Abstract
Porcine reproductive and respiratory syndrome (PRRS) virus (PRRSV), a positive-strand RNA virus that belongs to the Arteriviridae family of Nidovirales, has been identified as the causative agent of PRRS. Nsp1α is the amino (N)-terminal protein in a polyprotein encoded by the PRRSV genome and is reported to be crucial for subgenomic mRNA synthesis, presumably by serving as a transcription factor. Before functioning in transcription, nsp1α proteolytically releases itself from nsp1β. However, the structural basis for the self-releasing and biological functions of nsp1α remains elusive. Here we report the crystal structure of nsp1α of PRRSV (strain XH-GD) in its naturally self-processed form. Nsp1α contains a ZF domain (which may be required for its biological function), a papain-like cysteine protease (PCP) domain with a zinc ion unexpectedly bound at the active site (which is essential for proteolytic self-release of nsp1α), and a carboxyl-terminal extension (which occupies the substrate binding site of the PCP domain). Furthermore, we determined the exact location of the nsp1α self-processing site at Cys-Ala-Met180↓Ala-Asp-Val by use of crystallographic data and N-terminal amino acid sequencing. The crystal structure also suggested an in cis self-processing mechanism for nsp1α. Furthermore, nsp1α appears to have a dimeric architecture both in solution and as a crystal, with a hydrophilic groove on the molecular surface that may be related to nsp1α's biological function. Compared with existing structure and function data, our results suggest that PRRSV nsp1α functions differently from other reported viral leader proteases, such as that of foot-and-mouth disease.
Porcine reproductive and respiratory syndrome virus (PRRSV) is the pathogenic agent of the swine disease bearing the corresponding name (PRRS), also known as blue-ear pig disease. This economically devastating, pandemic disease causes reproductive failure in breeding stock and respiratory tract illness in young pigs. Initially referred to as a “mystery swine disease” and a “mystery reproductive syndrome,” it was first reported in 1987 in North America (VR-2332 strain) (1) and central Europe (Lelystad Virus/LV strain) (26). Although the European and North American PRRSV strains cause similar clinical symptoms, they represent two distinct major viral genotypes, with their genomes diverging by approximately 40% (30). The simultaneous emergence of two very different genotypes on different continents created a veil of mystery around the origin of the virus (15, 24) and presents a challenge to developing vaccines against PRRSV. Perhaps because PRRS does not appear to pose an immediate, direct threat to human health (22), it has not received sufficient attention worldwide. Nevertheless, since it was first reported in China in 2006, it has affected 22 of the 33 Chinese provinces and has had far-reaching effects on the Chinese swine industry and related economy. The PRRS disease similarly poses a real threat to the worldwide swine industry.
PRRSV, a small enveloped RNA virus, is a positive-strand RNA virus and belongs to the genus Arterivirus, family Arteriviridae, and order Nidovirales. Three other members of the Arterivirus genus have been reported, namely Equine arteritis virus (EAV), Simian hemorrhagic fever virus (SHFV), and Lactate dehydrogenase-elevating virus (LDV). The single-stranded, positive-sense RNA genome of PRRSV is about 15 kb in size and encodes nine open reading frames (ORFs), including two large ones, ORF1a and ORF1b. While the smaller ORFs encode structural proteins, the two large ORFs encode two multidomain replicase polyproteins, pp1a and pp1b (37). Both the pp1a and pp1b polyproteins are processed extensively by ORF1a-encoded viral proteases, yielding a number of nonstructural proteins (nsps) (Fig. 1A). In particular, nsp1α, nsp1β, and nsp2 release themselves from pp1a first, and then nsp4 (a 3C like protease) cleaves each of the remaining nsp's from the polyproteins, including polymerase (RdRp/nsp9), helicase (Hel/nsp10), and Xenopus laevis homolog poly(U)-specific endoribonuclease (N/nsp11) (5, 10, 11, 37). These resulting mature nsps form a replication/transcription complex essential for viral RNA synthesis (6, 27).
FIG. 1.
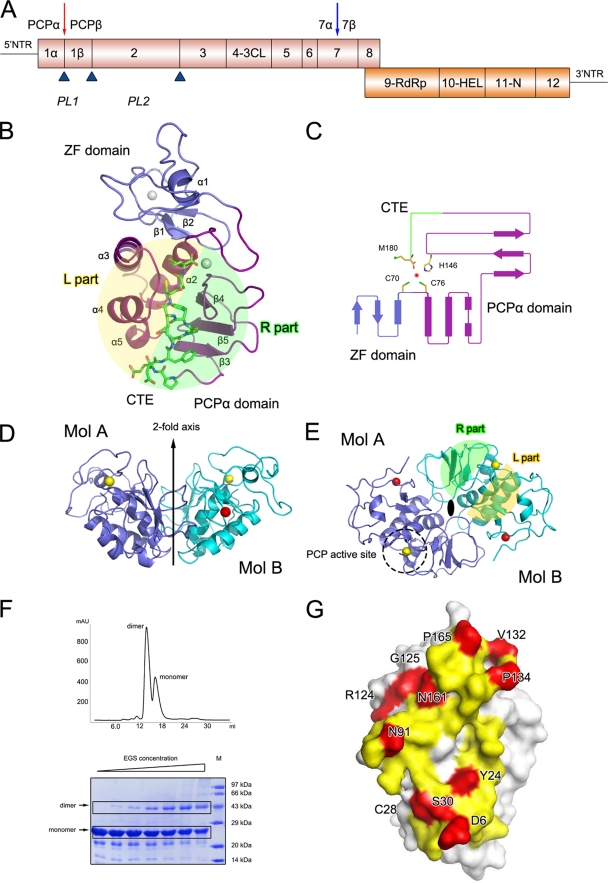
Crystal structure of PRRSV nsp1α. (A) Diagrammatic representation of the PRRSV genome. ORF1a and ORF1b are shown as magenta and orange bars. Putative nsp's are labeled. The predicted or reported PCPα, PCPβ, and PCP2 cleavage sites are represented as upward-pointing blue triangles. The Nsp1α cleavage site is highlighted by a red downward arrow, and the potential subdivision of PRRSV nsp7 suggested by the study of EAV nsp7 (34) is marked by a blue downward arrow. The named or putative signature motifs of nsp's are labeled at their positions in the PRRSV genome, including PCP (PL1), cysteine protease (PL2), serine/3C protease (3CL), polymerase (RdRp), helicase (Hel), and Xenopus laevis homolog poly(U)-specific endoribonuclease (N). (B) The monomer fold. The N-terminal ZF domain, C-terminal PCPα domain, and CTE are represented as slate and purple ribbons, and green sticks, respectively. (C) Topology diagram. The color scheme is the same as that used in panel A. The α-helices and β strands are shown as bars and arrows, respectively. The