Abstract
Infection with many mammalian orthoreovirus (MRV) strains results in shutoff of host, but not viral, protein synthesis via protein kinase R (PKR) activation and phosphorylation of translation initiation factor eIF2α. Following inhibition of protein synthesis, cellular mRNAs localize to discrete structures in the cytoplasm called stress granules (SGs), where they are held in a translationally inactive state. We examined MRV-infected cells to characterize SG formation in response to MRV infection. We found that SGs formed at early times following infection (2 to 6 h postinfection) in a manner dependent on phosphorylation of eIF2α. MRV induced SG formation in all four eIF2α kinase knockout cell lines, suggesting that at least two kinases are involved in induction of SGs. Inhibitors of MRV disassembly prevented MRV-induced SG formation, indicating that viral uncoating is a required step for SG formation. Neither inactivation of MRV virions by UV light nor treatment of MRV-infected cells with the translational inhibitor puromycin prevented SG formation, suggesting that viral transcription and translation are not required for SG formation. Viral cores were found to colocalize with SGs; however, cores from UV-inactivated virions did not associate with SGs, suggesting that viral core particles are recruited into SGs in a process that requires the synthesis of viral mRNA. These results demonstrate that MRV particles induce SGs in a step following viral disassembly but preceding viral mRNA transcription and that core particles are themselves recruited to SGs, suggesting that the cellular stress response may play a role in the MRV replication cycle.
The nonfusogenic mammalian orthoreovirus (MRV) is a member of the Reoviridae family of segmented double-stranded RNA (dsRNA) viruses. The genome of MRV consists of 10 segments of dsRNA contained within a nonenveloped, multilayered protein capsid. During entry into cells, the outermost MRV capsid layer is removed by endosomal proteases, creating intermediate subvirion particles (ISVPs). ISVPs undergo an additional conformational change, resulting in a particle (ISVP*) that is capable of penetration of the endosomal membrane. Coincident with cellular membrane disruption, the inner capsid, or core, is released into the cytoplasm (11). The core particle, which contains the viral polymerase, guanylyltransferase, and methyltransferase enzyme activities, transcribes mRNAs corresponding to each of the 10 viral genes in the cytoplasm (14, 20). MRV mRNAs are different from cellular mRNAs in that they do not contain a 3′ poly(A) tail but do have an m7GpppN cap structure on their 5′ end (6). As infection proceeds, distinct viral structures, termed viral factories (VFs), form in the cytoplasm primarily through the action of the nonstructural protein μNS, which constitutes the structural matrix of the factories (8, 10). Core particles, viral proteins, newly synthesized viral mRNA, and dsRNA are localized within VFs, suggesting that transcription, replication, and assembly of progeny viral core particles occur within these structures (7, 8, 10, 35, 36).
Infection with MRV has been shown to induce phosphorylation of the α subunit of the translation initiation factor eIF2 (39, 45, 47, 55), a modification that inhibits host cell translation initiation by preventing the formation of the ternary complex (eIF2/GTP/tRNAiMet) (reviewed in references 42 and 44). In the case of MRV infection, phosphorylation of eIF2α is associated with the activation of protein kinase R (PKR) (55). PKR activation and subsequent eIF2α phosphorylation, together with the interferon-regulated 2′,5′-oligoadenylate synthetase RNase L system, are necessary for MRV-induced host cell translation shutoff (48). MRV mRNA continues to be translated following shutoff of host cell translation, although the mechanism for escape is not well understood. Moreover, studies have indicated that MRV replication may benefit from this aspect of the cellular response to infection by demonstrating that MRV replication is more efficient in the presence of PKR and phosphorylatable eIF2α (47).
An additional level of cellular translation regulation has recently been identified. Treatment of cells with drugs that inhibit various aspects of protein translation (32, 38) or application of other external stresses (such as nutrient starvation, heat shock, oxidative stress, or viral infection) that lead to the phosphorylation of eIF2α by specific kinases (PKR, PKR-like endoplasmic reticulum kinase [PERK], general control nonderepressible kinase [GCN], or heme-regulated inhibitor kinase [HRI]) is sufficient to trigger the formation of distinct structures in the cytoplasm termed stress granules (SGs) (reviewed in references 3 and 24). SGs sequester stalled 43S preinitiation complexes (including 40S ribosomes, mRNAs, and many translation initiation factors) in a translationally inactive complex to hold non-stress-related cellular translation in check. Once the stress is removed, SGs are disassembled and the translational material is released for reuse (reviewed in reference 24). The translational silencing proteins T-cell intracellular antigen 1 (TIA-1) and TIA-1 related protein (TIAR) play a role in SG aggregation. These proteins contain three N-terminal RNA recognition motifs and a C-terminal domain that resembles the aggregation domain of prion proteins (21). The N terminus of TIA-1/TIAR presumably binds to mRNA through its RNA recognition domains and sequesters the mRNAs and associated translation initiation factors and small ribosomal subunits into SGs by auto-aggregation of the prion-like C-terminal prion-related domain of TIA-1/TIAR (reviewed in reference 3). Additional proteins, including Ras-GAP SH3 domain binding protein (G3BP) (51), tristetraprolin (TTP), fragile X mental retardation-related protein (FMRP), and many others, have been implicated in SG formation (reviewed in reference 1).
Many viruses alter the normal course of SG induction during their replication cycle. For poliovirus, viral infection induces transient SG formation at an early phase of infection (2 to 6 h postinfection [p.i.]) and later disrupts SGs by cleaving SG component G3BP with poliovirus 3C proteinase (54). Semliki Forest virus infection results in the shutoff of host protein synthesis largely due to activation of the cellular stress response via phosphorylation of eIF2α. Semliki Forest virus infection also induces transient SG formation during the early phase of infection (2 to 5 h p.i.). Later, SGs are dispersed by an unknown mechanism (34). West Nile virus disrupts SG formation by utilizing SG effecter proteins TIA-1 and TIAR during RNA replication within viral replication complexes (17). A previous study reported that MRV infection induces SGs in a strain-specific manner at late times p.i. (19.5 h p.i) that correlates with the extent of host translation shutoff and eIF2α phosphorylation (47). Whether SGs are hurdles that must be overcome during viral translation has not yet been fully determined.
In this study, we found that cells infected with MRV transiently form SGs at an early phase of infection (2 to 6 h. p. i.), which dissociate as infection progresses. eIF2α phosphorylation was found to be necessary for MRV induction of SGs; however, a single eIF2α kinase necessary for this phosphorylation was not identified. Utilizing known pharmaceutical inhibitors of virus disassembly and translation as well as UV-inactivated virus, we identified the steps in the viral life cycle that are necessary or dispensable for SG induction. Finally, we show that MRV core particles are recruited to SGs in a manner that depends on synthesis of viral mRNA.
MATERIALS AND METHODS
Cells and reagents.
CV-1 (African green monkey kidney fibroblast), HeLa (human cervical cancer cell), mouse embryonic fibroblast (MEF), PKR−/−, PERK−/−, GCN−/−, HRI−/−, MEFS51S/S51S, and MEFS51A/S51A cells were maintained in Dulbecco's modified essential medium (Invitrogen) containing 10% fetal calf serum (Atlanta Biologicals) and penicillin-streptomycin (100 IU/ml) (Mediatech). DU-145 cells were maintained in Eagle's modified essential medium (Invitrogen) containing 10% fetal calf serum (Atlanta Biologicals) and penicillin-streptomycin (100 IU/ml). Spinner-adapted L929 cells were maintained in Joklik's minimal essential medium (Sigma-Aldrich) containing 2% fetal calf serum, 2% fetal bovine serum (HyClone Laboratories), 2 mM l-glutamine (Mediatech), and penicillin-streptomycin (100 IU/ml) (Mediatech). The primary antibodies used in immunofluorescence and immunoblotting assays were as follows: goat polyclonal anti-TIA-1 antibodies (sc-1751), goat polyclonal anti-TIAR antibodies (sc-1749) (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-eIF2α antibodies (A300-721A), rabbit polyclonal anti-eIF3 antibodies (A300-376A) (Bethyl Laboratories), rabbit polyclonal anti-eIF4G antibodies (no. 2498), rabbit polyclonal anti-eIF4E antibodies (no. 9742), and rabbit polyclonal anti-phospho-eIF2α (Ser51) antibodies (no. 9721) (Cell Signaling Technologies). Mouse monoclonal antibody (7F4) against MRV structural protein λ2, rabbit polyclonal anti-μNS antiserum, and rabbit polyclonal anti-MRV core antiserum have previously been described (9, 53). Rabbit antiserum against DCP1a has been described (26). The secondary antibodies used in immunofluorescence and immunoblotting experiments were as follows: Alexa 488- or Alexa 594-conjugated donkey anti-mouse immunoglobulin G (IgG) antibodies, Alexa 488-conjugated donkey anti-goat IgG antibodies, Alexa 350- or Alexa 594-conjugated donkey anti-rabbit IgG antibodies (Invitrogen), and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody (Invitrogen). Sodium arsenite (SA) (Sigma-Aldrich) was used at a final concentration of 0.5 mM. E-64 (Roche) was used at a final concentration of 2 mM. Ammonium chloride (NH4Cl) was used at a final concentration of 2 mM. Puromycin (Invivogen) was used at final concentration of 0.1 mg/ml.
