Abstract
The coronaviruses mouse hepatitis virus type 2 (MHV-2) and severe acute respiratory syndrome coronavirus (SARS-CoV) utilize proteases to enter host cells. Upon receptor binding, the spike (S) proteins of both viruses are activated for membrane fusion by proteases, such as trypsin, present in the environment, facilitating virus entry from the cell surface. In contrast, in the absence of extracellular proteases, these viruses can enter cells via an endosomal pathway and utilize endosomal cathepsins for S protein activation. We demonstrate that the MHV-2 S protein uses multistep conformational changes for membrane fusion. After interaction with a soluble form of the MHV receptor (CEACAM1a), the metastable form of S protein is converted to a stable trimer, as revealed by mildly denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Liposome-binding assays indicate that the receptor-bound virions are associated with the target membrane through hydrophobic interactions. The exposure of receptor-bound S protein to trypsin or cathepsin L (CPL) induces the formation of six-helix bundles (6HB), the final conformation. This trypsin- or CPL-mediated conversion to 6HB can be blocked by a heptad repeat peptide known to block the formation of 6HB. Although trypsin treatment enabled receptor-bound MHV-2 to enter from the cell surface, CPL failed to do so. Interestingly, consecutive treatment with CPL and then chlorpromazine enabled a portion of the virus to enter from cell surface. These results suggest that trypsin suffices for the induction of membrane fusion of receptor-primed S protein, but an additional unidentified cellular factor is required to trigger membrane fusion by CPL.
Enveloped viruses enter cells through fusion of their envelope with the cellular membrane. A number of enveloped viruses utilize proteases to activate their surface glycoproteins for fusion. There are two major mechanisms for protease activation of viral glycoproteins. For many viruses, such as human immunodeficiency virus, influenza virus, and Nipah virus, the protease (e.g., furin, trypsin, or cathepsin) makes a simple proteolytic cut in the glycoprotein during biogenesis, separating the receptor-binding and fusion subunits. This cleavage converts the precursor glycoprotein to its fusion-competent state, which is presented on the virion surface. Alternatively, for other viruses such as ebolavirus and severe acute respiratory syndrome coronavirus (SARS-CoV), the proteases cleave the viral glycoprotein and induce its conformational changes during viral entry, following receptor binding and/or endocytosis (5, 13, 20, 23). However, the mechanism underlying this second situation has not been fully characterized.
The 200-kDa spike (S) proteins of murine coronavirus mouse hepatitis viruses (MHV) are class I fusion proteins that are generally cleaved into S1 and S2 subunits by cellular proteases during biogenesis. The N-terminal S1 subunit forms the surface knoblike structure of the spike, which is responsible for receptor binding, and the C-terminal membrane-anchored S2 subunit forms the stem, which is important for virus-cell membrane fusion (4). Similar to the S protein of SARS-CoV, the S protein of MHV type 2 (MHV-2) is not cleaved during biogenesis. However, regions corresponding to S1 and S2 are retained in these molecules.
The conformational changes of MHV-2 and SARS-CoV S proteins are thought to be similar to the class I fusion proteins human immunodeficiency virus GP160, influenza hemagglutinin (HA), and other coronaviral S proteins. One of the most important features of the protein is the conserved heptad repeat (HR) regions (HR-C and HR-N), which play an essential role in virus-cell fusion activities. In the fusion process, HR-N forms an interior trimeric coiled-coil structure to which the three HR-Cs bind in an antiparallel fashion, resulting in the formation of a six-helix bundle (6HB). This structure brings viral and cellular membranes into close proximity to facilitate membrane fusion. Synthetic short peptides corresponding to the HR (HR-P) regions of class-I fusion proteins have been shown to block the interaction of HR-N and HR-C complexes, resulting in the inhibition of a number of viral infections (27).
In the entry of SARS-CoV, Simmons et al. reported that two steps of conformational changes are necessary for final invasion of cells (22). The first step takes place after binding to the receptor, and the subsequent step is driven by cleavage of the S protein by protease. This mechanism was predicted by studies of viral infection using intervirion interactions; however, structural analyses of the conformational changes of S protein were not performed. In the present study, we examined the conformational changes of MHV-2 S in terms of the structural changes and revealed that, in agreement with the findings for SARS-CoV, MHV-2 S also requires at least two steps of conformational changes for fusion triggering. However, the last stage of MHV-2 entry, envelope-endosome membrane fusion, seems to be different from that of SARS-CoV. SARS-CoV entry from endosomes can be facilitated by cathepsin L (CPL) alone (3, 22), while another unidentified factor together with CPL is probably critical for the final virus-endosome membrane fusion of MHV-2. We propose a possible mechanism for the entry of MHV-2, which may also be utilized by other enveloped viruses that require endosomal cathepsins.
MATERIALS AND METHODS
Cells, viruses, and soMHVR.
DBT cells was maintained in Dulbecco modified Eagle medium (DMEM) low glucose (Gibco) supplemented with 5% fetal bovine serum. MHV-2 was propagated in DBT cells as previously reported, and the supernatants of culture fluids, DMEM containing 10% tryptose phosphate broth (Difco), were used for experiments. The soluble MHV receptor (soMHVR) was produced by using a recombinant baculovirus and purified as previously described (12).
Western blot analysis.
