothesize a more general role for Hox proteins in DNA replication origin function, which is that of facilitating pre-RC complex reassembly during G1 phase at multiple origins and possibly that of origin activation. Considering their low intrinsic DNA-binding specificity and their binding preference for AT-rich sequences, HOX transcription factors appear in fact to possess ideal properties for the regulation of replication initiation at multiple origins.
The geminin-Hox antagonism and DNA replication.
Geminin was identified both as an inhibitor of DNA replication relicensing, as well as a regulator of patterning processes (reviewed in reference 48). In Xenopus geminin has been shown to be an inducer of neuralization, while in Drosophila and Medaka fish it has been shown to be involved in eye development through its antagonism with the Six3 homeodomain protein (12). Geminin has been found in addition to interact with the SWI/SNF chromatin remodeling complex protein Brg1 (54) and with the Polycomb group protein Scmh1, thereby participating in the epigenetic regulation of Hox gene expression (39). Finally, geminin has been shown to interact with a number of Hox proteins and to inhibit their DNA-binding and hence their transcriptional activation functions (39). The geminin-interaction region within Hox proteins has been mapped to the homeodomain, providing a biochemical basis for the geminin-induced inhibition of HOX DNA-binding (39). Our results define an unforeseen function for the inhibition of HOX protein activity by geminin, which is that of blocking the HOX-mediated stimulation of origin licensing. We show that the antagonistic action of geminin on HOXD13 is exerted at the level of DNA replication origin function. Indeed, geminin overexpression was found to inhibit HOXD13 binding at origins in a dose-dependent manner. The observations that the HOXD13 homeodomain region is apparently sufficient to mediate both protein-protein interactions with CDC6 and with geminin (39; the present study) and that geminin interferes with the HOXD13-CDC6 interaction suggests that geminin, in addition to inhibiting HOX DNA-binding, also blocks interaction with CDC6, thus exerting a twofold control on HOX activity at replication origins.
The geminin-HOX antagonism is apparently reciprocal. Geminin has been shown to be degraded as the cells exit from mitosis, to be absent during G1 phase, and to accumulate again during S, G2, and M phases (41). Moreover, the overexpression of a nondegradable form of geminin or the targeted disruption of the geminin destruction box has been shown to inhibit DNA replication and to cause the accumulation of cells in G1 phase (55, 64). We found that HOXD13 expression relieves the geminin-induced accumulation of cells in G1, indicating that HOX proteins could potentially antagonize the licensing-inhibiting function of geminin. HOXD13 expression, however, did not interfere with the normal relicensing block of cells during S and G2/M phases, since HOXD13-enforced overexpression did not cause significant relicensing nor DNA damage checkpoint activation in HEK293 cells, suggesting that HOXD13 can antagonize geminin licensing inhibition only during the G1 phase or that HOXD13 is prevented from interfering with the geminin-mediated relicensing block during the S and G2/M phases of the cell cycle. In fact, we observed a substantial decrease of HOXD13 binding at origins during the S and G2/M phases, indicating that a specific mechanism(s), or more likely geminin itself, actually prevent HOXD13 from binding to replication origins in these phases. The differential binding behavior of HOXD13 between replication origins and the promoter of the EphA7 transcriptional target gene during S phase could depend on additional factors, such as overall stability of the pre-RC and/or chromatin context, that would render the binding of HOXD13 to origins locally more sensitive to variations of geminin concentration during the cell cycle.
The activity of HOX proteins at replication origins most likely precedes that of geminin, since the geminin protein, unlike HOXD13, is not present during G1 phase and accumulates at significant levels only at the beginning of the S phase (41, 63). HOX proteins would therefore be available during G1 phase to promote pre-RC binding to origin DNA. Subsequently, during early S phase, increasing levels of geminin would sequester both HOX proteins and Cdt1, thereby blocking reinitiation of DNA replication until geminin becomes inactivated during mitosis (41). We thus propose that the physiological role of the geminin-HOX antagonism at replication origins is to counteract the HOX-mediated stimulation of origin licensing and thus to prevent a potential HOX-induced relicensing during S, G2, and early M phases of the cell cycle.
