Abstract
Apoptosis is an important antivirus defense. To define the poorly understood pathways by which invertebrates respond to viruses by inducing apoptosis, we have identified replication events that trigger apoptosis in baculovirus-infected cells. We used RNA silencing to ablate factors required for multiplication of Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV). Transfection with double-stranded RNA (dsRNA) complementary to the AcMNPV late expression factors (lefs) that are designated as replicative lefs (lef-1, lef-2, lef-3, lef-11, p143, dnapol, and ie-1/ie-0) blocked virus DNA synthesis and late gene expression in permissive Spodoptera frugiperda cells. dsRNAs specific to designated nonreplicative lefs (lef-8, lef-9, p47, and pp31) blocked late gene expression without affecting virus DNA replication. Thus, both classes of lefs functioned during infection as defined. Silencing the replicative lefs prevented AcMNPV-induced apoptosis of Spodoptera cells, whereas silencing the nonreplicative lefs did not. Thus, the activity of replicative lefs or virus DNA replication is sufficient to trigger apoptosis. Confirming this conclusion, AcMNPV-induced apoptosis was suppressed by silencing the replicative lefs in cells from a divergent species, Drosophila melanogaster. Silencing replicative but not nonreplicative lefs also abrogated AcMNPV-induced shutdown of host protein synthesis, suggesting that virus DNA replication triggers inhibition of host biosynthetic processes and that apoptosis and translational arrest are linked. Our findings suggest that baculovirus DNA replication triggers a host cell response similar to the DNA damage response in vertebrates, which causes translational arrest and apoptosis. Pathways for detecting virus invasion and triggering apoptosis may therefore be conserved between insects and mammals.
DNA and RNA viruses are potent inducers of apoptosis. This host suicide response is an effective antiviral strategy because the destruction of virus-infected cells can significantly diminish virus yields and affect pathogenicity. Supporting the significance of apoptosis as an antiviral defense in invertebrates, insect viruses carry diverse genes that suppress apoptosis and facilitate virus multiplication (reviewed in references 5, 6, and 14). Importantly, the apoptotic response may also influence the competency of insects, including mosquitoes, to function as vectors of human viral pathogens (16, 56). Thus, a better understanding of the molecular mechanisms by which viruses trigger apoptosis in invertebrates and how pathogenesis is affected will provide a critical framework for control of insect-vectored diseases.
The baculoviruses provide an attractive model for defining the mechanisms by which insect viruses trigger apoptosis and thus influence virus dissemination and pathogenesis within an animal. These large, double-stranded DNA viruses cause widespread apoptosis during infection of insects, which include the larvae of moths and butterflies (order Lepidoptera) (5, 6, 14, 49). The baculoviruses encode potent antiapoptotic proteins that counter the host cell response by suppressing premature cell death and thus promoting prolific multiplication. In particular, baculovirus inhibitor-of-apoptosis (IAP), P35, and P49 inhibit the activation or activity of the death proteases known as caspases and thus prevent apoptosis, even in diverse organisms, including mammals (6, 14, 39). In the absence of virus-encoded apoptotic suppressors, baculovirus production and spread within infected larvae is severely restricted (4). Thus, in permissive insects, apoptosis serves to limit virus reproduction and pathology and can be considered an effective innate immune response (5, 7).
The best-studied model for virus-induced apoptosis in invertebrates is the prototype baculovirus Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV). In the absence of antiapoptotic genes, this 134-kb genome DNA virus induces widespread caspase-dependent apoptosis of permissive Spodoptera frugiperda (SF21) cells. This host response severely limits AcMNPV yields and destroys the infected cell by a rapid process that involves chromatin condensation, DNA fragmentation, plasma membrane blebbing, and cytolysis (8, 9, 19, 33). Although virus interaction with host cell receptors is insufficient to trigger apoptosis (25), the virus genes or replication events directly responsible for apoptosis are unknown.
Inhibition of AcMNPV DNA synthesis either by using the DNA polymerase inhibitor aphidicolin or temperature-sensitive AcMNPV mutations reduces virus-induced apoptosis (10, 25). In addition, the activation of host cell caspases coincides with the initiation of viral DNA synthesis (25, 26). These findings suggested that the apoptotic signal is linked to virus DNA replication. However, it was unclear whether DNA replication is the direct trigger or if virus DNA synthesis is indirectly involved because it promotes late multiplicative events, which could trigger apoptosis. The AcMNPV immediate-early protein IE1 is also necessary for virus-induced apoptosis (46). Because this transcriptional activator is required for viral DNA replication and replication-dependent late gene expression (43, 46, 48), it may trigger apoptosis by initiating DNA synthesis or promoting downstream multiplicative events. Alternatively, IE1 may directly activate host pro-death genes, perturb the cell cycle, or inhibit host biosynthetic pathways, all of which are sufficient to cause apoptosis (reviewed in references 18, 24, 29, 30, and 57).
To distinguish the replication events responsible for baculovirus-induced apoptosis, we used RNA silencing to ablate the factors required for AcMNPV multiplication in permissive, apoptosis-sensitive Spodoptera cells. RNA silencing is an effective means by which to selectively reduce viral or cellular gene expression during infection of invertebrates (13, 35, 46, 51). An important advantage to our approach was the capacity to evaluate the function of individual genes during replication of fully infectious virus after normal receptor-mediated entry. Thus, RNA silencing can be more informative than other approaches, such as the overexpression of specific viral genes in the absence of other contributing genes. We used here RNA silencing as a unique means to distinguish the proapoptotic contributions of virus DNA replication from those of viral late gene expression. By selectively silencing components of the AcMNPV-encoded RNA polymerase that is responsible for late and very late virus transcription (reviewed in references 14 and 41), the effects of DNA replication on the host cell were disengaged from those involving late virus gene expression.
We report here that silencing genes essential for AcMNPV DNA replication, defined as replicative late expression factors (lefs), suppressed AcMNPV-induced apoptosis. In contrast, silencing genes that are selectively required for virus late gene expression, defined as nonreplicative lefs, had no effect on virus-induced apoptosis. Thus, the activities of the AcMNPV replicative lefs or events associated with virus DNA replication provide the critical apoptotic signal in the infected cell, including those of the distantly related species Drosophila melanogaster. The replicative but not the nonreplicative lefs also contributed to the inhibition host protein synthesis, which is characteristic of AcMNPV infection. Our study therefore suggests that analogous to certain DNA viruses of vertebrates, the baculoviruses trigger a DNA damage and translational arrest response that culminates in apoptosis of the host cell.
MATERIALS AND METHODS
Cells.
Spodoptera frugiperda IPLB-SF21 cells (55) were propagated in TC100 growth medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone Laboratories). Drosophila melanogaster Schneider DL-1 cells (44) were propagated in Schneider's growth medium (Invitrogen) supplemented with 15% heat-inactivated FBS.
Viruses.
Wild-type (wt) l-1 strain AcMNPV (28) and AcMNPV recombinants wt/lacZ (p35+ polh− lacZ+), vΔ35K/lacZ (p35− polh− lacZ+), vP35 (p35− polh− ie-1prm-p35+ lacZ+), and vOpIAP (p35− polh− ie-1prm-Op-iap+ lacZ+) were described previously (19, 27, 60). The very late promoter of the polyhedrin gene (polh) directs expression of lacZ of recombinant wt/lacZ. The promoter of the immediate-early gene ie-1 directs expression of p35 and Opiap3 from the polh locus of recombinants vP35 and vOpIAP, respectively. For inoculation, extracellular budded virus in TC100 plus 10% FBS was added to SF21 and DL-1 monolayers using 10 and 20 PFU/cell, respectively. After rocking the samples for 1 h at room temperature, the inoculum was replaced with FBS-supplemented growth medium, and the cells were incubated at 27°C.
Plasmids.
pBluescript K/S+ (Invitrogen) plasmids containing portions of the enhanced green fluorescence protein gene (egfp) or the AcMNPV genes gp64 or ie-1 used as templates for in vitro transcription reactions were described previously (46). To generate plasmids with AcMNPV genes p143 and lef-3, a 1,200-bp SacI fragment from plasmid pRESTA-Acp143ORF and a 1,575-bp XbaI-XhoI fragment from pRESTB-Aclef3 (kindly provided by Eric Carstens, Queen's University), respectively, were inserted into pBluescript K/S+. All other baculovirus genes or fragments thereof were cloned by PCR amplification of the AcMNPV genome (GenBank accession number NC_001623). The full-length open reading frames (ORFs) of lef-1, lef-2, lef-11, p47, and lef-9 or a fragment of the ORF of dnapol (ORF nucleotides 61 to 862), lef-8 (ORF nucleotides 427 to 1,838), and pp31 (ORF nucleotides 94 to 837) were each inserted into pBluescript K/S+. The genomic sequences used to generate lef-specific double-stranded RNA (dsRNA) were selected so as not to overlap the ORFs of other AcMNPV lefs with two exceptions. Our ie-1/ie-0-specific dsRNA silenced both ie-1 and ie-0 due to the overlapping nature of the spliced ie-0 and unspliced ie-1 mRNAs (46). The lef-11-specific dsRNA overlapped the pp31 RNA transcript by 136 nucleotides and was expected to ablate pp31 expression; synthesis of PP31 during infection was reduced in lef-11 dsRNA-transfected cells (data not shown). Our pp31-specific dsRNA did not overlap lef-11.
dsRNA transfections.
