Abstract
The alphavirus Semliki Forest virus (SFV) uses a membrane fusion reaction to infect host cells. Fusion of the virus and cell membranes is triggered by low pH in the endosome and is mediated by the viral membrane protein E1. During fusion, E1 inserts into the target membrane, trimerizes, and refolds into a hairpin conformation. Formation of the E1 homotrimer is critical to membrane fusion, but the mechanism of trimerization is not understood. The crystal structure of the postfusion E1 trimer shows that an aspartate residue, D188, is positioned in the central core trimer interface. D188 is conserved in all reported alphavirus E1 sequences. We tested the contribution of this amino acid to trimerization and fusion by replacing D188 with alanine (D188A) or lysine (D188K) in an SFV infectious clone. These mutations were predicted to disrupt specific interactions at this position and/or change their pH dependence. Our results indicated that the D188K mutation blocked SFV fusion and infection. At low pH, D188K E1 inserted into target membranes but was trapped as a target membrane-inserted monomer that did not efficiently form the stable core trimer. In contrast, the D188A mutant was infectious, although trimerization and fusion required a lower pH. While there are extensive contacts between E1 subunits in the homotrimer, the D188K mutant identifies an important “hot spot” for protein-protein interactions within the core trimer.
In an aqueous environment, phospholipid bilayers are stable structures that do not spontaneously fuse. Fusion is inhibited by high energy barriers that oppose both the initial mixing of the outer membrane leaflets and the subsequent merger of the inner leaflets that forms a stable fusion pore joining the two membrane compartments (18). Enveloped viruses have developed efficient strategies to promote fusion between the virus and cellular membranes to deliver the viral genome into the host cell (reviewed in references 19 and 40). Such virus-membrane fusion reactions are mediated by transmembrane fusion proteins in the virus envelope. These proteins rearrange from a metastable prefusion conformation to a stable target membrane-inserted postfusion conformation. The energy released by these conformational changes acts to induce local bending of the membranes, destabilizing the bilayers and lowering the activation energy for fusion (8, 18). The refolding from the prefusion to the postfusion form is initiated by specific triggering mechanisms, such as receptor and/or coreceptor interactions or exposure to low pH in the endocytic pathway.
Virus membrane fusion proteins have been grouped into several classes based on shared structural features (23, 24). The postfusion structures of class I fusion proteins, exemplified by the influenza virus hemagglutinin and human immunodeficiency virus type 1 gp41, contain a central α-helical coiled-coil domain. The class II fusion proteins, exemplified by alphavirus E1 and flavivirus E, contain primarily β-sheet structures. The class III proteins, such as vesicular stomatitis virus (VSV) G protein, herpes simplex virus gB, and baculovirus gp64, contain features of both class I and class II (the structures of viral fusion proteins are reviewed in references 2, 19, 23, and 40). However, while their structural features differ, all virus fusion proteins whose postfusion structures have been defined mediate fusion through a common mechanism. The fusion proteins insert into the target membrane via a hydrophobic fusion peptide or loop and then refold into a final postfusion structure that is a stable trimeric “hairpin” in which the fusion peptide/loop and transmembrane (TM) domains are at the same end of a rod-like trimer. Refolding into this hairpin is critical in mediating membrane fusion, and fusion can be blocked by the inhibition of hairpin formation (11, 19, 20, 40).
The class II fusion proteins of the alphaviruses and flaviviruses have strikingly similar structures (reviewed in references 19, 20, 30, and 36). The E1 protein of alphaviruses and the E protein of flaviviruses are oriented tangentially to the virus membrane and are elongated molecules composed of β-sheets. Each protein contains a central domain I (DI) from which two long extensions form the finger-like domain II (DII) with the fusion loop at its distal tip. A linker region connects DI to domain III (DIII), which has an immunoglobulin-like fold. DIII is followed by the C-terminal stem region and TM domain. A distinguishing feature of these proteins is that their prefusion forms are dimeric and must undergo an oligomeric rearrangement to a trimer to mediate fusion. During this process, the DII fusion loop inserts into the target membrane, the protein trimerizes, and DIII and the stem region fold back toward the fusion loop to generate the trimeric postfusion hairpin (3, 16, 29) (Fig. 1A). Trimer formation and fusion are triggered by virus exposure to low pH during endocytic uptake. The key interactions that must be broken in the prefusion state and those required for formation of the trimer are largely uncharacterized. Recent evidence has identified a conserved DIII histidine residue as a pH sensor during dissociation of the flavivirus E protein homodimer (12).
FIG. 1.
E1 D188 conservation in alphaviruses and location in the E1 homotrimer. (A) Side view of the structure of the postfusion SFV E1 homotrimer (Protein Data Bank accession number 1RER) (16). DI is shown in red, DII in yellow, DIII in blue, the DI-DIII linker in black, the stem in purple, and the fusion loop (FL) at the tip of DII in green. D188 in the gh loop in the core trimer interface is represented in a space-filling model in gray. (B) Comparison of the amino acid sequences of the alphavirus E1 gh loop, residues 179 to 196 (SFV E1 numbering). Residues that are identical to the SFV sequence are represented as dots, and D188 is shown in red. The alphaviruses are SFV, Barmah Forest virus (BFV), eastern equine encephalitis virus (EEEV), Igbo Ora virus (IOV), Middelburg virus (MIDV), Ndumu virus (NDUV), salmon pancreas disease virus (SPDV), sleeping disease virus (SDV), chikungunya virus (CHIKV), o'nyong-nyong virus (ONNV), Ross River virus (RRV), Trocara virus (TROV), Venezuelan equine encephalitis virus (VEEV), Aura virus (AURAV), Sindbis virus (SIN), western equine encephalitis virus (WEEV), HbB17, and M1. (C) View looking into the core trimer interface, oriented with the fusion loops pointing at the viewer. The three E1 monomers are shown in red, blue, and green. D188 is represented in yellow as a stick drawing, with oxygen and nitrogen atoms drawn in red and blue, respectively.