Virions and ISVPs.
MRV stocks (T1L, T2J, and T3DC) were obtained from Max Nibert (Harvard Medical School, Boston, MA). The superscript C in T3DC is used to differentiate the T3D strain used in this study from a T3D strain (T3DN) that has previously been shown to differ in M1 gene sequence, factory morphology, and μ2 ubiquitination phenotype (37, 43). Purified virions were prepared as described previously (12) and stored in virion buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 10 mM MgCl2) at 4°C. ISVPs were prepared as previously described (12, 41). Titers of purified virus were determined by a standard MRV plaque assay using L929 cells (19). UV light-inactivated virions were prepared as follows. MRV virions were diluted in virion buffer to give a final titer (5.0 × 109 PFU/ml), 0.9 ml/well, in six-well cell culture plates. The viral solution was exposed to UV light with an intensity of 1.0 J/cm2. The resulting virions were shown to retain viral entry by an immunofluorescence assay and additionally confirmed to be completely deficient in virus replication by a plaque assay.
Infection.
Cells (1.0 × 105 or 2.0 ×105) were seeded onto 12-well cell culture plates or 35-mm cell culture dishes the day before infection. Cells were infected with MRV virions, ISVPs, or UV-inactivated virions at 10, 100, or 1,000 PFU/cell as indicated for each experiment. Viral particles were diluted in phosphate-buffered saline (PBS) (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4 [pH 7.5]) containing 2 mM MgCl2 and adsorbed to cells for 1 h, at which point cells were overlaid with Dulbecco's modified essential medium and incubated at 37°C until harvested.
Immunofluorescence assay.
Cells were seeded on six-well (9.6-cm2) dishes containing 18-mm-diameter coverslips at a density of 2 × 105 cells/well and then incubated overnight at 37°C. At the indicated times p.i., cells were fixed at room temperature for 10 min with 2% paraformaldehyde in PBS and then washed three times with PBS. Fixed cells were permeabilized by incubation with 0.2% Triton X-100 in PBS for 5 min and then washed three times with PBS. Samples were blocked for 10 min with 2% bovine serum albumin in PBS. Primary and secondary antibodies were diluted in 2% bovine serum albumin in PBS. After being blocked, cells were incubated for 1 h with primary antibody, washed three times with PBS, and then incubated for an additional hour with secondary antibody. Immunostained cells were washed a final three times with PBS and mounted on slides with ProLong reagent with or without DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Invitrogen). Immunostained samples were examined with a Zeiss Axiovert 200 inverted microscope equipped with fluorescence optics. Confocal images were taken with a Leica SP5 X confocal microscope. For each field selected, a total of 35 0.1-μm serial sections were taken horizontally. Images were prepared using Photoshop and Illustrator software (Adobe Systems).
Immunoblotting assay.
Cells (2.0 × 105) were seeded on 35-mm dishes the day before infection and then infected with viable and UV light-inactivated MRV virions as indicated. Infected cells were harvested at different times p.i. and lysed with 100 μl lysis buffer (200 mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 1% NP-40). Protein concentrations for each sample were measured by EZQ protein quantitation reagent (Invitrogen), and equivalent amounts of protein from each sample were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to nitrocellulose by electroblotting in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol [pH 8.3]). Nitrocellulose containing transferred proteins was blocked for 15 min with 5% nonfat skim milk in Tris-buffered saline (20 mM Tris, 137 mM NaCl [pH 7.6]) containing 0.1% Tween-20 (TBS-T) and then incubated overnight with primary antibodies in TBS-T containing 1% milk. Blots were washed three times for 15 min each with TBS-T, followed by a 4-h incubation with HRP-conjugated secondary antibodies in TBS-T containing 1% milk. Blots were washed a final three times and exposed to Western Lightning Plus enhanced chemiluminescence substrate (Perkin Elmer). Images were collected using a ChemiDoc XRS camera (Bio-Rad), and protein bands were quantified using Quantity-One software (Bio-Rad). At least two independent experiments were performed, and the results for a representative experiment are shown.
SG quantification.
Infected cells were immunostained at 2 or 4 h p.i. as indicated for each experiment. More than 10 fields were randomly chosen and counted, with at least 200 cells counted for each sample slide. MRV core-positive cells were recorded as infected cells, and SG-containing cells were counted from within the infected cell samples. The percentage of SG-containing cells in infected and uninfected cells was calculated, and Pearson's chi-square (χ2) test was applied to examine if there was a significant difference in SG formation between infected cells and noninfected cells. At least two independent experiments were performed, and the results for a representative experiment are shown.
RESULTS
MRV infection induces cytoplasmic structures containing SG marker proteins at early times p.i.
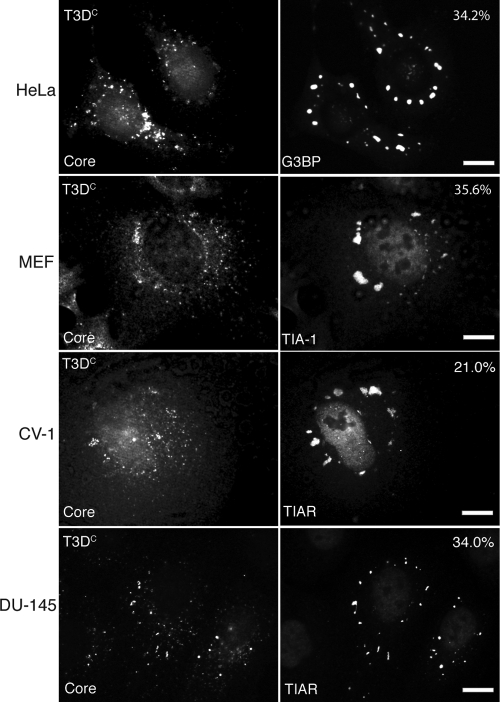
A previous study found that infections with MRV strains c87, c8, and T3D induce SGs to differing degrees in the DU-145 prostate cancer cell line at late times p.i. (19.5 h p.i.) (47). To further explore the ability of MRV to induce SGs, we infected HeLa, CV-1, MEF, and DU-145 cells with purified virions from MRV strains T1L, T2J, and T3DC. Mock-infected cells and cells treated with the known SG inducer SA served as negative and positive controls (data not shown). SA induces SGs in 98% of treated cells through activation of HRI and phosphorylation of eIF2α (33). Cells were infected with virus at 100 PFU/cell or 1,000 PFU/cell and then examined throughout infection for cytoplasmic granules containing SG marker proteins by using immunofluorescence microscopy. At early times p.i. (2 to 6 h), we found increased numbers of T2J- and T3DC-infected cells which contained cytoplasmic granules positive for the G3BP, TIA-1, or TIAR SG marker protein, compared to the level for mock-infected cells (Fig. 1 and data not shown). We quantified the number of SG-containing cells obtained following T3DC virion infection (1,000 PFU/cell, 4 h p.i.). We found that 34.2% of infected HeLa cells, 21.0% of infected CV-1 cells, 35.6% of infected MEF cells, and 34.0% of infected DU-145 cells contained putative SGs but that no uninfected HeLa, CV-1, or DU-145 cells and only 0.4% of uninfected MEF cells contained putative SGs. Pearson's chi-square (χ2) test (P < 0.005) showed that infection of these four cell types with MRV T3DC virions at 1,000 PFU/cell induces SG formation in a significantly increased number of cells compared to the level for noninfected cells at early times p.i.