Single-stranded RNA was synthesized by using in vitro transcription reactions (Ampliscribe T3 and T7 kits; Epicentre) with linearized pBluescript K/S+ plasmids as a template. Complementary RNAs were heated to 65°C and cooled 1°C per min to generate dsRNA. SF21 and DL-1 cells were transfected with a dsRNA-liposome mix as described previously (46). Visual inspection revealed that the dsRNAs tested here had no obvious effect on cell viability.
Immunoblots.
Infected or mock-infected cells were collected by centrifugation, lysed in 1% sodium dodecyl sulfate (SDS)-1% β-mercaptoethanol, and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). After protein transfer, the nitrocellulose membranes were incubated with the following antisera diluted as indicated in parentheses: polyclonal anti-IE1 (1:10,000) (40), monoclonal AcV5 anti-GP64 (a gift from Gary Blissard, Cornell University) (1:500) (20), and mouse monoclonal anti-actin (1:5,000 dilution) (BD Biosciences). Signal development was conducted as described previously (46). Films were scanned at 300 dpi by using an Epson TWAIN Pro scanner and prepared by using Adobe Photoshop CS2 and Adobe Illustrator CS2.
β-Galactosidase assays.
SF21 monolayers were infected with AcMNPV wt/lacZ (multiplicity of infection [MOI] = 10) 24 h after dsRNA transfection. The cells were collected 48 h later, washed, and lysed in Tropix buffer (Galacto-Light Plus kit; Applied Biosystems). The β-galactosidase activity was measured according to the manufacturer's instructions and is reported as the average activity ± the standard deviation determined from triplicate infections.
Cell survival assays.
Intact, nonapoptotic SF21 or DL-1 cells were counted after inoculation with AcMNPV recombinant vΔ35K or vOpIAP, respectively, by using a phase-contrast microscope (Axiovert 135TV; Zeiss) and IP Lab Spectrum P software (BD Biosciences) as described previously (21). The mean ± the standard deviation of surviving cells was determined from six nonoverlapping fields of view from triplicate infections and normalized to that of uninfected cells transfected with control egfp dsRNA.
Quantitation of AcMNPV DNA.
At the indicated times after infection, cells were collected, washed, and suspended in 10 mM Tris (pH 8.0)-1 mM EDTA. SDS and proteinase K were added to 0.2% and 0.1 mg per ml, respectively. After 4 h at 37°C, the mixture was phenol-chloroform extracted and treated with 120 μg of RNase A/ml. Nucleic acid was precipitated with ethanol and suspended in 10 mM Tris (pH 8.0)-1 mM EDTA. Quantitative real-time PCR was performed using 50-μl reactions containing DNA extracted from the equivalent of 25 SF21 cells or 50 DL-1 cells in 1× Taq buffer A (Promega), 5.5 mM MgCl2, 90 nM SuperROX reference dye (Biosearch Technologies), 200 nM deoxynucleoside triphosphates, 100 nM DNA probe, and 1 U of Taq polymerase (Promega). Because the content of Spodoptera frugiperda cellular DNA does not increase during AcMNPV infection (1), nucleic acid samples were normalized for cell equivalents by using the sfiap gene from Spodoptera (22) (provided by John Reed, Burnham Institute) as a genome standard for SF21 cells. Likewise, the β-actin gene from Drosophila (15) (provided by Paul Ahlquist, University of Wisconsin-Madison) was used as a DL-1 genome standard. AcMNPV DNA from purified extracellular budded virus and linearized plasmids containing sfiap or β-actin were used to generate the standard curves used for quantitation. The PCR cycling conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 56°C for 1 min on a real-time PCR machine (7900HT; Applied Biosystems). The following oligonucleotides were used for primers and probes: sfiap, forward primer 5′-ATTGGAAGAACCATGACGTACCCT-3′ (50 nM), reverse primer 5′-ACGTATTCTCGACCCTTCACCAAT-3′ (300 nM), and probe 5′-TET-ACAACACGCAAGGTGGTTTGACCGTTGC-TAMRA-3′; Drosophila β-actin, forward primer 5′-TGACCGACTACCTGATGAAGATCC-3′ (300 nM), reverse primer 5′-GCAACATAGCACAGCTTCTCCTTG-3′ (300 nM), and probe 5′-FAM-TTTCACCACCACCGCTGAGCGTGAAAT-TAMRA-3′; and AcMNPV HindIII-T genome fragment, forward primer 5′-ATTTAACATCGGGCGTGTTAGCTT-3′ (900 nM), reverse primer 5′-TCGCTTTCTAACGTGTTGTCTGAAT-3′ (900 nM), and probe 5′-FAM-CGATTTTGCCATAGCCACACGACGCCT-TAMRA-3′.
Protein radiolabeling.
At the indicated times after infection, the growth medium above SF21 monolayers was replaced with phosphate-buffered saline (pH 6.2) (28) containing 200 μCi of Trans35S-label (1,175 Ci/mmol, methionine at 70%; cysteine at ≤15%; MP Biomedical, LLC)/ml. After 1 h at 27°C, the cells were dislodged, collected by centrifugation, and lysed with 1% SDS-1% β-mercaptoethanol. The lysates were subjected to SDS-PAGE and autoradiography.
RESULTS
RNA silencing of AcMNPV lefs prevents very late gene expression.
To define the virus genes and replication events involved in triggering baculovirus-induced apoptosis, we used dsRNA-mediated silencing to selectively knock down the expression of AcMNPV genes required for essential replicative processes. A principal advantage to this approach is the capacity to evaluate gene function during replication initiated by normal receptor-mediated entry of fully infectious virus (46). By generating gene-specific dsRNA, which was used to transfect cultured cells 24 h prior to inoculation, we ablated viral proteins necessary for AcMNPV DNA replication and late gene expression. To this end, we generated dsRNAs complementary to AcMNPV ie-1/ie-0, p143, lef-1, lef-2, lef-3, lef-11, and dnapol, which are designated replicative lefs because each gene is required for DNA replication and late gene expression in transient-transfection assays (Table 1). Similarly, we synthesized dsRNAs complementary to p47, lef-8, lef-9, and pp31, which are designated as nonreplicative lefs because each is required for late gene expression but not virus DNA replication (Table 1). In particular, we chose p47, lef-8, and lef-9 because they are components of the AcMNPV RNA polymerase complex responsible for late viral transcription (14, 41). As such, ablation of the late RNA polymerase was expected to suppress late and very late multiplicative events.
TABLE 1.