Hox proteins and cell division during development.
During development, generation of form must necessarily rely on the amount of cells fated to be part of a given structure. Hox transcription factors have been implicated in the control of cell proliferation that underlies morphogenetic processes (16). A clear example is, in effect, offered by the Hoxd13 gene, whose targeted inactivation in mice causes a phenotype that was interpreted as being the consequence of defects in the proliferation of limb mesenchymal cells (11, 14). The relevant role played by Hox proteins in cell proliferation is moreover testified by their involvement in oncogenesis (6). HOXD13, in particular, was found to cause myeloid malignancies as a fusion with the NUP98 gene (1). Although our results do not exclude that HOXD13 and/or other Hox gene products may regulate cell cycle progression via canonical transcriptional mechanisms, they suggest that HOXD13 and/or other Hox proteins exert a control on cell division by promoting G1/S transition and DNA replication initiation via an additional mechanism, based on their direct action at replication origins.
Supplementary Material
Acknowledgments
We thank Anindya Dutta for the kind gift of Myc-tagged Orc2 and Cdc6 expression constructs and Carol Imbriano for sharing anti-NF-YB antibodies. We also thank Chiara Vecchi and Antonello Pietrangelo for help with the quantitative real-time PCR analysis.
This study was supported by grants (to V.Z.) from the Italian Association for Cancer Research, the Italian Telethon Foundation, and the Italian Ministry of University and Research.
Footnotes
Published ahead of print on 24 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abramovich, C., N. Pineault, H. Ohta, and R. K. Humphries. 2005. Hox genes: from leukemia to hematopoietic stem cell expansion. Ann. N. Y. Acad. Sci. 1044:109-116. [DOI] [PubMed] [Google Scholar]
- 2.Brylawski, B. P., P. D. Chastain II, S. M. Cohen, M. Cordeiro-Stone, and D. G. Kaufman. 2007. Mapping of an origin of DNA replication in the promoter of fragile X gene FMR1. Exp. Mol. Pathol. 82:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capellini, T. D., G. Di Giacomo, V. Salsi, A. Brendolan, E. Ferretti, D. Srivastava, V. Zappavigna, and L. Selleri. 2006. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development 133:2263-2273. [DOI] [PubMed] [Google Scholar]
- 4.Caronia, G., F. R. Goodman, C. M. McKeown, P. J. Scambler, and V. Zappavigna. 2003. An I47L substitution in the HOXD13 homeodomain causes a novel human limb malformation by producing a selective loss of function. Development 130:1701-1712. [DOI] [PubMed] [Google Scholar]
- 5.Chiang, C. M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cillo, C., A. Faiella, M. Cantile, and E. Boncinelli. 1999. Homeobox genes and cancer. Exp. Cell Res. 248:1-9. [DOI] [PubMed] [Google Scholar]
- 7.Cobb, J., and D. Duboule. 2005. Comparative analysis of genes downstream of the Hoxd cluster in developing digits and external genitalia. Development 132:3055-3067. [DOI] [PubMed] [Google Scholar]
- 8.Cocker, J. H., S. Piatti, C. Santocanale, K. Nasmyth, and J. F. Diffley. 1996. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379:180-182. [DOI] [PubMed] [Google Scholar]
- 9.Comelli, L., L. Marchetti, D. Arosio, S. Riva, G. Abdurashidova, F. Beltram, and A. Falaschi. 2009. The homeotic protein HOXC13 is a member of human DNA replication complexes. Cell Cycle 8:454-459. [DOI] [PubMed] [Google Scholar]
- 10.Cvetic, C., and J. C. Walter. 2005. Eukaryotic origins of DNA replication: could you please be more specific? Semin. Cell Dev. Biol. 16:343-353. [DOI] [PubMed] [Google Scholar]