MHV-2 S protein was analyzed by Western blotting as described previously (12). Briefly, each sample was boiled with sample buffer containing 0.5% sodium dodecyl sulfate (SDS), separated by using a 3 to 10% gradient gel with 0.1% SDS (SDS-polyacrylamide gel electrophoresis ), and transferred to a membrane. S proteins on the membrane were reacted with anti-S1 (11F) and anti-S2 (10G) monoclonal antibodies (MAbs) (21) or anti-S2A (25) and subsequently with anti-mouse (for 11F and 10G) or anti-rabbit (for anti-S2A) immunoglobulin G labeled with horseradish peroxidase.
S protein oligomer assay.
The step 1 conformational changes of S protein were detected by mildly denaturing SDS-PAGE. A 9-μl portion of MHV-2 was mixed with 1 μl (10 μg/ml) of soMHVR (12). For step 1 incubations, samples were warmed to 37°C for 20 min, treated with sample buffer containing 0.1% SDS, and incubated for 5 min at room temperature. Samples were analyzed by Western blotting as described above.
Proteinase K digestion assay.
The step 2 conformational changes of the S protein were examined by a proteinase digestion assay as previously described (12). MHV-2 and soMHVR were mixed and incubated as described above. For two-step incubations, samples were adjusted to pH 7.4 or 5.0 with 40 mM HEPES-40 mM MES (morpholineethanesulfonic acid) and mixed with 1 μg of trypsin/ml or 18 μg of CPL (Calbiochem)/ml. After a 20-min incubation at 37°C, 1 μl of 1 M Tris (pH 8.0) was added to neutralize low-pH-treated samples, and samples were treated with 1 μg of proteinase K/ml at 4°C for 20 min. The digested S proteins were analyzed by Western blotting as described above.
Liposome-binding assay.
Lipids, l-phosphatidylcholine (PC; egg; Avanti-Polar Lipids), l-phosphatidylethanolamine (PE; egg; Avanti-Polar Lipids), sphingomyelin (Sph; brain; Avanti-Polar Lipids), and cholesterol (Chol; Sigma Chemical Co.) were mixed in a 1:1:1:1.5 molar PC-PE-Sph-Chol ratio, dried under N2 gas in a glass tube, and lyophilized overnight. After the addition of phosphate-buffered saline (pH 7.2; PBS), the lipid suspension was vortexed and then extruded 25 times through a 0.4-μm-pore-size Nuclepore filter in an Avanti-Mini-Extruder. Liposomes (5.6 mM lipid on the basis of the input lipid) were stored at 4°C and used within 1 week. A liposome-binding assay was performed as described previously (6, 28), with minor modifications. Then, 4 μl of MHV-2 and 15 μl of liposomes were incubated with 1 μM soMHVR in a final volume of 20 μl of PBS at 37°C for 20 min. The samples were overlaid on a discontinuous sucrose gradient consisting of 300 μl of 20% sucrose, 300 μl of 30% sucrose, and 30 μl of 40% sucrose in a 700-μl Ultra-Clear centrifuge tube (Beckman). After centrifugation at 40,000 rpm for 1 h at 4°C, 200-, 250-, and 200-μl fractions were drawn from the air-fluid interface. Viral RNAs were isolated from the fractions with the addition of an equal volume of Isogen (Nippon Gene), and 50 μg of yeast tRNA (Sigma) was added as a carrier. Samples were then subjected to real-time PCR to quantify the amount of genomic RNA as described below.
Virus entry assay.
DBT cells in 96-well culture plates were treated with DMEM containing 200 μM bafilomycin A1 (DMEM+Baf) at 37°C for 30 min and then chilled on ice for 10 min. Approximately 105 PFU of virus in DMEM+Baf was used to infect 105 cells on ice. After a 30-min adsorption, the virus was removed, and infected cells were treated for 10 min with various concentrations of proteases in DMEM+Baf prewarmed to room temperature. Trypsin was used as a positive control for inducing fusion at the plasma membrane. After the protease was removed, the cells were cultured in DMEM+Baf at 37°C for 5 h. Viral RNAs were isolated from cells with the addition of 200 μl of Isogen. Real-time PCR was performed to estimate the amounts of newly synthesized mRNA7 as described below.
Quantitative estimation of viral RNA by real-time PCR.
Real-time reverse transcription-PCR to estimate the amount of viral RNA was performed as described previously for SARS-CoV mRNA detection (13), with slight modifications for MHV-2 detection. To quantify the amounts of mRNA7, the target sequence was set at the inside of MHV-2 N gene. To detect the amplified fragments, we used hybridization probes labeled with fluorescent dye, 5′-GCTCCTCTGGAAACCGCGCTGGTAATGG-3′ (3′ fluorescein isothiocyanate labeled) and 5′-ATCCTCAAGAAGACCACTTGGGCTGACCAAACC-3′ (5′ LCRed640 labeled). For amplification of the fragment from viral RNA, we used the pair of oligonucleotides 5′-TGTCTTTTGTTCCTGGGCA-3′ (MHV-2 mRNA7 forward) and 5′-CAAGAGTAATGGGGAACCA-3′ (MHV-2 mRNA7 reverse). Finally, for amplification of the fragment from mRNA7, we used the pair of oligonucleotides 5′-GTACGTACCCTTTCTACTC-3′ (MHV-2 leader) and 5′-CAAGAGTAATGGGGAACCA-3′ (MHV-2 mRNA7 reverse). These oligonucleotides were synthesized according to the MHV-2 N gene sequence. PCR analysis was performed under the following conditions (room temperature, 61°C for 20 min; PCR, 95°C for 30 s; 40 cycles for 95°C for 5 s, 55°C for 15 s, and 72°C for 13 s). The amount of virus in cells was calculated from the calibration line as described previously (13).