residues that correspond to PCPα activity are shown as sticks. (D and E) Side and top views of the nsp1α homodimer. The two monomers are colored in slate and cyan, respectively. The zinc atoms in the ZF domains and PCP domains are shown as red and golden spheres, respectively. The twofold axis, PCP active site, and L/R parts are marked or highlighted. (F) Gel filtration and cross-linking results for nsp1α. In the gel filtration of nsp1α (top), 0.5-ml nsp1α solution at a concentration of 2 mg/ml was loaded on a Superdex 200 column (GE Healthcare) and eluted with a buffer containing 50 mM MES (pH 6.0), 1 M NaCl, and 50 mM DTT. The EGS cross-linking result is also shown (bottom). The EGS concentration was used at 0 mM, 0.315 mM, 0.625 mM, 1.25 mM, 2.5 mM, 5.0 mM, and 10.0 mM, shown from left to right. Recombinant nsp1α was diluted to 5 mg/ml in phosphate buffered saline (pH 7.0), 1 M NaCl, and 1 mM DTT. The sizes of the molecular mass markers (in thousands) are indicated on the right. (G) Dimer interface of an nsp1α subunit. Residues that contribute to the dimer interaction are colored yellow for parts that are buried (solvent inaccessible) and red for parts that are involved in close contacts (<3.6 Å).
Located at the N terminus of pp1a, PRRSV nsp1 is processed into two multifunctional proteins, nsp1α and nsp1β, each of which contains a papain-like cysteine protease (PCP) domain essential for self-release from the polyprotein. In particular, nsp1α contains an amino-terminal zinc finger (ZF) and the PCPα protease domain, while nsp1β contains PCPβ (3, 25, 32, 33). Nsp1β is responsible for the cleavage between nsp1β and the downstream protein nsp2 at the cleavage site Trp-Tyr-Gly203↓Ala-Gly-Lys (numbered according to the nsp1β amino acid sequence), while the exact cleavage site of nsp1α has not yet been identified (16). In principle, such a self-processing event may occur either intra- or intermolecularly. A previous study of the crystal structure of the foot-and-mouth disease virus (FMDV) leader protease (Lpro), which is a PCP Lpro, suggests that in cis self-processing may be favored (8, 9). However, such a process remains speculative for PRRSV nsp1. Although nsp1α of PRRSV shares low amino acid sequence identity with papain (≤15%), the characteristic residues of papain-like proteases are generally conserved in the primary sequences of nsp1α proteins of all reported arteriviruses, except EAV. Moreover, it has been shown that loss of PCPα activity totally abolishes the synthesis of subgenomic (sg) mRNA in PRRSV but has no effect on its genome replication, suggesting a role for nsp1α in transcription (31, 33). Furthermore, the homologue of PRRSV nsp1 in EAV was found to bind with p100, a transcription coactivator and thus affect sg mRNA synthesis (32).
To understand the PRRSV Lpro processing mechanisms and to identify potential drug targets against the PRRS disease, we carried out structure-function studies of the nsp1α protein. We report here the three-dimensional structure of nsp1α of PRRSV (strain XH-GD; GenBank accession no. EU624117) and discuss its relationship with the papain superfamily. Moreover, we experimentally determined the exact self-processing site between nsp1α and -β. The crystallographic and biological data indicated a dimeric architecture for nsp1α, with a hydrophilic groove on the molecular surface that could be related to the biological function of nsp1α. In light of these available data, we also propose a probable mechanism of self-processing and substrate recognition for nsp1α.
MATERIALS AND METHODS
Protein expression, purification, and characterization.
The primers 5′-CCG GAA TTC ATG TCT GGG ATA CTT GAT CG-3′ and 5′-CCG CTC GAG TTA ACC GTA CCA CTT ATG ACT GC-3′ were used to amplify the full-length nsp1 gene from the cDNA of the PRRSV XH-GD strain (EU624117). The primers included EcoRI and XhoI restriction sites (shown in bold). The nsp1 gene was amplified using PCR and then cloned into the pET-28a vector (Invitrogen), and the sequence of the insert was verified by DNA sequencing. The recombinant plasmid was transformed into Escherichia coli strain BL21(DE3). Transformed cells were then cultured at 37°C in LB medium containing 50 μg/ml kanamycin. When the culture density reached an A600 of 0.6, induction with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was performed, and cell growth continued for an additional 12 to 14 h at 16°C. Cells were harvested by centrifugation, then resuspended in lysis buffer (20 mM 2-(N-morpholino)ethanesulfonic acid [MES; pH 6.5], 500 mM NaCl, 10% [vol/vol] glycerol, and 50 mM imidazole), and the cell lysing was facilitated by sonication. The lysate was centrifuged at 20,000 × g for 30 min to remove cell debris. The supernatant was applied to a Ni2+ chelating column (1 ml Ni2+-NTA agarose), and nonspecifically bound proteins were washed off with the lysis buffer. Presumably, self-processing occurred before the purification. The N-terminal His-tagged self-processed nsp1α protein was eluted with an eluting buffer (20 mM MES [pH 6.5], 500 mM NaCl, 10% [vol/vol] glycerol, and 200 mM imidazole). The purity of the eluted protein was estimated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis to be greater than 95% and was transferred into a storage buffer of 50 mM MES (pH 6.0), 1 M NaCl, and 50 mM dithiothreitol (DTT).