The alphavirus Semliki Forest virus (SFV) has been extensively studied as a membrane fusion system (reviewed in reference 20), and the pre- and postfusion structures of the SFV E1 protein have been determined (16, 24, 33). The prefusion form of E1 is associated with the E2 TM protein on the virus surface. Low pH triggers the dissociation of the E2/E1 dimer. E1 then inserts into the target bilayer via the fusion loop, thus bridging the viral and target membranes via an “extended intermediate.” The membrane-inserted E1 then refolds to form the postfusion hairpin and mediate membrane fusion.
Trimerization of E1 happens quickly, as the entire fusion process is complete within seconds at 37°C (4). Both trimerization and fusion are inhibited by an E1 fusion loop mutation (22, 25) and by the inclusion of zinc during in vitro fusion reactions (9). Addition of exogenous DIII proteins during low-pH treatment specifically blocks alphavirus and flavivirus fusion (26). DIII proteins act by binding and trapping a trimeric E1 intermediate and preventing the foldback of endogenous DIII to form the hairpin. Thus, a variety of evidence demonstrates that formation of the final postfusion E1 trimer is required for alphavirus fusion and infection.
Despite our detailed structural and functional knowledge of the pre- and postfusion states, however, little is known about the dynamic process of E1 trimer formation. Recently, we demonstrated that a truncated form of SFV E1 containing only DI and DII forms trimers with pH dependencies and properties similar to those of full-length E1 (34). This result suggests that important interactions in DI/DII drive the formation of the core trimer. Examination of the SFV homotrimer structure revealed that the aspartic acid residues at position 188 in DII are located 2.5 Å from each other in the trimer interface (Fig. 1A and C). Alphavirus E1 D188 is located in a conserved region of E1 and is itself completely conserved, even in the more distantly related fish alphaviruses (Fig. 1B). During crystallization trials with SFV E1, it was observed that the addition of the heavy atom holmium results in better diffraction (16). The trimer structure revealed that holmium is bound by the three D188 residues in the central trimer interface. These three aspartates lie within hydrogen-bonding distance of each other, although this positioning could be affected by the presence of holmium in the three-dimensional structure (16). We hypothesized that holmium could stabilize the trimer by substituting for interactions that are functionally important in the trimer interface region and that involve D188. Together, the sequence conservation, the location in the central trimer interface, and the stabilizing effect of holmium binding suggest a potential role for D188 in the formation or stabilization of the E1 trimer.
Here, we conducted a mutagenesis study to address the contributions of D188 to trimerization and fusion. A mutation of D188 to alanine shifted the pH dependence of fusion to more acidic pH. Mutation of D188 to lysine blocked virus fusion and infection. The lysine mutant defined a fusion intermediate in which monomeric E1 was inserted into the target bilayer but the “prehairpin” extended trimer was not formed.
(The data in this paper are from a thesis to be submitted by C. Y. Liu in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University, Bronx, NY.)
MATERIALS AND METHODS
Cells.
BHK-21 cells were maintained at 37°C in BHK medium (Dulbecco's modified Eagle's medium containing 5% fetal bovine serum, 10% tryptose phosphate broth, 100 U penicillin/ml, and 100 μg streptomycin/ml).
Construction of SFV E1-D188K and E1-D188A mutant infectious clones.
Mutagenesis was based on the subgenomic DG-1 plasmid as a template, as previously described (5, 27), using Pfu Turbo DNA polymerase (Stratagene, Inc., La Jolla, CA) to introduce D188K and D188A into DG-1. NsiI/SpeI fragments containing the mutations were subcloned into the pSP6-SFV4 infectious clone (wt-ic) (28) to generate D188K-ic and D188A-ic. Two independent clones of each mutant were generated from two separate PCRs and were sequenced to verify the introduction of specific mutations and the absence of other mutations in E1. In vitro transcription was used to generate viral RNAs, which were electroporated into BHK cells for further analysis (28). Initial experiments used both clones of each mutant to confirm the phenotype; subsequent experiments were performed with one clone of each mutant.
Infectivity assays.
Primary and secondary infections were evaluated by mixing electroporated cells with nonelectroporated cells at a 1:20 ratio and plating them on 22-mm-square coverslips for 2 h at 37°C. The medium was then replaced with BHK medium with or without 20 mM NH4Cl, and the cells were incubated overnight at 37°C. The cells were fixed in methanol, stained with a polyclonal antibody to E1/E2, and evaluated by fluorescence microscopy (22).
Growth curves were performed by quantitating virus release from electroporated BHK cells. Alternatively, BHK cells were infected with virus for 1 h at 37°C at a multiplicity of infection (MOI) of 0.05 PFU/cell, washed, and incubated for the indicated times. Progeny virus titers were determined by plaque assay on BHK cells.
Protein expression and virus assembly.
Cell surface expression of viral envelope proteins was characterized by fluorescence microscopy of electroporated cells fixed with paraformaldehyde and stained with monoclonal antibodies (MAbs) to the E1 or E2 protein (21). To evaluate virus assembly, pulse-chase experiments were performed on electroporated cells essentially as previously described (5). After electroporation, the cells were incubated for 6 h at 37°C, labeled with 100 μCi/ml [35S]methionine and [35S]cysteine (Promix; Amersham Life Sciences) for 30 min at 37°C, and chased in the absence of radiolabel. Media and cell lysates were immunoprecipitated with a polyclonal antibody to E1/E2. Medium samples were precipitated in the absence of detergent to retrieve whole virus particles in the media. The samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Fusion assays.