RESULTS
The first step in conformational change in the MHV S protein is induced by receptor binding.
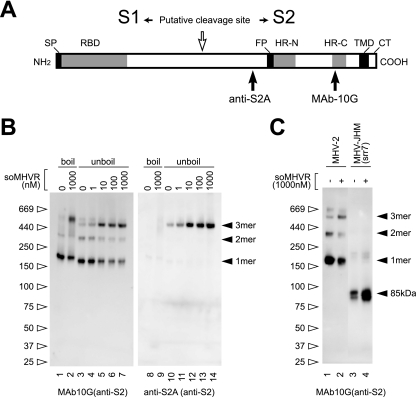
To visualize the first step in conformational changes in MHV-2 S, an S protein oligomer assay was performed using two S2-specific antibodies: MAb 10G, which recognizes HR-C, and anti-S2A, which is antiserum produced by immunizing rabbits with a synthetic peptide that encompasses the S2A epitope and recognizes the region near the fusion peptide (19, 25), as depicted in Fig. 1A. As shown in Fig. 1B, a major band at 200 kDa, as well as 380-kDa and 500-kDa bands, was detected by MAb 10G when the virus was incubated without soMHVR (Fig. 1B, lane 3). When the virus was incubated with increasing concentrations of soMHVR, the major band was shifted to 500 kDa in a concentration-dependent manner (lane 4 to 7). This 500-kDa band is thought to be a trimer of the S protein induced by soMHVR. The dimer and trimer formations of S protein detected by SDS-PAGE were also reported in a study of SARS-CoV (24), as was dimerization of S1 subunits by receptor binding in a study of MHV (10). For SARS-CoV, the major band of S protein was detected as a stable trimer without receptor interaction. However, in the present study, the MHV-2 S protein exhibited a trimer band whose mobility is similar to the SARS-CoV S trimer only after interaction with soMHVR. Furthermore, boiling the sample failed to reduce the amount of the 500-kDa band, suggesting that the trimerization is established in a stable manner (lane 2). Interestingly, the anti-S2A antibody detected the 500-kDa band in unboiled samples (lanes 10 to 14) but not in boiled samples. This indicates that S2A recognizes native S protein that has undergone conformational changes exposing the fusion peptide after binding to soMHVR. These results collectively suggest that the MHV-2 S protein undergoes step 1 conformational changes after interaction with soMHVR, from the metastable form to a stable trimer, which results in the exposure of the fusion peptide. Furthermore, since the conformational changes of srr7 S protein have been well documented (12), we compared the MHV-2 S trimer to the cleaved type S protein bearing the MHV-JHM-srr7 and found that soMHVR induced its 6HB. As shown in Fig. 1C, soMHVR induced the formation of an 85-kDa srr7 S2 subunit reactive to the MAb 10G but failed to induce a trimer (lane 4). This result suggests that the srr7 S protein is not arrested at the intermediate stage as a trimer after receptor binding.
FIG. 1.
Schematic structure and step 1 conformational change of MHV-2 S protein induced by receptor binding. (A) The S protein has an N-terminal signal peptide (SP), a receptor-binding domain (RBD), a fusion peptide (FP), two heptad repeats (HR-N and HR-C), a transmembrane domain (TMD), and a cytoplasmic tail (CT). The putative cleavage site in the central region is identified by a white arrow. Epitopes recognized by the two antibodies (MAb 10G and anti-S2A) are depicted. (B) MHV-2 virions were incubated with various concentrations of soMHVR at 37°C for 30 min. The samples were boiled (lanes 1, 2, 8, and 10) or left unboiled (lanes 3 to 7 and lanes 10 to 14) and analyzed by Western blotting with the indicated antibody. (C) Precleaved and uncleaved S proteins of MHV-JHM-srr7 and MHV-2 were incubated in the presence or absence of soMHVR to compare the trimer formation. The samples were analyzed by native PAGE and Western blotting with the indicated antibody.
Binding of S protein to soMHVR facilitates the interaction of MHV with liposomes.
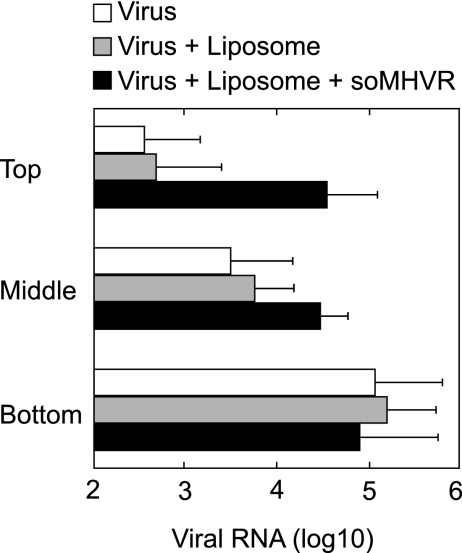
We next examined whether the fusion peptide in S protein interacts with target membranes by using a liposome binding assay. We first tried to detect liposome binding of the virions by Western blotting, as has been done with other viruses (6, 28), but we could not detect virus binding. We therefore performed this binding assay using real time-PCR, which is more sensitive at detecting a small amount of virus. In this assay, virus and liposomes are mixed in the presence or absence of receptor, overlaid on a sucrose step gradient, and centrifuged. If the virus binds the liposomes, it floats to the top of the gradient with the liposomes (28). Otherwise, it is precipitated at the bottom of the tube. As shown in Fig. 2, in the absence of receptor the majority of the viral RNA was detected in the bottom fraction, and only a small amount was detected in the middle and top fractions. However, incubation of MHV-2 with soMHVR and liposomes resulted in high colocalization of MHV-2 with the liposomes in the top fraction of the gradient. This figure is shown in a log scale and implies that ca. 50% of the virus was attached to the liposomes. This value is not unexpectedly low since some coronaviruses contain a few projections of S protein that could fail to bind soMHVR. Together with the result shown in Fig. 1B, this indicates that the interaction of MHVR with MHV-2 S protein induces conformational changes in S that expose the fusion peptide and result in an interaction of S with the target cell membrane.