Requirement for AcMNPV replicative and nonreplicative lefs
| Virus gene | Requirement fora: |
Gene function/homologyb | ||
|---|---|---|---|---|
| DNA replication | Late gene expression | Virus apoptosis | ||
| ie-1/ie-0 | + | + | + | Transcription activator, binds DNA origins |
| p143 | + | + | + | Helicase, binds DNA, ATPase |
| lef-1 | + | + | − | DNA primase activity, LEF2 association |
| lef-2 | + | + | + | LEF1 primase accessory factor |
| lef-3 | + | + | + | Single-strand DNA binding, P143 association |
| dnapol | + | + | + | DNA polymerase, 3′→5′ exonuclease |
| lef-11 | + | + | +/− | Unknown function |
| p47 | − | + | − | Late RNA polymerase subunit |
| lef-8 | − | + | − | Late RNA polymerase subunit |
| lef-9 | − | + | − | Late RNA polymerase subunit |
| pp31 | − | + | − | Binds DNA, virogenic stroma association |
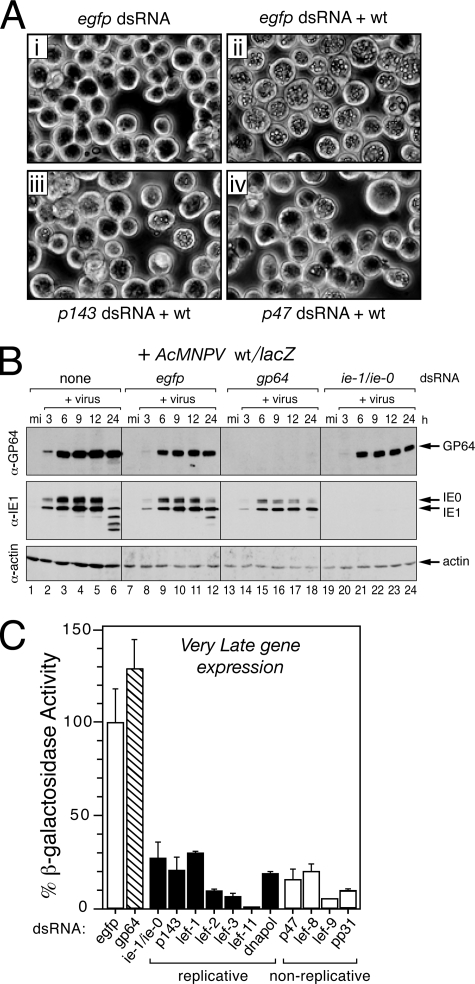
We first showed that dsRNA had no deleterious effects. Transfection with control dsRNA, including egfp-specific dsRNA, did not affect the viability or morphology of mock-infected SF21 cells (Fig. 1Ai). Moreover, upon infection with wild-type AcMNPV, occluded virus particles accumulated in ≥95% of egfp dsRNA-transfected cells, as expected (Fig. 1Aii). The synthesis of essential AcMNPV proteins, including envelope fusion protein GP64 and transactivators IE1 and IE0, was comparable to that in untreated cells (Fig. 1B, compare lanes 2 to 6 and lanes 8 to 12). Thus, control dsRNA had a minimal effect on synthesis of AcMNPV proteins or replication events. In contrast, gp64-specific dsRNA ablated GP64 synthesis without affecting transactivators IE1 or IE0 (Fig. 1B, lanes 14 to 18). Likewise, ie-1/ie-0 dsRNA, which is complementary to the overlapping lefs ie-1 and ie-0 (46), ablated these early proteins without affecting GP64 (Fig. 1B, lanes 20 to 24). Thus, gene-specific dsRNA was effective in knockdown of AcMNPV gene products during infection, as shown previously (46).
FIG. 1.
Silencing of AcMNPV lefs blocks very late gene expression. (A) Polyhedra accumulation. SF21 monolayers were transfected with the indicated dsRNAs and inoculated 24 h later with wild-type (+wt) AcMNPV (MOI = 10). Representative photographs (×500 magnification) taken 48 h after infection are shown. (B) Intracellular levels of AcMNPV early proteins. SF21 cells were transfected and then mock infected (mi) or infected with AcMNPV recombinant wt/lacZ as described for panel A. Whole-cell lysates prepared at the indicated times (in hours) after infection were subjected to immunoblot analysis by using anti-GP64 (top), anti-IE1 (middle), and anti-actin (bottom); IE1 and its larger splice variant IE0 were detected by anti-IE1. (C) Very late polh gene expression. SF21 cells were transfected with the indicated dsRNAs and infected with wt/lacZ (MOI = 10) in which the polh promoter directs lacZ expression. Gene expression was quantified by measuring β-galactosidase in cell extracts prepared 48 h after infection. The values reported are the average β-galactosidase activity ± the standard deviation from duplicate infections normalized to that of wt/lacZ-infected cells transfected with control egfp-specific dsRNA. The results of a representative experiment are shown.
Upon transfection of SF21 cells, dsRNA specific for each of the replicative lefs reduced accumulation of occluded virus produced by wild-type AcMNPV. Occluded virus levels were comparable to that effected by p143-specific dsRNA (Fig. 1Aiii and data not shown), in which virus was evident in only 5 to 10% of the cells and in lower quantities on a per cell basis. Likewise, occluded virus was reduced to comparably low levels by dsRNAs specific to the nonreplicative lefs, including that for p47 (Fig. 1Aiv). To quantify the inhibitory effect of lef silencing on very late gene expression, we monitored β-galactosidase production in dsRNA-treated cells infected with AcMNPV recombinant wt/lacZ, in which the very late polh promoter directs expression of a lacZ reporter. Very late expression of lacZ in cells transfected with dsRNA specific to replicative lefs ie-1/ie-0, p143, lef-1, lef-2, lef-3, lef-11, or dnapol was reduced from 4- to 20-fold compared to that of cells transfected with control egfp dsRNA (Fig. 1C). Transfection with dsRNA specific to each of the nonreplicative lefs p47, lef-8, lef-9, or pp31 reduced very late expression to similar levels. In contrast, dsRNA specific to gp64 had no effect. We concluded that RNA silencing of replicative and nonreplicative lefs was sufficient to suppress AcMNPV late gene expression.
Replicative lef silencing blocks replication of AcMNPV DNA.
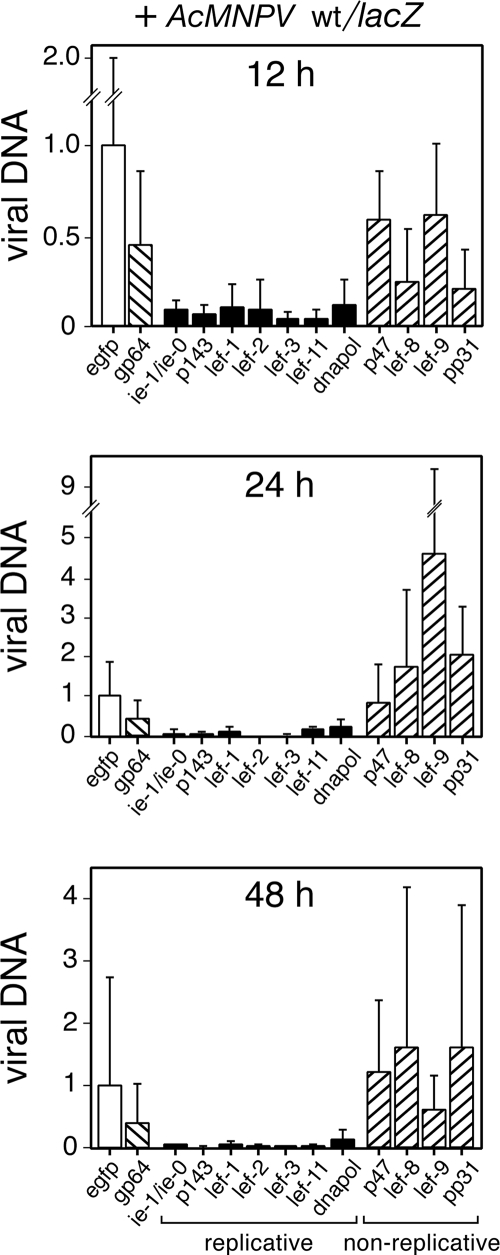
To determine the contribution of the lefs to virus DNA replication during infection, we silenced each lef and monitored virus DNA accumulation by using quantitative real-time PCR. In the absence of dsRNA, newly synthesized AcMNPV DNA was detected in SF21 cells as early as 6 h after infection, whereupon it increased through 12 h and peaked thereafter (Fig. 2A). Verifying the reliability of our PCR assay, this pattern of virus DNA accumulation is typical of L-1 AcMNPV infection of permissive SF21 cells (25, 43). Importantly, the kinetics of viral DNA synthesis in nonpermissive Drosophila cells was comparable (Fig. 2B), as described below. By 12 h after infection, dsRNA specific for the replicative lefs ie-1/ie-0, p143, lef-1, lef-2, lef-3, lef-11, and dnapol reduced accumulation of viral DNA by more than 10-fold compared to that by control egfp dsRNA (Fig. 3). Similar reductions were observed at 24 and 48 h. In contrast, levels of viral DNA in cells transfected with dsRNAs specific to the nonreplicative lefs p47, lef-8, lef-9, or pp31 were as high or higher than that in cells transfected with dsRNA specific for egfp or gp64. Virus DNA accumulation in lef-8- and pp31-silenced cells was reduced at 12 h after infection, but not 24 or 48 h (Fig. 3). These findings were confirmed in Drosophila cells, whereupon lef-8 and pp31 silencing had no effect on AcMNPV DNA levels (see below). We concluded that silencing the replicative lefs blocked or reduced virus DNA accumulation and thus established their role in AcMNPV DNA replication during infection. Moreover, the nonreplicative lefs p47, lef-8, lef-9, and pp31 have little or no effect on virus DNA synthesis.
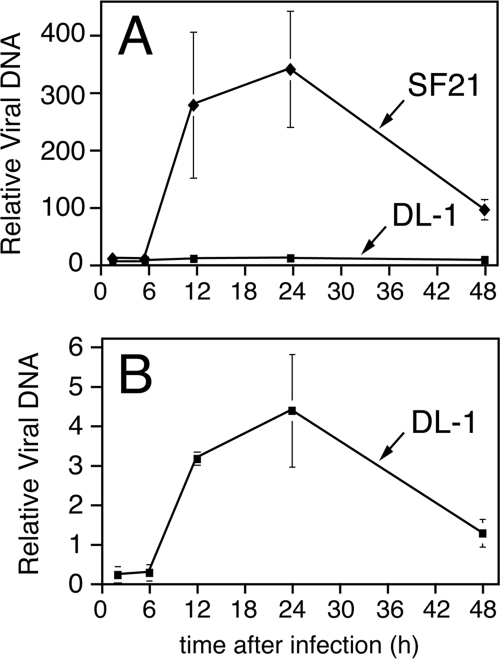
FIG. 2.