To determine the native cleavage site between nsp1α and nsp1β, we designed a construct of full-length nsp1 with both N- and C-terminal His tags in a pET-28a vector. The new construct contained a thrombin cleavage site between the N-terminal His tag and nsp1. After loading the samples to a Ni2+ chelating column (1 ml Ni2+-NTA agarose) and washing them with 20 ml MCAC buffer (20 mM MES, pH 6.5; 500 mM NaCl; and 10% [vol/vol] glycerol) containing 50 mM imidazole, thrombin was added to the column and incubated 12 h at 4°C to remove the autocleft nsp1α while holding the nsp1β protein on the column through the C-terminal His tag. Both nsp1α and nsp1β were collected for analysis.
Crystallization.
Crystallization of nsp1α was performed at 18°C using the hanging-drop vapor-diffusion method. A series of crystallization grids was prepared by mixing ∼15 mg/ml nsp1α in the storage buffer with an equal volume of the reservoir solution containing 100 mM Tris-HCl (pH 8.5) and 25% (vol/vol) tert-butanol. Single crystals with a size of 50 by 50 by 100 μm and suitable for data collection appeared in 15 days and were soaked in a cryo-protectant solution consisting of the reservoir solution and 10% (vol/vol) glycol. Crystals were flash-frozen in liquid nitrogen and then transferred into a dry nitrogen stream at 100 K for X-ray diffraction data collection.
X-ray data collection, processing, and structure determination.
Multiwavelength anomalous dispersion (MAD) data sets using the bound zinc were collected for native nsp1α at 100 K using a Mar555 flat panel detector on the 1W2 beamline at Beijing Synchrotron Radiation Facility (BSRF). Data were processed and scaled using the program package MOSFLM (19). Two zinc ions per asymmetric unit were identified using the MAD data sets with the program SHELXD (28), with a partial occupancy over 0.9 and a CCall/CCweak score of 58.55/41.42. Initial phases were obtained using the computer program SOLVE (29) and were subsequently improved by using RESOLVE (29) to a mean overall figure of merit of 0.48. The electron density map at this stage was traceable with RESOLVE (29), and the automated model-building resulted in multiple peptide fragments. Manual model-building with the program O (12) and initial refinement with CNS software (2) generated a starting model for further refinement. The final model building and refinement were performed with the programs COOT (4) and Refmac (23). Solvent molecules were located from stereochemically reasonable peaks in the σA-weighted Fo-Fc difference electron density map. The quality of the final refined model was verified using the program PROCHECK (18). Final refinement statistics are summarized in Table 1. Structural figures were drawn with the program PyMOL (W. DeLano, Delano Scientific, San Carlos, CA).
TABLE 1.
Statistics of data collection and refinement
| Parameter | Zn peak value(s) | Zn edge value(s) |
|---|---|---|
| Data statistics | ||
| Cell parameters | a = b = 51.0 Å | |
| c = 154.6 Å | ||
| α = β = γ = 90° | ||
| Space group | P43212 | |
| Wavelength used (Å) | 1.2821 | 1.2827 |
| Resolution range (Å) | 50 (2.5)-2.4 | 50 (2.5)-2.4c |
| No. of all reflections | 61,067 | 60,801 |
| No. of unique reflections | 8,281 | 8,263 |
| Completeness of data (%) | 95.3 (95.1) | 99.5 (99.9) |
| Average I/σ(I) | 35.3 (1.3) | 35.7 (1.2) |
| Rmergea (%) | 6.5 (41.9) | 6.6 (43.6) |
| Refinement statistics | ||
| No. of reflections used [σ(F) > 0] | 8,269 | |
| Rworkb (%) | 22.3 | |
| Rfreeb (%) | 25.7 | |
| RMSD bond distance (Å) | 0.012 | |
| RMSD bond angle (°) | 2.070 | |
| No. of nonsolvent atoms | 1,367 | |
| No. of solvent atoms | 52 | |
| Avg overall B value (Å2) | 57.2 | |
| Ramachandran plot (excluding Pro and Gly) | ||
| No. of residues in most favored regions (%) | 115 (81.0) | |
| No. of residues in additionally allowed regions (%) | 22 (15.5) | |
| No. of residues in generously allowed regions (%) | 5 (3.5) |
Rmerge = ΣhΣl [pipe] Iih − <Ih> [pipe]/ΣhΣI <Ih>, where <Ih> is the mean of multiple observations Iih of a given reflection h.
Rwork = Σ‖Fp(obs)[pipe] − [pipe]Fp(calc)‖/Σ[pipe]Fp(obs) [pipe]; Rfree is an R factor for a selected subset (5%) of reflections that was not included in prior refinement calculations.
Numbers in parentheses are corresponding values for the highest-resolution shell (2.5 to 2.4 Å).
dValues for wavelength, resolution, number of all reflections, number of unique reflections, completeness, average I/σ(I), and Rmerge are given in two distinct groups: data determined at the Zn peak and data determined at the Zn edge. RMSD, root mean square deviation.
Cross-linking gel assay.
Recombinant nsp1α was diluted to 5 mg/ml in phosphate buffered saline (pH 7.0), 1 M NaCl, and 1 mM DTT. Ethylene glycol disuccinate di(N-succinimidyl) ester (EGS) was dissolved in dimethyl sulfoxide to a concentration of 25 mM and then added to 20 μl of protein sample, with final EGS concentrations of 0, 0.315, 0.625, 1.25, 2.5, 5.0, and 10.0 mM individually. After the mixture was incubated at room temperature for 15 min, the reaction was quenched for 10 min by adding 1 M Tris-HCl (pH 7.5) to a final concentration of 50 mM. An equal volume of 2× SDS-polyacrylamide gel electrophoresis sample buffer was added to the reaction mixture, and a small amount was analyzed on a 15% SDS polyacrylamide gel.
Protein structure accession number.
Coordinates and structure factors for the crystal structure of PRRSV nsp1α were deposited with the Protein Data Bank (PDB accession no. 3IFU).
RESULTS AND DISCUSSION
3D structure of the PRRSV nsp1α monomer.