FIG. 2.
Hydrophobic interactions of receptor-primed MHV-2 with liposome. Virions incubated in the absence of liposomes and soMHVR (□) or in the presence of liposomes only (░⃞) or both liposomes and soMHVR (▪) were overlaid onto the top fraction of a step gradient and spun at 40,000 rpm for 1 h. The top, middle, and bottom fractions were drawn from the air-fluid interface, and viral RNAs were isolated from the fractions. Viral genomic RNA was quantified by real-time RT-PCR. The vertical line extending to the right of the bar indicates the standard deviation of four different experiments.
Interaction with the receptor induces changes in S that expose the trypsin recognition site.
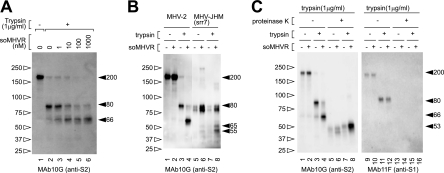
It has been well documented that trypsin treatment can activate the membrane fusion potential of MHV-2 S protein after receptor binding (18). Therefore, we next examined the fragments of S protein produced by trypsin digestion. As shown in Fig. 3A, S protein was cleaved from 200 kDa (lane 1) to 80 kDa (lane 2) by trypsin in the absence of soMHVR. However, a clear band of 66 kDa was detected when the virus was incubated with more than 1 nM soMHVR (lanes 3 to 6). The amount of 66-kDa protein increased according to the increase in concentration of added soMHVR. These results indicate that receptor binding induces the step 1 conformational changes of S protein that expose the trypsin target region and allow trypsin-mediated activation of the fusion machinery. In addition, the liposome-binding ability of MHV-2 was not enhanced by the trypsin treatment after the addition of soMHVR (data not shown). Furthermore, we compared the cleaved S proteins of srr7 to MHV2 S, as shown in Fig. 3B; an 80-kDa srr7 S2 subunit was induced by adding soMHVR (lanes 6 and 8), and 65- and 55-kDa bands became detectable after treatment of soMHVR and trypsin (lane 8) as we previously reported (12).
FIG. 3.
Step 2 conformational change of MHV-2 S protein is induced by trypsin. (A) soMHVR concentration-dependent conformational changes induced by trypsin. Virions were incubated with various concentrations of soMHVR (lanes 3 to 6) or PBS (lanes 1 and 2), and then the samples were digested with trypsin (lanes 2 to 6). (B) Comparison of the trypsin-digested S fragment between MHV-2 and MHV-JHM-srr7. Samples were treated with 1 μM soMHVR and 1 μg of trypsin/ml as in panel A. (C) Analysis of conformational changes induced by trypsin by proteinase K digestion. Virions were incubated in the absence (lanes 2, 4, 6, 8, 10, 12, 14, and 16) or presence (lanes 1, 3, 5, 7, 9, 11, 13 and 15) of soMHVR. The samples were digested with trypsin (lanes 3, 4, 7, 8, 11, 12, 15, and 16) and then further digested with proteinase K (lanes 5 to 8 and lanes 13 to 16) and analyzed by SDS-PAGE and Western blotting with the indicated antibody.
Both trypsin and CPL can induce six-helix bundle formation.
We hypothesized that the 66-kDa form of S protein induced by soMHVR and trypsin represented the fusogenic 6HB conformation, since trypsin treatment induces cell-cell fusion in S-expressing cells. 6HB conformations are characterized by the presence of a protease-resistant core. To test this hypothesis, the 66-kDa form of S protein was further digested with proteinase K, and the proteinase K-resistant fraction was detected. As shown in Fig. 3C, a clear proteinase K-resistant 53-kDa band was detected when the virus was incubated with soMHVR and then exposed to trypsin (lane 8), but this band was greatly reduced and more disperse when the virus was incubated without soMHVR or trypsin (lanes 5 to 7). The 53-kDa protein was revealed to be a constituent of 6HB, since it was not detected when treated with HR-P (see Fig. 5, compare lane 5 with lane 6). HR-P is believed to inhibit the formation of 6HB by blocking the binding of HR-C with HR-N and consequently no apparent band corresponding to 6HB is detected after treatment with proteinase K. In contrast, the S1 subunit detected with MAb 11F was cleaved to 100 kDa by trypsin and was lost by further digestion by proteinase K (Fig. 3C, lanes 9 to 16).
FIG. 5.