The ability of virus particles to fuse with the cell plasma membrane was evaluated by prebinding wild-type (wt) or mutant virus particles to BHK cells on ice for 90 min. The samples were treated at the indicated pH for 1 min at 37°C and incubated at 28°C overnight in BHK medium containing 20 mM NH4Cl to prevent secondary infection. Infected cells were quantitated by fluorescence microscopy using a polyclonal antibody to E1/E2 (37).
To assess E1's cell-cell fusion activity, BHK cells were electroporated with viral RNA, mixed with unelectroporated BHK cells (1:20), plated on 22-mm-square coverslips at 37°C for 2 h, and cultured at 28°C overnight. The cells were then treated with medium at the indicated pH for 3 min at 37°C and cultured for 3 h at 28°C to allow E1/E2 expression in the fused cells. Expressing cells were stained with a polyclonal antibody to E1/E2, and nuclei were stained with propidium iodide. The cells were evaluated by fluorescence microscopy, and the fusion index was calculated as follows: 1 − (cells/nuclei) (25).
Production of radiolabeled virus.
BHK-21 cells were electroporated with viral RNA, plated at 37°C for 4 h in BHK medium, and labeled with 100 μCi/ml [35S]methionine for 12 h at 37°C. The 12-h labeling was used to avoid selecting for revertants of the lethal D188K mutation. Independent experiments showed that 7 to 10 days of culture of infected BHK cells were required to generate viable revertants of D188K (C. Y. Liu, unpublished results). Labeled virus was harvested and purified on sucrose gradients, all as previously described (5).
Fusion loop exposure.
Exposure of the fusion loop was detected as previously described using MAb E1f, which maps to residues 85 to 95 in the fusion loop (15). In brief, radiolabeled virus was treated at the indicated pH for 5 min at 37°C, adjusted to neutral pH, and immunoprecipitated with MAb E1f, a polyclonal antibody to E1/E2, to obtain total protein or with nonspecific antibody I1 to VSV G protein. The E1f-bound virus particles were washed three times in a detergent-free buffer to remove free antibody and then washed once with detergent-containing buffer to retain only E1 that was directly bound to MAb E1f.
Virus-liposome association.
Radiolabeled virus was mixed with 0.2 mM liposomes (egg phosphatidylcholine, egg phosphatidylethanolamine, sphingomyelin, and cholesterol in a ratio of 1:1:1:1.5) and treated at the indicated pH for 30 s at 37°C. Samples were adjusted to pH 8 and mixed with sucrose to a final concentration of 40% (wt/vol) sucrose, layered over a 60% (wt/vol) sucrose cushion, and overlaid with 25% (wt/vol) sucrose and then 5% (wt/vol) sucrose. The sucrose solutions were in 50 mM Tris, pH 8.0, 100 mM NaCl. The gradients were centrifuged in a TLS-55 rotor for 2 h at 50,000 rpm at 4°C and separated into seven fractions, and the radioactivity was quantitated (5).
E2/E1 dimer dissociation and E1 trimer formation.
Radiolabeled virus was mixed with 0.8 mM liposomes (dioledylphosphatidylcholine, dioleoylphosphatidylethanolamine, sphingomyelin, and cholesterol in a ratio of 1:1:1:1.5), incubated at the indicated pH for 5 min at 20°C (conditions previously shown to produce maximal trimers), and adjusted to pH 7.0. To evaluate E2/E1 heterodimer dissociation, samples were digested with 200 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone-trypsin (Sigma) for 10 min on ice, conditions that selectively digest monomeric E2 (10). The reactions were stopped by the addition of a threefold excess of soybean trypsin inhibitor. Control samples were incubated with premixed trypsin and soybean trypsin inhibitor.
Generation of the SDS-resistant E1 homotrimer was assayed by solubilization of aliquots of the samples with SDS sample buffer at 30°C for 5 min (39). Parallel aliquots were solubilized at 95°C and used to determine the total E1. For each pH, the amount of E1 migrating in the homotrimer position at 30°C was divided by the total E1 in the 95°C lane to determine the fraction of E1 that was SDS resistant. Generation of the trypsin-resistant E1 trimer was monitored by digestion with trypsin as described above but using sample incubation at 37°C for 10 min. Acid conformation-specific epitope exposure was assayed by immunoprecipitation with MAb E1a-1 in the presence of detergent and compared with samples retrieved with polyclonal antibody to E1 and E2 (1, 21). The efficiency of trimer retrieval by MAb E1a-1 was affected by the multiple immunoprecipitation washes. All samples were analyzed by SDS-PAGE and phosphorimaging.
DIII binding.
Formation of the E1 core trimer was tested by its interaction with exogenous DIII protein (26). In brief, radiolabeled virus was prebound to BHK cells on ice and then treated at the indicated pH for 1 min at 37°C in the presence of 2 μM His-DIII with the stem (His-DIIIS). The cells were then lysed; immunoprecipitated with a MAb to the His tag, a polyclonal antibody to E1/E2, or an irrelevant MAb to VSV G; and analyzed by SDS-PAGE and phosphorimaging.
RESULTS
Generation of SFV E1 D188A and D188K mutants.
