Abstract
Drug resistance is an important cause of antiretroviral therapy failure in human immunodeficiency virus (HIV)-infected patients. Mutations in the protease render the virus resistant to protease inhibitors (PIs). Gag cleavage sites also mutate, sometimes correlating with resistance mutations in the protease, but their contribution to resistance has not been systematically analyzed. The present study examines mutations in Gag cleavage sites that associate with protease mutations and the impact of these associations on drug susceptibilities. Significant associations were observed between mutations in the nucleocapsid-p1 (NC-p1) and p1-p6 cleavage sites and various PI resistance-associated mutations in the protease. Several patterns were frequently observed, including mutations in the NC-p1 cleavage site in combination with I50L, V82A, and I84V within the protease and mutations within the p1-p6 cleavage site in combination with D30N, I50V, and I84V within the protease. For most patterns, viruses with mutations both in the protease and in either cleavage site were significantly less susceptible to specific PIs than viruses with mutations in the protease alone. Altered PI resistance in HIV-1 was found to be associated with the presence of Gag cleavage site mutations. These studies suggest that associated cleavage site mutations may contribute to PI susceptibility in highly specific ways depending on the particular combinations of mutations and inhibitors. Thus, cleavage site mutations should be considered when assessing the level of PI resistance.
Combination antiretroviral treatment regimens containing protease inhibitors (PIs) often do not completely and durably suppress human immunodeficiency virus type 1 (HIV-1) infection, resulting in therapy failure. Selective drug pressure of competitive PIs has resulted in an accumulation of combinations of mutations within both Gag and GagProPol polyproteins that render the virus highly resistant to multiple drugs (2, 23, 36). In addition, there is an increasing incidence of new infections with viruses that are already drug resistant (3).
Drug resistance provides an escape mechanism for the virus by lowering the affinity of target enzymes for inhibitors while still maintaining efficient processing, thus leading to therapy failure (14, 35). This paradox may be explained by using the concept of a “substrate envelope,” which states that substrate specificity is based not on a particular amino acid sequence but on a conserved shape (34). The consensus volume of the majority of the substrates defines the substrate envelope. Although most of the volume of any given inhibitor also fits within this envelope, particular inhibitor atoms occasionally protrude beyond the envelope to contact surrounding protease residues. Mutations at these residues preferentially impact inhibitor binding over substrate recognition and cleavage (16). Often, these resistance-associated mutations reduce the catalytic efficiency of the protease, resulting in immature or noninfectious viruses (5). To compensate for this loss in efficiency, secondary mutations develop within the protease (24). In addition, mutations develop within the cleavage sites, complementing the changes in the resistant protease. All genotype-based algorithms used to predict the susceptibility of the patient virus to PIs assess only mutations within the protease (12, 15, 24). However, resistance-associated mutations in Gag, especially within the cleavage sites, are often associated with protease mutations (1, 10, 11, 17, 21, 40) and may be important for resistance testing.
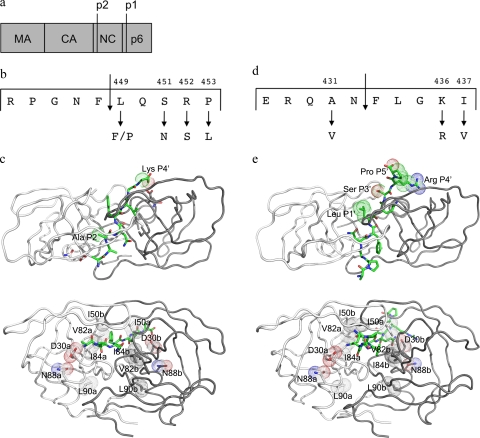
Gag A431V, within the nucleocapsid-p1 (NC-p1) cleavage site, was one of the first cleavage site mutations to be associated with the V82A protease mutation (40), and more recently, I437V was also shown to be associated with 82 (25). Association was reported between mutations within the p1-p6 cleavage site and the I50V protease mutation (18). In earlier studies (17), we showed an association between the occurrence of mutations within the p1-p6 cleavage site and the signature protease mutations of nelfinavir (NFV) resistance, D30N/N88D (26, 29). The present study focuses on the primary resistance-associated mutations at residues 30, 50, 82, 84, 88, and 90 in the protease and their association with mutations within the Gag cleavage sites, NC-p1 and p1-p6 (Fig. 1a to e).
FIG. 1.
(a) Overview of Gag showing the constituent proteins. (b) Details of the NC-p1 cleavage site. Also shown are two residues that mutate frequently. (c) Two different views of the crystal structure of HIV-1 protease complexed with NC-p1 cleavage site (1TSU) (33). The substrate is shown in green, and the two monomers are shown in white and gray. (Top) The top view showing van der Waals surfaces on cleavage site residues that mutate frequently. (Bottom) The side view with van der Waals surfaces on protease residues that are the focus of this study. (d) Details of the p1-p6 cleavage site. Frequently occurring mutations are shown. (e) Two different views of the crystal structure of HIV-1 protease complexed with p1-p6 cleavage site (1KJF) (34). Shown are the substrate (green) and the two monomers (white and gray). (Top) The top view showing van der Waals surfaces on cleavage site residues that mutate frequently. (Bottom) The side view with van der Waals surfaces on protease residues that are the focus of this study. Images were generated using PyMol (8).
To further study the impact of these associations, the susceptibility of viruses to various PIs was examined by mining a large database of linked genotypes and phenotypic measures of viral susceptibility to antiretroviral drugs. Several of these patterns significantly alter the virus susceptibility to PIs, indicating that the evolution of drug resistance not only occurs within the protease but also involves multiple regions of Gag and GagProPol polyproteins in an interdependent manner.
MATERIALS AND METHODS
Sequence database.
HIV-1 protease (amino acids 1 to 99), reverse transcriptase (RT) (amino acids 1 to 305), and Gag (amino acids 418 to 500) sequences were obtained from samples submitted to Monogram Biosciences for routine phenotype and genotype drug resistance testing. Viral sequences from infected patients were determined by the GeneSeq HIV assay (31). A data set of 39,152 subtype B sequences from the Monogram Biosciences database was used. Wild-type (WT) viruses, i.e., those that show none of the known resistance mutations in either protease or RT, were excluded. RT mutations that were included are M41L, K65R, D67N, T69X, K70ER, L74X, V75A/M/S/T, A98G, L100I, K101P, K103N/S, V106A/M, Y115F, Q151M, Y181X, M184X, Y188X, G190X, L210W, T215F/Y, K219X, P225X, F227X, M230L, and P236L (X stands for any mutation). Protease mutations that were included are L10X, K20X, L23X, L24X, D30X, V32X, L33X, M46X, I47X, G48X, I50X, I54X, A71X, G73X, L76X, V82X, I84X, N88X, and L90X. Only one sample per patient was included. Gag sequence alignments were performed using the profile hidden Markov model software HMMER (http://hmmer.janelia.org). The profile hidden Markov model was previously constructed from 1,152 gag sequences (data not shown). Amino acid positions were numbered through the alignment with the HXB2 sequence. All statistical analyses were performed using the R software environment version 2.4.1 (http://www.r-project.org).