Kinetics of intracellular AcMNPV DNA accumulation is similar in lepidopteran and dipteran cells. Spodoptera SF21 and Drosophila DL-1 monolayers were inoculated (MOI = 10) with AcMNPV recombinants wt/lacZ and vP35, respectively; these viruses encode caspase inhibitor p35 under the control of the early p35 or ie-1 promoters, respectively, which prevents apoptosis of the infected cell. At the indicated times after inoculation, cells were collected, lysed, and extracted for total DNA. (A) Intracellular AcMNPV DNA was quantified by using real-time PCR. Values reported are the quantities of virus DNA ± the standard deviation at the indicated times for triplicate plates and normalized to that detected in mock-infected cells. Nucleic acid samples were normalized for cell equivalents by measuring SF21 and DL-1 genomic DNA content by using the Spodoptera sfiap and Drosophila β-actin genes, respectively, as standards. (B) High-resolution depiction of panel A to highlight the lower levels of AcMNPV DNA accumulation in DL-1 cells.
FIG. 3.
AcMNPV DNA synthesis is blocked upon silencing of the replicative lefs. SF21 monolayers were transfected with the indicated dsRNAs and infected 24 h later with AcMNPV wt/lacZ (MOI = 10). At 12, 24, and 48 h after infection, the cells were collected, lysed, and extracted for total DNA. AcMNPV DNA levels were quantified by real-time PCR as described in Fig. 2. Values are reported as the level of viral DNA ± the standard deviation from six different plates compared to that of control egfp dsRNA-transfected, wt/lacZ-infected cells. Solid and cross-hatched bars depict values obtained upon transfection of dsRNA specific for replicative and nonreplicative lefs, respectively.
Replicative but not nonreplicative lefs contribute to AcMNPV-mediated inhibition of host protein synthesis.
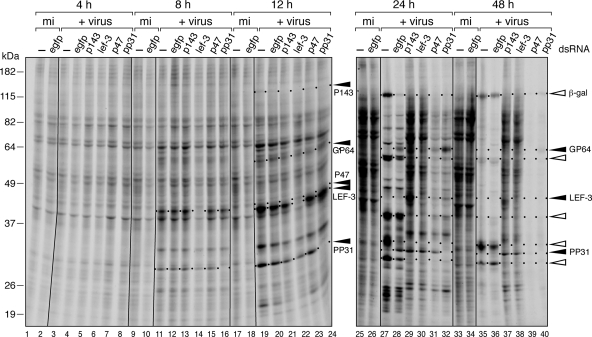
To verify the effects of gene silencing on virus replicative events, we monitored the temporal synthesis of viral proteins. Early, late, and very late phases of AcMNPV protein synthesis are readily distinguished by radiolabeling (8, 9, 19, 33). We therefore labeled dsRNA-transfected cells with [35S]methionine-cysteine for 1-h periods after infection with AcMNPV wt/lacZ. As expected, dsRNA specific to p143, lef-3, p47, and pp31 ablated the protein product of each gene as indicated by the absence of these proteins at 8 and 12 h after infection (Fig. 4, lanes 11 to 16 and lanes 19 to 24). Upon silencing these individual genes, there was little or no effect on the early pattern (4 and 8 h) of virus and host proteins compared to that of cells transfected with control egfp dsRNA (lanes 1 to 16). In contrast, p143- and lef-3-silenced cells synthesized no detectable late (24 h) or very late (48 h) viral proteins (lanes 29 to 30 and lanes 37 and 38). Rather, these cells continued to synthesize several early viral proteins then. Upon silencing the late RNA polymerase subunit gene p47 or nonreplicative pp31, late and very late virus proteins were reduced or eliminated (lanes 31 to 32 and lanes 39 and 40). By comparison, untreated or egfp dsRNA-treated cells exhibited normal or near normal synthesis of late and very late proteins (lanes 27 to 28 and lanes 35 and 36), including polh promoter-directed β-galactosidase. We concluded that silencing the replicative and nonreplicative lefs was sufficient to block the transition from early to late phases of infection.
FIG. 4.
AcMNPV-mediated inhibition of host protein synthesis is prevented upon silencing of replicative lefs. SF21 monolayers were transfected with the indicated dsRNAs and infected 24 h later with AcMNPV wt/lacZ (MOI = 10). The cells were radiolabeled for 1 h with [35S]methionine-cysteine, collected at the indicated times (in hours), and lysed. Protein samples from equal cell numbers were subjected to SDS-PAGE and autoradiography. Early and late/very late virus proteins are denoted by closed and open arrows, respectively; unlabeled arrows denote unknown late virus proteins. Protein size standards (in kilodaltons) are indicated to the left. The two autoradiographs (4 to 12 h and 24 to 48 h) represent two contiguous gels of a representative experiment.
At late times after AcMNPV infection, host protein synthesis declines dramatically, whereas late viral proteins accelerate. Although the mechanisms and baculovirus genes responsible for this host shutdown are unknown (for a review, see reference 49), the response is common among diverse viruses. AcMNPV inhibition of host protein synthesis was readily apparent in untreated and egfp dsRNA-transfected cells by 24 and 48 h (Fig. 4, lanes 27 to 28 and lanes 35 and 36). In contrast, cells transfected with dsRNA specific to replicative lefs p143 and lef-3 exhibited near-normal levels of host protein synthesis, as indicated by efficient radiolabeling of nonvirus proteins at the same times (lanes 29 to 30 and lanes 37 and 38). Silencing the envelope fusion protein gene gp64, another early gene, had no effect on the inhibition of host protein synthesis (data not shown). Thus, silencing these replicative lefs was sufficient to suppress virus-mediated inhibition of host protein synthesis. In contrast, host protein synthesis inhibition was unaffected upon silencing the nonreplicative lefs p47 and pp31 (Fig. 4, lanes 31 to 32 and lanes 39 and 40). By 48 h, few if any host proteins were radiolabeled in either p47- or pp31-silenced cells (lanes 39 to 40) compared to p143- and lef-3-silenced cells (lanes 37 and 38). Thus, these nonreplicative lefs failed to contribute to AcMNPV-mediated shutoff of host protein synthesis. We concluded that whereas both replicative and nonreplicative lefs are required for the transition to late stages of infection, only the replicative lefs are required for virus-mediated translational arrest. This finding is consistent with viral DNA replication acting as the signal for host protein synthesis shutdown.
Replicative but not nonreplicative lefs are required for AcMNPV-induced apoptosis.
AcMNPV-mediated apoptotic signaling requires ie-1 (46). However, it was unclear whether the proapoptotic activity of ie-1 is due to its promotion of downstream virus events, including virus DNA replication, late gene expression, or both. Here, by selectively silencing components of the AcMNPV late stage RNA polymerase, it was possible to independently evaluate the contribution of virus DNA replication and late virus gene expression to apoptotic signaling.