FIG. 1.
Infection with MRV induces SG formation at early times p.i. HeLa (first row), MEF (second row), CV-1 (third row), and DU-145 (fourth row) cells were infected with MRV T3DC virions (1,000 PFU/cell). At 4 h p.i., cells were fixed and immunostained with rabbit anti-MRV core polyclonal antiserum (left column) and mouse monoclonal antibody against G3BP (first row, right column), goat anti-TIA-1 (second row, right column), or goat anti-TIAR (third and fourth rows, right column) polyclonal antibodies, followed by Alexa 594-conjugated donkey anti-rabbit IgG and Alexa 488-conjugated donkey anti-mouse IgG or donkey anti-goat IgG. More than 200 infected cells were counted on each slide, and the percentage of infected cells containing SG-like granules at the time of fixation is indicated. Scale bars = 10 μm.
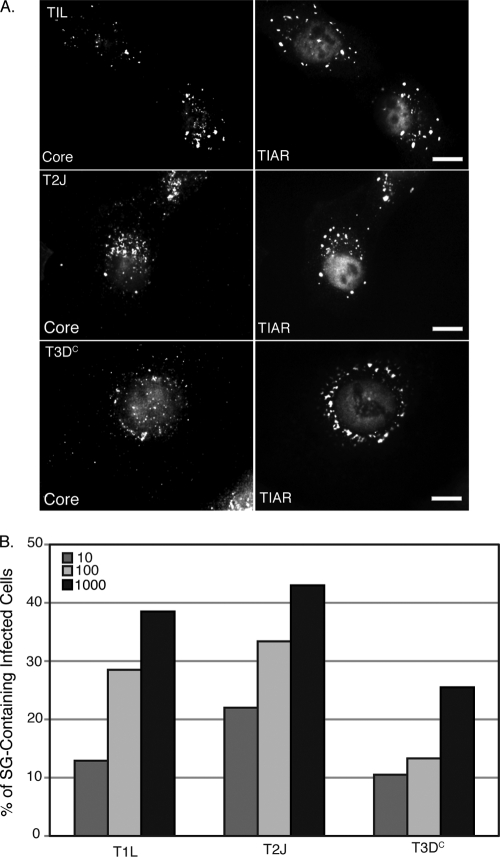
Unlike for T2J and T3DC virion-infected cells, we did not observe putative SGs in T1L virion-infected cells, even when the number of PFU/cell was increased to 10,000 (data not shown). We hypothesized that this was due to delayed entry of T1L virions into these cells on the basis of a large decrease in cells containing viral core particles at early times p.i. as measured by an immunofluorescence assay using antibodies specific for MRV core particles (data not shown). In order to clarify this, we repeated these experiments using T1L, T2J, and T3DC ISVPs, which are predicted to directly enter the cell through membrane penetration. Similar to our findings with T2J and T3DC virions, all T1L, T2J, and T3DC ISVPs induced cytoplasmic structures that stained with SG marker proteins in HeLa, CV-1, and MEF cells at early times p.i. (Fig. 2A and data not shown). This suggests that the inability of T1L virions to induce SGs is based on a deficiency in virus entry in these cell types. Furthermore, MRV ISVPs induce SGs in a dose-dependent manner (Fig. 2B), with the number of SG-positive cells increasing with the number of viral particles used to infect the cells. We found that at later times in virus infection (8 to 24 h p.i.), as viral protein translation increased and VFs became the prominent structures within the cytoplasm, SGs were no longer present in any tested cell type infected with 1, 10, 100, or 1,000 PFU/cell with T1L, T2J, or T3DC, unlike for a previous report that MRV induces SGs at late times p.i. (see Fig. 5B and 6, top right panel; data not shown) (47).
FIG. 2.
Infection with MRV ISVPs induces SG formation in a dose-dependent manner. (A) HeLa cells were infected with T1L ISVPs (top row), T2J ISVPs (middle row) or T3DC ISVPs (bottom row) (1,000 PFU/cell). At 2 h p.i., cells were fixed and immunostained with rabbit anti-MRV core antiserum (left column) and goat polyclonal anti-TIAR antibodies (right column), followed by Alexa 594-conjugated donkey anti-rabbit IgG and Alexa 488-conjugated donkey anti-goat IgG. Scale bars = 10 μm. (B) Cells were infected with MRV ISVPs (T1L, T2J, and T3DC) at 10, 100, or 1,000 PFU/cell and fixed and immunostained as in panel A. More than 200 infected cells were counted in each treatment to calculate the percentage of infected cells containing SG-like granules at the time of fixation.
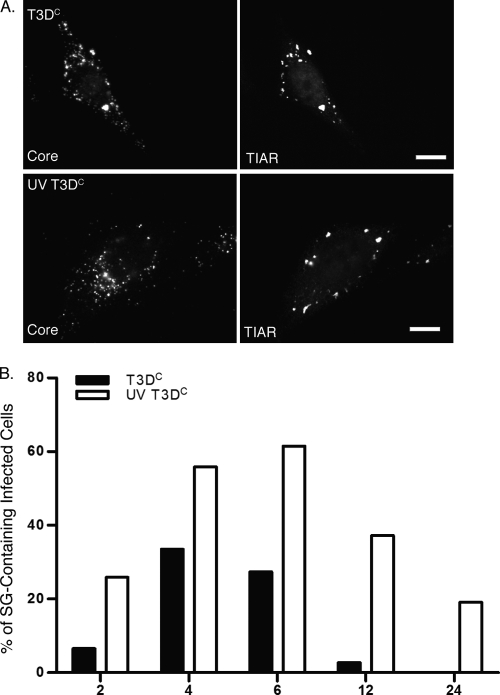
FIG. 5.
UV-inactivated MRV virions induce SGs. (A) HeLa cells were infected with untreated (top row) or UV-inactivated (bottom row) T3DC virions (1,000 PFU/cell). Cells were fixed at 4 h p.i. and immunostained with rabbit anti-MRV core polyclonal antiserum (left column) and goat anti-TIAR antibodies (right column), followed by Alexa 594-conjugated donkey anti-rabbit IgG and Alexa 488-conjugated donkey anti-goat IgG. (B) HeLa cells were infected, fixed, and immunostained as in panel A at 2, 4, 6, 12, and 24 h p.i. The percentage of infected cells containing SGs was calculated at each time point as described in Materials and Methods.
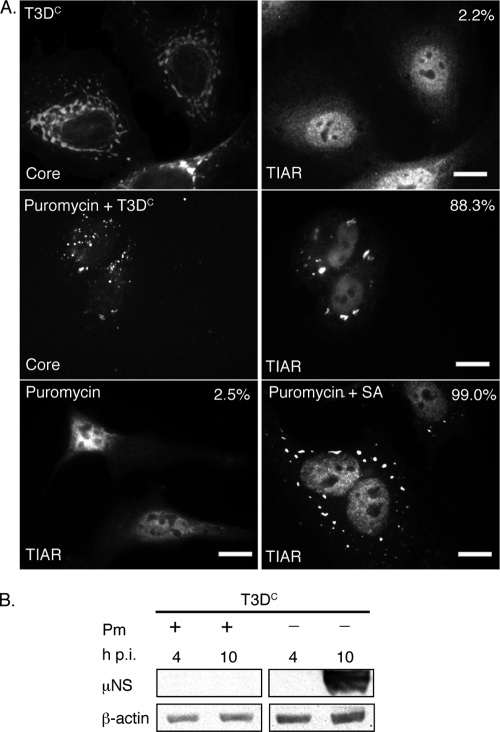
FIG. 6.