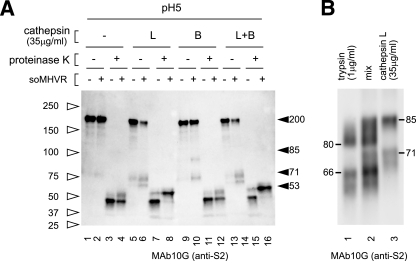
S Inhibition of conformational changes by HRP. All samples were treated with soMHVR and then mixed with HRP (lanes 2, 4, 6, 8, 10, and 12) or PBS (lanes 1, 3, 5, 7, 9, and 11). The samples were treated with trypsin (lanes 3 to 6) or CPL (lanes 9 to 12) and then with proteinase K (lanes 5, 6, 11, and 12) or PBS (lanes 1 to 4 and lanes 7 to 10).
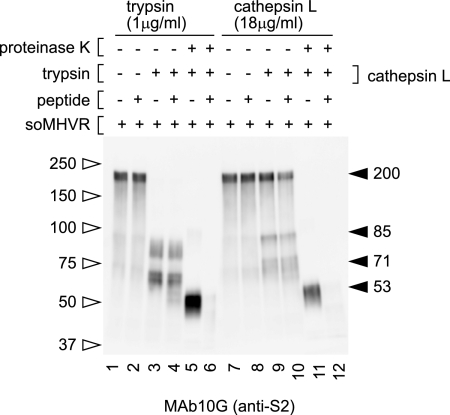
It has been reported that CPL inhibitors specifically inhibit SARS-CoV S-mediated infection (22), and exogenous CPL induces cell-cell fusion in S expressing cells (3). Similarly, Qiu et al. reported the importance of CPL and cathepsin B (CPB) for infection by MHV-2 (18). Therefore, we next tested whether CPL or CPB digestion could induce the 6HB formation of the MHV-2 S protein. When MHV-2 was pretreated with soMHVR, CPL digested S protein from 200 kDa to a 71-kDa species and a slightly smaller species (Fig. 4A, lane 6), while in the absence of soMHVR CPL digestion of S protein was greatly reduced (Fig. 4A, lane 5). An 85-kDa band of S2 was also detected after treatment with trypsin or CPL in some experiments. (Fig. 5, lanes 3 and 9). To examine the molecular mass of the fragments generated by treatment with trypsin and CPL, MHV-2 was mixed with soMHVR and then digested with each protease (Fig. 4B, lanes 1 and 3). Samples treated with trypsin or CPL were mixed and loaded into a single lane of an SDS-PAGE gel to visualize any differences in mobility. As shown in Fig. 4B lane 2, the major band produced by CPL was detected at 71 kDa, a slightly larger size than the 66-kDa band produced by trypsin.
FIG. 4.
Step 2 conformational change of MHV-2 S protein is induced by cathepsins. (A) Analysis of conformational changes induced by CPL or CPB. Virions incubated with soMHVR were digested with CPL (lanes 5, 6, 7, and 8), CPB (lanes 9, 10, 11, and 12), or CPL plus CPB (lanes 13, 14, 15, and 16) and digested with proteinase K (lanes 3, 4, 7, 8, 11, 12, 15, and 16). (B) Comparison of cleaved fragments between trypsin and CPL treated S protein after interaction with soMHVR. Samples digested with trypsin or CPL as shown in Fig. 3C, lane 4, and Fig. 4A, lane 6, were loaded in lanes 1 and 3, respectively. A mixture of trypsin and CPL treated S protein was loaded in lane 2.
CPB also digested 200-kDa S to an 85-kDa and a 71-kDa species in the presence of soMHVR (Fig. 4A, lane 10). Combined digestion with CPB plus CPL produced bands similar to those produced by treatment with CPL alone (Fig. 4A, compare lanes 6 and 14). When the virus was further exposed to proteinase K after incubation with soMHVR and treatment with CPL, a clear band of 53 kDa was detected, as shown in Fig. 4A, lane 8. This 53-kDa band was the same size as that produced by trypsin and proteinase K that appeared to be a component of 6HB, since this band was not detected after treatment with HRP, as shown in Fig. 5, lane 12. A 45-kDa proteinase K-resistant band was detected when MHV-2 was incubated at pH 5.0 in the presence or absence of CPL (Fig. 4A, lanes 3, 4, 7, 11, 12, and 15). This indicates that the 45-kDa form is induced by low pH, but this does not relate to fusion activity since MHV-2-infected cells fail to develop syncytia at low pH (18). These results suggest that trypsin or CPL treatment alone is sufficient to induce 6HB formation of the S protein after interaction with MHVR.
CPL induces 6HB formation but not membrane fusion.
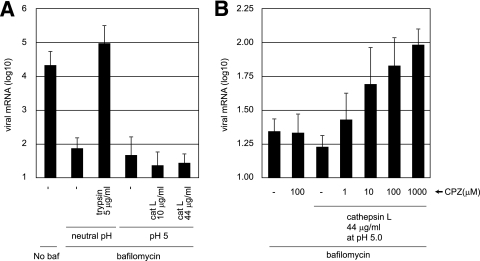
Although the 6HB formation of MHV-2 S protein could be induced by CPL cleavage, we failed to induce the direct cell-cell fusion or hemifusion of MHV-2-infected cells by CPL or CPB, detected by staining the cells with octadecyl rhodamine B (R18) and calcein blue, under the various conditions tested (data not shown). Therefore, we used a real-time PCR-based entry assay to detect fusion, which is more sensitive than the detection of syncytium formation. If CPL could trigger membrane fusion by S, MHV-2 would be able to enter cells directly from the cell surface when MHV-2 bound to its receptor was treated with CPL. As shown in Fig. 6A, 5 μg of trypsin/ml extensively facilitated the viral entry into bafilomycin-treated cells, demonstrating that trypsin activates the fusogenicity of S protein and induces fusion of the virus envelope and plasma membrane. In contrast, CPL failed to facilitate MHV-2 infection. These findings support the hypothesis that CPL treatment is not sufficient to facilitate membrane fusion.