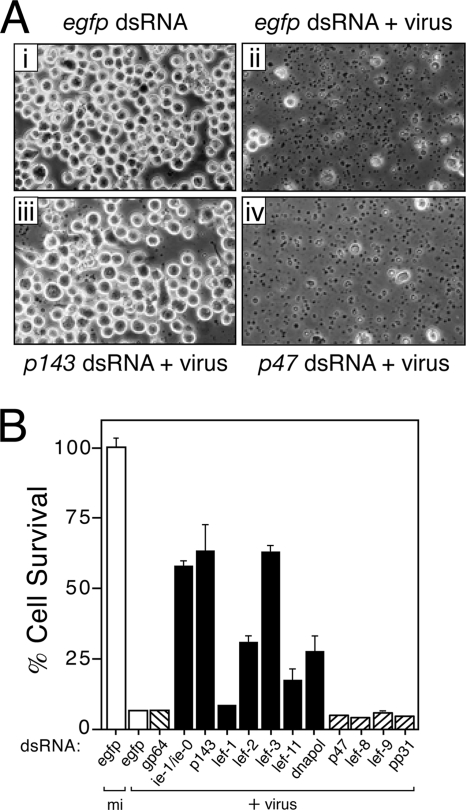
To this end, we monitored the level of virus-induced apoptosis in RNA-silenced SF21 cells by using the AcMNPV mutant vΔ35K/lacZ (p35− polh− lacZ+), which lacks the caspase inhibitor p35 and thus cannot prevent caspase-mediated apoptosis. Transfection of control egfp dsRNA had no effect on the capacity of vΔ35K/lacZ to trigger widespread apoptosis in SF21 cells (Fig. 5A, compare panels i and ii). By 24 h after infection, apoptotic blebbing and cytolysis encompassed >90% of the culture, which was comparable to that in the absence of dsRNA (46). Proteolytic processing of the Spodoptera effector caspase, Sf-caspase-1, and the usual appearance of intracellular caspase activity confirmed that cell death was by apoptosis (data not shown). In contrast, p143-specific dsRNA prevented vΔ35K/lacZ-induced apoptosis (Fig. 5Aiii). Quantitation indicated that >60% of these cells survived through 24 h (Fig. 5B), which was comparable to that of cells treated with ie-1/ie-0 dsRNA (46). Because p143-silenced cells synthesized early viral proteins (except P143) at normal levels (Fig. 4), the lack of apoptosis was not due to inefficient infection. Virus-induced apoptosis was also reduced upon silencing the replicative lefs lef-3, lef-2, dnapol, and lef-11, in order of decreasing effectiveness (Fig. 5B); cell survival was 2.5- to 10-fold higher than that of cells transfected with control egfp or gp64 dsRNA. Interestingly, lef-1 had the smallest contribution to virus-induced apoptosis in SF21 cells (Fig. 5B). Nonetheless, lef-1 was essential for virus-induced apoptosis in Drosophila cells (see below). We concluded that replicative lefs are required for AcMNPV-induced apoptosis but that they differ in their proapoptotic activities. It is noteworthy that when silenced individually none of the replicative lefs restored survival of vΔ35K/lacZ-infected cells to 100%. We attributed this effect to either incomplete silencing that allowed very low levels of DNA replication or to other early proapoptotic events not associated with virus DNA replication, as suggested previously (25).
FIG. 5.
AcMNPV-induced apoptosis is blocked upon silencing of replicative but not nonreplicative lefs. (A) Apoptotic cytolysis. SF21 monolayers were transfected with the indicated dsRNAs and inoculated (+virus) 24 h later with apoptosis-inducing AcMNPV p35-deletion mutant vΔ35K/lacZ (MOI = 10). Representative photographs (×250 magnification) taken 26 h after infection are shown. (B) Cell survival. Intact SF21 cells that were mock-infected (mi) or infected (+virus) 26 h earlier with vΔ35K/lacZ as described for panel A were counted by using computer-aided microscopy. Values shown represent the percent ± the standard deviation of surviving cells averaged for three independent plates and compared to that of mock-infected, egfp dsRNA-transfected cells. The results of a representative experiment are shown.
When SF21 cells were transfected with dsRNA specific to nonreplicative lefs p47, lef-8, lef-9, or pp31, they remained sensitive to AcMNPV-induced apoptosis (Fig. 5B). For each of the nonreplicative lefs, <5% of the cells survived infection with vΔ35K/lacZ. Virus-induced apoptotic blebbing and cytolysis were extensive, as typified by p47-silenced cells (Fig. 5Aiv). Because the conditions for silencing the nonreplicative lefs were identical to those that prevented late gene expression (Fig. 1 and 4), we concluded that silencing was achieved here. These findings indicated that the nonreplicative lefs are not required for virus-induced apoptosis and therefore suggested for the first time that late virus multiplicative events contribute little to apoptotic signaling.
Replicative lefs are required for AcMNPV DNA replication in nonpermissive dipteran cells.
To further test the role of viral DNA replication in triggering apoptosis, we determined the effect of lef ablation on AcMNPV-induced apoptosis of DL-1 cells, a Drosophila cell line that is highly responsive to RNA-mediated gene silencing (27, 46, 47). DL-1 cells fail to support productive infection by AcMNPV but are highly sensitive to apoptosis induced by inoculation of AcMNPV mutants that lack apoptotic suppressors (27, 34, 37, 60). Thus, early virus events that include DNA replication may be sufficient to trigger apoptosis (46). To define the kinetics of AcMNPV DNA replication in DL-1 cells, we quantified intracellular viral DNA. Real-time PCR detected virus DNA by 6 h after inoculation (Fig. 2B). This DNA increased through 12 and 24 h and then declined. At peak accumulation, virus DNA was ∼80-fold lower than that of permissive SF21 cells on a per-cell basis (Fig. 2A). Nonetheless, the kinetics of virus DNA accumulation was strikingly similar in both cell lines and argued that viral DNA synthesis is an active process in Drosophila cells.
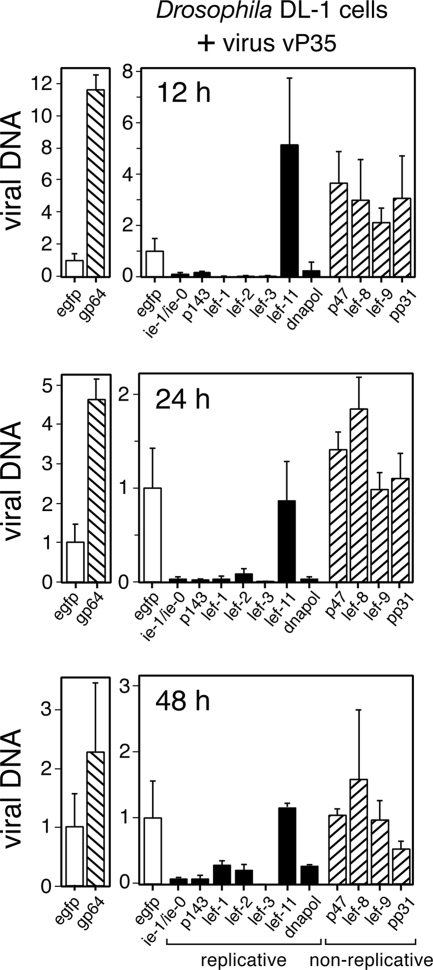
Active DNA synthesis was confirmed by demonstrating that replicative lef silencing decreased AcMNPV DNA accumulation in DL-1 cells (Fig. 6). Transfection with dsRNA specific to the replicative lefs ie-1/ie-0, p143, lef-1, lef-2, lef-3, and dnapol reduced virus DNA accumulation at 12, 24, and 48 h after inoculation by ca. 10- to 30-fold compared to that in the presence of egfp dsRNA. Only lef-11 dsRNA failed to affect virus DNA accumulation (Fig. 6). In contrast to the replicative lefs, dsRNAs specific to the nonreplicative lefs p47, lef-8, lef-9, and pp31 had no effect. Interestingly, virus DNA accumulation was highest in gp64-silenced cells. We concluded that the replicative lefs, with the exception of lef-11, are required for virus DNA synthesis in Drosophila cells. Conversely, the nonreplicative lefs are dispensable.
FIG. 6.
AcMNPV DNA synthesis in DL-1 cells is blocked upon silencing of replicative lefs. DL-1 monolayers were transfected with the indicated dsRNAs and infected 24 h later with AcMNPV vP35 (MOI = 10). Intracellular virus DNA was quantified by real-time PCR as described in Fig. 2. Values are reported as the level of viral DNA ± the standard deviation from four different plates relative to that of control egfp dsRNA-transfected, vP35-infected cells. Solid and cross-hatched bars depict values obtained upon transfection of dsRNA specific for replicative and nonreplicative lefs, respectively.
Replicative lefs are required for AcMNPV-induced apoptosis in DL-1 cells.
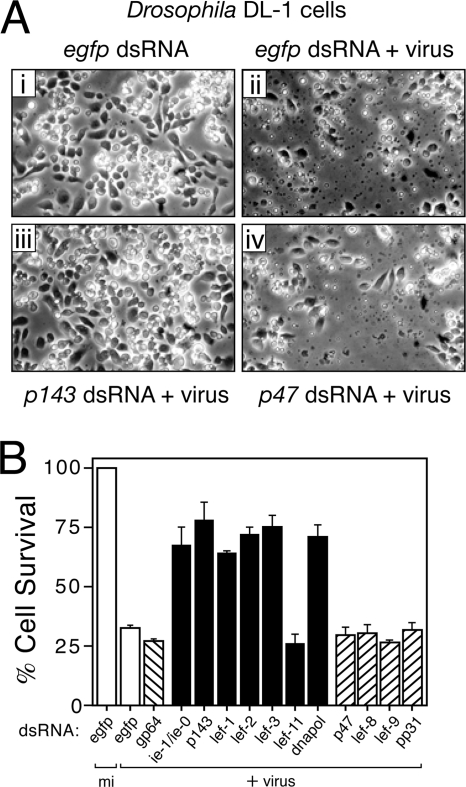
Having established that replicative lefs are required for virus DNA synthesis, we next tested their contribution to virus-induced apoptosis. To this end, we used AcMNPV recombinant vOpIAP, which lacks p35 but encodes Op-iap3 that fails to block apoptosis in Drosophila (27, 58, 60). Op-iap3 prevents apoptosis in permissive SF21 cells and thus facilitated production of high-titered vOpIAP stocks used here. As expected, vOpIAP caused widespread apoptosis in DL-l cells transfected with control egfp dsRNA (Fig. 7Aii); by 24 h after inoculation, >75% of these cells underwent membrane blebbing and cytolysis, which is the result of Drosophila caspase activation (46). The absence of cytolysis of mock-infected cells transfected with the same dsRNA indicated that apoptosis was virus mediated (Fig. 7Ai). In contrast, transfection with replicative p143-specific dsRNA suppressed virus-induced apoptosis (Fig. 7Aiii); >75% of these cells survived virus inoculation through 24 h, which was a level comparable to that of ie-1-silenced cells (Fig. 7B). Likewise, dsRNA specific to replicative lef-1, lef-2, lef-3, or dnapol increased cell survival. Only lef-11-specific dsRNA failed to suppress apoptosis (Fig. 7B). Because only those replicative lefs that contributed to virus-induced apoptosis were also necessary for AcMNPV DNA synthesis (Fig. 6), we concluded that viral DNA synthesis triggers apoptosis in Drosophila cells.