Puromycin inhibits viral translation but does not prevent MRV induction of SGs. (A) HeLa cells were untreated (first row) or pretreated with 0.1 mg/ml puromycin (second row) for 1 h and then infected with MRV T3DC virions (1,000 PFU/cell) for 1 h. New medium containing 0.1 mg/ml puromycin was added to cells following infection (second row). At 10 h p.i., cells were fixed and immunostained with rabbit anti-MRV core polyclonal antisera (first and second rows, left column) and goat anti-TIAR polyclonal antibodies (first and second rows, right column), followed by Alexa 594-conjugated donkey anti-rabbit IgG and Alexa 488-conjugated donkey anti-goat IgG. Uninfected HeLa cells were treated with 0.1 mg/ml puromycin (third row, left and right columns), incubated (left column) or treated with 0.5 mM SA for 1 h (right column) after 9 h puromycin treatment, and then fixed and stained with goat anti-TIAR polyclonal antibodies followed by Alexa 488-conjugated donkey anti-goat IgG. Following immunostaining, the percentage of infected cells containing SGs (first and second rows) or total cells containing SGs (third row) was quantified as described in Materials and Methods. Percentages of SG-containing cells are indicated. Scale bars = 10 μm. (B) HeLa cells were infected with MRV T3DC virions (1,000 PFU/cell) with or without 0.1 mg/ml puromycin (Pm) as indicated, and at 4 and 10 h p.i., cells were lysed, and proteins were separated by SDS-PAGE, followed by immunoblotting using anti-μNS polyclonal antiserum or anti-β-actin polyclonal antibodies. Proteins were detected using HRP-conjugated goat anti-rabbit IgG, followed by chemiluminescence imaging.
MRV-induced SGs are structurally similar to SGs induced by eIF2α phosphorylation.
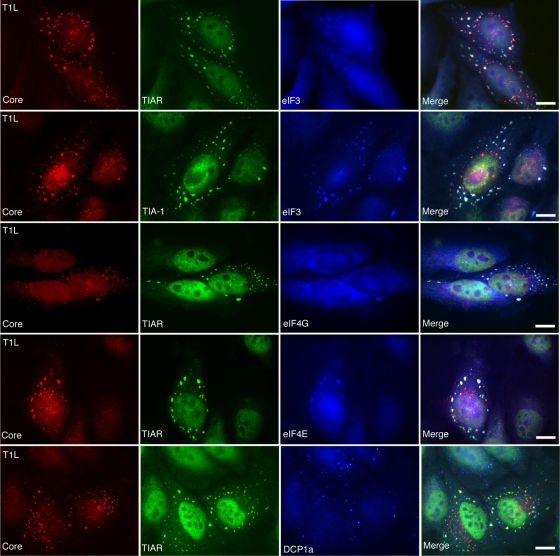
Because there are a growing number of cytoplasmic structures that form in cells in response to different stresses (reviewed in reference 3), we further characterized SGs induced by MRV infection to determine if they contained components, such as translation initiation factors (eIF4G, eIF4E, and eIF3), that are characteristic of SGs induced by phosphorylation of eIF2α and other types of protein synthesis inhibition (25). HeLa cells were infected with ISVPs (T1L, T2J, and T3DC) or virions (T2J and T3DC) with 1,000 PFU/cell. At 2 h p.i. or 4 h p.i., cells were fixed for three-color immunofluorescence using mouse antibody (7F4) against viral core protein λ2 to visualize infected cells; goat antibodies against TIAR or TIA-1 to visualize SGs; and rabbit antibodies against eIF4G, eIF4E, or eIF3 to visualize translation initiation factors. Mock-infected cells and SA-treated cells were used as negative and positive controls for SG formation (data not shown). The SG effecter protein TIAR (or TIA-1) colocalized in independent experiments with eIF3 (Fig. 3, first and second rows), eIF4G (Fig. 3, third row), and eIF4E (Fig. 3, fourth row) in the granules that formed in MRV-infected cells. As a negative control for SGs, we examined the localization of DCP1a, a marker for processing bodies (26), and found that this protein did not localize in the structures induced by MRV infection and was found instead in small separate structures that are likely processing bodies (Fig. 3, fifth row). These results indicate that MRV-induced SGs are structurally similar to those induced by inhibition of protein translation initiation and eIF2α phosphorylation.
FIG. 3.
MRV-induced SGs contain translation initiation factors eIF3, eIF4G, and eIF4E but not P body component DCP1a. HeLa cells were infected with T1L ISVPs (1,000 PFU/cell) for 2 h and then fixed and immunostained with mouse anti-λ2 monoclonal antibody 7F4 (Core) (left column); goat anti-TIA-1 or goat anti-TIAR antibodies (second column); and rabbit anti-eIF3, eIF4G, and eIF4E antibodies or DCP1a antisera (third column); followed by Alexa 594-conjugated donkey anti-mouse IgG, Alexa 488-conjugated donkey anti-goat IgG, and Alexa 350-conjugated donkey anti-rabbit IgG. Merged images are shown in the right column. Scale bars = 10 μm.
Viral uncoating is required for MRV induction of SGs.
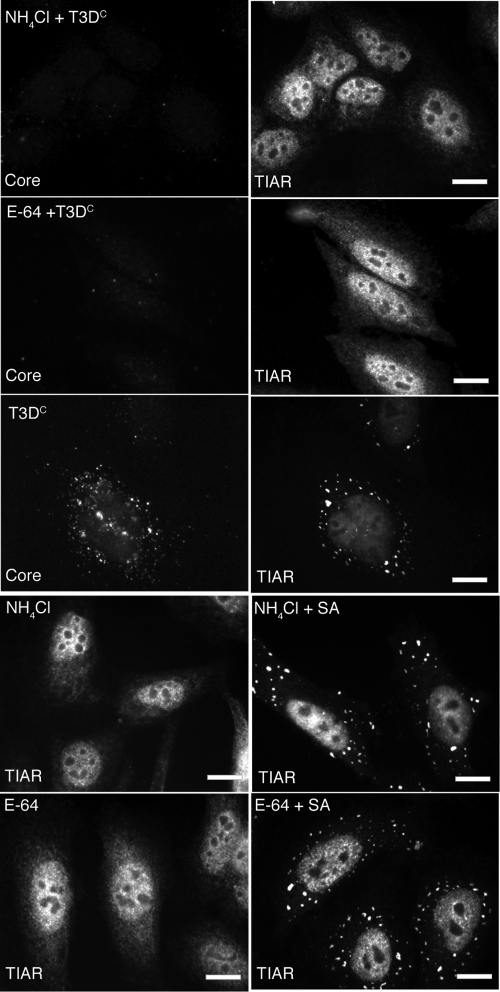
To unravel the steps in MRV infection that are necessary for induction of SGs, we examined the effects of previously described pharmaceutical inhibitors of viral uncoating (ammonium chloride and E-64) on the ability of MRV infection to induce SGs. It has previously been shown that these drugs are able to block viral uncoating by changing the pH of endosomes (ammonium chloride) or blocking endosomal protease activity (E-64) (5, 49). HeLa cells were pretreated with 2 mM ammonium chloride or 2 mM E-64 for 4 h preceding infection. Following this incubation, cells were infected with T3DC virions (1,000 PFU/cell), incubated for 1 h at room temperature, and then retreated with ammonium chloride or E-64 at the above-mentioned concentrations. At 4 h p.i. and 8 h p.i., cells were fixed and stained with antibodies to detect viral cores and SGs. In contrast to what was found for untreated, infected cells that contain both core particle staining and SGs (Fig. 4, third row), neither core particle staining nor SGs were observed in the ammonium chloride- or E-64-treated, infected cells at 4 h p.i. (Fig. 4, first and second rows). Control experiments indicate that neither ammonium chloride nor E-64 induces SGs or prevents SG formation induced by SA (Fig. 4, fourth and fifth rows). At 8 h p.i., MRV virions were still unable to uncoat and induce SGs (data not shown). These results suggest that viral uncoating is required for SG induction.
FIG. 4.