D188 is completely conserved among the reported sequences of alphavirus E1 proteins (Fig. 1B). Its location in the central trimer interface and the stabilizing effects of its interaction with holmium suggest that D188 may play a role during E1 trimerization. We tested the function of D188 by constructing infectious SFV clones in which D188 was mutated to alanine (D188A) or lysine (D188K). Mutation to alanine tested the potential importance of protonation of D188 during low-pH-triggered fusion and disrupted any specific chemical interactions of D188. Mutation to lysine not only disrupted such interactions, but also replaced aspartate's neutral charge (at low pH) or negative charge with a bulky positive charge. Mutant viral RNAs were transcribed from the infectious clones and electroporated into BHK cells for phenotypic evaluation.
Initial characterization of the D188A mutant.
We first tested the production of infectious D188A virus particles. BHK cells were electroporated with wt or mutant RNA, mixed with nonelectroporated cells, and incubated at 37°C, and primary and secondary infections were evaluated. Cells electroporated with either RNA produced virus that efficiently infected neighboring cells (data not shown). The growth kinetics of the wt and D188A were similar when tested by electroporation of BHK cells with comparable amounts of infectious RNA, followed by plaque assay of the growth media (data not shown). To test for small differences in the growth kinetics, wt and D188A virus stocks were collected at 13 h postelectroporation and used to infect BHK cells at low multiplicity (MOI = 0.05) (Fig. 2). D188A SFV showed growth kinetics similar to those of wt SFV, indicating that the mutation did not significantly disrupt the infection cycle. Immunofluorescence experiments confirmed that comparable levels of mutant and wt E1 and E2 proteins were present at the plasma membranes of BHK cells electroporated with the respective RNAs (data not shown). Pulse-chase experiments showed that mutant virus particles were produced with kinetics and efficiency comparable to those of wt virus particles (data not shown). Thus, the D188A mutation did not significantly affect the SFV life cycle.
FIG. 2.
Growth properties of wt and D188A mutant SFV. BHK cells were infected with wt or D188A SFV at an MOI of 0.05 for 1 h at 37°C. The cells were washed and incubated for the indicated times at 37°C, and the progeny virus in the medium was quantitated by plaque assay. D188A-1 and -2 represent two independent clones of the mutant virus.
D188A membrane fusion activity.
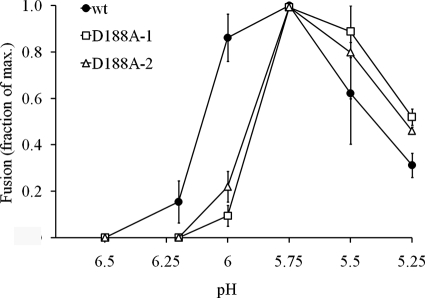
We used the fusion of virus particles with the cell plasma membrane to compare the pH requirements for wt and D188A SFV. Wt or mutant virus particles were prebound to BHK-21 cells on ice and treated at low pH for 1 min at 37°C to trigger virus fusion and infection. The pH threshold for wt SFV fusion activity was ∼6.2, while no fusion was observed at this pH value for D188A (Fig. 3). The pH threshold for D188A SFV was shifted to ∼6.0, a pH at which mutant fusion was only ∼15% of maximal while that of the wt virus was ∼85%. Thus, although D188A SFV could efficiently infect cells, there was a shift in the pH required to trigger virus fusion.
FIG. 3.
Virus-membrane fusion activities of wt and D188A mutant SFV. Wt and D188A virus particles were prebound to BHK cells on ice. Virus fusion with the plasma membrane was then induced by treatment for 1 min at 37°C with medium of the indicated pH. The cells were cultured overnight in medium containing NH4Cl to prevent secondary infection, and the infected cells were quantitated by immunofluorescence. The data are shown as the fraction of maximal fusion and represent the average of two experiments; the bars indicate the range. D188A-1 and -2 represent two independent clones of the mutant virus.
Low-pH-dependent conformational changes in D188A.
Several established assays were used to test the effects of the D188A mutation on the conformational changes observed during virus fusion (reviewed in reference 20). The first step in fusion that is biochemically detectable is the low-pH-dependent dissociation of the E2/E1 heterodimer, a process that results in increased sensitivity of the E2 protein to mild trypsin digestion (10). Radiolabeled wt or D188A mutant viruses were resistant to trypsin digestion after incubation in buffer at pH 7.0 (Fig. 4A). Incubation at pH 6.2 or below allowed comparable trypsin digestion of wt and mutant E2 proteins. Thus, the dissociation of the E2/E1 heterodimer was similar in wt and D188A SFV, suggesting that the decreased fusion of D188A at pH 6.2 and 6.0 was due to effects following dimer dissociation.
FIG. 4.
Low-pH-induced conformational changes in wt and D188A mutant SFV. Radiolabeled wt or D188A mutant virus particles were mixed with liposomes, treated at the indicated pH for 5 min at 20°C, adjusted to neutral pH, and analyzed for conformational changes as follows and as described in Materials and Methods. (A) Dissociation of the E2/E1 dimer. Samples were digested with trypsin at 0°C and analyzed by SDS-PAGE. The trypsin-resistant E2 at each pH point was compared to the total E2 in control samples. (B) Trypsin-resistant E1 trimer formation. Samples were digested with trypsin at 37°C and analyzed by SDS-PAGE, and the trypsin-resistant E1 homotrimer was quantitated compared to control samples. (C) SDS-resistant E1 trimer formation. Samples were solubilized in sample buffer at 30°C and analyzed by SDS-PAGE, and SDS-resistant E1 was quantitated as described in Materials and Methods. All data represent the averages of two experiments; the bars indicate the range.