FIG. 7.
AcMNPV-induced apoptosis of DL-1 cells is blocked upon silencing of replicative but not nonreplicative lefs. (A) Apoptotic cytolysis. DL-1 monolayers were transfected with the indicated dsRNAs and inoculated (+virus) 24 h later with AcMNPV recombinant vOpIAP (MOI = 10), which lacks p35 and causes vigorous apoptosis in DL-1 cells (27, 58, 60). Representative photographs (×250 magnification) taken 24 h after infection are shown. (B) Cell survival. Intact DL-1 cells that were mock-infected (mi) or infected (+virus) 24 h earlier with vOpIAP as described for panel A were counted by using computer-aided microscopy. Values shown represent the percent ± the standard deviation of surviving cells averaged for three independent plates and normalized to that of mock-infected, egfp dsRNA-transfected cells. The results of a representative experiment are shown.
The AcMNPV nonreplicative lefs p47, lef-8, lef-9, and pp31 had little or no effect on vOpIAP-induced apoptosis of DL-1 cells (Fig. 7B). The reduced survival of these cells was comparable to those transfected with control egfp or gp64 dsRNAs. Moreover, the level of virus-induced membrane blebbing and cytolysis was comparable to that of control egfp dsRNA-treated cells (Fig. 7A). Although these findings suggested that the nonreplicative lefs fail to contribute to virus-induced apoptosis, it is relevant to note that AcMNPV late and very late gene expression is greatly reduced or absent in DL-1 cells (37). Thus, in the absence of late multiplicative events it is unlikely that silencing of nonreplicative lefs would have an observed effect.
DISCUSSION
By using RNA silencing to selectively knock down expression of essential genes required for baculovirus multiplication, we report here that AcMNPV replicative lefs, but not nonreplicative lefs, are required to trigger apoptosis in cells derived from different orders of insects (Lepidoptera and Diptera). Our study indicates that baculovirus DNA replication activities or events, rather than virus late gene expression, are responsible for host apoptosis. Importantly, RNA silencing of replicative but not nonreplicative lefs abrogated AcMNPV-induced shutoff of host protein synthesis, suggesting that virus DNA replication also triggers inhibition of host biosynthetic processes during infection. Thus, the shutoff of host protein synthesis and apoptotic signaling may be linked. Our findings are consistent with a model in which baculovirus DNA replication triggers a host cell response, like that occurring during DNA damage or cell cycle perturbations, which cause translational arrest and apoptosis in vertebrates. Thus, pathways for sensing virus invasion and triggering apoptosis appear to be conserved between insects and mammals.
Different roles for AcMNPV lefs during permissive infection.
The lefs were originally defined by transient-transfection assays as factors required for baculovirus late gene expression and were subsequently assigned as replicative and nonreplicative on the basis of their role in transient DNA replication assays (41). As determined by gene-specific RNA silencing, we report here that AcMNPV lefs ie-1/ie-0, p143, lef-1, lef-2, lef-3, lef-11, and dnapol (Table 1) contribute to virus DNA replication and late gene expression during permissive AcMNPV infection (Fig. 1 and 3). Our data confirm the critical roles of ie-1/ie-0, p143, lef-11, and dnapol as demonstrated by conditional lethal mutations or gene knockouts (31, 32, 48, 53). The roles of lef-1, lef-2, and lef-3 for DNA replication, and thus late gene expression, during infection are established here. We also demonstrated that IE1 is required for AcMNPV DNA replication during infection (Fig. 3 and 6). In addition to its transactivation potential for replicative genes, IE1 likely has a direct role in AcMNPV DNA synthesis (D. Taggart and P. Friesen, unpublished data). AcMNPV nonreplicative lefs had little or no effect in virus DNA replication but were required for late gene expression (Fig. 1 and 3). Thus, our study confirmed the critical roles of p47 and pp31 for late gene expression (3, 59) and demonstrated definitive roles for RNA polymerase subunits lef-8 and lef-9 during infection.
RNA silencing is effective because it selectively ablates gene products and thereby abolishes gene function. Here, we verified the loss of viral proteins IE1, IE0, and EFP GP64 by immunoblot analysis of silenced cells (Fig. 1B). Ablation of P143, LEF-3, P47, and PP31 was confirmed by protein radiolabeling during infection (Fig. 4). Due to low-level synthesis or antisera unavailability, we have not confirmed the depletion of LEF-1, -2, -8, -9, -11, or DNA polymerase. However, because of the striking inhibition of AcMNPV multiplicative processes effected by dsRNAs specific to their genes (Fig. 1, 3, and 6), we concluded that significant silencing was accomplished.
Induction of apoptosis by AcMNPV DNA replication.
Previous studies have suggested that baculovirus DNA synthesis or late gene expression, which is dependent on virus DNA replication, triggers apoptosis during infection (10, 25). By preventing late gene expression through silencing nonreplicative lefs comprising the AcMNPV late stage RNA polymerase (p47, lef-8, and lef-9), we demonstrated here that late gene expression is not required for virus-induced apoptosis of Spodoptera cells (Fig. 4). Conversely, ablation of six of seven AcMNPV replicative lefs, including ie-1/ie-0, p143, lef-2, lef-3, lef-11, and dnapol, arrested virus-induced apoptosis. Because silencing of multiple, independent targets required for virus DNA replication had the identical effect of suppressing virus-induced apoptosis, it is likely that one or more activities constituting the DNA replication process is responsible for triggering apoptosis. Not ruled out is the possibility that a late virus gene, expressed independent of the late-stage RNA polymerase but dependent on virus DNA replication, is responsible for apoptosis. However, this possibility is unlikely because dsRNA-mediated ablation of pp31 was as ineffective in preventing AcMNPV-induced apoptosis as that of the late RNA polymerase subunits (Fig. 4). The pp31 product is a DNA-binding protein not associated with the late RNA polymerase but required for late gene expression (41).
It is noteworthy that among the replicative lefs, ablation of ie-1/ie-0, p143, or lef-3 had the strongest suppressive effect on AcMNPV-induced apoptosis (Fig. 4). This difference could be attributed to enhanced sensitivity of these lefs to RNA silencing. Alternatively, it is possible that the replicative lefs differ in their proapoptotic activities. In particular, IE1 may upregulate expression of the other replicative lefs through its transactivator properties, as well as initiate viral DNA synthesis by direct binding to origins of virus DNA replication and recruiting factors of the replication complex (D. Taggart and P. Friesen, unpublished data). LEF-3 and P143, which are interdependent for nuclear import, may also provide required early functions for DNA synthesis, including virus DNA unwinding and formation of the replication complex (reviewed in reference 52). Thus, ablation of any one of these replicative factors may have a potent negative effect at the earliest stage of apoptotic signaling. Interestingly, lef-1 was required for AcMNPV DNA synthesis, but not virus-induced apoptosis in Spodoptera cells (Fig. 1 and 4). In contrast, lef-1 was required for both activities in Drosophila cells. Although additional studies are required to define the mechanisms by which lef-1 and the other lefs promote virus DNA replication and trigger apoptosis (see below), this finding strengthens the premise that the replicative lefs have nonequivalent proapoptotic activities.
AcMNPV DNA replication-induced apoptosis in Drosophila.