Ammonium chloride and E-64 prevent MRV induction of SGs. HeLa cells were pretreated with 2 mM ammonium chloride (NH4Cl) (first row) or 2 mM E-64 (second row) or left untreated (third row) for 4 h and then infected with MRV T3DC virions (1,000 PFU/cell). At 4 h p.i., cells were fixed and immunostained with rabbit anti-MRV core polyclonal antisera (first, second, and third rows, left column) and goat anti-TIAR polyclonal antibodies (first, second, and third rows, right column), followed by Alexa 594-conjugated donkey anti-rabbit IgG and Alexa 488-conjugated donkey anti-goat IgG. Uninfected HeLa cells were treated with 2 mM ammonium chloride (fourth row) or 2 mM E-64 (fifth row), incubated (fourth and fifth rows, left column) or additionally treated for 1 h with 0.5 mM SA (fourth and fifth rows, right column), and then fixed and stained with goat anti-TIAR polyclonal antibodies followed by Alexa 594-conjugated donkey anti-rabbit IgG. Scale bars = 10 μm.
UV-inactivated MRV virions induce SGs.
In order to examine whether gene expression is involved in SG induction, we examined the ability of UV-inactivated virions to induce SGs. T3DC virions were inactivated by UV light treatment as described in Materials and Methods. HeLa cells were infected with untreated T3DC virions or UV-treated T3DC virions, and at 0, 2, 4, 6, 12, and 24 h p.i., cells were fixed and immunostained to detect viral core particles and SGs. We found that similar to untreated virions (Fig. 5A, first row), UV-inactivated virions were able to enter cells and to induce SGs (Fig. 5A, second row). Additionally, we quantified the number of SGs that were induced by active versus inactive viruses over time (Fig. 5B). Interestingly, UV-inactivated virions induced higher percentages of SG-containing cells throughout MRV infection than viable MRV-infected cells (Fig. 5B). In fact, by 12 h p.i., less than 5% of cells infected with active MRV contained SGs, whereas nearly 40% of cells infected with UV-inactivated virus still contained SGs at this time. By 24 h p.i., no actively infected cells contained SGs, while 20% of cells infected with UV-inactivated MRV still contained SGs. These results suggest that SG induction is mainly due to viral entry events. Moreover, because cells containing SGs appear to rapidly decrease as viral mRNA is transcribed and translated in actively infected cells but remain present in a large number of cells infected with virus that is unable to express protein, these results also suggest that viral gene expression may be involved with SG disruption as viral infection progresses. The reason for the decline over time of SG-containing cells following infection with UV-inactivated virus remains to be determined.
Translation is dispensable for SG induction but required for SG disassembly in MRV-infected cells.
To further examine the role of viral mRNA transcription and translation in MRV induction of SGs, we examined MRV induction of SGs in the presence of a known pharmaceutical inhibitor of translation (puromycin). Puromycin is a general inhibitor of cellular and viral protein translation that inhibits translation elongation by forming methionyl-puromycin (31). HeLa cells were treated with puromycin for 1 h preceding infection. Following this incubation, cells were infected with T3DC virions at 1,000 PFU/cell and incubated for 1 h at room temperature, at which point cells were retreated with puromycin at the above-mentioned concentration. At 10 h p.i., cells were fixed and stained with antibodies to detect viral cores and SGs. In untreated, infected cells at 10 h p.i., rabbit anti-core antiserum stained VFs, and only 2.2% of infected cells formed SGs (Fig. 6A, first row), confirming our previous observations that SGs dissipate as MRV infection proceeds. In puromycin-treated, infected cells, no VFs were present as a result of puromycin translation inhibition, however, 88.3% of cells contained SGs (Fig. 6A, second row) at this time. In uninfected, puromycin-treated cells, only 2.5% of cells formed SGs (Fig. 6A, third row, left). Similar to previously reported data (50), puromycin treatment did not prevent SA induction of SGs, and 99.0% of SA-treated cells contained SGs (Fig. 6A, third row, right). Immunoblots against MRV nonstructural protein μNS confirmed no MRV protein synthesis in the puromycin-treated, infected cells at 10 h p.i., while untreated cells contained significant levels of μNS synthesis (Fig. 6B). These results suggest that viral translation is not required for MRV induction of SGs. They further suggest, similar to our results with UV-treated virions, that when viral protein synthesis is inhibited, the SGs that form in response to MRV infection are not disrupted. This again implicates a role for MRV protein expression in SG disruption as viral infection proceeds from early to late times.
eIF2α phosphorylation correlates with, and is required for, MRV induction of SGs.
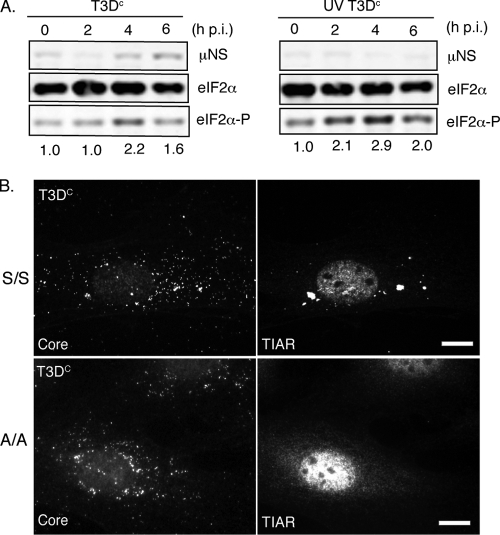
Many lines of evidence suggest that eIF2α phosphorylation is sufficient to induce SGs (29); however, a number of drugs and small molecules that can induce SGs independently of eIF2α phosphorylation have been described (38). These include the small molecule NSC119893, which inhibits the interaction between eIF2α and Met-tRNA (38), and the drugs pateamine (15) and 15D-PGJ2 (28), which bind and inhibit translation initiation factor eIF4A. To determine if MRV induction of SGs correlates with phosphorylation of eIF2α, we examined the relative levels of phosphorylated eIF2α compared to total cellular eIF2α during MRV infection at times in which SGs were present. HeLa cells were infected with untreated or UV-treated T3DC virions, and at 0, 2, 4, and 6 h p.i., samples were collected. Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies against MRV nonstructural protein μNS, total eIF2α, and phosphorylated eIF2α (Fig. 7A). These experiments show that the levels of phosphorylated eIF2α increase in both active virus-infected cells (Fig. 7A, left panel) and UV-inactivated virus-infected cells (Fig. 7A, right panel) at times when SGs are routinely found in MRV-infected cells. This suggests MRV-induced SG formation may occur as a result of eIF2α phosphorylation.
FIG. 7.
eIF2α phosphorylation correlates with, and is required for, MRV induction of SGs. (A) HeLa cells were infected with untreated or UV-inactivated T3DC virions (1,000 PFU/cell), and cells were harvested at 0, 2, 4, and 6 h p.i. Following cell lysis, proteins were separated by SDS-PAGE and immunoblotted using rabbit anti-μNS polyclonal antiserum, rabbit anti-eIF2α polyclonal antibodies, and rabbit anti-phospho-eIF2α polyclonal antibodies. Proteins were detected and quantified using HRP-conjugated goat anti-rabbit antibodies, followed by chemiluminescence imaging. Levels of phosphorylated eIF2α relative to total eIF2α at each time point were calculated and normalized to the level seen at time zero and are indicated below the panel for each time point. (B) MEFS51S/S51S (S/S) (first row) and MEFS51A/S51A (A/A) (second row) cells were infected with MRV T3DC virions (1,000 PFU/cell). At 4 h p.i., cells were fixed and immunostained with rabbit anti-MRV core antiserum (left column) and goat anti-TIAR polyclonal antibodies (right column), followed by Alexa 594-conjugated donkey anti-rabbit IgG and Alexa 488-conjugated donkey anti-goat IgG. Scale bars = 10 μm.
In order to examine whether phosphorylation of eIF2α is required for MRV induction of SGs, we utilized a cell line, MEFS51A/S51A, in which the eIF2α gene is genetically altered such that the serine residue at amino acid 51 is changed to an alanine. This results in loss of the phosphorylation event that is necessary to inhibit ternary complex formation (46). MEFS51A/S51A cells are unable to form SGs following SA treatment, which activates eIF2α phosphorylation through HRI, but are able to form SGs following treatment with NSC11893 (38), pateamine A (15), or 15D-pGJ2 (28). Wild-type MEFS51S/S51S and mutant MEFS51A/S51A cells were infected with ISVPs (T1L, T2J, or T3DC) or virions (T2J or T3DC) at 1,000 PFU/cell. At 2 h p.i. or 4 h p.i., cells were fixed and stained for immunofluorescence with antiserum against MRV cores to visualize infected cells and with antibodies against TIAR to visualize SGs. In these experiments, although we counted various numbers of SG-containing cells in infected wild-type MEFS51S/S51S cells (ranging from 13% to 54%, depending on virus strain and particle type used), we did not detect any infected cells containing SGs in the mutant MEFS51A/S51A cells (Fig. 7B and data not shown). These findings suggest that SG induction by MRV occurs through a pathway that requires eIF2α phosphorylation.