In order to obtain naturally self-processed nsp1α, we expressed the full-length nsp1 of PRRSV (i.e., residues Met1 to Gly383 of the pp1a polyprotein) in Escherichia coli, and the recombinant nsp1α protein that was self released from downstream nsp1β was purified. The crystal structure of the 19-kDa nsp1α (i.e., residues Met1 to Met180) was determined using the MAD method and refined at a 2.4-Å resolution, resulting in a final Rwork value of 22.5% (Rfree = 27.1%). The crystal form belongs to the space group P43212, and there is one nsp1α monomer per asymmetric unit, with a Matthews coefficient of 2.6 Å3/Da, corresponding to a 51% solvent content (21). The nsp1α monomer possesses a compact, elliptical structure with dimensions of 50 by 35 by 30 Å, consisting of five β strands and five α-helices. It can be divided into three parts (Fig. 1B): the N-terminal ZF domain (Met1 to Glu65), the PCP domain (PCPα domain, Pro66 to Gln166), and the C-terminal extension (CTE; Arg167 to Met180). The overall refined structure was of good quality (Table 1).
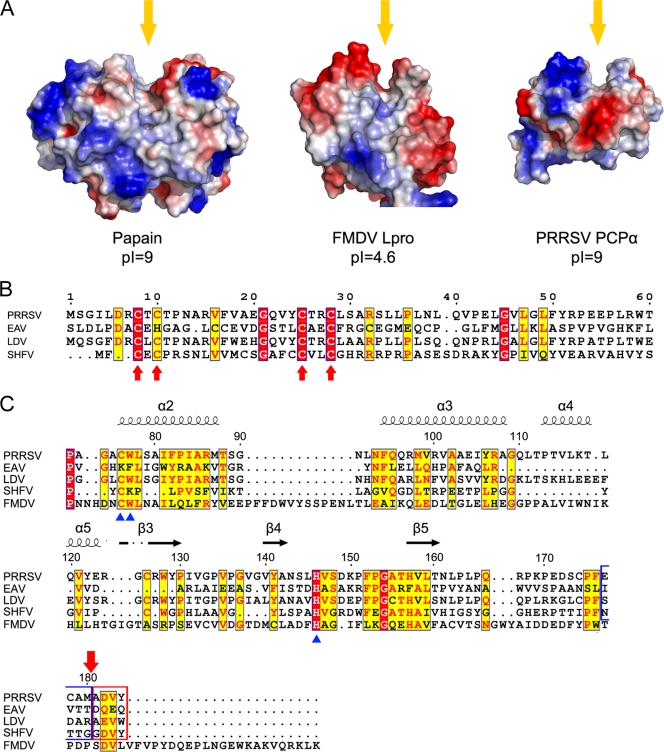
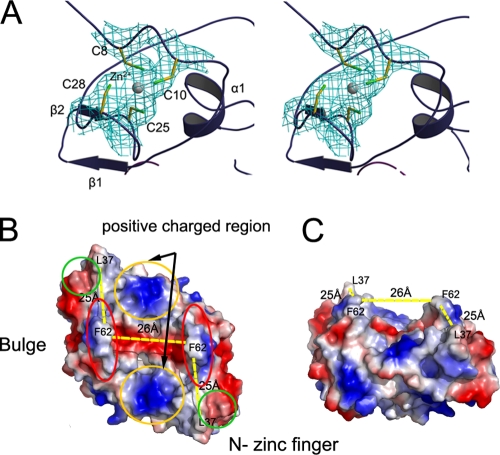
The N-terminal ZF domain consisted mainly of “random” coils plus two short antiparallel β strands (β1 and β2) and a short α-helix (α1) (Fig. 1C). A previous bioinformatics study and computer modeling predicted that the N-terminal ZF domain of nsp1α belongs to the 4-Cys (C4) ZF superfamily (20, 33). Our crystal structure of nsp1α confirmed this prediction and further revealed that the topology of the ZF domain is generally similar to that of the ββα ZF family (Fig. 1C), which includes over 1,000 known transcription factors (20). As a protease domain is not common in transcription factors, the ZF domain of nsp1α could explain the previously reported (16) effect of nsp1α on sg mRNA synthesis in PRRSV. Moreover, the homologue of nsp1α in EAV was demonstrated to be unnecessary for replication, but crucial for transcription, and several mutations in the putative N-terminal ZF domain of EAV nsp1 selectively abolished transcription of certain mRNAs (31). The conservation of the ZF domain among PRRSV, LDV, SHFV, and EAV suggests that their N-terminal ZF domains play similar roles in Arterivirus (see Fig. 4B) (31).
FIG. 4.
Structural and sequence comparisons of nsp1α homolog proteins. (A) Comparison of surface electrostatic potential of the PCP catalytic centers of papain (PDB accession no. 9PAP), FMDV Lpro (PDB accession no. 1QOL), and PRRSV PCPα (residues Pro66 to Gln166). Yellow arrows indicate the substrate binding grooves. (B) Sequence alignment of the N-terminal domains from PRRSV, EAV, LDV, and SHFV. Key residues of the C4 motif are marked with red arrows. Residue numbers at the top refer to the PRRSV sequence. (C) Sequence alignment of the C-terminal domains from PRRSV, EAV, LDV, SHFV, and FMDV Lpro. Key residues for the proteolytic activity of PCPs are marked with blue triangles at the bottom. Secondary-structure elements of PRRSV nsp1α are marked on top of the alignment; α-helices and β strands are presented as curves and arrows, respectively. Four residues upstream and downstream of the cleavage site of PRRSV nsp1α are highlighted with blue and red frames, and the cleavage site is marked with a red arrow on the top. The residue numbers on the top refer to the PRRSV sequence. The sequences were aligned with CLUSTALW (17), and the alignment was drawn with ESPript (7).
The PCPα domain of PRRSV nsp1α has a typical papain fold (13), which is a compact global structure consisting of sequentially connected left (L) and right (R) parts in a so-called standard orientation (9) (Fig. 1B). The L subdomain of PCPα consists of four α-helices (α2 to α5), while the R subdomain is formed by three antiparallel β strands (β3 to β5). Residue Cys76, which is located at the N-terminal end of the longest helix, α2, in the L subdomain, and His146, which is located in the turn connecting the two longest β strands, β4 and β5, in the R subdomain, face each other at the L-R interface and form the catalytic center of the PCPα domain.