FIG. 6.
Entry of MHV-2 from the cell surface is facilitated by protease and CPZ. (A) Effect of proteases on MHV-2 entry into DBT cells treated with bafilomycin. DBT cells were treated with bafilomycin for 30 min and infected with MHV-2 at a multiplicity of infection of 1. The cells were treated with trypsin or CPL and cultured in the presence of bafilomycin for a further 5 h. The amount of viral mRNA7 was measured quantitatively by real-time PCR. Cells not treated with bafilomycin or cells treated with bafilomycin but not with protease were used as controls. (B) Effect of CPZ on MHV-2 entry after treatment with CPL. DBT cells were treated with bafilomycin, infected with MHV-2, and treated with CPL as described for panel A. Immediately after CPL treatment, various concentrations of CPZ (pH 5.0) were treated for 3 min, and cells were cultured in the presence of bafilomycin for a further 5 h. The amount of viral mRNA7 was measured as described for panel A. Cells not treated with CPL or cells treated with CPZ but not with CPL were used as controls. baf, bafilomycin.
The failure of CPL to induce viral envelope-plasma membrane fusion could be explained if CPL-treated virus is arrested at the stage of hemifusion. If this is the case, then subsequent fusion pore formation could be triggered by treatment with chlorpromazine (CPZ), a membrane-permeable cationic drug that lowers the energy requirement for forming fusion pores (15). To test the ability of CPZ to induce fusion, virus was adsorbed onto the surface of bafilomycin-treated cells, treated with CPL, and then treated with CPZ for only 3 min. As shown in Fig. 6B, virus entry was facilitated by treatment with CPZ in a concentration-dependent manner. However, virus entry into the CPZ-treated cells was only sixfold higher than in untreated cells, which is a small enhancement compared to that caused by trypsin. This weak enhancement by CPZ could be attributed to the small degree of cleavage and 6HB formation achieved by CPL (Fig. 4A, lanes 6 and 8) compared to the more efficient cleavage by trypsin (Fig. 3C, lanes 4 and 8). The cytotoxicity of CPZ could also be responsible for the weak enhancement. Nevertheless, these results suggest that membrane fusion cannot be induced by CPL alone and that another unidentified factor that functions like CPZ must cooperate with CPL to trigger fusion of the MHV-2 envelope and the plasma membrane.
DISCUSSION
Soluble forms of receptor proteins are useful in the research of conformational changes of coronavirus S glycoproteins. However, the soluble SARS-CoV receptor (soACE2) prepared in our laboratory does not work satisfactorily for these experiments. This failure could be attributed to the previously reported insufficient neutralization activity of soACE2 compared to the clear-cut neutralization seen with soMHVR (8). Complete neutralization of pseudotyped virus bearing SARS-S was not obtained even with a high concentration of soACE2 (>200 nM), whereas 10 nM soMHVR (CEACAM1a) completely neutralized the MHV-JHM strain (16). Under these conditions, conformational changes in the S protein of SARS-CoV were not induced by soACE2 (data not shown). Proteolytic digestion by endosomal cathepsins has been described to be involved in the entry of four different enveloped viruses: ebolavirus, SARS-CoV, MHV-2, and human coronavirus 229E (5, 9, 18, 22). Cathepsins are thought to be utilized by the ebolavirus glycoprotein to generate its fusion-competent state (20), but the receptor for this virus has not been identified, and therefore a soluble form of the receptor is not available. The soluble form of aminopeptidase N, the receptor of 229E, has also not been well documented for these analyses. Therefore, the combination of MHV-2 and soMHVR is currently the only system available for the investigation of the protease-induced conformational changes in a viral glycoprotein. Our previous study using soMHVR has enabled the detection of 6HB of the MHV S protein, which is the final product of conformational changes (12). In the present study, we demonstrate the multistep conformational changes undergone by MHV-2 S protein to induce fusion. A multistep mechanism for fusion activation has also been proposed for SARS-CoV using an intervirion fusion assay (22).
Receptor-induced step 1 conformational changes.
A two-step fusion activation mechanism was first reported for the avian sarcoma leukosis virus (ASLV) envelope protein, Env (17). The first step involves receptor-induced conformational changes in Env at neutral pH. The changes lead to exposure of the fusion peptide so that Env inserts its fusion peptide into the cell surface membrane. The next step involves low pH activation of Env, which results in completion of the fusion reaction in an acidic endosomal compartment, following virus uptake and endosomal trafficking. Detailed analysis of conformational changes in Env identified at least five conformational states during activation (11). Similarly, the data in our study indicate that the activation events of MHV-2 S protein can also be divided into two steps, a receptor-triggered membrane-binding step and a protease-activated step. In various viruses, such as SARS-CoV, the possibility has been reported that a prehairpin intermediate forms a stable trimer by HR-Ns (7). In the present study, we show that receptor binding induces a stable trimer that likely exists in a prehairpin conformation. This structure seems to be maintained when S protein is cut by proteases such as CPL in the endosome. Interestingly, the anti-S2A antibody bound only to the native intermediate of S that had undergone step 1 conformational changes after receptor binding. This suggests that the S2A site is exposed by receptor-induced (step 1) conformational changes out of the spike globule, which is normally not accessible to antibodies against S2A. Since the intermediate structure of S protein is not yet well studied, analysis of MHV-2 S protein with this particular antibody will aid the study of this intermediate conformation.