We next compared E1 homotrimer formation in wt and D188A mutant SFV. The postfusion SFV E1 homotrimer is resistant to trypsin digestion and to dissociation by SDS sample buffer at 30°C. We used these methods to quantitate E1 trimers following low-pH treatment of radiolabeled wt and D188A mutant virus. SDS- and trypsin-resistant E1 trimers formed efficiently after treatment of wt SFV at pH 6.2 or below (Fig. 4B and C). In contrast, although the D188A mutant generated E1 trimers, treatment at pH 6.0 or below was required, and trimerization appeared to be less efficient than that of wt SFV. The difference in D188A trimer efficiency observed between samples tested by trypsin resistance (Fig. 4B) and SDS resistance (Fig. 4C) suggested a decrease in trimer stability to SDS, as previously observed for other E1 mutants (7). The more acidic fusion threshold of D188A SFV thus correlates with the more acidic pH required to generate the E1 homotrimer. The overall reduction in trimerization suggests that D188A trimers form less efficiently and/or have reduced stability.
Initial characterization of the D188K mutant.
BHK cells were electroporated with wt or mutant RNA, mixed with nonelectroporated cells, and incubated at 37°C in medium in the presence or absence of NH4Cl to block secondary infection. In the presence of NH4Cl, similar numbers of primary infected cells were observed for wt SFV and the D188K mutant (Fig. 5). In the absence of NH4Cl, the wt virus spread to infect essentially every cell while D188K showed no signs of secondary infection. The growth media were also collected at 14 h postelectroporation, and the titer on BHK cells was determined. The wt-infected cells produced titers of 107 to 108 PFU/ml, while the D188K-infected cells produced no plaques (data not shown).
FIG. 5.
Secondary infection by wt and D188K mutant SFV. BHK cells were electroporated with wt SFV or D188K mutant RNA. The cells were then mixed with nonelectroporated cells and cultured overnight at 37°C in the presence or absence of NH4Cl to detect primary versus secondary infection, respectively. Infected cells were visualized by immunofluorescence microscopy using a polyclonal antibody to E1 and E2. Shown is a representative example of three experiments.
The lack of secondary infection by D188K virus could be due to defects in virus protein biogenesis or in particle assembly. Indirect immunofluorescence showed comparable expression of the E1 and E2 proteins on the surfaces of wt- and D188K virus-infected cells, suggesting correct folding and transport through the secretory pathway (data not shown). Pulse-chase experiments were used to quantitate the release of virus particles into the chase medium (5, 10). The efficiency of virus production and the electrophoretic mobility and stoichiometry of the viral proteins in virus particles were comparable for wt SFV and the D188K mutant (Fig. 6, 3 h, lanes M). Electron microscopy studies showed that the morphology of budding virus particles was also similar in wt and mutant viruses (data not shown). Together, these results indicate that the D188K mutant-infected cells produce virus particles but that these mutant particles are unable to mediate infection of host cells.
FIG. 6.
Assembly of wt and D188K mutant SFV. BHK cells were electroporated with wt SFV or D188K mutant RNA, incubated for 6 h at 37°C, pulse labeled with [35S]methionine and [35S]cysteine, and chased at 37°C in the absence of radiolabel for 0 or 3 h. Cell lysates (C) and media (M) were immunoprecipitated with a polyclonal antibody to E1/E2. Medium samples were immunoprecipitated in the absence of detergent to permit retrieval of intact virus particles. Samples were analyzed by SDS-PAGE and fluorography. The positions of the viral proteins p62, E2, E1, and capsid are indicated on the left. Shown is a representative example of two experiments.
D188K mutant membrane fusion activity.
We directly tested the membrane fusion activity of the D188K mutant E1 protein by assaying its ability to mediate cell-cell fusion. BHK cells were electroporated with wt or mutant RNA and cultured overnight at 28°C to produce abundant expression of the E1 and E2 proteins at the cell surface. The cells were then treated briefly with buffers at pH values ranging from 4 to 10.5 to induce fusion between cells, and the number of nuclei in the E1/E2-expressing cells was determined (Fig. 7). At all pH points, virtually no cell-cell fusion was seen above background for the D188K mutant, whereas efficient fusion was seen for wt virus-infected cells at pH 6.2 and below. Thus, the D188K mutant is blocked at the fusion step of the viral entry pathway.
FIG. 7.
Cell-cell fusion activities of wt and D188K mutant SFV. BHK cells were electroporated with wt or D188K mutant RNA, mixed with nonelectroporated cells, and incubated overnight at 28°C to permit abundant E1 and E2 expression at the cell surface. The cells were treated for 3 min at 37°C to trigger cell-cell fusion. Polykaryon formation was quantitated and expressed as a fusion index, as detailed in Materials and Methods. Shown are the averages of two or three experiments, with the bars indicating the range.
Initial response of the D188K mutant to low pH.
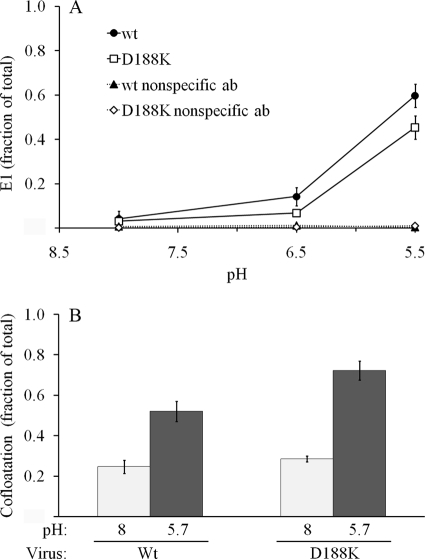
We used purified radiolabeled wt SFV and D188K mutant virus to assay the initial conformational changes induced upon exposure of virus to low pH. Dissociation of the E2/E1 dimer and exposure of the E1 fusion loop were monitored using monoclonal antibody E1f, which maps to residues 85 to 95 of the fusion loop (15, 17). The pH dependence and efficiency of fusion loop exposure were similar for wt SFV and the D188K mutant (Fig. 8A). Thus, both the initial E2/E1 heterodimer interaction that masks the fusion loop and its low-pH-induced dissociation were comparable in the two viruses.