Covariation analysis.
Sequences were analyzed with respect to the presence or absence of the following primary mutations within the protease, either individually or in combination: D30N, I50V, I50L, V82A, I84V, N88D, and L90M. Samples with mixtures at particular positions and mutations other than those specified were excluded. The corresponding NC-p1 and p1-p6 cleavage sites were screened for mutations relative to HXB2. The association of cleavage site mutations with a specific protease mutation was computed as the proportion of the total number of sequences with that particular protease mutation. These proportions were compared to the case when the cleavage site mutations occurred in the absence of any primary protease mutation. The significance of the association was tested by using the chi-square test (R function “chisq.test”). Adjustments were made for multiple testing using the Bonferroni method. Corrected P values of <0.05 indicated statistical significance.
RC.
Relative fitness of the virus was determined with the replication capacity (RC) and expressed as a percentage relative to that of a reference virus, NL4-3 (7). RCs for viruses including the protease mutations of interest with and without Gag cleavage site mutations were extracted and analyzed. The difference in mean RC was tested using the Mann-Whitney test (R function “wilcoxon.test”). Adjustments were made for multiple testing using the Bonferroni method. Corrected P values of <0.05 indicated statistical significance.
Drug susceptibilities.
Virus susceptibilities to amprenavir (APV), atazanavir (ATV), indinavir (IDV), lopinavir (LPV), nelfinavir (NFV), ritonavir (RTV), saquinavir (SQV), and tipranavir (TPV) were determined with the PhenoSense assay (Monogram Biosciences, South San Francisco, CA) (27, 31). Results are expressed as the fold change (FC) in 50% inhibitory concentration (IC50) of the patient virus relative to that of the drug-sensitive reference, NL4-3. FC distribution for groups of samples containing certain protease mutations with and without Gag cleavage site mutations were extracted and analyzed. Samples with mixtures at particular positions and mutations other than those specified were considered to not meet the profile and were excluded. The difference in median IC50 FC was tested using the Mann-Whitney test (R function “wilcoxon.test”). Adjustments were made for multiple testing using the Bonferroni method. Corrected P values of <0.05 indicated statistical significance.
Additional validation studies.
To help to ensure the validity of our studies, the following additional studies were performed. (i) The average number of secondary protease mutations in viruses with or without particular Gag mutations was computed and the difference was evaluated using the t test. (ii) Sequences with and without Gag mutations in the absence of protease mutations were extracted from the database. The difference in mean FC in IC50s to various PIs was evaluated using the t test. (iii) Two sets of Gag sequences (418 to 500) were aligned. The first group consists of non-WT samples that were used for the susceptibility analyses, and the second group consists of sequences that were WT for protease. The mutation frequency at each Gag position was calculated for both sets, and the frequency difference between the two groups was plotted.
RESULTS
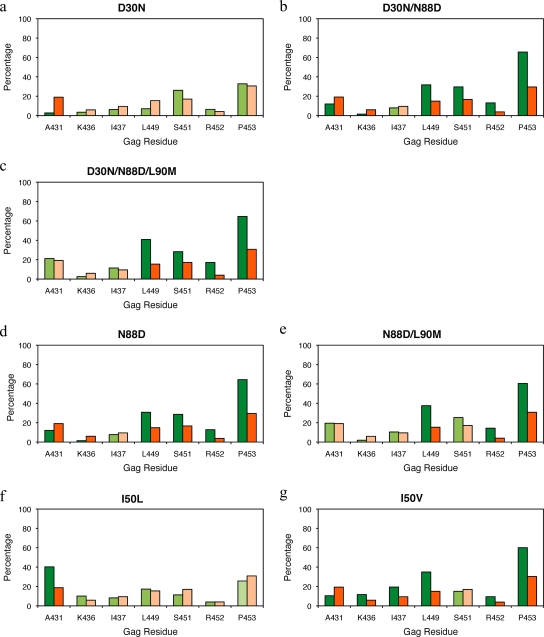
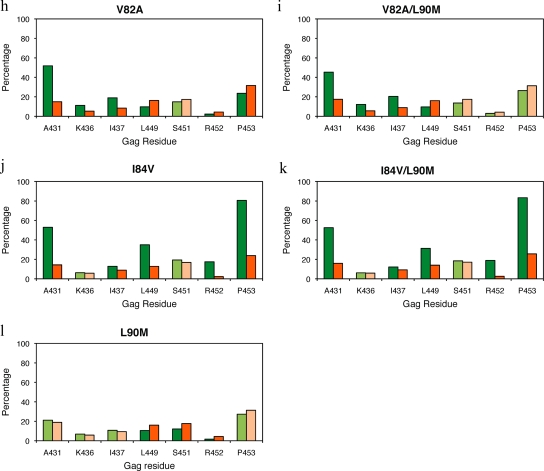
The NC-p1 and p1-p6 cleavage sites (Fig. 1b and d) are essential in the critical final steps of viral budding (37) and maturation. To examine the mutational patterns in these two key cleavage sites and their association with HIV-1 protease mutations, data was extracted from 39,152 HIV-1, subtype B viral sequences from infected patients. These sequences had a median of one PI-associated protease mutation (range, zero to eight). Sequences were examined for associated mutations between D30N, D30N/N88D(/L90M), I50V/L, V82A(/L90M), I84V(/L90M), and L90M primary drug-resistance mutations in protease and mutations within Gag in either the NC-p1 or the p1-p6 cleavage site. Gag A431 within the NC-p1 cleavage site was the most common site for associated mutations. The A431V mutation was observed to associate with the V82A protease mutation, as previously reported (10, 40). Mutations at Gag 431 also associated with either I50L or I84V protease mutations. Mutations at Gag 436, another site within NC-p1, associated with the V82A protease mutation. The Gag I437V mutation within NC-p1, recently shown to enhance PI resistance (25), was observed in this study to significantly associate with both V82A and I84V protease mutations. Previously, we observed that mutations at the p1-p6 cleavage site associate with the D30N/N88D NFV-resistance protease mutations (17). In the present study, p1-p6 mutations were also observed to associate with N88D alone. In addition, p1-p6 cleavage site mutations associated with either I50V or I84V protease mutations, independently. The particular patterns of mutations within the p1-p6 cleavage site varied specifically depending on the particular protease mutation. D30N/N88D and I84V protease mutations associated with mutations at Gag 449, 451, 452, and 453, while I50V protease mutation associated with mutations at Gag 449, 452, and 453. Thus, associated mutations were observed in the NC-p1 and/or the p1-p6 cleavage site in the presence of all the primary active site resistance mutations.