The structure of the 14-residue CTE region of PRRSV nsp1α corresponds to unambiguous electron densities in experimentally MAD-phased 2Fo-Fc maps. The average B factor of this domain was 62 Å2, comparable with the overall average B factor of 58 Å2. This CTE peptide inserts into a putative substrate binding site of the PCPα domain from the same protein subunit. This observation suggests that CTE likely serves as a natural substrate for PCPα through intramolecular interactions, implying an in cis self-processing mode for nsp1α. At the amino acid sequence level, CTE is a characteristic feature of all nsp1α proteins of the Arteriviridae family, yet it has not been shown in a 3D structure before for any of the family members.
In the crystal structure of PRRSV nsp1α, we identified two zinc ions associated with each nsp1α subunit, one for the ZF domain and the other for the PCPα domain. In fact, the MAD signals from the zinc ions were successfully used to determine the initial phases of the crystal structure (Table 1). The zinc binding to nsp1α was further confirmed by atomic absorption spectrum and extended X-ray absorption fine structure spectra. In the 3D structure, both the N- and C-terminal zinc ions are tetrahedrally coordinated with Cys8, Cys10, Cys25, and Cys28 and with Cys70, Cys76, His146, and Met180, respectively. The N-terminal zinc coordination is consistent with the bioinformatics prediction of the C4 type of ZFs and was previously found to be related to sg mRNA synthesis (16). In contrast, the C-terminal-bound zinc ion is the first one among the reported 3D structures of papain-like proteases, and its precise biological role remains unclear (Fig. 2A and 3A). Unlike the conserved ZF domain among the other Arterivirus genus members, key residues observed forming the zinc coordination in the PCP domain of EAV vary from those in SHFV, LDV, and PRRSV, consistent with an earlier observation that the PCP-like domain of EAV nsp1 lacks PCP activity due to the absence of the catalytic cysteine. This further suggests that SHFV and LDV share similar zinc binding modes and PCPα activities (Fig. 4C) (33).
FIG. 2.
Functional implications of the N-terminal ZF domain in nsp1α. (A) Tetrahedral coordination of the zinc ion. The zinc ion and four coordinated cysteine residues are covered with a 2Fo-Fc electron density map phased with a MAD data set and contoured at 1.0 σ (cyan). (B and C) Top and side views of the hydrophilic groove on the molecular surface of the nsp1α homodimer.
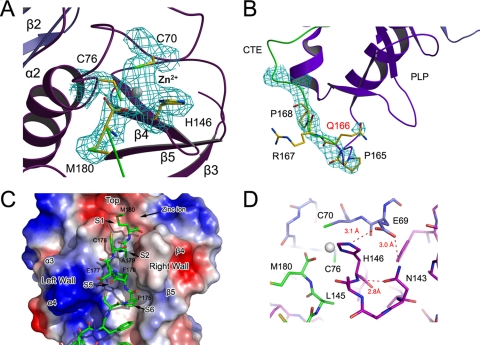
FIG. 3.
Active site of the C-terminal PCPα domain and its interaction with CTE. (A) Bound zinc ion and its coordination residues. The structural model is overlapped with a 2Fo-Fc electron density map phased with a MAD data set and contoured at 1.0 σ. (B) Uncleaved Glu166-Arg167 peptide bond. The 2Fc-Fc electron density map, contoured at 1.0 σ, shows inconsistency with a previously predicted cleavage site. (C) CTE binding to PCPα. PCPα is shown as a molecular surface model colored according to electrostatic potential (red for negatively charged regions and blue for positively charged regions). The CTE is shown as stick models, green for carbon, red for oxygen, and blue for nitrogen atoms. The zinc ion, key residues, and major binding pockets are labeled. (D) Hydrogen bond network around His146. Residues are colored according to their domain association (blue for ZF domain, purple for PCPα, and green for CTE). Hydrogen bonds are shown as red dashed lines and labeled with the distance.
PCPα active site bound with a zinc ion.
As a conserved structural feature of the papain superfamily, the active site of PRRSV nsp1α contains the catalytic residues Cys76 and His146 facing each other at the L-R interface. His146 of the His-Cys catalytic dyad is stabilized by an elaborate hydrogen bond network that includes Glu69 and Asn143, similar to two asparagines in other reported PCPs (Fig. 3D). The Oδ1 and Nδ2 atoms of Asn143 contacted the backbone amide group of His146 and the Oɛ2 atom of Gln69 at 2.8 Å and 3.0 Å, respectively. The Oɛ1 atom of Gln69 further stabilized the imidazole side chain of His146 via a 3.1-Å hydrogen bond with the Nɛ1 atom. Through this hydrogen bond network, the imidazole ring of His146 was kept in a favorable orientation relative to the cysteine residue, stabilizing the thiolate-imidazolium ion pair.
Consistent with this active site description, the position of the very C-terminal residue of CTE, Met180, likely represents the S1 site of the proteolytic reaction. Its carboxylate oxygen atoms are positioned close to the active site Cys76 and are associated with well-defined electron density. One of its carboxylate oxygen atoms (with a B factor of 51.4 Å2) is located in the putative oxyanion hole (formed by residues Ser71 to Ala75) and forms hydrogen bonds with both the main-chain carboxyl oxygen of Gly74 (2.8 Å) and the amide nitrogen of Cys76 (2.8 Å), whereas the second carboxylate oxygen (with a B factor of 51.6 Å2) coordinates the metal ion of the catalytic site (2.1 Å).