FIG. 8.
Fusion loop exposure and E1-membrane interaction of wt and D188K mutant SFV. (A) Radiolabeled viruses were treated at the indicated pH for 5 min at 37°C in the absence of target membranes. Samples were then adjusted to neutral pH, immunoprecipitated with MAb E1f against the fusion loop or with an irrelevant MAb (nonspecific ab), and quantitated by SDS-PAGE and phosphorimaging. (B) Radiolabeled viruses were mixed with liposomes, incubated at the indicated pH for 5 min at 37°C, and adjusted to neutral pH. Liposome-bound virus was separated by sucrose gradient flotation. Shown are the averages of two experiments; the bars indicate the range.
During fusion, the dissociation of the heterodimer is followed by the insertion of the E1 fusion loop into the target membrane. We assayed this step by following the low-pH-induced association of radiolabeled virus with target liposomes (Fig. 8B). Both wt SFV and the D188K mutant comigrated with liposomes in a sucrose flotation gradient after treatment at pH 5.7. Similar low-pH dependence and efficiency of membrane interaction were observed for the two viruses.
D188K E1 homotrimer formation.
The data to this point indicated that the block in D188K fusion and infection was at a step following the insertion of the E1 fusion loop into the target membrane. We therefore tested the abilities of radiolabeled wt SFV and the D188K mutant to form the target membrane-inserted E1 homotrimer (Fig. 9A). The assays were based on the resistance of this postfusion E1 conformation to dissociation by SDS and to trypsin digestion (as in Fig. 4B and C) and on its recognition by the acid conformation-specific MAb E1a-1 (20). In each case, the results demonstrated efficient low-pH-induced conversion of the wt E1, while essentially no D188K virus E1 homotrimers were detectable using any of these methods (Fig. 9A).
FIG. 9.
E1 trimer formation in wt and D188K mutant SFV. (A) Radiolabeled viruses were mixed with liposomes and treated at the indicated pH for 5 min at 20°C. Aliquots of the samples were analyzed for SDS-resistant-trimer formation, trypsin-resistant-trimer formation, or reactivity with the acid conformation-specific MAb E1a-1, as detailed in Fig. 4 and Materials and Methods. Shown are the averages of two experiments; with the bars representing the range. Recovery of the trimer by immunoprecipitation with MAb E1a-1 is less efficient than its detection by SDS or trypsin resistance. (B) Interaction of viral E1 with exogenous DIII protein. Radiolabeled wt or D188K mutant SFV was prebound to BHK cells on ice and treated for 1 min at 37°C with buffer at the indicated pH and containing 2 μM His-DIIIS protein. The cells were lysed, and aliquots of the samples were immunoprecipitated with a polyclonal antibody to E1/E2 to recover total E1, an antibody to the His tag (His), and an irrelevant MAb (ns). The data shown are the averages and standard deviations of three independent experiments.
Since D188K E1 interacted with target membranes but did not refold to the final stable E1 homotrimer, we asked whether the mutant protein could form the “prehairpin,” or extended, trimer (19, 20). This is a trimeric intermediate in which the fusion loop has inserted into the target membrane and a core trimer has formed but DIII has not folded back to form the final hairpin structure. We showed in earlier studies that exogenously added DIII proteins can inhibit membrane fusion by stably binding to such a trimeric intermediate (26) and that a truncated form of E1 consisting of DI and DII can form a membrane-inserted core trimer that efficiently binds DIII (34). We therefore used the binding of exogenous DIII to test whether the D188 residue is important in the formation of the initial extended trimer versus the refolding of this trimer into the final SDS- and trypsin-resistant hairpin. Purified radiolabeled wt and D188K viruses were bound to BHK cells on ice and treated at pH 7 or 5.7 in the presence of exogenous DIII proteins (Fig. 9B). In agreement with our previous results (26), wt E1 protein was efficiently bound by DIII during low-pH treatment. In contrast, DIII binding to the D188K E1 protein was low after treatment at either neutral or low pH. Thus, E1 D188 is an important site in the initial formation of the stable core trimer. Although D188K E1 inserts into the target membrane, the mutation inhibits formation of the extended E1 trimer, leading to a block in virus fusion and infection.
DISCUSSION
Effects of mutations of SFV E1 D188.
The alphavirus fusion reaction is initiated by the low-pH-dependent dissociation of the E2/E1 heterodimer and the exposure of the E1 fusion loop (15, 20, 38). E1 then inserts into the target membrane and trimerizes. The available data suggested that the initial step in trimerization was the formation of a “prehairpin” extended trimer of E1, followed by foldback of DIII and membrane fusion. The existence of such an extended trimer is supported by the ability of exogenous DIII to trap a trimer intermediate, suggesting the presence of E1 trimers with “empty” DIII binding sites (26). In addition, truncated E1 DI/DII proteins can form a stable membrane-inserted core trimer at low pH in the absence of DIII (34). The mechanism of trimerization and the critical interactions within such extended trimers were unknown. We hypothesized that E1 D188 might play a role, based on its sequence conservation and location within the trimer interface region on DII and the stabilizing effect of the D188-holmium interaction on trimer crystals. Here, our studies of the D188A and D188K mutants revealed an important site of interactions in the central trimer and highlighted the role of a monomeric prehairpin E1 intermediate in the SFV fusion reaction.