These patterns of associations between cleavage site mutations and particular primary protease resistance mutations were further analyzed in terms of RC and drug susceptibility. The effects of additional protease mutations (positions 10, 20, 23, 24, 32, 33, 46, 47, 48, 54, 71, 73, and 76) have not been excluded, as attempts to exclude them limited the sample size and did not significantly alter the observed associations. However, additional control studies, as detailed below, were performed to understand other factors that may likely contribute to alterations in phenotypic susceptibility to various inhibitors in addition to the role of Gag mutations.
Groups of viruses with particular protease mutations in the presence and absence of Gag mutations were compared for the number of secondary protease mutations (see Table S1 in the supplemental material). With the exception of viruses with V82A/L90M mutations, all viruses with other protease mutations in combination with Gag A431V had significantly more secondary mutations in the protease. Of these, only viruses with D30N and I50V mutations did not show increased FCs. Viruses with V82A and V82A/L90M mutations in combination with a mutation at Gag 449 had slightly more secondary protease mutations and showed higher FCs in IC50 than viruses without the Gag 449 mutation. Viruses with mutations at Gag 452 had an increased number of secondary mutations when in combination with the I84V, I84V/L90M, V82A, and V82A/L90M protease mutations and also showed significant FCs. A similar trend was observed in viruses that had mutations at Gag 453 in combination with V82A and V82A/L90M protease mutations.
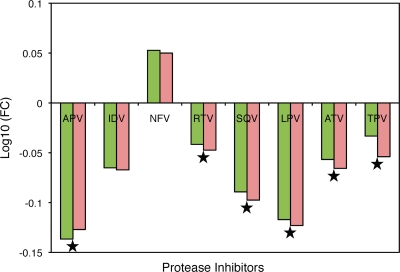
Alterations in phenotypic susceptibilities to PIs as a result of various Gag cleavage site mutations in the absence of protease mutations were analyzed (Fig. 2). With the exception of IDV and NFV, there were small but statistically significant increases in susceptibilities to various PIs as a result of cleavage site mutations alone in the absence of any resistance-associated protease mutations. However, both groups of viruses were found to be highly susceptible to all PIs except to NFV, where a slight reduction in susceptibility was observed.
FIG. 2.
PI susceptibility of viruses with and without Gag mutations in the absence of protease resistance mutations. Mean FCs in IC50 are shown on a log scale on the y axis, and the various PIs tested against are shown on the x axis. Green bars represent viruses with Gag mutations, and pink bars represent viruses without Gag mutations. Stars indicate significant differences in susceptibilities of the viruses to a particular PI. P values of ≤0.05 are significant.
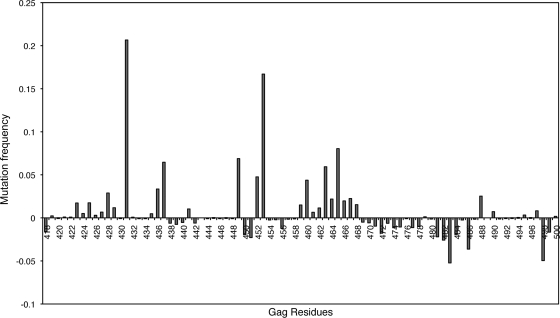
Mutation frequencies within Gag were higher in the case of non-WT samples that were used in the phenotypic susceptibility analyses compared to WT sequences (Fig. 3). Specifically, Gag 431 and 453 mutated much more frequently in non-WT samples than in WT sequences. Increased mutational frequencies were also observed at Gag 436, 437, 449, and 452 in the case of non-WT samples compared to WT samples. Small increases in mutation frequencies were also observed in the PTAP region in non-WT samples.
FIG. 3.
Differential mutation rates within Gag from viruses with and without protease resistance mutations. Mutation frequencies are shown on the y axis, and Gag residues numbering 418 to 500 are shown on the x axis.
RCs.
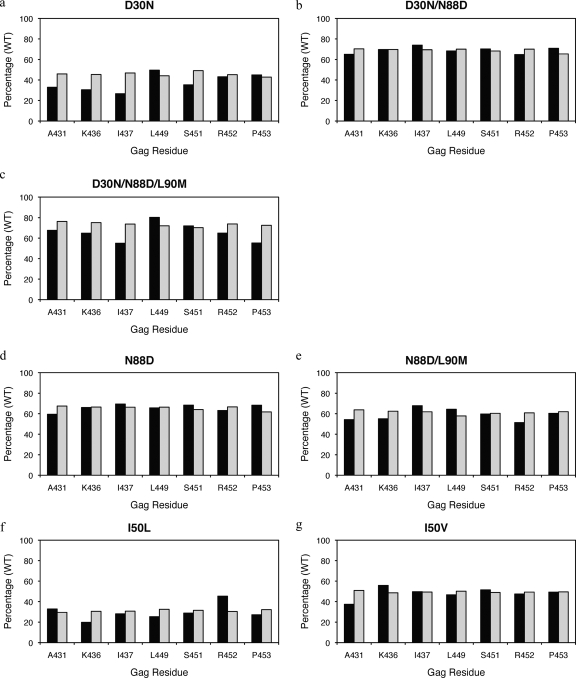
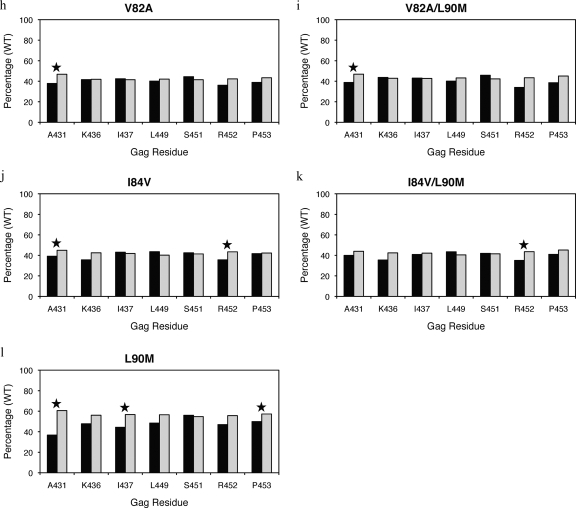
In this study, the fitness of groups of viruses with particular protease mutations in the presence and absence of Gag mutations were compared to a reference virus. Interestingly, in most instances, viruses with Gag mutations in combination with particular protease mutations did not improve fitness compared to viruses with mutations in the protease alone (Fig. 4 and see Table S2 in the supplemental material). In fact, in the few cases where significant differences were observed, Gag mutations actually decreased fitness compared to viruses that have protease mutations in the absence of Gag mutations. Both V82A and V82A/L90M viruses had lower RCs in combination with mutations at Gag 431 compared to those without this mutation (Fig. 4h and i). Similarly, a mutation at Gag 431 also lowered viral fitness in the presence of I84V protease mutation (Fig. 4j). In addition, viruses with I84V or I84V/L90M protease mutations in combination with mutations at Gag 452 also had lower RCs compared to viruses with the protease mutation alone (Fig. 4j and k). Viruses with an L90M protease mutation in combination with mutations at Gag 431, 437, or 453 were less fit than viruses with an L90M mutation alone (Fig. 4l).