At least two eIF2α kinases are involved in MRV induction of SGs.
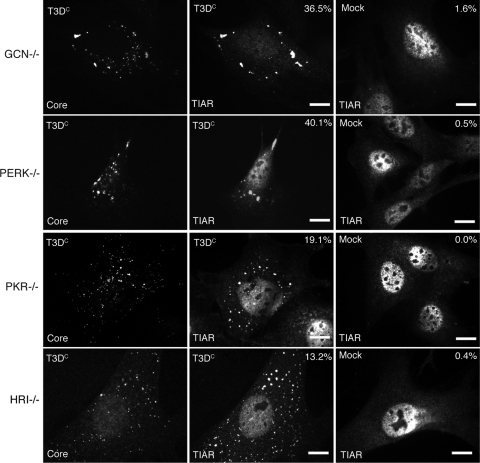
Because our data suggested that eIF2α phosphorylation is required for SG formation at early times in infection, we were interested in further identifying the cellular pathway involved in MRV induction of SGs. Four kinases that phosphorylate eIF2α in response to different cellular stresses have been identified. These include PKR, which is activated by viral infection; PERK, which is activated by protein misfolding in the endoplasmic reticulum; GCN, which is activated by nutrient deprivation; and HRI, which is activated by oxidative stress (reviewed in reference 3). We utilized a knockout cell line for each of the four eIF2α kinases (PKR−/−, PERK−/−, HRI−/−, and GCN−/−) to determine if knockout of any individual stress kinase results in loss of SG formation following MRV infection. Each cell line was infected with T3DC at 1,000 PFU/cell, and at 4 h p.i., cells were fixed and immunostained to visualize viral cores and SGs. We found that each individual knockout cell line was able to form SGs in response to MRV infection with 36.5% GCN−/− cells, 40.1% PERK−/− cells, 19.1% PKR−/− cells, and 13.2% HRI−/− cells containing SGs (Fig. 8, left and middle columns). Only a few cells contained SGs in these cell lines when they were mock infected (Fig. 8, right column). These results indicate that at least two eIF2α kinases can be activated to phosphorylate eIF2α by MRV infection at early times p.i. and suggest that the signaling pathway leading to SG induction by MRV infection is more complex than activation of a single eIF2α kinase.
FIG. 8.
MRV induces SG formation in eIF2α kinase knockout cell lines. GCN−/− (first row), PERK−/− (second row), PKR−/− (third row), or HRI−/− (fourth row) cells were infected with T3DC virions (1,000 PFU/cell) (left and middle columns) or mock infected (right column). At 4 h p.i., cells were fixed and immunostained with rabbit anti-MRV core antiserum (left column) and goat anti-TIAR polyclonal antibodies (middle and right columns), followed by Alexa 594-conjugated donkey anti-rabbit IgG and Alexa 488-conjugated donkey anti-goat IgG. More than 200 infected cells were counted on each slide, and the percentage of infected cells containing SG-like granules at the time of fixation is indicated. Scale bars = 10 μm.
Viral core particles are recruited to SGs in a transcription-dependent manner.
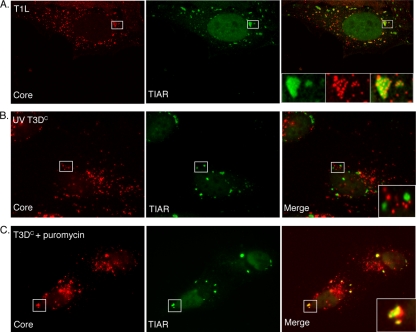
In our study, we found that in MRV-infected, SG-containing cells, a portion of viral core particles localized to SGs at early times p.i. (Fig. 1, 2, 3, 4, 5, 6, 7, and 8). This colocalization was variable depending on cell type and experiment; however, in each experiment, core particles were visualized in SGs. In order to confirm these findings, we examined MRV core and SG localization by using confocal microscopy. HeLa cells were infected with T1L ISVPs, and at 2 h p.i., cells were fixed and stained with antibodies against MRV cores and TIAR to visualize core particle localization relative to SGs. Confocal images confirmed that many, but not all, viral core particles are intensely localized in SGs induced by MRV (Fig. 9A). To determine whether gene expression is necessary for the localization of viral cores to SGs, we examined the localization of viral core particles in UV-inactivated, infected cells and puromycin-treated, infected cells. UV irradiation is expected to prevent viral transcription (and in our studies, we found no evidence of protein translation or replication following UV treatment of particles) (Fig. 7A and data not shown). Puromycin is not expected to prevent viral transcription but is expected to prevent viral translation (and in our studies, we did not detect viral protein translation following puromycin treatment) (Fig. 6B) (31). In these experiments, we found that in cells infected with UV-inactivated viruses, no viral core particles colocalized in SGs (Fig. 9B). However, in the puromycin-treated, infected cells, although not all viral core particles localized to SGs, by 10 h p.i, almost all cells formed SGs which contained viral core particles (Fig. 9C). These findings suggest that the localization of viral core particles to SGs may be mediated by newly synthesized viral mRNA. The absence of complete colocalization of core particles with SGs is likely a result of two issues. First, virion preparations always contain a portion of viral particles defective in viral transcription, and second, some cores are likely not actively synthesizing viral mRNA at the moment when cells were fixed for immunostaining. While viral core particles may be targeted to SGs via viral mRNA, it has yet to be determined whether viral mRNA, like cellular mRNA, is translationally silent when cores are sequestered in SGs.
FIG. 9.
MRV core particles colocalize with SGs in a manner dependent on viral gene expression. (A) HeLa cells were infected with MRV T1L ISVPs (1,000 PFU/cell). At 2 h p.i., cells were fixed and stained with rabbit anti-MRV core antiserum (left) and goat anti-TIAR antibodies (middle), followed by Alexa 594-conjugated donkey anti-rabbit IgG and Alexa 488-conjugated donkey anti-goat IgG. A merged image is shown (right). Confocal images were taken at 0.1-μm-slice intervals using a Leica SP5 X confocal microscope. The boxed regions in each image were amplified and are shown in insets in the merged image. (B) HeLa cells were infected with UV-inactivated T3DC virions (1,000 PFU/cell). At 4 h p.i., cells were fixed and stained with rabbit anti-MRV core antiserum (left) and goat anti-TIAR antibodies (middle). A merged image is shown (right). The boxed region in the merged image was amplified and is shown in the inset. (C) HeLa cells were pretreated with 0.1 mg/ml puromycin for 1 h, incubated with T3DC virions (1,000 PFU/cell) for 1 h, and then retreated with puromycin for an additional 10 h, at which point cells were fixed and stained with rabbit anti-MRV core antiserum (left) and goat anti-TIAR polyclonal antibodies (middle). A merged image is shown (right). The boxed region in the merged image was amplified and is shown in the inset.
DISCUSSION
Viral entry is required for MRV induction of SGs.
SG formation is a defensive mechanism used by host cells in response to many types of external or internal stresses (2). The size and number of SGs within cells are dependent on the intensity of different stresses. For example, SA induces SGs in a dose-dependent manner, with a lower dose of SA inducing fewer and smaller SGs, and vice versa (2). In these studies, we also found that the number of cells containing MRV-induced SGs is dependent on the levels of core particle entry into the cytoplasm. ISVPs from all prototype viral strains were able to induce SG formation in a dose-dependent manner, with increasing numbers of PFU/cell resulting in increasing numbers of SG-containing cells. In fact, with the use of ISVPs, a significant number of SGs could be induced at 10 PFU/cell (Fig. 2B). It is important to note that our experiments quantifying numbers of SG-containing, infected cells are complicated by the fact that viral infection and SG formation are both dynamic processes. Because cells and viral infections are not synchronized in these experiments, fixation for immunofluorescence represents only a snapshot of a population of cells at one moment in time. It is likely that infected cells will be at different stages of these dynamic processes at this moment and that the percentage of cells in which MRV induces SGs is much higher than what is indicated in these experiments. In fact, we found that when MRV gene expression was inhibited by UV inactivation (Fig. 5) or by puromycin treatment (Fig. 6), much higher percentages of SG-containing infected cells were observed in these assays. MRV produces 200 to 3,000 PFU in each infected cell (40). All of these viruses are likely released simultaneously into the surrounding cellular matrix by cell lysis, making it likely that the conditions used in our experiments are biologically relevant.