- 11.Davis, A. P., and M. R. Capecchi. 1996. A mutational analysis of the 5′ HoxD genes: dissection of genetic interactions during limb development in the mouse. Development 122:1175-1185. [DOI] [PubMed] [Google Scholar]
- 12.Del Bene, F., K. Tessmar-Raible, and J. Wittbrodt. 2004. Direct interaction of geminin and Six3 in eye development. Nature 427:745-749. [DOI] [PubMed] [Google Scholar]
- 13.de Stanchina, E., D. Gabellini, P. Norio, M. Giacca, F. A. Peverali, S. Riva, A. Falaschi, and G. Biamonti. 2000. Selection of homeotic proteins for binding to a human DNA replication origin. J. Mol. Biol. 299:667-680. [DOI] [PubMed] [Google Scholar]
- 14.Dolle, P., A. Dierich, M. LeMeur, T. Schimmang, B. Schuhbaur, P. Chambon, and D. Duboule. 1993. Disruption of the Hoxd-13 gene induces localized heterochrony leading to mice with neotenic limbs. Cell 75:431-441. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Sola, D., C. Y. Ying, C. Grandori, L. Ruggiero, B. Chen, M. Li, D. A. Galloway, W. Gu, J. Gautier, and R. Dalla-Favera. 2007. Non-transcriptional control of DNA replication by c-Myc. Nature 448:445-451. [DOI] [PubMed] [Google Scholar]
- 16.Duboule, D. 1995. Vertebrate Hox genes and proliferation: an alternative pathway to homeosis? Curr. Opin. Genet. Dev. 5:525-528. [DOI] [PubMed] [Google Scholar]
- 17.Favier, B., and P. Dolle. 1997. Developmental functions of mammalian Hox genes. Mol. Hum. Reprod. 3:115-131. [DOI] [PubMed] [Google Scholar]
- 18.Feng, D., Z. Tu, W. Wu, and C. Liang. 2003. Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res. 63:7356-7364. [PubMed] [Google Scholar]
- 19.Ferraresi, R., L. Troiano, E. Roat, E. Lugli, E. Nemes, M. Nasi, M. Pinti, M. I. Fernandez, E. L. Cooper, and A. Cossarizza. 2005. Essential requirement of reduced glutathione (GSH) for the anti-oxidant effect of the flavonoid quercetin. Free Radic. Res. 39:1249-1258. [DOI] [PubMed] [Google Scholar]
- 20.Gabellini, D., I. N. Colaluca, H. C. Vodermaier, G. Biamonti, M. Giacca, A. Falaschi, S. Riva, and F. A. Peverali. 2003. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 22:3715-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacca, M., L. Zentilin, P. Norio, S. Diviacco, D. Dimitrova, G. Contreas, G. Biamonti, G. Perini, F. Weighardt, S. Riva, et al. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. USA 91:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez, M., and F. Antequera. 1999. Organization of DNA replication origins in the fission yeast genome. EMBO J. 18:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grande, A., B. Piovani, A. Aiuti, S. Ottolenghi, F. Mavilio, and G. Ferrari. 1999. Transcriptional targeting of retroviral vectors to the erythroblastic progeny of transduced hematopoietic stem cells. Blood 93:3276-3285. [PubMed] [Google Scholar]
- 24.Hateboer, G., A. Wobst, B. O. Petersen, L. Le Cam, E. Vigo, C. Sardet, and K. Helin. 1998. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol. 18:6679-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herbig, U., C. A. Marlar, and E. Fanning. 1999. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol. Biol. Cell 10:2631-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 27.Hyrien, O., C. Maric, and M. Mechali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994-997. [DOI] [PubMed] [Google Scholar]
- 28.Keller, C., E. M. Ladenburger, M. Kremer, and R. Knippers. 2002. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 277:31430-31440. [DOI] [PubMed] [Google Scholar]
- 29.Khochbin, S., A. Chabanas, and J. J. Lawrence. 1988. Early events in murine erythroleukemia cells induced to differentiate: variation of the cell cycle parameters in relation to p53 accumulation. Exp. Cell Res. 179:565-574. [DOI] [PubMed] [Google Scholar]
