Abstract
Polyomaviruses are a growing family of small DNA viruses with a narrow tropism for both the host species and the cell type in which they productively replicate. Species host range may be constrained by requirements for precise molecular interactions between the viral T antigen, host replication proteins, including DNA polymerase, and the viral origin of replication, which are required for viral DNA replication. Cell type specificity involves, at least in part, transcription factors that are necessary for viral gene expression and restricted in their tissue distribution. In the case of the human polyomaviruses, BK virus (BKV) replication occurs in the tubular epithelial cells of the kidney, causing nephropathy in kidney allograft recipients, while JC virus (JCV) replication occurs in the glial cells of the central nervous system, where it causes progressive multifocal leukoencephalopathy. Three new human polyomaviruses have recently been discovered: MCV was found in Merkel cell carcinoma samples, while Karolinska Institute Virus and Washington University Virus were isolated from the respiratory tract. We discuss control mechanisms for gene expression in primate polyomaviruses, including simian vacuolating virus 40, BKV, and JCV. These mechanisms include not only modulation of promoter activities by transcription factor binding but also enhancer rearrangements, restriction of DNA methylation, alternate early mRNA splicing, cis-acting elements in the late mRNA leader sequence, and the production of viral microRNA.
Polyomaviruses comprise a family of small nonenveloped DNA tumor viruses which have small, circular, double-stranded DNA genomes, have been isolated from many species of mammals and birds, and are characterized by a very limited host range with respect to the species that they can productively infect (48). We will focus mainly on three primate viruses, simian vacuolating virus 40 (SV40), BK virus (BKV), and JC virus (JCV), not only because they have taught us important lessons about eukaryotic molecular biology but also because BKV and JCV cause important human diseases. SV40 was discovered almost 50 years ago (115) and was the first primate polyomavirus to be described. SV40 differs significantly from the previously discovered mouse polyoma virus (111) in that it does not possess a middle T antigen in the early region and it expresses an accessory regulatory protein, agnoprotein, which is encoded in the late region. The species of origin of SV40 is the rhesus macaque, and the virus was discovered as a contaminant in early batches of polio vaccine. While the polio vaccinations at that time likely infected many people, the prevalence of SV40 infections in humans today has been debated (36, 122). In 1971, two bona fide human polyomaviruses were discovered. BKV, also known as polyomavirus BK, was first isolated by Gardner et al. (37) by culture in Vero cells from the urine of a patient receiving immunosuppressive therapy following kidney transplantation. BKV is widespread throughout the human population around the world, with more than 70% of individuals testing serologically positive (59). Primary infection is thought to occur during childhood and is usually subclinical. However, BKV can rarely reactivate from latency under conditions of severe immunosuppression to cause nephropathy. Importantly, kidney transplant recipients who receive highly immunosuppressive drugs may develop BKV-associated nephropathy, and this is a leading cause of allograft failure (45, 77). JCV, also known as polyomavirus JC, was first isolated from brain tissue of a patient with the central nervous system demyelinating disease progressive multifocal leukoencephalopathy (PML) by Padgett et al. (85). Brain tissue was used to inoculate primary cultures derived from the human fetal brain. The virus was then successfully isolated from these long-term cultures, which consisted mainly of glial cells (85). This was the first direct evidence for a neurotropic virus associated with PML, and now JCV is the proven causative agent of PML. PML occurs mainly in individuals with highly suppressed immune system function, especially those with human immunodeficiency virus (HIV) infection/AIDS (47, 56), and involves productive infection of both oligodendrocytes and astrocytes, as judged by the production of viral capsid protein observed by immunohistochemistry and virions observed by electron microscopy (24, 73).
In tissue culture, JCV growth is restricted largely to primary human fetal glial cells. The expression of JCV early mRNA depends on tissue-specific factors found in both human and rodent glial cells, while in the presence of JCV T antigen, viral DNA replication requires a species-specific factor, perhaps a component of DNA polymerase, which is found only in primate cells (28). JCV can transform cells in culture and cause tumors in experimental animals. Furthermore, the detection of the JCV genome in a variety of human tumors raises the possibility that JCV may be associated with human tumors (reviewed in reference 24).