SG induction by MRV requires viral uncoating but not viral gene transcription and translation.
Previous studies showed that MRV infection induces phosphorylation of eIF2α at late times p.i., likely through activation of PKR kinase during viral replication, resulting in SG formation (47). However, in our study, we tested several cell lines (HeLa, L929, MEF, CV-1, Cos-7, and DU-145) and found that no SGs formed in cells containing the large, perinuclear VFs that form in infected cells at late times in infection (12 to 24 h p.i.). We consistently visualized a small percentage of SG-containing, infected cells expressing very low levels of viral protein when we examined SGs at late times p.i. (Fig. 5B and 6A; also Q. Qin and C. L. Miller, unpublished data). These findings suggest that as MRV infection continues, MRV-induced SGs are dispersed. We cannot fully explain the difference in our data from the published report indicating that MRV-infected cells contain SGs at late times p.i. We have tested more than 10 cell lines (this study and data not shown), and the presence of SGs at early, but not late, times in MRV infection does not appear to be cell type specific. Moreover, while we have demonstrated that the induction of SGs following MRV infection is dependent on number of PFU/cell, the absence of SGs in cells at late times in infection is not dependent on number of PFU/cell, as cells expressing high levels of MRV protein following infection with 1, 10, 100, or 1,000 PFU/cell also do not contain SGs at later times p.i. (Fig. 5B and 6A and data not shown). It is also interesting that a recent report has shown that the highly related rotavirus also does not contain SGs at late times in infection, even though eIF2α is phosphorylated at these times (39).
In order to understand the mechanism of SG induction by MRV, we chose some well-defined pharmaceutical inhibitors to block specific steps of viral infection to identify which step is required for SG induction. We found that viral uncoating is required for SG formation on the basis of the fact that treatment of cells with ammonium chloride and E-64, which inhibit the cleavage and subsequent conformation changes of outer capsid proteins (σ3 and μ1) in the endosome, prevents the formation of SGs following MRV infection (Fig. 4). We also found that viral transcription and translation are not required for SG formation on the basis of the facts that UV-inactivated MRV virions and ISVPs still retain their ability to induce SGs (Fig. 5A) and cells treated with puromycin are still able to form SGs (Fig. 6A) even though viral translation is completely inhibited. These findings suggest that MRV induction of SGs is mainly dependent on viral entry and precedes viral protein synthesis and replication.
Phosphorylation of eIF2α is a key factor for SG formation in response to MRV infection during viral entry.
SG formation is a complicated process, and the mechanism involved in the formation and dispersal of these structures is poorly understood. Formation of SGs can be induced by phosphorylation of eIF2α (3), inhibiting the formation of ternary complex (eIF2-GTP-tRNAiMet) or interfering with translation initiation factors (eIF4A [15], eIF4B, eIF4H, or PABP [38]). Phosphorylation of eIF2α appears to play an important role in SG induction by some but not all viruses (18, 34). This is not surprising, given the fact that many viruses activate PKR, including alphavirus (52), reovirus (47), rotavirus (39), and influenza viruses (22). Herpes simplex virus 1 activates PERK as well as PKR (13). Activated PKR or PERK phosphorylates eIF2α on Ser 51, which leads to the failure of ternary complex formation (eIF2-GTP-tRNAiMet), leading to SG formation (25). Our data show that phosphorylation of eIF2α is required for SG formation following MRV infection (Fig. 7). However, PKR does not appear to be the only kinase that phosphorylates eIF2α during MRV infection. We found that MRV infection induces SGs in a PKR−/− cell line as well as other eIF2α kinase knockout cell lines (GCN−/−, PERK−/−, and HRI−/−) (Fig. 8). We speculate, based on this data, that MRV infection can induce eIF2α phosphorylation through a number of pathways. It has previously been shown that MRV infection activates the unfolded protein response, which involves PERK (47), raising the possibility that both PKR and PERK may play a role in MRV induction of SGs.
The recruitment of viral cores in SGs may be a consequence of pathogen-host coevolution.
During MRV-induced SG formation, we observed many viral core particles localized in SGs (Fig. 9A). Our data suggest viral mRNA transcription is necessary for core localization to SGs. How newly synthesized viral mRNA might mediate core particle localization to SGs is unknown. Following the release of core particles into the cytoplasm, they do not further disassemble (56). Viral core particles directly transcribe viral mRNA from within the core, and newly synthesized viral mRNA is released from λ2-formed turrets as mRNA synthesis proceeds (56). We propose that the process of SG formation is triggered as a result of phosphorylation of eIF2α during viral entry. It is possible that during SG assembly, some major component(s) of SGs, such as TIAR, TIA-1 (27), G3BP (51), FMRP (16), or Stau 1 (50), plays a role in sequestering viral mRNA and its associated core particles into SGs. SGs sequester and translationally silence mRNA. Some stress-induced transcripts, such as ATF4, GCN4, and Hsp70, are selectively translated when SGs are present in cells (24), although the molecular features that distinguish between constitutive and stress-induced transcripts are not well understood. For West Nile virus, TIAR and TIA-1 interact with the 3′ stem-loop of the complementary minus-strand RNA and facilitate virus replication. This finding suggests that viruses can either escape SG arrest or take advantage of SGs. Whether MRV translation is inhibited by SGs is undetermined, although, in every case in this study, MRV infection continued following SG formation.
Our data suggest that SGs are diminished as viral proteins accumulate (Fig. 5B and 6A, first panel, and data not shown), suggesting that SGs are disassembled as infection proceeds. It is unlikely that the dissolution of SGs seen in MRV-infected cells relieves the host cell translation shutoff seen following infection with some strains of MRV, because in our experiments, SGs were absent at late times p.i. in cells infected with both viral strains that induce host translational shutoff (T2J) and those that do not induce host cell translational shutoff (T3DC). However, SG disruption may be necessary for synthesis of viral protein in the translational shutoff environment. MRV induction and disruption of SGs and subsequent escape from the translation inhibition that is a consequence of this induction could be the consequence of coevolution between MRV and the host cell.
SGs and MRV factories.
SG formation depends on the microtubule network (23). The deacetylase HDAC6 was proposed to coordinate the formation of SGs by mediating the motor protein-driven movement of SG components along microtubules (30). During MRV infection, the viral core protein, μ2, and the VF structural matrix protein, μNS, are associated with the microtubule network. While the association of μ2 with microtubules plays a role in VF morphology, the association of μNS with microtubules appears to be important for the development of MRV factories from small structures scattered throughout the cytoplasm to larger perinuclear structures (10, 43). MRV core particles are embedded in microtubule-associated VFs (4, 8) and associate with μNS, both in vitro and in vivo (9, 10). The fact that μNS binds to viral cores and uses the microtubule network to facilitate VF formation, coupled with our findings that viral core particles colocalize with microtubule-associated SGs at very early times p.i., suggests that there may be a link between SGs and VF formation. The possibility that MRV VFs may nucleate around core-containing MRV-induced SGs through interactions between μNS, viral cores, and the microtubule network will be interesting to examine in future studies.
Acknowledgments
We thank Paul Anderson and Nancy Kedersha for wild-type MEF cells. We thank Randall Kaufman for MEFS51S/S51S and MEFS51A/S51A cells. We thank David Ron for GCN−/− and PERK−/− cells. We thank Scott Kimball for HRI−/− and PKR−/− cells. We thank Max Nibert for MRV core and μNS polyclonal antisera as well as monoclonal cell lines expressing λ2 antibodies (7F4). We thank Jens Lykke-Andersen for the rabbit antiserum against DCP1a.