FIG. 4.
RCs of viruses with primary drug-resistant protease mutations (a to l) and associated NC-p1 and p1-p6 cleavage site mutations. RCs of viruses tested are shown as a percentage of the WT reference virus (100%) on the y axis, and on the x axis are Gag residues within NC-p1 and p1-p6 that mutate frequently. Closed bars represent viruses with both the particular protease and cleavage site mutations present. Open bars represent viruses with only the particular protease mutations present. Stars indicate a significant difference in RCs for a particular set. P values of ≤0.05 are significant.
D30N.
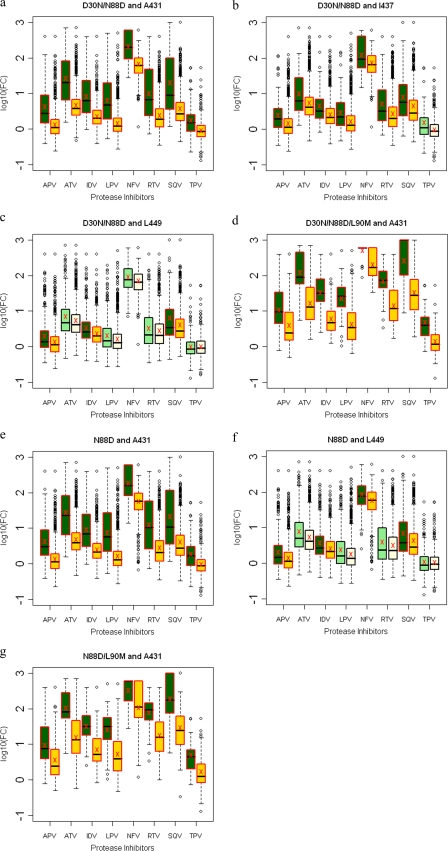
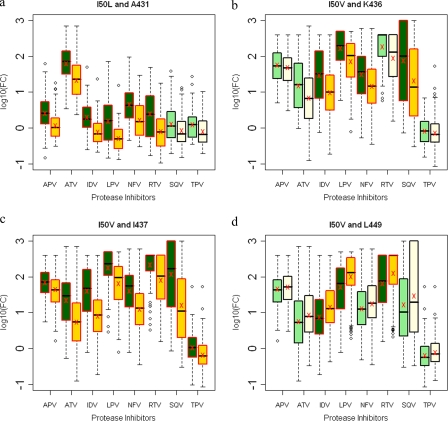
D30N is a signature mutation for NFV often in combination with N88D or occasionally N88D/L90M (22, 29). D30N, D30N/N88D, and N88D protease mutations were not associated with mutations in NC-p1 (Fig. 5a, b, and d). However, there was a significant decrease in phenotypic susceptibilities to all PIs in viruses where an A431V mutation occurred in combination with D30N/N88D(/L90M), N88D, or N88D/L90M (Fig. 6a, d, and g). No corresponding changes in PI sensitivity were seen with mutations at Gag 436, whereas mutations at Gag 437 in combination with D30N/N88D protease mutations resulted in viruses with reduced sensitivity to APV, ATV, IDV, LPV, NFV, RTV, and SQV (Fig. 6b). In contrast, as we have previously seen, D30N/N88D protease mutations were significantly associated with mutations within the p1-p6 cleavage site where 32, 30, and 66% of sequences had mutations at Gag 449, 451, and 453, respectively, whereas only 15, 17, and 30% of sequences had mutations at these positions in the absence of D30N/N88D (Fig. 5b and see Table S3 in the supplemental material), confirming results of our earlier studies (17). Although not as frequent, Gag R452 was also observed to mutate in an associated manner with D30N/N88D (Fig. 5b). A similar trend was observed in D30N/N88D/L90M viruses (Fig. 5c). Viruses with D30N/N88D in combination with mutations at Gag 449 showed small changes in FC across APV, IDV, and SQV (Fig. 6c). Mutations within p1-p6 are also associated with the N88D and N88D/L90M protease mutations (Fig. 5d and e), and decreased sensitivity to APV, IDV, NFV, SQV, and TPV was observed in viruses with a Gag 449 mutation in combination with N88D (Fig. 6f). Thus, D30N/N88D and N88D protease mutations associate with mutations within p1-p6, in contrast with mutations in NC-p1, although both combinations modulate susceptibilities to the PIs.
FIG. 5.
Associated mutations within the NC-p1 and p1-p6 cleavage sites with primary resistance-associated mutations in the protease at residues 30, 50, 82, 84, 88, and 90 (a to l). The frequencies of cleavage site mutations are shown as percentages on the y axis. On the x axis are the amino acid residues (Gag numbering) within the NC-p1 and p1-p6 cleavage sites. Shown are cleavage sites that mutate in the presence of the primary protease mutation (green) and cleavage sites that mutate in the absence of the given protease mutations (orange). Dark green and dark orange indicate significant association of mutations. P values of ≤0.05 are significant.
FIG. 6.
Box plots (ratio of median FCs on a log scale; y axis) showing susceptibilities of NFV-resistant viruses to various PIs (x axis) in the presence (green boxes) or absence (yellow boxes) of associating mutations within the NC-p1 and p1-p6 cleavage sites. The boxes represent the interquartile range between the first and third quartile. The upper and lower lines represent 1.5 times the upper and lower quartile limits. The black line represents the median, and the red X represents the mean of the distribution. Outliers are shown as open circles. Dark green and gold boxes indicate a significant effect of associating mutations within Gag cleavage sites on PI susceptibilities. Shown are susceptibilities of viruses with mutation D30N/N88D with and without mutations at Gag 431 (a), Gag 437 (b), and Gag 449 (c); susceptibilities of viruses with mutation D30N/N88D/L90M with and without mutations at Gag 431 (d); susceptibilities of viruses with mutation N88D with and without mutations at Gag 431 (e) and Gag 449 (f); and susceptibilities of viruses with mutation N88D/L90M with and without mutations at Gag 431 (g). P values of ≤0.05 are significant.
I50V and I50L.