The proapoptotic activities of the replicative lefs were confirmed by our finding that they are also required for AcMNPV-induced apoptosis in Drosophila DL-1 cells (Fig. 7). Although dipteran DL-1 cells are nonpermissive for AcMNPV, inoculation with mutants that lack antiapoptotic genes causes widespread apoptosis that is caspase dependent (27, 37, 58, 60). Quantitative PCR revealed that although total accumulation of AcMNPV DNA is significantly lower than that in permissive Spodoptera cells, the kinetics of viral DNA synthesis were comparable (Fig. 2). With the exception of lef-11, all of the replicative lefs were required for viral DNA synthesis in DL-1 cells (Fig. 6); the nonreplicative lefs were dispensable. We concluded that the mechanics of AcMNPV DNA synthesis are similar in both Drosophila and Spodoptera. Each of the replicative lefs required for viral DNA synthesis also contributed to AcMNPV-induced apoptosis (Fig. 7). This finding suggested that AcMNPV triggers apoptosis by comparable mechanisms in dipteran and lepidopteran species. Thus, the pathway by which insects detect DNA virus entry and respond by apoptosis may be conserved. Our study also endorses the use of Drosophila as an advantageous model system for defining mechanisms of DNA virus-induced apoptosis.
Host protein synthesis arrest by AcMNPV replicative lefs.
Inhibition of host protein synthesis is common among viruses (reviewed in references 11 and 45). Baculoviruses induce a dramatic but poorly understood shutoff of host protein synthesis that parallels late and very late gene expression (49). It is unclear whether this shutoff is a virus-based strategy to selectively facilitate virus protein synthesis or whether it is a host-mediated response to establish an antiviral state. We discovered here that silencing AcMNPV replicative lefs prevented virus-induced translational arrest of host proteins (Fig. 4 and data not shown); nonreplicative lefs had no effect. Because ablation of independent replicative lef targets had the same effect, we concluded that virus DNA replication activities rather than late gene functions trigger the inhibitory response.
Our finding that host translational arrest and apoptosis are triggered by the same baculovirus replication events raises the possibility that both processes are linked and represent host responses. Shutoff of host protein synthesis can trigger apoptosis by blocking replenishment of short-lived antiapoptotic proteins, including the invertebrate IAP proteins that are required for cell survival (18, 24). For example, nodavirus-mediated depletion of the principal IAP of Drosophila, DIAP1, can trigger apoptosis of Drosophila DL-1 cells upon infection (47). Our preliminary studies indicated that DIAP1 and SfIAP, the Spodoptera cellular IAP, are also depleted upon AcMNPV infection (R. Cerio, K. Schultz, Rianna Vandergaast, and P. Friesen, unpublished data). In mammals, protein synthesis inhibition often precedes cell cycle arrest and apoptosis that is mediated by tumor suppressor p53, which is activated upon virus infection or DNA damage (12). In Drosophila, DNA damage-induced activation of Dmp53 upregulates proapoptotic genes, including reaper, that antagonize DIAP1 antiapoptotic activity and inhibit global protein synthesis (reviewed in references 50 and 54). In Spodoptera, baculovirus DNA replication-triggered protein synthesis arrest may provide supplementary contributions toward apoptosis. Thus, analogous to the large herpesviruses in which multiple genes or events cause apoptosis (17, 38), baculoviruses may also present multiple apoptotic triggers (25). Further study is required to distinguish the potential roles of host lepidopteran genes in translational arrest and apoptosis during baculovirus infection.
Mechanisms of baculovirus DNA replication-induced apoptosis.
The induction of unscheduled DNA synthesis during infection or the detection of replicating viral DNA as damaged DNA are potent proapoptotic signals in vertebrates (reviewed in references 29, 30, and 57). Because the replication of DNA virus genomes often requires active host DNA synthetic machinery, pathogens such as adenovirus, simian virus 40, and human papillomavirus induce cell cycle progression that pushes cells into S phase and as a consequence triggers the DNA damage apoptosis pathways. Thus, to promote multiplication, these viruses encode factors that inactivate p53 and block apoptosis (reviewed in references 29, 30, and 57).
DNA damage and unscheduled DNA synthesis are sufficient to trigger apoptosis in invertebrates, including Drosophila (2, 36, 54). The highly conserved DNA repair machinery of Drosophila is capable of recognizing DNA breaks and single-stranded DNA, which activates Dmp53 and upregulates proapoptotic factors including Reaper (2, 36). Representing various intermediates during virus DNA replication, double-stranded DNA ends or stretches of single-stranded DNA are recognized as damaged DNA (30). It is unknown how the large (>120 kb), circular DNA genome of baculoviruses is replicated (reviewed in references 39 and 52). Several replication strategies have been proposed, including rolling circle and recombination-mediated mechanisms. Each of these pathways involves the generation of replication intermediates that should be recognized by the host repair machinery as damaged DNA and thereby trigger cell cycle arrest and apoptosis. Indeed, AcMNPV replication causes cell cycle perturbations (1, 23, 42). Thus, the pathways by which apoptosis are triggered in DNA virus-infected cells may be conserved between insects and mammals. Additional studies should uncover both novel and conserved mechanisms by which baculovirus DNA replication is detected by the host insect cell and triggers apoptosis.
Acknowledgments
We thank Eric Carstens (Queen's University) for the gift of the p143 and lef-3 plasmids, John Reed (Burnham Institute) for sfiap plasmid, Paul Ahlquist (University of Wisconsin-Madison) for the β-actin plasmid, and Gary Blissard (Cornell University) for GP64 monoclonal antibody. We also thank Justin Wetter for helpful discussions.
This study was supported in part by Public Health Service grants AI25557 and AI40482 from the National Institute of Allergy and Infectious Diseases (P.D.F.).
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Braunagel, S. C., R. Parr, M. Belyavskyi, and M. D. Summers. 1998. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology 244:195-211. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky, M. H., B. T. Weinert, G. Tsang, Y. S. Rong, N. M. McGinnis, K. G. Golic, D. C. Rio, and G. M. Rubin. 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 24:1219-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carstens, E. B., A. L. Lu, and H. L. Chan. 1993. Sequence, transcriptional mapping, and overexpression of p47, a baculovirus gene regulating late gene expression. J. Virol. 67:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke, T. E., and R. J. Clem. 2003. In vivo induction of apoptosis correlating with reduced infectivity during baculovirus infection. J. Virol. 77:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clem, R. J. 2007. Baculoviruses and apoptosis: a diversity of genes and responses. Curr. Drug Targets 8:1069-1074. [DOI] [PubMed] [Google Scholar]
- 6.Clem, R. J. 2001. Baculoviruses and apoptosis: the good, the bad, and the ugly. Cell Death Differ. 8:137-143. [DOI] [PubMed] [Google Scholar]
- 7.Clem, R. J. 2005. The role of apoptosis in defense against baculovirus infection in insects. Curr. Top. Microbiol. Immunol. 289:113-129. [DOI] [PubMed] [Google Scholar]
- 8.Clem, R. J., M. Fechheimer, and L. K. Miller. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388-1390. [DOI] [PubMed] [Google Scholar]
- 9.Clem, R. J., and L. K. Miller. 1993. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J. Virol. 67:3730-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clem, R. J., and L. K. Miller. 1994. Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell. Biol. 14:5212-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemens, M. J. 2005. Translational control in virus-infected cells: models for cellular stress responses. Semin. Cell Dev. Biol. 16:13-20. [DOI] [PubMed] [Google Scholar]
- 12.Constantinou, C., and M. J. Clemens. 2007. Regulation of translation factors eIF4GI and 4E-BP1 during recovery of protein synthesis from inhibition by p53. Cell Death Differ. 14:576-585. [DOI] [PubMed] [Google Scholar]
- 13.Flores-Jasso, C. F., V. J. Valdes, A. Sampieri, V. Valadez-Graham, F. Recillas-Targa, and L. Vaca. 2004. Silencing structural and nonstructural genes in baculovirus by RNA interference. Virus Res. 102:75-84. [DOI] [PubMed] [Google Scholar]
- 14.Friesen, P. D. 2007. Insect viruses, p. 707-736. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 15.Fyrberg, E. A., B. J. Bond, N. D. Hershey, K. S. Mixter, and N. Davidson. 1981. The actin genes of Drosophila: protein coding regions are highly conserved but intron positions are not. Cell 24:107-116. [DOI] [PubMed] [Google Scholar]