- 30.Kilstrup_Nielsen, C., M. Alessio, and V. Zappavigna. 2003. PBX1 nuclear export is regulated independently of PBX-MEINOX interaction by PKA phosphorylation of the PBC-B domain. EMBO J. 22:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinoshita, Y., and E. M. Johnson. 2004. Site-specific loading of an MCM protein complex in a DNA replication initiation zone upstream of the c-MYC gene in the HeLa cell cycle. J. Biol. Chem. 279:35879-35889. [DOI] [PubMed] [Google Scholar]
- 32.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 33.Knosp, W. M., V. Scott, H. P. Bachinger, and H. S. Stadler. 2004. HOXA13 regulates the expression of bone morphogenetic proteins 2 and 7 to control distal limb morphogenesis. Development 131:4581-4592. [DOI] [PubMed] [Google Scholar]
- 34.Kohzaki, H., and Y. Murakami. 2005. Transcription factors and DNA replication origin selection. Bioessays 27:1107-1116. [DOI] [PubMed] [Google Scholar]
- 35.Krumlauf, R. 1994. Hox genes in vertebrate development. Cell 78:191-201. [DOI] [PubMed] [Google Scholar]
- 36.Ladenburger, E. M., C. Keller, and R. Knippers. 2002. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 22:1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau, E., C. Zhu, R. T. Abraham, and W. Jiang. 2006. The functional role of Cdc6 in S-G2/M in mammalian cells. EMBO Rep. 7:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei, H., H. Wang, A. H. Juan, and F. H. Ruddle. 2005. The identification of Hoxc8 target genes. Proc. Natl. Acad. Sci. USA 102:2420-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo, L., X. Yang, Y. Takihara, H. Knoetgen, and M. Kessel. 2004. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature 427:749-753. [DOI] [PubMed] [Google Scholar]
- 40.Manfredini, R., R. Zini, S. Salati, M. Siena, E. Tenedini, E. Tagliafico, M. Montanari, T. Zanocco-Marani, C. Gemelli, T. Vignudelli, A. Grande, M. Fogli, L. Rossi, M. E. Fagioli, L. Catani, R. M. Lemoli, and S. Ferrari. 2005. The kinetic status of hematopoietic stem cell subpopulations underlies a differential expression of genes involved in self-renewal, commitment, and engraftment. Stem Cells 23:496-506. [DOI] [PubMed] [Google Scholar]
- 41.McGarry, T. J., and M. W. Kirschner. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93:1043-1053. [DOI] [PubMed] [Google Scholar]
- 42.Mendez, J., and B. Stillman. 2003. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays 25:1158-1167. [DOI] [PubMed] [Google Scholar]
- 43.Morgan, E. A., S. B. Nguyen, V. Scott, and H. S. Stadler. 2003. Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadia. Development 130:3095-3109. [DOI] [PubMed] [Google Scholar]
- 44.Nishitani, H., and Z. Lygerou. 2002. Control of DNA replication licensing in a cell cycle. Genes Cells. 7:523-534. [DOI] [PubMed] [Google Scholar]
- 45.Ouyang, P. 1998. An in vitro model to study mesenchymal-epithelial transformation. Biochem. Biophys. Res. Commun. 246:771-776. [DOI] [PubMed] [Google Scholar]
- 46.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen, B. O., C. Wagener, F. Marinoni, E. R. Kramer, M. Melixetian, E. Lazzerini Denchi, C. Gieffers, C. Matteucci, J. M. Peters, and K. Helin. 2000. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14:2330-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pitulescu, M., M. Kessel, and L. Luo. 2005. The regulation of embryonic patterning and DNA replication by geminin. Cell Mol. Life Sci. 62:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rijli, F. M., and P. Chambon. 1997. Genetic interactions of Hox genes in limb development: learning from compound mutants. Curr. Opin. Genet. Dev. 7:481-487. [DOI] [PubMed] [Google Scholar]