I50, positioned at the flap tips of the protease, plays an essential role in substrate recognition and cleavage by locking down the active site (20, 34). This key residue did not mutate in response to the first-generation PIs; however, subsequent PIs result in I50 mutating in a drug-dependent manner. The I50L mutation results in severe resistance to ATV but causes hypersusceptibility to most other PIs (4, 39), whereas the I50V mutation results in resistance to APV and, to a lesser extent, DRV (9, 28, 30). The I50L protease mutation was observed to associate with mutations at Gag 431 within the NC-p1 cleavage site (Fig. 5f). Previous studies showed that L449 and P453 within the p1-p6 cleavage site mutated in a manner associated with the I50V protease mutation (18, 19), which was also observed here (Fig. 5g). Less frequently, mutations at Gag 452 were also associated with the I50V mutation. In addition, the I50V protease mutation was also associated with mutations at K436 and I437 within the NC-p1 cleavage site (Fig. 5g). Altered susceptibilities to various drugs were observed with both I50L and I50V protease mutations in the presence of mutations in NC-p1 and p1-p6 cleavage sites, respectively. Viruses with the I50L mutation, a signature mutation of ATV resistance, often show hypersusceptibility to other PIs (38). Viruses with the I50L mutation in combination with Gag A431V lose this hypersusceptibility and show decreased susceptibility to APV, ATV, IDV, LPV, NFV, and RTV (Fig. 7a). Viruses with the I50V mutation showed significantly reduced susceptibilities to IDV, LPV, NFV, and SQV in combination with the K436R mutation (Fig. 7b) and showed reduced susceptibility to all PIs in combination with mutations at Gag 437 (Fig. 7c), whereas viruses with the I50V mutation in combination with Gag L449F showed increased susceptibilities to IDV, LPV, and especially to RTV (Fig. 7d). Thus, the impact of mutations at I50 is modulated by the addition of particular cleavage site mutations.
FIG. 7.
Box plots (ratio of median FCs on a log scale; y axis) showing susceptibilities of I50V and I50L viruses to various PIs (x axis) in the presence (green boxes) or absence (yellow boxes) of associating mutations within the NC-p1 and p1-p6 cleavage sites. The boxes represent the interquartile range between the first and third quartile. The upper and lower lines represent 1.5 times the upper and lower quartile limits. The black line represents the median, and the red X represents the mean of the distribution. Outliers are shown as open circles. Dark green and gold boxes indicate a significant effect of associating mutations within Gag cleavage sites on PI susceptibilities. Shown are susceptibilities of viruses with mutation I50L with and without mutations at Gag 431 (a) and susceptibilities of viruses with mutation I50V with and without mutations at Gag 436 (b), Gag 437 (c), and Gag 449 (d). P values of ≤0.05 are significant.
V82A.
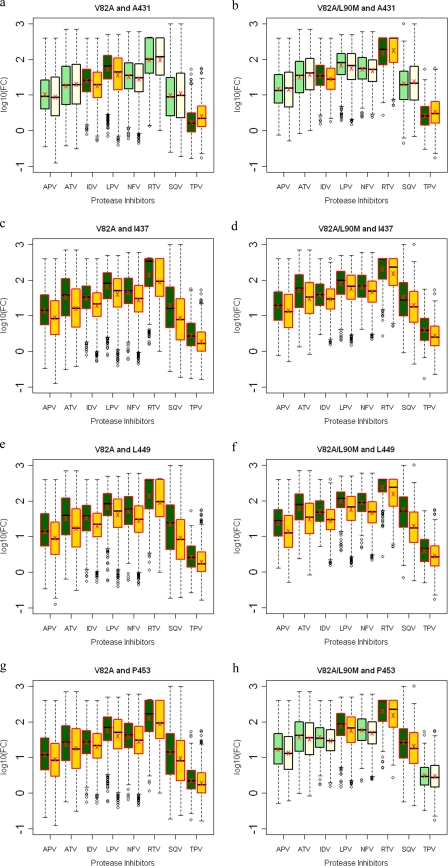
The V82A protease mutation has been well documented to associate with Gag A431V (33, 40). Fifty-two percent of viruses have this combination of mutations, whereas only 15% have the A431V mutation in the absence of the V82A protease mutation (Fig. 5h and see Table S3 in the supplemental material). Those viruses with both mutations exhibited lower susceptibilities to IDV and LPV but were hypersusceptible to TPV (Fig. 8a). However, a significant increase in susceptibility to RTV and TPV, to a lesser extent, was observed in viruses with the combination of V82A/L90M and Gag A431V mutations (Fig. 8b). Mutations at Gag 436 and 437 within the NC-p1 were also associated with both V82A and V82A/L90M protease mutations (Fig. 5h and i). Mutations at Gag 436 were observed to associate with the V82A protease mutation, where Gag 436 mutated in 11% and 12% of the viruses in the presence and only 5% and 6% in the absence of V82A and V82A/L90M, respectively (see Table S3 in the supplemental material). However, no associated changes in susceptibilities were observed. The Gag 437 mutations, which occurred 19% and 20% of the time with V82A and V82A/L90M, respectively, resulted in reduced sensitivity to all PIs (Fig. 8c and d). Mutations within the p1-p6 site were significantly not associated with either a V82A or V82A/L90M protease mutation. Only 10% of viruses had a Gag L449F mutation and a V82A or V82A/L90M protease mutation as opposed to 16% that had mutations only in the cleavage site (Fig. 5h and i and see Table S3 in the supplemental material). Mutations at Gag 449 in combination with V82A or V82A/L90M decreased susceptibilities to all PIs tested (Fig. 8e and f). Mutations at Gag 453 with V82A viruses also showed significantly decreased susceptibilities to all PIs (Fig. 8g), whereas in combination with V82A/L90M viruses, decreased susceptibilities were observed with LPV, RTV, and SQV (Fig. 8h). Thus, with a few exceptions, mutations within the NC-p1 and p1-p6 cleavage sites, often modulated by L90M, are seen to decrease the susceptibilities of V82A protease-containing viruses.
FIG. 8.
Box plots (ratio of median FCs on a log scale; y axis) showing susceptibilities of viruses with V82A and V82A/L90M mutations to various PIs (x axis) in the presence (green boxes) or absence (yellow boxes) of associating mutations within the NC-p1 and p1-p6 cleavage sites. The boxes represent the interquartile range between the first and third quartile. The upper and lower lines represent 1.5 times the upper and lower quartile limits. The black line represents the median, and the red X represents the mean of the distribution. Outliers are shown as open circles. Dark green and gold boxes indicate a significant effect of associating mutations within Gag cleavage sites on PI susceptibilities. Susceptibilities are shown as follows: viruses with mutation V82A with and without mutations at Gag 431 (a), viruses with mutation V82A/L90M with and without mutations at Gag 431 (b), viruses with mutation V82A with and without mutations at Gag 437 (c), viruses with mutation V82A/L90M with and without mutations at Gag 437 (d), viruses with mutation V82A with and without mutations at Gag 449 (e), viruses with mutation V82A/L90M with and without mutations at Gag 449 (f), viruses with mutation V82A with and without mutations at Gag 453 (g), and viruses with mutation V82A/L90M with and without mutations at Gag 453 (h). P values of ≤0.05 are significant.
I84V.