The D188A mutant was able to grow as efficiently as wt SFV, although the mutant's fusion activity required a somewhat lower pH. This fusion effect correlated with a shift in the pH threshold for E1 trimerization, while E2/E1 dimer dissociation was unchanged. These results suggest that the possible role of D188 is preserved, although not completely, when alanine is substituted. Because of the size difference between the side chains of these two amino acids, it is possible that a polar water molecule may enter the trimer interface of D188A and maintain the central trimer interactions. Alternatively, it is interesting that although aspartic acid carries a negative charge at neutral pH, if it becomes protonated during fusion at low pH it will be uncharged. As the alanine side chain is also uncharged, this feature could explain why the alanine mutation does not significantly affect SFV fusion and infection.
The D188K mutant was blocked in fusion and infection and was negative in all of our assays of E1 trimerization. Dimer dissociation and E1 fusion loop membrane insertion were unaltered. Our previous studies showed that exogenous DIII proteins bind only to the groove between two E1 subunits in the core trimer and not to E1 monomers (26, 34). Here, we demonstrated that DIII did not efficiently interact with E1 from the D188K mutant, suggesting that the D188K block occurred early in trimerization and prevented the formation of the initial extended trimers. Thus, the presence of the positively charged lysine side chain destabilized the trimer interface to such an extent that other interactions were not able to mediate the formation of a stable core trimer.
The role of aspartate residues in other virus fusion reactions.
The flavivirus E protein is structurally similar to alphavirus E1 in both its pre- and postfusion conformations. An analogous aspartic acid is located in the central trimer interface of the E proteins from the flaviviruses dengue virus and tick-borne encephalitis (TBE) virus (3, 29). This aspartate (D193 in the TBE virus E protein numbering) is oriented similarly to D188 of SFV and is strongly, although not completely, conserved among the flavivirus E proteins. The distance between the three D193 residues in the flavivirus trimer structures is 7.1 Å to 8.5 Å, significantly farther apart than D188 in the SFV trimer interface (where the orientation of the D188 side chain may be affected by the presence of holmium). Interestingly, in the TBE virus trimer, D193 on one E protein appears to interact with the main chain of the adjacent E protein, perhaps contributing to the stability of the core trimer (3). While the overall mechanisms of fusion protein refolding are similar in the alphaviruses and flaviviruses, further studies will clearly be required to compare critical interactions during trimerization.
The class III fusion protein VSV G reversibly transitions between pre- and postfusion trimeric states (13). The protonation and neutralization of a key aspartate residue, D268, located in the trimeric interface of the postfusion trimer, helps to trigger this conformational switch (31, 32). D268 appears to be protonated in the postfusion trimer even though the structure was solved at pH 7, suggesting that the pKa of D268 is higher than in an exposed aqueous environment (31). These studies thus illustrate important aspects of the acidic side chain and its modulation by the local environment.
Mechanism of the D188 mutations.
The alphavirus E1 postfusion trimer is considerably more stable than the prefusion monomer (14) and is resistant to dissociation by SDS treatment (39). This trimer stability is mediated by multiple contacts both in the central trimer interface and in the interaction of DIII and the stem with the core trimer (16). We used a software program (Ligand-Protein Contacts [35]) to analyze the central trimer interface (data not shown). This analysis indicated that the majority of interface contacts are hydrophilic interactions between side chains and the main chain backbone. Given the extent of these contacts, it is rather surprising that changing aspartic acid to lysine caused such a drastic fusion block, suggesting that this region is a “hot spot” for critical protein-protein interactions within the central trimer interface. The loss of stabilizing interactions, the addition of repulsive forces, or a combination of these features presumably explains why D188K is unable to trimerize. We note that a previous point mutation in the E1 fusion loop, glycine 91 to aspartate (G91D), was also found to block fusion by blocking trimer formation (22, 25). Similarly to the D188K mutant, the G91D mutant binds to target membranes but does not form a biochemically detectable homotrimer. As no interchain interactions are observed in this region of the E1 trimer, G91D presumably blocks trimerization at a step distinct from D188K. Other evidence suggests that the DII region containing D188 is important in trimerization and fusion. This region is the location of several mutations that affect SFV cholesterol dependence and trimer stability (7) and is also the site of a series of mutations that rescue the lethal fusion block mutant E1 H230A (5, 6). Together, these results support an important role for this region of E1 in trimerization and fusion.
How might D188 be involved in the formation of the core trimer? The presence of three negatively charged aspartic acid side chains would presumably cause repulsion in the trimer center, resulting in a block in trimerization similar to that caused by the D188K mutation. Thus, we hypothesize that during trimer formation at low pH, D188 must be largely neutralized, eliminating this repulsive force. In fact, holmium has a charge of +3, and thus, holmium binding would help to neutralize the three D188 residues in the trimer interface, which could explain its observed effect on trimer stability. What could be the physiological means of neutralizing D188? The pKa of aspartic acid in free solution is ∼3 to 4, while E1 trimerization occurs efficiently at a pH of ∼6. However, the local environment of D188 could increase its pKa so that it is protonated and neutralized during fusion at mildly acidic pH. Alternatively, the negative charge of D188 could be neutralized by the formation of a hydrogen bond or salt bridge with another residue in the trimer interface. Analyses of D188K revertants and further studies of the trimer interface interactions will help to clarify the role of D188 in alphavirus fusion.