As mentioned above, the zinc ion bound in the PCP domain active site is a novel feature of PRRSV nsp1α that has not previously been reported in any other crystal structure of the papain superfamily. This metal ion was first identified as a Zn2+ ion during X-ray diffraction data collection and was later confirmed by ICP-MS performed with an ICP-MS XII instrument (Thermo). In this experiment, 95 nmol zinc ion was found in a 1-mg (∼50-nmol) protein sample of PRRSV nsp1α. This suggests that there is one more Zn2+ ion in PRRSV nsp1α in addition to the well-known one associated with the ZF domain. Moreover, in an anomalous difference Fourier map that was calculated using the diffraction data collected at the zinc absorption edge (λ = 1.2821 Å), two distinct peaks were observed in each nsp1α monomer at a contour level as high as 6.5 standard deviations and thus were assigned as Zn2+ ions in the subsequent structure determination. To verify the importance of this Zn2+ ion to proteolytic activity, we synthesized a fluorescent substrate MCA-ECAMADVY-Lys(Dnp)-Lys-NH2 and measured the proteolytic activity of nsp1α in both the absence and presence of EDTA. However, the native full-length PRRSV nsp1α was totally inactive, which is not surprising, considering the above-mentioned CTE binding mode (also see below). In addition, all our attempts to construct nsp1α variants of varied C-terminal truncations to restore the activity of PCP failed, as none of them could be expressed in a soluble form. These negative results unfortunately prevented us from directly examining the proteolytic activity of PRRSV nsp1α and further investigating the effect of zinc ion on PCPα activity. Nevertheless, considering that there was no Zn2+ ion artificially added during the purification and crystallization of nsp1α, our crystal structure and ICP-MS data unambiguously suggest a native zinc binding site at the PCPα active site, which is quite novel among the reported crystal structures of papain-like protease. Its precise biological role needs to be further investigated.
Homodimer assembly of PRRSV nsp1α.
The equilibrium between the monomer and dimer of PRRSV nsp1α was first observed in gel filtration chromatography during purification of nsp1α, even in the presence of 1 M NaCl (Fig. 1F). Subsequently, the results of EGS cross-linking and analytical ultracentrifugation also showed that nsp1α exists as a homodimer in solution and indicated that such a dimer remains associated even at high salt concentrations (Fig. 1F).
Consistently, two monomers formed a homodimer via the only available crystallographic twofold axis in the crystal form (Fig. 1D and E). This dimer possesses a hydrophilic, half-cylinder-shaped, symmetrical, open channel with a curvature diameter of ∼26 Å and length of ∼50 Å on its molecular surface (Fig. 2B and C). Major contacts between the two subunits were provided by several hydrophobic residues of the PCPα domain, especially long loops connecting α1 and α2, β3 and β4, and β5 and CTE, while minor contributions came from residues of the ZF domain, including the region connecting β1 and β2 and some coil regions near the peptide N terminus. The two putative protease active sites of each homodimer are on opposite sides and exposed to solvent, permitting independent function if they are not blocked by CTE (also see below). The relatively hydrophobic contact surface between the two monomers was 2,400 Å2 (25% of total) from each monomer, consistent with the strong dimerization in solution (Fig. 1G). In addition, we constructed several dimer-breaking point mutations but failed to express them as soluble proteins in E. coli, presumably because of unfavorable solvent-exposed hydrophobic surfaces. Together, both the structural and solution data suggest that the nsp1α homodimer is likely a biologically functional unit. In sharp contrast, a similar investigation of FMDV reveals that its corresponding protease Lpro functions as a monomer when it proteolytically cleaves the host cell protein eukaryotic initiation factor eIF4G (8, 9). Although the PCPα domain of PRRSV nsp1α shares high structural similarity with FMDV Lpro, the N-terminal ZF domain is absent from the latter. Therefore, the additional structural features, including the ZF domain and dimeric architecture of PRRSV nsp1α, may bear important functions beyond the protease activity.
Identifying the autocleavage site.
The crystal structure of PRRSV nsp1α showed the autocleavage site during nsp1 processing. PRRSV nsp1α releases itself from the parental polypeptide chain by a cleavage between its own C terminus and the N terminus of nsp1β, the downstream protein in the polyprotein peptide. During the early stages of model building, we built an initial model of nsp1α from Met1 to Gln166 into the experimental electron density map. However, unambiguous and consecutive electron density was found for an additional peptide C-terminal to Gln166. By carefully analyzing the primary sequence, we were able to fit 14 more residues (Arg167 to Met180) into this extra piece of electron density; the final model contained residues Met1 to Met180. Moreover, in an unbiased Fo-Fc difference Fourier map, the C-terminal residue Met180 had excellent electron density in the vicinity of the PCPα catalytic center. Meanwhile, the peptide between Gln166 and Arg167 was covered with clear continuous electron density (Fig. 3B). These results suggest that the nsp1α peptide ends exactly at Met180 and that the CTE region is an intact part of nsp1α instead of an isolated ligand or substrate.
To further verify the biological relevance of the above structural observation, we constructed a full-length nsp1 fused with both an N- and C-terminal His tag and a thrombin cleavage site between the N-terminal His tag and nsp1. After thrombin cleavage and self-processing, nsp1α and nsp1β were separated, and we were able to purify the nsp1β protein using affinity chromatography and to analyze its N-terminal amino acid sequence. The result showed that the N-terminal sequence of nsp1β begins with Ala181-Val-Asp-Ala-Tyr. In addition, the purified nsp1α was analyzed by mass spectroscopy methods, and the result showed a molecular weight of 19,900 for the nsp1α peptide, consistent with cleavage after Met180. Moreover, residues in the downstream region of this self-cleavage site in PRRSV, LDV, and SFHV are relatively conserved, and they correspond to the N terminus of nsp1β after cleavage. Notably, residues in EAV corresponding to this conserved nsp1α cleavage site also show some recognizable similarity with other family members, although EAV nsp1α lacks proteolytic activity because of an inactive PCP activity center (33) (Fig. 4C). Thus, it is likely that most Arterivirus members share a self-processing site similar to that of nsp1α. Taking all these data together, we conclude that the exact native nsp1α autocleavage site is after Met180 in the sequence of Cys-Ala-Met180↓Ala-Asp-Val (Fig. 3A).
Interaction between PCPα and CTE.