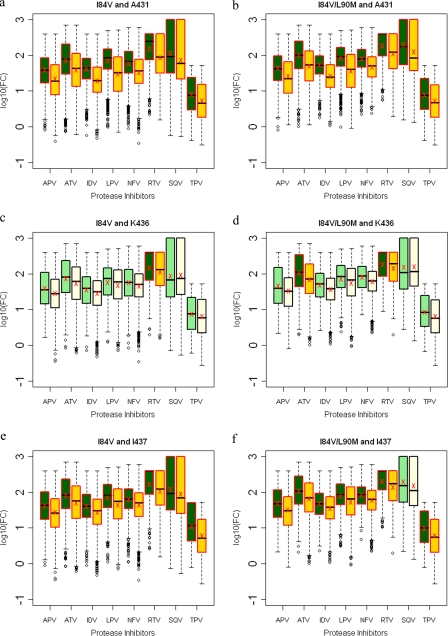
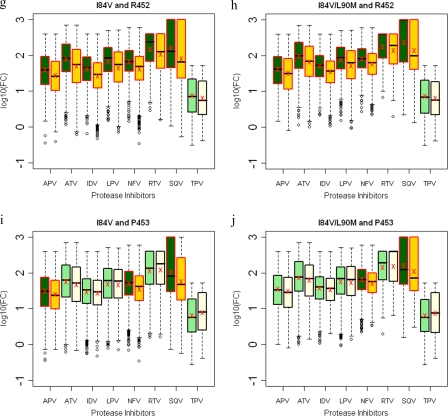
Several Gag cleavage site mutations result in viruses with significant decreases in susceptibilities when combined with the I84V protease mutation. Within NC-p1, mutations at Gag 431 occurred in 53% of the viruses in the presence of I84V or I84V/L90M mutations compared to 14% and 16%, respectively, in the absence of these protease mutations (Fig. 5j and k and see Table S3 in the supplemental material). Decreased susceptibilities to all PIs were observed when mutations at Gag 431 occurred with either I84V alone or I84V/L90M protease mutations (Fig. 9a and b). Even though no associations were observed, mutations at Gag 436 in combination with I84V resulted in decreased susceptibility to RTV, whereas in combination with I84V/L90M, there was decreased susceptibility to ATV and RTV (Fig. 9c and d). Mutations at I437 within the NC-p1 were also associated with these protease mutations (Fig. 5j and k). In the case of viruses with the I84V mutation and mutations at Gag 437, a significant reduction for all PIs was observed (Fig. 9e), whereas with viruses with the I84V/L90M mutation, reduced sensitivity was observed for all PIs except SQV (Fig. 9f). A significant association of mutations was also observed between I84V and mutations within the p1-p6 cleavage site. Mutations at Gag 449 and 453 were the most common, and mutations at Gag 452 were less frequent. Gag 449 mutations associated with I84V and I84V/L90M in 35% and 31% of the viruses, respectively, compared to 13% and 44%, respectively, in the absence of I84V and I84V/L90M mutations (Fig. 5j and k and see Table S3 in the supplemental material). Mutations at Gag 452 were also associated, to a lesser extent, with both I84V and I84V/L90M protease mutations (Fig. 5j and k). No corresponding modulations in drug susceptibilities were observed in viruses with either the I84V or I84V/L90M mutation and with mutations at Gag 449. Significant decreases in susceptibilities were observed with APV, ATV, IDV, LPV, NFV, and especially RTV and SQV, when I84V or I84V/L90M associated with mutations at Gag 452 (Fig. 9g and h). Viruses with mutations at Gag 453 showed reduced susceptibilities to APV, NFV, and SQV in combination with I84V (Fig. 9i), whereas with I84V/L90M reduced susceptibilities were observed with NFV and SQV (Fig. 9j). Thus, several Gag cleavage site mutations, in combination with I84V, modulate the level of susceptibility to various PIs.
FIG. 9.
Box plots (ratio of median FCs on a log scale; y axis) showing susceptibilities of I84V and I84V/L90M viruses to various PIs (x axis) in the presence (green boxes) or absence (yellow boxes) of associating mutations within the NC-p1 and p1-p6 cleavage sites. The boxes represent the interquartile range between the first and third quartile. The upper and lower lines represent 1.5 times the upper and lower quartile limits. The black line represents the median, and the red X represents the mean of the distribution. Outliers are shown as open circles. Dark green and gold boxes indicate a significant effect of associating mutations within Gag cleavage sites on PI susceptibilities. Susceptibilities are shown as follows: viruses with mutation I84V with and without mutations at Gag 431 (a), viruses with mutation I84V/L90M with and without mutations at Gag 431 (b), viruses with mutation I84V with and without mutations at Gag 436 (c), viruses with mutation I84V/L90M with and without mutations at Gag 436 (d), viruses with mutation I84V with and without mutations at Gag 437 (e), viruses with mutation I84V/L90M with and without mutations at Gag 437 (f), viruses with mutation I84V with and without mutations at Gag 452 (g), viruses with mutation I84V/L90M with and without mutations at 452 (h), viruses with mutation I84V with and without mutations at Gag 453 (i), and viruses with mutation I84V/L90M with and without mutations at Gag 453 (j). P values of ≤0.05 are significant.
L90M.
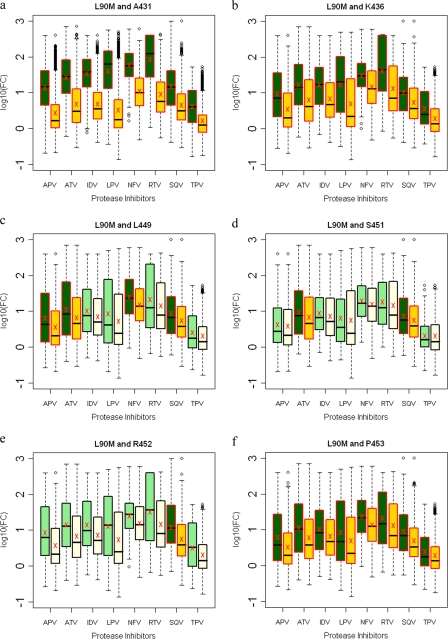
L90M protease mutation is most commonly associated with other primary PI-resistant protease mutations, leading to an increase in resistance. Mutations within both NC-p1 and p1-p6 cleavage sites were not associated when an L90M mutation occurred independently of other primary protease mutations (Fig. 5l). Mutations at Gag 431 and 436 decreased susceptibilities to all PIs in combination with the L90M protease mutation (Fig. 10a and b). Decreased susceptibilities to APV, ATV, NFV, and SQV were observed with mutations at Gag 449, to ATV and SQV with mutations at Gag 451, to SQV with mutations at Gag 452, and to all PIs with mutations at Gag 453 when they occurred in combination with the L90M protease mutation (Fig. 10c to f). Thus, decreased sensitivity to various PIs was observed in combination with mutations in both NC-p1 and p1-p6 cleavage sites.
FIG. 10.