Protease-induced step 2 conformational changes.
It was reported previously and shown in the present study that treatment of MHV-2 S protein with trypsin cleaves S2 to an 80-kDa form (18), similar to what is seen with trypsin treatment of SARS-CoV S (3). However, unlike with SARS-CoV, this cleavage event is not responsible for cell entry of MHV-2 because it occurs in the absence of receptor. In the present study, the receptor-primed form of S was cut into 66- or 71-kDa fragments of S2 by trypsin or CPL, respectively. Moreover, 66- and 71-kDa forms of S2 could form 6HB, which is thought to be the minimal size containing a fusion peptide, two HRs, and a transmembrane domain. Similarly, we and Belouzard et al. recently reported that cleavage at 120 amino acids downstream of the predicted cleavage site conferred fusion activity to the SARS-CoV S2 subunit (2, 26).
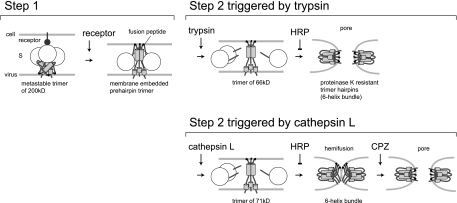
The results of our study have led to the model presented in Fig. 7, in which receptor binding and trypsin cleavage act sequentially to form distinct conformations of MHV-2 S protein that mediate virus-cell membrane fusion. Our study clearly shows that trypsin induces the complete membrane fusion of MHV-2 S protein, being consistent with SARS-CoV fusion events. However, CPL fails to induce complete membrane fusion, which is different from the observation reported for SARS-CoV S-mediated fusion (3, 22). This distinct feature of trypsin and CPL could be attributed to the small difference in the size of the residual S2 after trypsin versus CPL digestion, as revealed by detailed electrophoresis analysis (Fig. 4B). Two different groups have reported that membrane fusion is induced by CPL in SARS-CoV infection. Simmons et al. observed S protein-mediated membrane fusion by an intervirion fusion assay. Two different pseudotyped viruses were used in this assay: one with both SARS-CoV S and ASLV Env and the other expressing ACE2 and encoding luciferase. These pseudotypes were mixed, and fusion was induced by CPL; the fused pseudotypes were then allowed to infect cells expressing the ASLV receptor. Finally, infection was monitored by measuring the expression of luciferase as a result of membrane fusion between these two different pseudotype virions in vitro (22). However, ASLV can also utilize an endosomal pathway for entry. Therefore, even if the intervirion membrane fusion by S protein of SARS-CoV was not complete, i.e., arrested at the hemifusion stage, it could be completed by an unidentified factor in the endosomal environment resulting in the expression of luciferase. Bosch et al. reported significantly smaller cell-cell fusions induced in S-expressing Vero-E6 cells treated with CPL compared to the robust fusion induced by trypsin (3). However, we failed to detect SARS-CoV fusion events by CPL treatment alone, either by a cell-cell fusion assay or by the more sensitive virus-cell fusion assay used in the present study.
FIG. 7.
Model of the two-step conformational changes of MHV-2 S protein. After receptor interaction at the cell surface, MHV-2 S protein forms a stable trimer from its metastable state, and the fusion peptide is exposed and inserted into the target membrane (step 1). Upon treatment with trypsin, the receptor-primed form of MHV-2 S protein is digested to a 65-kDa fragment, which forms a stable 6HB to make a fusion pore (step 2). However, upon treatment with CPL, S protein cannot make a fusion pore even when 6HB formation is induced. CPZ treatment facilitates pore formation of CPL-primed S protein, allowing delivery of the viral core into the cytoplasm.
Our study shows that trypsin and CPL both induce similar conformational changes in S protein from a prehairpin intermediate to a folded hairpin structure. However, while membrane fusion events were completed in two steps when treated with trypsin, they were not executed in two steps when treated with CPL. The effect of trypsin could correspond to the step 2 conformational changes of ASLV Env observed in a low-pH environment. Figure 4B indicates that the CPL cleavage site is likely more N-terminal than the trypsin cleavage site. It is likely that this difference presents an obstacle to conformational changes in S. It is possible that if the CPL-initiated 6HB formation of S protein was not complete, additional cleavage by proteinase K could contribute to the 6HB formation seen in the present study. In any case, these results show that CPL can trigger conformational changes in S protein that at least make an intermediate to achieve 6HB formation. In addition, although the S protein could form a hairpin structure after CPL treatment, it failed to induce membrane fusion. This state of S protein seems to be similar to the cold-arrested stage of ASLV Env (1). Even if the 6HB of Env is formed at a low pH, the membrane fusion event fails to proceed past hemifusion at a low temperature. Incubation at physiological temperature is critical for making a membrane pore (1).
It was reported that a mutant of influenza HA that is arrested at hemifusion can complete fusion with the aid of CPZ, a reagent that binds to the membrane and promotes formation of the fusion pore (15). In the present study, we observed that CPZ can also bring about pore formation in MHV-2-infected cells treated with CPL. However, the effect was not complete, likely due to the incomplete cleavage of S by CPL. This finding suggests that the membrane fusion event induced by CPL is arrested at a hemifusion stage, presumably because the energy of the S protein was not enough to pull and mix the membranes and another factor is necessary to stimulate S protein to overcome the energy barrier of conversion from hemifusion to complete fusion. Interestingly, it was also reported that the ebolavirus glycoprotein is activated by CPL, but it is not the final trigger to induce membrane fusion. Another, as-yet-unidentified factor is predicted to cooperate with CPL for membrane fusion (20). We speculate that another protease may cut away at the N-terminal part of the CPL fragment of S protein and allow the conversion from hemifusion to complete membrane fusion.