This work was supported by NIH NIAID grant K22 AIO65496 to C.L.M. Other assistance to C.L.M. was provided by the Roy J. Carver Charitable Trust; the Office of the Dean, College of Veterinary Medicine; the Office of Biotechnology; and the Office of the Provost, Iowa State University.
Footnotes
Published ahead of print on 26 August 2009.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141-150. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, P., and N. Kedersha. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babiss, L. E., R. B. Luftig, J. A. Weatherbee, R. R. Weihing, U. R. Ray, and B. N. Fields. 1979. Reovirus serotypes 1 and 3 differ in their in vitro association with microtubules. J. Virol. 30:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baer, G. S., and T. S. Dermody. 1997. Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 71:4921-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Both, G. W., S. Lavi, and A. J. Shatkin. 1975. Synthesis of all the gene products of the reovirus genome in vivo and in vitro. Cell 4:173-180. [DOI] [PubMed] [Google Scholar]
- 7.Broering, T. J., M. M. Arnold, C. L. Miller, J. A. Hurt, P. L. Joyce, and M. L. Nibert. 2005. Carboxyl-proximal regions of reovirus nonstructural protein μNS necessary and sufficient for forming factory-like inclusions. J. Virol. 79:6194-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broering, T. J., J. Kim, C. L. Miller, C. D. Piggott, J. B. Dinoso, M. L. Nibert, and J. S. Parker. 2004. Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 78:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broering, T. J., J. S. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandran, K., and M. L. Nibert. 2003. Animal cell invasion by a large nonenveloped virus: reovirus delivers the goods. Trends Microbiol. 11:374-382. [DOI] [PubMed] [Google Scholar]
- 12.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, G., Z. Feng, and B. He. 2005. Herpes simplex virus 1 infection activates the endoplasmic reticulum resident kinase PERK and mediates eIF-2α dephosphorylation by the γ134.5 protein. J. Virol. 79:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleveland, D. R., H. Zarbl, and S. Millward. 1986. Reovirus guanylyltransferase is L2 gene product λ2. J. Virol. 60:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang, Y., N. Kedersha, W. K. Low, D. Romo, M. Gorospe, R. Kaufman, P. Anderson, and J. O. Liu. 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870-32878. [DOI] [PubMed] [Google Scholar]
- 16.Dolzhanskaya, N., G. Merz, and R. B. Denman. 2006. Oxidative stress reveals heterogeneity of FMRP granules in PC12 cell neurites. Brain Res. 1112:56-64. [DOI] [PubMed] [Google Scholar]
- 17.Emara, M. M., and M. A. Brinton. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. USA 104:9041-9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esclatine, A., B. Taddeo, and B. Roizman. 2004. Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-rich RNAs. J. Virol. 78:8582-8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. σ1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuichi, Y., S. Muthukrishnan, J. Tomasz, and A. J. Shatkin. 1976. Mechanism of formation of reovirus mRNA 5′-terminal blocked and methylated sequence, m7GpppGmpC. J. Biol. Chem. 251:5043-5053. [PubMed] [Google Scholar]
- 21.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman, A. G., J. A. Smith, S. Balachandran, O. Perwitasari, S. C. Proll, M. J. Thomas, M. J. Korth, G. N. Barber, L. A. Schiff, and M. G. Katze. 2007. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J. Virol. 81:2221-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov, P. A., E. M. Chudinova, and E. S. Nadezhdina. 2003. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp. Cell Res. 290:227-233. [DOI] [PubMed] [Google Scholar]
- 24.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 25.Kedersha, N., S. Chen, N. Gilks, W. Li, I. J. Miller, J. Stahl, and P. Anderson. 2002. Evidence that ternary complex (eIF2-GTP-tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13:195-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fritzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, W. J., J. H. Kim, and S. K. Jang. 2007. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 26:5020-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimball, S. R., R. L. Horetsky, D. Ron, L. S. Jefferson, and H. P. Harding. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273-C284. [DOI] [PubMed] [Google Scholar]
- 30.Kwon, S., Y. Zhang, and P. Matthias. 2007. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21:3381-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin, D. H., D. Kyner, and G. Acs. 1972. Formation of a mammalian initiation complex with reovirus messenger RNA, methionyl-tRNA F, and ribosomal subunits. Proc. Natl. Acad. Sci. USA 69:1234-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazroui, R., R. Sukarieh, M. E. Bordeleau, R. J. Kaufman, P. Northcote, J. Tanaka, I. Gallouzi, and J. Pelletier. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol. Biol. Cell 17:4212-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McEwen, E., N. Kedersha, B. Song, D. Scheuner, N. Gilks, A. Han, J. J. Chen, P. Anderson, and R. J. Kaufman. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925-16933. [DOI] [PubMed] [Google Scholar]
- 34.McInerney, G. M., N. L. Kedersha, R. J. Kaufman, P. Anderson, and P. Liljestrom. 2005. Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, C. L., M. M. Arnold, T. J. Broering, C. Eichwald, J. Kim, J. B. Dinoso, and M. L. Nibert. 2007. Virus-derived platforms for visualizing protein associations inside cells. Mol. Cell. Proteomics 6:1027-1038. [DOI] [PubMed] [Google Scholar]
- 36.Miller, C. L., T. J. Broering, J. S. Parker, M. M. Arnold, and M. L. Nibert. 2003. Reovirus σNS protein localizes to inclusions through an association requiring the μNS amino terminus. J. Virol. 77:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, C. L., J. S. Parker, J. B. Dinoso, C. D. Piggott, M. J. Perron, and M. L. Nibert. 2004. Increased ubiquitination and other covariant phenotypes attributed to a strain- and temperature-dependent defect of reovirus core protein μ2. J. Virol. 78:10291-10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mokas, S., J. R. Mills, C. Garreau, M. J. Fournier, F. Robert, P. Arya, R. J. Kaufman, J. Pelletier, and R. Mazroui. 2009. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell 20:2673-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montero, H., M. Rojas, C. F. Arias, and S. Lopez. 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J. Virol. 82:1496-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 793-842. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 41.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pain, V. M. 1996. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 236:747-771. [DOI] [PubMed] [Google Scholar]
- 43.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preiss, T., and W. H. M. 2003. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 25:1201-1211. [DOI] [PubMed] [Google Scholar]
- 45.Samuel, C. E., R. Duncan, G. S. Knutson, and J. W. Hershey. 1984. Mechanism of interferon action. Increased phosphorylation of protein synthesis initiation factor eIF-2α in interferon-treated, reovirus-infected mouse L929 fibroblasts in vitro and in vivo. J. Biol. Chem. 259:13451-13457. [PubMed] [Google Scholar]
- 46.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 47.Smith, J. A., S. C. Schmechel, A. Raghavan, M. Abelson, C. Reilly, M. G. Katze, R. J. Kaufman, P. R. Bohjanen, and L. A. Schiff. 2006. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 80:2019-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, J. A., S. C. Schmechel, B. R. Williams, R. H. Silverman, and L. A. Schiff. 2005. Involvement of the interferon-regulated antiviral proteins PKR and RNase L in reovirus-induced shutoff of cellular translation. J. Virol. 79:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas, M. G., L. J. Tosar, M. A. Desbats, C. C. Leishman, and G. L. Boccaccio. 2009. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J. Cell Sci. 122:563-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tourriere, H., K. Chebli, L. Zekri, B. Courselaud, J. M. Blanchard, E. Bertrand, and J. Tazi. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823-831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Ventoso, I., M. A. Sanz, S. Molina, J. J. Berlanga, L. Carrasco, and M. Esteban. 2006. Translational resistance of late alphavirus mRNA to eIF2α phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 20:87-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White, J. P., A. M. Cardenas, W. E. Marissen, and R. E. Lloyd. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295-305. [DOI] [PubMed] [Google Scholar]
- 55.Yue, Z., and A. J. Shatkin. 1997. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 234:364-371. [DOI] [PubMed] [Google Scholar]
- 56.Zarbl, H., K. E. Hastings, and S. Millward. 1980. Reovirus core particles synthesize capped oligonucleotides as a result of abortive transcription. Arch. Biochem. Biophys. 202:348-360. [DOI] [PubMed] [Google Scholar]