Box plots (ratio of median FCs on a log scale; y axis) showing susceptibilities of viruses with the L90M mutation to various PIs (x axis) in the presence (green boxes) or absence (yellow boxes) of associating mutations within the NC-p1 and p1-p6 cleavage sites. The boxes represent the interquartile range between the first and third quartile. The upper and lower lines represent 1.5 times the upper and lower quartile limits. The black line represents the median, and the red X represents the mean of the distribution. Outliers are shown as open circles. Dark green and gold boxes indicate a significant effect of associating mutations within Gag cleavage sites on PI susceptibilities. Susceptibilities are shown as follows: viruses with mutation L90M with and without mutations at Gag 431 (a), viruses with mutation L90M with and without mutations at Gag 436 (b), viruses with mutation L90M with and without mutations at Gag 449 (c), viruses with mutation L90M with and without mutations at Gag 451 (d), viruses with mutation L90M with and without mutations at Gag 452 (e), and viruses with mutation L90M with and without mutations at Gag 453 (f).
DISCUSSION
NC-p1 and p1-p6 cleavage sites, two critical sites in Gag processing, were systematically analyzed for mutations associated with primary PI resistance mutations in the protease. For every single active site protease mutation studied, at least one, often novel, associated mutation was observed in either of these two critical cleavage sites. In this study, we show that a number of these cleavage site mutations are associated with drug-resistant mutations within the protease. While previous studies have shown increased resistance and/or improved fitness for some pairs of protease cleavage site mutations (1, 6, 10, 11, 17, 18, 21), our analyses reveal that even though only some of these alter fitness, a majority of these cleavage site mutations significantly modulate resistance, usually increasing it.
NC-p1 mutations occurred frequently at Gag 431, 436, and 437, often in combination with several highly resistant protease mutations. Earlier studies have shown that mutations at Gag 436 and 437 enhance Gag processing (6, 18, 25) and increase resistance, although the exact mechanism is not clearly understood. In our study, we observed that the K436R mutation increased resistance to certain PIs in combination with I50V, I84V, and I84V/L90M mutations in protease and that the I437V mutation increased resistance to all PIs with I50V, V82A and I84V mutations in protease. Gag 436 and 437 are farther from the catalytic D25 than Gag 431, thus making structural predictions of their potential mechanism of action less reliable. The A431V mutation results in a more efficient cleavage of the NC-p1 cleavage site irrespective of the protease variant (11) as a result of additional contacts with the protease (32). A recent study showed that in viruses from heavily treated patients, A431V also appeared to directly enhance resistance as a result of this improved NC-p1 processing (6). This nonspecific improvement in processing of the NC-p1 cleavage site is possibly why this mutation was observed frequently in combination with V82A/F, I84V, and I50L protease mutations in our study. Even in combination with the D30N protease mutation, which is not associated with A431V, an increased level of resistance to multiple PIs in the 12% of viruses that have this combination was observed. In the validation studies, unlike with most of the cleavage site mutations, generally more secondary mutations were observed in the protease in viruses with A431V (see Table S1 in the supplemental material), perhaps contributing to the increased resistance and warranting further study. Overall, our study demonstrates that mutations within NC-p1 result in higher levels of resistance when coupled with specific protease mutations.
Mutations within the p1-p6 cleavage site occurred frequently in combination with D30N, D30N/N88D, I84V, and I84V/L90M protease mutations, sometimes enhancing resistance to various PIs. However, mutations within this cleavage site are negatively associated with V82A and V82A/L90M, even while enhancing resistance. Mutations at Gag 449 and 453 alone, in the absence of protease mutations, were previously shown not to reduce susceptibility to PIs (18); however, mutated peptides corresponding to these cleavage sites are better substrates for a variety of protease variants (11, 18). L449F may enhance catalytic efficiency as a result of improved van der Waals contacts, as we and others have previously suggested (11, 17). The overall improved catalytic efficiency from mutations in p1-p6 in combination with specific protease mutations may be part of the potential mechanism by which the observed increase in resistance to various PIs occurs.
Other underlying causes for the observed increased levels of resistance were evaluated and seem not alter our conclusions. Systematic analysis of the impact of these associated NC-p1 and p1-p6 cleavage site mutations on fitness and susceptibility to PIs showed these processes not to be directly coupled. There were no significant improvements in viral fitness due to the cleavage site mutations, even as significant alterations in sensitivity to various PIs were observed. Overall, in resistant viruses, most of the changes in this region of Gag (amino acids 418 to 500) occurred within the cleavage sites, though increased mutation rates were also observed in the PTAP region (Fig. 3) but to a lesser extent. The number of secondary protease mutations observed in viruses with mutations in protease with and without Gag cleavage site mutations generally did not show a trend (see Table S1 in the supplemental material) except with mutations at Gag 431. Thus, the interdependency that leads to modulation of resistance appears to be specific between the mutations in protease and the mutations in the NC-p1 and p1-p6 cleavage sites.
This study strongly suggests that the association of mutations within Gag cleavage sites and protease contributes to PI susceptibility in an inhibitor-specific manner. Mutations within NC-p1 and p1-p6 cleavage sites, in combination with specific protease mutations, were observed to significantly modulate drug susceptibilities. Frequently, these specific combinations enhance PI resistance, while in a few cases increased susceptibility was observed. Modulated sensitivity to PIs due to these mutations may be the result of alterations in polyprotein processing and structural adaptations of both the protease and Gag proteins (6, 13, 18, 24, 25). In mutations in either NC-p1 or p1-p6, cases occur that make the cleavage site a better substrate, complementing the mutated protease even with protease mutations not associated with it and thereby causing increased levels of resistance. Further studies are warranted to determine the exact mechanisms and extent to which these cleavage site mutations contribute to PI resistance. The inclusion of Gag during genotypic or phenotypic resistance testing may lead to improved accuracy in the measurement of viral resistance and have direct implications for evaluating patients failing PI therapy.
Supplementary Material
Acknowledgments
We acknowledge Neil T. Parkin for his assistance in obtaining and evaluating the Monogram Biosciences Inc. data.
This research was supported by the National Institutes of Health (R01-GM65347).
Footnotes
Published ahead of print on 12 August 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bally, F., R. Martinez, S. Peters, P. Sudre, and A. Telenti. 2000. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease. AIDS Res. Hum. Retrovir. 16:1209-1213. [DOI] [PubMed] [Google Scholar]
- 2.Boden, D., and M. Markowitz. 1998. Minireview: resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 42:2775-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner, B. G., J. P. Routy, M. Petrella, D. Moisi, M. Oliveira, M. Detorio, B. Spira, V. Essabag, B. Conway, R. Lalonde, R. P. Sekaly, and M. A. Wainberg. 2002. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J. Virol. 76:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colonno, R., R. Rose, C. McLaren, A. Thiry, N. Parkin, and J. Friborg. 2004. Identification of I50L as the signature atazanavir (ATV)-resistance mutation in treatment-naive HIV-1-infected patients receiving ATV-containing regimens. J. Infect. Dis. 189:1802-1810. [DOI] [PubMed] [Google Scholar]
- 5.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dam, E., R. Quercia, B. Glass, D. Descamps, O. Launay, X. Duval, H. G. Krausslich, A. J. Hance, and F. Clavel. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 5:e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 8.DeLano, W. L. 2008. The PyMol molecular graphics system. DeLano Scientific, LLC, Palo Alto, CA. http://www.pymol.org.