- 50.Salsi, V., M. A. Vigano, F. Cocchiarella, R. Mantovani, and V. Zappavigna. 2008. Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. Dev. Biol. 317:497-507. [DOI] [PubMed] [Google Scholar]
- 51.Salsi, V., and V. Zappavigna. 2006. Hoxd13 and Hoxa13 directly control the expression of the EphA7 ephrin tyrosine kinase receptor in developing limbs. J. Biol. Chem. 281:1992-1999. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki, T., T. Sawado, M. Yamaguchi, and T. Shinomiya. 1999. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 19:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaefer, J. F., M. L. Millham, B. de Crombrugghe, and L. Buckbinder. 2003. FGF signaling antagonizes cytokine-mediated repression of Sox9 in SW1353 chondrosarcoma cells. Osteoarthritis Cartilage 11:233-241. [DOI] [PubMed] [Google Scholar]
- 54.Seo, S., A. Herr, J. W. Lim, G. A. Richardson, H. Richardson, and K. L. Kroll. 2005. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 19:1723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shreeram, S., A. Sparks, D. P. Lane, and J. J. Blow. 2002. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene 21:6624-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simmons, N. L. 1990. A cultured human renal epithelioid cell line responsive to vasoactive intestinal peptide. Exp. Physiol. 75:309-319. [DOI] [PubMed] [Google Scholar]
- 57.Stoeber, K., A. D. Mills, Y. Kubota, T. Krude, P. Romanowski, K. Marheineke, R. A. Laskey, and G. H. Williams. 1998. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 17:7219-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeda, D. Y., Y. Shibata, J. D. Parvin, and A. Dutta. 2005. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 19:2827-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor, E. R., and I. M. Morgan. 2003. A novel technique with enhanced detection and quantitation of HPV-16 E1- and E2-mediated DNA replication. Virology 315:103-109. [DOI] [PubMed] [Google Scholar]
- 60.Troiano, L., R. Ferraresi, E. Lugli, E. Nemes, E. Roat, M. Nasi, M. Pinti, and A. Cossarizza. 2007. Multiparametric analysis of cells with different mitochondrial membrane potential during apoptosis by polychromatic flow cytometry. Nat. Protoc. 2:2719-2727. [DOI] [PubMed] [Google Scholar]
- 61.Valerius, M. T., L. T. Patterson, D. P. Witte, and S. S. Potter. 2002. Microarray analysis of novel cell lines representing two stages of metanephric mesenchyme differentiation. Mech. Dev. 112:219-232. [DOI] [PubMed] [Google Scholar]
- 62.Williams, T. M., M. E. Williams, R. Kuick, D. Misek, K. McDonagh, S. Hanash, and J. W. Innis. 2005. Candidate downstream regulated genes of HOX group 13 transcription factors with and without monomeric DNA binding capability. Dev. Biol. 279:462-480. [DOI] [PubMed] [Google Scholar]
- 63.Wohlschlegel, J. A., B. T. Dwyer, S. K. Dhar, C. Cvetic, J. C. Walter, and A. Dutta. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290:2309-2312. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida, K., N. Oyaizu, A. Dutta, and I. Inoue. 2004. The destruction box of human geminin is critical for proliferation and tumor growth in human colon cancer cells. Oncogene 23:58-70. [DOI] [PubMed] [Google Scholar]
- 65.Zakany, J., and D. Duboule. 1999. Hox genes in digit development and evolution. Cell Tissue Res. 296:19-25. [DOI] [PubMed] [Google Scholar]
- 66.Zappavigna, V., D. Sartori, and F. Mavilio. 1994. Specificity of HOX protein function depends on DNA-protein and protein-protein interactions, both mediated by the homeo domain. Genes Dev. 8:732-744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.