- 16.Girard, Y. A., B. S. Schneider, C. E. McGee, J. Wen, V. C. Han, V. Popov, P. W. Mason, and S. Higgs. 2007. Salivary gland morphology and virus transmission during long-term cytopathologic West Nile virus infection in Culex mosquitoes. Am. J. Trop. Med. Hyg. 76:118-128. [PubMed] [Google Scholar]
- 17.Han, J. Y., S. A. Miller, T. M. Wolfe, H. Pourhassan, and K. R. Jerome. 2009. Cell type-specific induction and inhibition of apoptosis by herpes simplex virus type 2 ICP10. J. Virol. 83:2765-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hay, B. A., and M. Guo. 2006. Caspase-dependent cell death in Drosophila. Annu. Rev. Cell Dev. Biol. 22:623-650. [DOI] [PubMed] [Google Scholar]
- 19.Hershberger, P. A., J. A. Dickson, and P. D. Friesen. 1992. Site-specific mutagenesis of the 35-kilodalton protein gene encoded by Autographa californica nuclear polyhedrosis virus: cell line-specific effects on virus replication. J. Virol. 66:5525-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125:432-444. [DOI] [PubMed] [Google Scholar]
- 21.Hozak, R. R., G. A. Manji, and P. D. Friesen. 2000. The BIR motifs mediate dominant interference and oligomerization of inhibitor of apoptosis Op-IAP. Mol. Cell. Biol. 20:1877-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, Q., Q. L. Deveraux, S. Maeda, G. S. Salvesen, H. R. Stennicke, B. D. Hammock, and J. C. Reed. 2000. Evolutionary conservation of apoptosis mechanisms: lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc. Natl. Acad. Sci. USA 97:1427-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda, M., and M. Kobayashi. 1999. Cell-cycle perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology 258:176-188. [DOI] [PubMed] [Google Scholar]
- 24.Kornbluth, S., and K. White. 2005. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm). J. Cell Sci. 118:1779-1787. [DOI] [PubMed] [Google Scholar]
- 25.LaCount, D. J., and P. D. Friesen. 1997. Role of early and late replication events in induction of apoptosis by baculoviruses. J. Virol. 71:1530-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaCount, D. J., S. F. Hanson, C. L. Schneider, and P. D. Friesen. 2000. Caspase inhibitor P35 and inhibitor of apoptosis Op-IAP block in vivo proteolytic activation of an effector caspase at different steps. J. Biol. Chem. 275:15657-15664. [DOI] [PubMed] [Google Scholar]
- 27.Lannan, E., R. Vandergaast, and P. D. Friesen. 2007. Baculovirus caspase inhibitors P49 and P35 block virus-induced apoptosis downstream of effector caspase DrICE activation in Drosophila melanogaster cells. J. Virol. 81:9319-9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, H. H., and L. K. Miller. 1978. Isolation of genotypic variants of Autographa californica nuclear polyhedrosis virus. J. Virol. 27:754-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine, A. J. 2009. The common mechanisms of transformation by the small DNA tumor viruses: the inactivation of tumor suppressor gene products: p53. Virology 384:285-293. [DOI] [PubMed] [Google Scholar]
- 30.Lilley, C. E., R. A. Schwartz, and M. D. Weitzman. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15:119-126. [DOI] [PubMed] [Google Scholar]
- 31.Lin, G., and G. W. Blissard. 2002. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J. Virol. 76:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, A., and E. B. Carstens. 1991. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology 181:336-347. [DOI] [PubMed] [Google Scholar]
- 33.Manji, G. A., and P. D. Friesen. 2001. Apoptosis in motion. An apical, P35-insensitive caspase mediates programmed cell death in insect cells. J. Biol. Chem. 276:16704-16710. [DOI] [PubMed] [Google Scholar]
- 34.McIntosh, A. H., J. J. Grasela, and H. J. Popham. 2005. AcMNPV in permissive, semipermissive, and nonpermissive cell lines from Arthropoda. In Vitro Cell. Dev. Biol. Anim. 41:298-304. [DOI] [PubMed] [Google Scholar]
- 35.Means, J. C., I. Muro, and R. J. Clem. 2003. Silencing of the baculovirus Op-iap3 gene by RNA interference reveals that it is required for prevention of apoptosis during Orgyia pseudotsugata M nucleopolyhedrovirus infection of Ld652Y cells. J. Virol. 77:4481-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moon, N. S., L. Di Stefano, E. J. Morris, R. Patel, K. White, and N. J. Dyson. 2008. E2F and p53 induce apoptosis independently during Drosophila development but intersect in the context of DNA damage. PLoS genetics 4:e1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris, T. D., and L. K. Miller. 1993. Characterization of productive and non-productive AcMNPV infection in selected insect cell lines. Virology 197:339-348. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, M. L., and J. A. Blaho. 2007. Apoptosis during herpes simplex virus infection. Adv. Virus Res. 69:67-97. [DOI] [PubMed] [Google Scholar]
- 39.Okano, K., A. L. Vanarsdall, V. S. Mikhailov, and G. F. Rohrmann. 2006. Conserved molecular systems of the Baculoviridae. Virology 344:77-87. [DOI] [PubMed] [Google Scholar]
- 40.Olson, V. A., J. A. Wetter, and P. D. Friesen. 2001. Oligomerization mediated by a helix-loop-helix-like domain of baculovirus IE1 is required for early promoter transactivation. J. Virol. 75:6042-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passarelli, A. L., and L. A. Guarino. 2007. Baculovirus late and very late gene regulation. Curr. Drug Targets 8:1103-1115. [DOI] [PubMed] [Google Scholar]
- 42.Prikhod'ko, E. A., and L. K. Miller. 1998. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J. Virol. 72:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribeiro, B. M., K. Hutchinson, and L. K. Miller. 1994. A mutant baculovirus with a temperature-sensitive IE-1 transregulatory protein. J. Virol. 68:1075-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353-365. [PubMed] [Google Scholar]
- 45.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 46.Schultz, K. L., J. A. Wetter, D. C. Fiore, and P. D. Friesen. 2009. Transactivator IE1 is required for baculovirus early replication events that trigger apoptosis in permissive and nonpermissive cells. J. Virol. 83:262-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Settles, E. W., and P. D. Friesen. 2008. Flock house virus induces apoptosis by depletion of Drosophila inhibitor-of-apoptosis protein DIAP1. J. Virol. 82:1378-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, T. M., I. Huijskens, L. G. Willis, and D. A. Theilmann. 2005. The Autographa californica multiple nucleopolyhedrovirus ie0-ie1 gene complex is essential for wild-type virus replication, but either IE0 or IE1 can support virus growth. J. Virol. 79:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiem, S. M. 2009. Baculovirus genes affecting host function. In Vitro Cell. Dev. Biol. Anim. 45:111-126. [DOI] [PubMed] [Google Scholar]
- 50.Thomenius, M., and S. Kornbluth. 2006. Multifunctional reaper: sixty-five amino acids of fury. Cell Death Differ. 13:1305-1309. [DOI] [PubMed] [Google Scholar]
- 51.Valdes, V. J., A. Sampieri, J. Sepulveda, and L. Vaca. 2003. Using double-stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J. Biol. Chem. 278:19317-19324. [DOI] [PubMed] [Google Scholar]
- 52.Vanarsdall, A. L., V. S. Mikhailov, and G. F. Rohrmann. 2007. Baculovirus DNA replication and processing. Curr. Drug Targets 8:1096-1102. [DOI] [PubMed] [Google Scholar]
- 53.Vanarsdall, A. L., K. Okano, and G. F. Rohrmann. 2005. Characterization of the replication of a baculovirus mutant lacking the DNA polymerase gene. Virology 331:175-180. [DOI] [PubMed] [Google Scholar]
- 54.van den Heuvel, S., and N. J. Dyson. 2008. Conserved functions of the pRB and E2F families. Nat. Rev. 9:713-724. [DOI] [PubMed] [Google Scholar]
- 55.Vaughn, J. L., R. H. Goodwin, G. J. Tompkins, and P. McCawley. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13:213-217. [DOI] [PubMed] [Google Scholar]
- 56.Wang, H., C. D. Blair, K. E. Olson, and R. J. Clem. 2008. Effects of inducing or inhibiting apoptosis on Sindbis virus replication in mosquito cells. J. Gen. Virol. 89:2651-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weitzman, M. D., C. T. Carson, R. A. Schwartz, and C. E. Lilley. 2004. Interactions of viruses with the cellular DNA repair machinery. DNA repair. 3:1165-1173. [DOI] [PubMed] [Google Scholar]
- 58.Wright, C. W., J. C. Means, T. Penabaz, and R. J. Clem. 2005. The baculovirus antiapoptotic protein Op-IAP does not inhibit Drosophila caspases or apoptosis in Drosophila S2 cells and instead sensitizes S2 cells to virus-induced apoptosis. Virology 335:61-71. [DOI] [PubMed] [Google Scholar]
- 59.Yamagishi, J., E. D. Burnett, S. H. Harwood, and G. W. Blissard. 2007. The AcMNPV pp31 gene is not essential for productive AcMNPV replication or late gene transcription but appears to increase levels of most viral transcripts. Virology 365:34-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zoog, S. J., J. J. Schiller, J. A. Wetter, N. Chejanovsky, and P. D. Friesen. 2002. Baculovirus apoptotic suppressor P49 is a substrate inhibitor of initiator caspases resistant to P35 in vivo. EMBO J. 21:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]