- 9.Delaugerre, C., D. Mathez, G. Peytavin, H. Berthe, K. Long, T. Galperine, and P. de Truchis. 2007. Key amprenavir resistance mutations counteract dramatic efficacy of darunavir in highly experienced patients. AIDS 21:1210-1213. [DOI] [PubMed] [Google Scholar]
- 10.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feher, A., I. T. Weber, P. Bagossi, P. Baross, B. Mahalingam, J. M. Louis, T. D. Copeland, I. Y. Yorshin, R. W. Harrison, and J. Tozser. 2002. Effect of sequence polymorphism and drug resistance on two HIV-1 Gag processing sites. J. Biochem. 269:4114-4120. [DOI] [PubMed] [Google Scholar]
- 12.Gulnik, S. V., L. I. Suvorov, B. Liu, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 34:9282-9287. [DOI] [PubMed] [Google Scholar]
- 13.Ho, S. K., R. M. Coman, J. C. Bunger, S. L. Rose, P. O'Brien, I. Munoz, B. M. Dunn, J. W. Sleasman, and M. M. Goodenow. 2008. Drug-associated changes in amino acid residues in Gag p2, p7(NC), and p6(Gag)/p6(Pol) in human immunodeficiency virus type 1 (HIV-1) display a dominant effect on replicative fitness and drug response. Virology 378:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kantor, R., W. J. Fessel, A. R. Zolopa, D. Israelski, N. Shulman, J. G. Montoya, M. Harbour, J. M. Schapiro, and R. W. Shafer. 2002. Evolution of primary protease inhibitor resistance mutations during protease inhibitor salvage therapy. Antimicrob. Agents Chemother. 46:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan, A. H., S. F. Michael, R. S. Wehbie, M. F. Knigge, D. A. Paul, L. Everitt, D. J. Kempf, D. W. Norbeck, J. W. Erickson, and R. Swanstrom. 1994. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc. Natl. Acad. Sci. USA 91:5597-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, N. M., M. Prabu-Jeyabalan, E. A. Nalivaika, and C. A. Schiffer. 2004. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem. Biol. 11:1333-1338. [DOI] [PubMed] [Google Scholar]
- 17.Kolli, M., S. Lastere, and C. A. Schiffer. 2006. Co-evolution of nelfinavir-resistant HIV-1 protease and the p1-p6 substrate. Virology 347:405-409. [DOI] [PubMed] [Google Scholar]
- 18.Maguire, M. F., R. Guinea, P. Griffin, S. Macmanus, R. C. Elston, J. Wolfram, N. Richards, M. H. Hanlon, D. J. Porter, T. Wrin, N. Parkin, M. Tisdale, E. Furfine, C. Petropoulos, B. W. Snowden, and J. P. Kleim. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 76:7398-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maguire, M. F., S. MacManus, P. Griffin, C. Guinea, W. Harris, N. Richards, J. Wolfram, M. Tisdale, W. Snowden, and J. P. Kleim. 2001. Interaction of HIV-1 protease and gag gene mutations in response to amprenavir-selective pressure exerted in amprenavir-treated subjects—contribution of Gag p6 changes L449F and P453L. Antivir. Ther. 6(Suppl. 1):48. [Google Scholar]
- 20.Mahalingam, B., J. Louis, C. Reed, J. Adomat, J. Krouse, Y. Wang, R. Harrison, and I. Weber. 1999. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 263:238-245. [DOI] [PubMed] [Google Scholar]
- 21.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsuya, Y., M. A. Winters, W. J. Fessel, S. Y. Rhee, L. Hurley, M. Horberg, C. A. Schiffer, A. R. Zolopa, and R. W. Shafer. 2006. N88D facilitates the co-occurrence of D30N and L90M and the development of multidrug resistance in HIV type 1 protease following nelfinavir treatment failure. AIDS Res. Hum. Retrovir. 22:1300-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molla, A., G. R. Granneman, E. Sun, and D. J. Kempf. 1998. Recent developments in HIV protease inhibitor therapy. Antivir. Res. 39:1-23. [DOI] [PubMed] [Google Scholar]
- 24.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 25.Nijhuis, M., N. M. van Maarseveen, S. Lastere, P. Schipper, E. Coakley, B. Glass, M. Rovenska, D. de Jong, C. Chappey, I. W. Goedegebuure, G. Heilek-Snyder, D. Dulude, N. Cammack, L. Brakier-Gingras, J. Konvalinka, N. Parkin, H. G. Krausslich, F. Brun-Vezinet, and C. A. Boucher. 2007. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 4:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pai, V. B., and M. C. A. P. Nahata. 1999. Nelfinavir mesylate: a protease inhibitor. Ann. Pharmacother. 33:325-339. [DOI] [PubMed] [Google Scholar]
- 27.Parkin, N. T., N. S. Hellmann, J. M. Whitcomb, L. Kiss, C. Chappey, and C. J. Petropoulos. 2004. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:437-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Partaledis, J. A., K. Yamaguchi, M. Tisdale, E. E. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, et al. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patick, A., M. Duran, Y. Cao, D. Shugarts, M. Keller, E. Mazabel, M. Knowles, S. Chapman, D. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pazhanisamy, S., J. Partaledis, B. Rao, and D. Livingston. 1998. In vitro selection and characterization of VX-478 resistant HIV-1 variants. Adv. Exp. Med. Biol. 436:75-83. [DOI] [PubMed] [Google Scholar]
- 31.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prabu-Jeyabalan, M., E. A. Nalivaika, N. M. King, and C. A. Schiffer. 2004. Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease. J. Virol. 78:12446-12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prabu-Jeyabalan, M., E. A. Nalivaika, K. Romano, and C. A. Schiffer. 2006. Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate. J. Virol. 80:3607-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabu-Jeyabalan, M., E. A. Nalivaika, and C. A. Schiffer. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369-381. [DOI] [PubMed] [Google Scholar]
- 35.Rhee, S. Y., M. J. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 1997. Mutations in retroviral genes associated with drug resistance. Int. Antivir. News 5:129-142. [Google Scholar]
- 37.Sheng, N., S. C. Pettit, R. J. Tritch, D. H. Ozturk, M. M. Rayner, R. Swanstrom, and S. Erickson-Viitanen. 1997. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 71:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sista, P., B. Wasikowski, P. Lecocq, T. Pattery, and L. Bacheler. 2008. The HIV-1 protease resistance mutation I50L is associated with resistance to atazanavir and susceptibility to other protease inhibitors in multiple mutational contexts. J. Clin. Virol. 42:405-408. [DOI] [PubMed] [Google Scholar]
- 39.Yanchunas, J., Jr., D. R. Langley, L. Tao, R. E. Rose, J. Friborg, R. J. Colonno, and M. L. Doyle. 2005. Molecular basis for increased susceptibility of isolates with atazanavir resistance-conferring substitution I50L to other protease inhibitors. Antimicrob. Agents Chemother. 49:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.