The presence of CTE residues inside the substrate-binding pockets illustrates the natural form of substrate binding during self-processing. The last six residues of the CTE were in an extended conformation (Fig. 3C) similar to that observed in complexes of enzymes of the papain superfamily with their peptide-like inhibitors (35, 36). Three major sequence regions in PCPα contributed to the CTE binding. In Fig. 3D, the loop between helices α3 and α4 (Gly109 to Pro113) formed the left wall of the binding pocket, while the loop between strands β4 and β5 formed the right wall. The central helix, α2, plus some side chains from the two loops mentioned above formed the bottom of the binding pocket. The two walls sandwiched CTE tightly to stabilize its extended conformation, and the catalytic center was located at the top of the substrate binding pocket.
More details on the CTE side of the major interactions between CTE and PCPα were provided by residues Phe173 to Met180. Whereas Met180 occupied the S1 position as mentioned above, Ala179 occupied the S2 subsite. The PCPα loop regions that connect helices α3 and α4 (i.e., residues Gln110 to Thr112) and strands β4 and β5 (i.e., residues Leu145 to Val147) formed a narrow cleft shaping the S2 subsite, as in many other papain-like proteases (14). The putative P5 residue of CTE was surrounded by the hydrophobic side chains of Leu111, Leu116, Val142, Val147, and Trp128, and therefore, the corresponding S5 site of the PCPα domain was a mostly hydrophobic subsite with a vacancy volume of ∼350 Å3. The hydrophobic side chain of Phe176 from CTE was completely buried in this subsite. This hydrophobic subsite is conserved to some extent in many papain-like proteases (Fig. 4B). The S6 subsite was at the end of the binding pocket and was occupied by Pro175. Phe173 and Pro175 were additionally stabilized by Trp128 and His157 through π electron interaction. In addition, residues of CTE also established some main-chain hydrogen bonds with PCPα to stabilize its conformation (Fig. 3C and Table 2).
TABLE 2.
Hydrogen bonds between CTE and PCPα residues
| CTE |
PCPα |
Distance (Å) | ||
|---|---|---|---|---|
| Residue | Atom | Residue | Atom | |
| Met180 | N | Leu145 | O | 3.3 |
| Ala179 | O | Leu111 | N | 3.1 |
| N | O | 3.0 | ||
| C | Leu145 | O | 3.2 | |
| Cys178 | O (via Wat42) | Ser144 | O | 3.1 (2.6) |
| N (via Wat42) | 3.0 (2.6) | |||
| Sγ | Gln110 | Oɛ1 | 3.2 | |
| Glu177 | O | Thr112 | Oγ1 | 2.8 |
| Pro175 | O | Ser144 | Cα | 3.2 |
| Gly69 | N | Trp128 | O | 3.2 |
Nsp1α proteins of PRRSV, LDV, SHFV, and FMDV Lpro show recognizable sequence homology, including the residues involved in PCP activity (Fig. 4C). For example, besides the Cys76 and His146 catalytic dyad (PRRSV numbers) (14), Trp77, which shields the hydrogen bond between the dyad from bulk solvent, is also among the most conserved residues, indicating similar cleavage reaction mechanisms for these Lpros of the Arteriviridae and Picornaviridae viruses. Nevertheless, there are distinct differences between the crystal structures of PRRSV nsp1α and FMDV Lpro. In the crystal structure of FMDV Lpro, one of two CTEs in each crystallographic asymmetric unit is in an unbound and flexible state, while the other one is well stabilized by the PCP domain of an adjacent Lpro molecule. This observation illustrates that the contact between FMDV Lpro CTE and the PCP catalytic domain is not very stable in solution, which would be consistent with its alternative proteolytic activity toward the protein substrate eIF4G. In sharp contrast, the CTE peptide of PRRSV nsp1α is well stabilized by the PCP domain and does not appear to allow subsequent substrate replacement for any further protease activity after self-cleavage. This product inhibitor phenomenon suggests that the PCP domain of the Lpro from the Arteriviridae family is unlikely to have specific proteolytic activity toward a host cell protein; instead, the self-released nsp1α is likely to be involved in nonproteolytic activities, such as acting as a platform for the N-terminal ZF domain to play its role in sg mRNA synthesis (16).
A putative self-processing mechanism.
Although there is not yet sufficient biochemical data to firmly establish a model regarding the self-processing mechanism of nsp1α-like Lpros, several structural features of PRRSV nsp1α suggest that an in cis (intramolecular) processing mode may be favored in PRRSV nsp1α self-processing. First of all, in our crystal structure, the residues N-terminal to the cleavage site were well defined in the electron density map and were strongly stabilized by an intramolecular hydrogen bond network (Table 2). Notably, as mentioned above, one carboxylate oxygen atom of C-terminal Met180 pointed toward the “oxyanion hole” and was stabilized by the active site. This oxygen atom likely represents the actual product after peptide hydrolysis. In addition, as we mentioned above, the cleavage sites of the two subunits in the homodimer were on opposite sides from each other, with a linear distance of 45 Å. Considering the limited peptide length of the CTE region, it is unlikely that the two subunits perform self-processing by swapping their CTE peptides. Therefore, intramolecular self-processing seems the most probable mechanism for PRRSV nsp1 based on our current model. A similar conclusion was suggested by the crystallographic investigation of FMDV Lpro. Although an in trans binding of the CTE peptide was observed with the FMDV Lpro crystal structure, it was attributed to crystal packing, and an in cis cleavage mechanism is considered the most probable scenario in solution (9).
In order to explore the possibility of studying the quaternary structure of a full-length nsp1 (i.e., nsp1α-nsp1β) polyprotein, we constructed an active site point mutation, C76S, to abolish the PCPα protease activity. Although we attempted to express it in dozens of combinations of expression vectors and culture conditions, this mutation was always present only in inclusion bodies, which prompted us to speculate that accurate self-processing might be essential for the structural stability of nsp1α and nsp1β as well as their functions.
Conclusion.
PRRSV nsp1α is the Lpro of the PRRSV pp1a polyprotein. Our crystal structure of nsp1α shows that it consists of an N-terminal ZF domain, which could be related to its biological role as a transcription factor; a PCP domain, which can self-release nsp1α from the polyprotein chain; and a CTE, which is the result of proteolytic self-processing and illustrates part of the substrate binding mode. Moreover, the crystal structure of nsp1α shows a dimeric architecture, which provides a putative hydrophilic surface channel and may be related to some important biological functions of nsp1α. The crystallographic and ICP-MS results also show a novel zinc ion binding site at the active site of PCPα, which has not been reported before for any crystal structure of PCP. Furthermore, our crystal structure suggest that an in cis mode is the most likely mechanism of PCPα self-processing.