Acknowledgments
We thank all of the members of our laboratory for helpful discussions and comments on the manuscript. We also thank Kartik Chandran and the members of his laboratory for their helpful input. Gwen Taylor from our laboratory and the staff of the Einstein Analytical Imaging Facility made important contributions to the electron microscopy analysis. We thank Anuja Ogirala and Sonu Nanda for excellent technical assistance.
This work was supported by a grant to M.K. from the National Institute of Allergy And Infectious Diseases (R01-AI075647) and by Cancer Center Core Support Grant NIH/NCI P30-CA013330. C.Y.L. was supported in part through the Medical Scientist Training Program of the Albert Einstein College of Medicine (NIH T32 GM07288).
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print on 19 August 2009.
REFERENCES
- 1.Ahn, A., M. R. Klimjack, P. K. Chatterjee, and M. Kielian. 1999. An epitope of the Semliki Forest virus fusion protein exposed during virus-membrane fusion. J. Virol. 73:10029-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backovic, M., and T. S. Jardetzky. 2009. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bron, R., J. M. Wahlberg, H. Garoff, and J. Wilschut. 1993. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 12:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanel-Vos, C., and M. Kielian. 2004. A conserved histidine in the ij loop of the Semliki Forest virus E1 protein plays an important role in membrane fusion. J. Virol. 78:13543-13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanel-Vos, C., and M. Kielian. 2006. Second-site revertants of a Semliki Forest virus fusion-block mutation reveal the dynamics of a class II membrane fusion protein. J. Virol. 80:6115-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, P. K., C. H. Eng, and M. Kielian. 2002. Novel mutations that control the sphingolipid and cholesterol dependence of the Semliki Forest virus fusion protein. J. Virol. 76:12712-12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernomordik, L. V., and M. M. Kozlov. 2008. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 15:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corver, J., R. Bron, H. Snippe, C. Kraaijeveld, and J. Wilschut. 1997. Membrane fusion activity of Semliki forest virus in a liposomal model system: specific inhibition by Zn2+ ions. Virology 238:14-21. [DOI] [PubMed] [Google Scholar]
- 10.Duffus, W. A., P. Levy-Mintz, M. R. Klimjack, and M. Kielian. 1995. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J. Virol. 69:2471-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 12.Fritz, R., K. Stiasny, and F. X. Heinz. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudin, Y. 2000. Reversibility in fusion protein conformational changes. The intriguing case of rhabdovirus-induced membrane fusion. Subcell. Biochem. 34:379-408. [DOI] [PubMed] [Google Scholar]
- 14.Gibbons, D. L., A. Ahn, P. K. Chatterjee, and M. Kielian. 2000. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J. Virol. 74:7772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons, D. L., A. Ahn, M. Liao, L. Hammar, R. H. Cheng, and M. Kielian. 2004. Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J. Virol. 78:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbons, D. L., M.-C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320-325. [DOI] [PubMed] [Google Scholar]
- 17.Hammar, L., S. Markarian, L. Haag, H. Lankinen, A. Salmi, and H. R. Cheng. 2003. Prefusion rearrangements resulting in fusion peptide exposure in Semliki forest virus. J. Biol. Chem. 278:7189-7198. [DOI] [PubMed] [Google Scholar]
- 18.Harrison, S. C. 2005. Mechanism of membrane fusion by viral envelope proteins. Adv. Virus Res. 64:231-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison, S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian, M. 2006. Class II virus membrane fusion proteins. Virology 344:38-47. [DOI] [PubMed] [Google Scholar]
- 21.Kielian, M., S. Jungerwirth, K. U. Sayad, and S. DeCandido. 1990. Biosynthesis, maturation, and acid-activation of the Semliki Forest virus fusion protein. J. Virol. 64:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kielian, M., and F. A. Rey. 2006. Virus membrane fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 25.Levy-Mintz, P., and M. Kielian. 1991. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J. Virol. 65:4292-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao, M., and M. Kielian. 2005. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus-membrane fusion. J. Cell Biol. 171:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao, M., and M. Kielian. 2006. Functions of the stem region of the Semliki Forest virus fusion protein during virus fusion and assembly. J. Virol. 80:11362-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13-22. [DOI] [PubMed] [Google Scholar]
- 31.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 32.Roche, S., F. A. Rey, Y. Gaudin, and S. Bressanelli. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843-848. [DOI] [PubMed] [Google Scholar]
- 33.Roussel, A., J. Lescar, M.-C. Vaney, G. Wengler, G. Wengler, and F. A. Rey. 2006. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14:75-86. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez San Martin, C., H. Sosa, and M. Kielian. 2008. A stable prefusion intermediate of the alphavirus fusion protein reveals critical features of class II membrane fusion. Cell Host Microbe 4:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sobolev, V., A. Sorokine, J. Prilusky, E. E. Abola, and M. Edelman. 1999. Automated analysis of interatomic contacts in proteins. Bioinformatics 15:327-332. [DOI] [PubMed] [Google Scholar]
- 36.Stiasny, K., and F. X. Heinz. 2006. Flavivirus membrane fusion. J. Gen. Virol. 87:2755-2766. [DOI] [PubMed] [Google Scholar]
- 37.Vashishtha, M., T. Phalen, M. T. Marquardt, J. S. Ryu, A. C. Ng, and M. Kielian. 1998. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J. Cell Biol. 140:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahlberg, J. M., W. A. M. Boere, and H. Garoff. 1989. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 63:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahlberg, J. M., and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus. I: Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J. Cell Biol. 116:339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]