Alternatively, there is the possibility that endosomal membranes with a lipid composition different from that of the plasma membrane can be fused more easily with the viral envelope. A typical biological membrane is composed of hundreds of different lipids, and lipids can be delivered to a variety of organelles by selective partitioning in a series of sorting steps (14). Particularly, the cholesterol content of late endosomes and lysosomes is lower than that in the plasma membrane (21), which results in a higher flexibility of membranes in these organelles. In addition, many kinds of viral receptors on the plasma membrane are reported to localize to lipid rafts, which contain a heavy concentration of cholesterol and thus are not very flexible. Since virus carried into late endosomes encounters a more flexible membrane, virus-cell membrane fusion could be induced with lower energy. Another possibility is that clathrin depletion enhances the membrane fusion on the cell surface, since CPZ is also known as an inhibitor of clathrin-coated pit formation at the cell surface. Reduction of the netlike structure made of clathrin would result in a more flexible surface. Thus, it may be possible that CPL is able to induce virus-cell membrane fusion in the endosome but not on the plasma membrane, whereas trypsin can successfully induce cell-cell and virus-cell membrane fusion on the plasma membrane. Studies are in progress to examine this possibility using liposomes with different lipid compositions.
Acknowledgments
We thank B. J. Bosch and P. J. Rottier (Utrecht University) for providing the heptad repeat peptide and K. L. Schornberg, D. Dube, S. E. Delos, and J. M. White (University of Virginia) for valuable comments on this work.
This study was supported by a Ministry of Education, Culture, Sports, Science, and Technology grant and by grants from the Uehara Memorial Foundation and the Ichiro Kanehara Foundation.
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Barnard, R. J. O., D. Elleder, and J. A. T. Young. 2006. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane fusion. Virology 344:25-29. [DOI] [PubMed] [Google Scholar]
- 2.Belouzard, S., V. C. Chu, and G. R. Whittaker. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA 106:5871-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, B. J., W. Bartelink, and P. J. M. Rottier. 2008. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82:8887-8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch, B. J., R. van der Zee, C. A. M. de Haan, and P. J. M. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandran, K., N. J. Sullivan, U. Felbor, S. P. Whelan, and J. M. Cunningham. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damico, R. L., J. Crane, and P. Bates. 1998. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc. Natl. Acad. Sci. USA 95:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, Y., J. Liu, Q. Zheng, W. Yong, and M. Lu. 2006. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure 14:889-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukushi, S., T. Mizutani, M. Saijo, S. Matsuyama, N. Miyajima, F. Taguchi, S. Itamura, I. Kurane, and S. Morikawa. 2005. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 86:2269-2274. [DOI] [PubMed] [Google Scholar]
- 9.Kawase, M., K. Shirato, S. Matsuyama, and F. Taguchi. 2008. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 83:712-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewicki, D. N., and T. M. Gallagher. 2002. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 277:19727-19734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama, S., S. E. Delos, and J. M. White. 2004. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 78:8201-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuyama, S., and F. Taguchi. 2002. Receptor-induced conformational changes of murine coronavirus spike protein. J. Virol. 76:11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuyama, S., M. Ujike, S. Morikawa, M. Tashiro, and F. Taguchi. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA 102:12543-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxfield, F. R., and T. E. McGraw. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121-132. [DOI] [PubMed] [Google Scholar]
- 15.Melikyan, G. B., S. Lin, M. G. Roth, and F. S. Cohen. 1999. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell 10:1821-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miura, H. S., K. Nakagaki, and F. Taguchi. 2004. N-terminal domain of the murine coronavirus receptor CEACAM1 is responsible for fusogenic activation and conformational changes of the spike protein. J. Virol. 78:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 18.Qiu, Z., S. T. Hingley, G. Simmons, C. Yu, J. Das Sarma, P. Bates, and S. R. Weiss. 2006. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 80:5768-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Routledge, E., R. Stauber, M. Pfleiderer, and S. G. Siddell. 1991. Analysis of murine coronavirus surface glycoprotein functions by using monoclonal antibodies. J. Virol. 65:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schornberg, K., S. Matsuyama, K. Kabsch, S. Delos, A. Bouton, and J. White. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulze, H., T. Kolter, and K. Sandhoff. 2008. Principles of lysosomal membrane degradation: cellular topology and biochemistry of lysosomal lipid degradation. Biochim. Biophys. Acta 1793:674-683. [DOI] [PubMed] [Google Scholar]
- 22.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 102:11876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 101:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song, H. C., M.-Y. Seo, K. Stadler, B. J. Yoo, Q.-L. Choo, S. R. Coates, Y. Uematsu, T. Harada, C. E. Greer, J. M. Polo, P. Pileri, M. Eickmann, R. Rappuoli, S. Abrignani, M. Houghton, and J. H. Han. 2004. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 78:10328-10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taguchi, F., and Y. K. Shimazaki. 2000. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J. Gen. Virol. 81:2867-2871. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe, R., S. Matsuyama, K. Shirato, M. Maejima, S. Fukushi, S. Morikawa, and F. Taguchi. 2008. Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J. Virol. 82:11985-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]