Acknowledgments
We thank Liao M. of South China Agricultural University for kindly providing the PRRSV XH-GD genome.
This work was supported by the National Natural Science Foundation of China (grant no. 30870486), 863 Project (China) (grant no. 2006AA02A322), the National Major Projects (grant no. 2009ZX09311-001), the Protein Studies Project (China) (grant no. 2006CB10901), the Ministry of Science and Technology (MOST) 973 Project (grant no. 2006CB806503 and 2007CB914304), and the PSA II Project from MOST and KNAW (grant no. 2008AA000238).
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Benfield, D., J. Collins, S. Dee, P. Halbu, H. Joo, and K. Lager. 1999. Porcine reproductive and respiratory syndrome, p. 201-232. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of the swine, 8th ed. Iowa State University Press, Ames, IA.
- 2.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.den Boon, J. A., E. J. Snijder, E. D. Chirnside, A. A. de Vries, M. C. Horzinek, and W. J. Spaan. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 65:2910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 5.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 8.Guarne, A., B. Hampoelz, W. Glaser, X. Carpena, J. Tormo, I. Fita, and T. Skern. 2000. Structural and biochemical features distinguish the foot-and-mouth disease virus leader proteinase from other papain-like enzymes. J. Mol. Biol. 302:1227-1240. [DOI] [PubMed] [Google Scholar]
- 9.Guarne, A., J. Tormo, R. Kirchweger, D. Pfistermueller, I. Fita, and T. Skern. 1998. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 17:7469-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han, J., Y. Wang, and K. S. Faaberg. 2006. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 122:175-182. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 13.Kamphuis, I. G., J. Drenth, and E. N. Baker. 1985. Thiol proteases. Comparative studies based on the high-resolution structures of papain and actinidin, and on amino acid sequence information for cathepsins B and H, and stem bromelain. J. Mol. Biol. 182:317-329. [DOI] [PubMed] [Google Scholar]
- 14.Kamphuis, I. G., K. H. Kalk, M. B. Swarte, and J. Drenth. 1984. Structure of papain refined at 1.65 Å resolution. J. Mol. Biol. 179:233-256. [DOI] [PubMed] [Google Scholar]
- 15.Kapur, V., M. R. Elam, T. M. Pawlovich, and M. P. Murtaugh. 1996. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J. Gen. Virol. 77:1271-1276. [DOI] [PubMed] [Google Scholar]
- 16.Kroese, M. V., J. C. Zevenhoven-Dobbe, J. N. Bos-de Ruijter, B. P. Peeters, J. J. Meulenberg, L. A. Cornelissen, and E. J. Snijder. 2008. The nsp1alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 89:494-499. [DOI] [PubMed] [Google Scholar]
- 17.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski, R., M. MacArthur, D. Moss, and J. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 19.Leslie, A. 1995. MOSFLM users guide. MRC Laboratory of Molecular Biology, Cambridge, United Kingdom.
- 20.Luscombe, N. M., S. E. Austin, H. M. Berman, and J. M. Thornton. 2000. An overview of the structures of protein-DNA complexes. Genome Biol. 1:REVIEWS001.1-REVIEWS001.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491-497. [DOI] [PubMed] [Google Scholar]
- 22.Meng, X. J., P. S. Paul, P. G. Halbur, and I. Morozov. 1995. Sequence comparison of open reading frames 2 to 5 of low and high virulence United States isolates of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 76:3181-3188. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 24.Nelsen, C. J., M. P. Murtaugh, and K. S. Faaberg. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 73:270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oleksiewicz, M. B., E. J. Snijder, and P. Normann. 2004. Phage display of the Equine arteritis virus nsp1 ZF domain and examination of its metal interactions. J. Virol. Methods 119:159-169. [DOI] [PubMed] [Google Scholar]
- 26.Paton, D. J., I. H. Brown, S. Edwards, and G. Wensvoort. 1991. ‘Blue ear’ disease of pigs. Vet. Rec. 128:617. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheldrick, G. M. 2008. A short history of SHELX. Acta Crystallogr. A 64:112-122. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiel, H. J., G. Meyers, R. Stark, N. Tautz, T. Rumenapf, G. Unger, and K. K. Conzelmann. 1993. Molecular characterization of positive-strand RNA viruses: pestiviruses and the porcine reproductive and respiratory syndrome virus (PRRSV). Arch. Virol. 7(Suppl.):41-52. [DOI] [PubMed] [Google Scholar]
- 31.Tijms, M. A., D. D. Nedialkova, J. C. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2007. Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease. J. Virol. 81:10496-10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tijms, M. A., and E. J. Snijder. 2003. Equine arteritis virus non-structural protein 1, an essential factor for viral subgenomic mRNA synthesis, interacts with the cellular transcription co-factor p100. J. Gen. Virol. 84:2317-2322. [DOI] [PubMed] [Google Scholar]
- 33.Tijms, M. A., L. C. van Dinten, A. E. Gorbalenya, and E. J. Snijder. 2001. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. USA 98:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 87:3473-3482. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto, A., K. Tomoo, M. Doi, H. Ohishi, M. Inoue, T. Ishida, D. Yamamoto, S. Tsuboi, H. Okamoto, and Y. Okada. 1992. Crystal structure of papain-succinyl-Gln-Val-Val-Ala-Ala-p-nitroanilide complex at 1.7-Å resolution: noncovalent binding mode of a common sequence of endogenous thiol protease inhibitors. Biochemistry 31:11305-11309. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto, D., K. Matsumoto, H. Ohishi, T. Ishida, M. Inoue, K. Kitamura, and H. Mizuno. 1991. Refined x-ray structure of papain.E-64-c complex at 2.1-Å resolution. J. Biol. Chem. 266:14771-14777. [DOI] [PubMed] [Google Scholar]
- 37.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]