GENOMIC ORGANIZATION
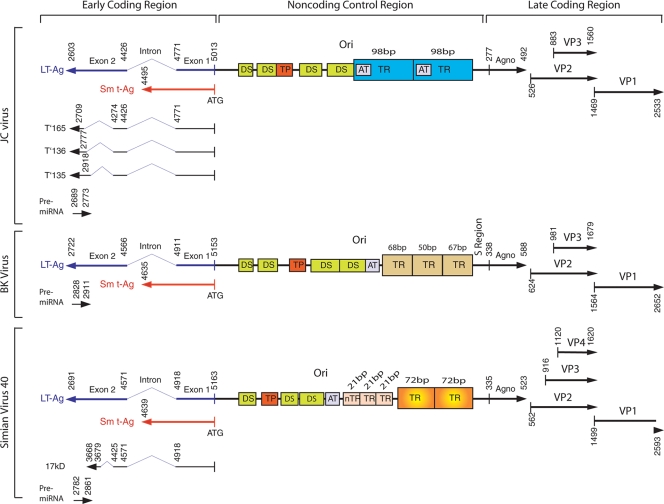
For each of the three viruses, study has focused mainly on a prototypical or reference strain of virus. For SV40, this is strain 776; for JCV, it is the Mad-1 strain; and for BKV, it is the Dunlop strain. In 1978, the complete nucleotide sequence of the circular 5.2-kb DNA genome of SV40 strain 776 was elucidated (31, 95), making it one of the first genomes for which complete sequence information had been described. The genome of the Dunlop strain of BKV (106) and the Mad-1 strain of JCV (33) have also been described. For each of the three viruses, the genome is approximately the same size and has the same organization as that of the others (Fig. 1). The viral genome is divided into two protein-coding regions that are transcribed in opposite directions starting from a common noncoding control region (NCCR). The NCCR constitutes a bidirectional regulatory region in that it contains promoter/enhancer elements for the viral early and late genes and also contains the origin of viral DNA replication. During lytic infection, transcription of these regions is temporally regulated (48). Prior to the initiation of DNA synthesis, only one region is transcribed (the early region). Once DNA replication is initiated, transcription of the other region (the late region) begins. In cells which are not permissive for viral replication, only the early region is expressed and cells may become transformed. The early region encodes large T antigen and small t antigen, which impel the cell cycle into S phase, allowing DNA replication to occur. Large T antigen also binds to the origin of viral replication located in the NCCR and cooperates with DNA polymerase and other cell DNA replication factors to initiate DNA synthesis. The early proteins, large T antigen and small t antigen, are also responsible for the oncogenic properties of these small DNA tumor viruses, i.e., their abilities to transform cells in culture and cause tumors in laboratory animals (reviewed in references 49 and 124). In the case of SV40, it has been reported that the early region also encodes a third protein, with a molecular weight of 17 kDa (dubbed tiny-t or 17kT protein), which is expressed during SV40 infection from an alternatively spliced third SV40 early mRNA and consists of 135 amino acids (130). Similarly, an additional early region product, with an apparent molecular weight of 17 to 20 kDa, has been reported for BKV expressed from an alternatively spliced early mRNA termed truncTAg (3). BKV truncTAg shares its first 133 amino acids with the N terminus of large T antigen and has three new amino acids at the C terminus translated from a different reading frame. In the case of JCV, the early coding region also encodes three additional early proteins, T′135, T′136, and T′165, which are expressed during the lytic infection of cells and are generated by alternative splicing (119). The T′ proteins are functionally different from full-length large T antigen as revealed by their abilities to differentially interact with the retinoblastoma family of tumor suppressor proteins and to alter their phosphorylation status (12, 13, 120). A mutagenesis analysis of splice sites in the early region involved in T′ protein production revealed that the T′ proteins enhance T-antigen-mediated viral DNA replication (88). Thus, while the early region of the primate polyomaviruses encodes two major products (large T and small t), there are subtle differences in the nature of the minor splice products between the viruses; the importance of these differences has not been fully explored.
FIG. 1.
Comparison of the genomes of JCV, BKV, and SV40. The circular genomes of the three polyomaviruses are shown as a linear schematic diagram (not to scale) with the NCCR at the center flanked by the coding regions—the early region on the left, which is transcribed from right to left, and the late region on the right, which is transcribed left to right. The early region of each virus encodes two primary regulatory proteins, large T antigen (LT-Ag) and small t antigen (Sm t-Ag). JCV and SV40 early regions also encode additional regulatory proteins. JCV encodes T′135, T′136, and T′165 (119), and SV40 encodes an additional 17-kDa protein (130). The late region of each virus, on the other hand, encodes three structural capsid proteins (VP1, VP2, and VP3). SV40 has also recently been shown to encode an additional very late protein, VP4, which functions in virus-mediated cell lysis (21). The late coding region of each virus also encodes a regulatory protein known as agnoprotein (Agno). The far 3′ region of LT-Ag of each virus was shown to encode pre-miRNAs that give rise to regulatory miRNAs (107, 112). Also shown in the NCCR are regions with dyad symmetry (DS), true palindromes (TP), poly(A/T) tract (AT), tandem repeats (TR), nontandem repeats (nTR), and the origin of viral DNA replication (Ori). Numbering for each virus is based on the Mad-1 strain of JCV (GenBank accession no. NC_001699, formerly J02226), the Dunlop strain of BKV (NC_001538, formerly J02038), and the 776 strain of SV40 (NC_001669, formerly J02400).
The late region encodes the three capsid proteins, VP1, VP2, and VP3, as well as a small highly basic accessory protein, known as agnoprotein, which has several functions during the viral life cycle and, at least in the case of JCV, has also been found to perturb several cellular functions (55). Recently, a fifth late protein has been reported for SV40 and has been dubbed VP4 or very late viral protein (21). Like VP3, the synthesis of VP4 is initiated from a downstream AUG start codon within the mRNA for VP2. VP4 is expressed 24 h later than the other late proteins at the time of cell lysis and is not incorporated into virions. Rather, VP4 appears to be involved in cell lysis, since coexpression of VP3 and VP4 in Escherichia coli causes bacterial lysis (21). Interestingly, this AUG codon is conserved in BKV and JCV.
NCCR
The bidirectional NCCR controls the transcription of both the early and late promoters and also contains the origin of replication, which regulates the initiation of viral DNA synthesis. It is defined as the region between the ATG start codon for T antigen and the start of the agnogene region, which encodes agnoprotein. A comparison of the NCCRs of JCV, BKV, and SV40 is shown in Fig. 1. The early proximal side of the NCCR is highly conserved between different strains of the same virus, contains the origin of viral DNA replication, and almost never undergoes rearrangement. Also contained within this region are dyad symmetry elements and palindromes. The late proximal side of the NCCR contains the repetitive enhancer elements and undergoes rearrangements, including mutations, deletions, and duplications, that account for most of the differences between different strains of the same virus, as will be discussed in detail below.
In the case of the SV40 NCCR, there is a TATA box just upstream from the start site for the early transcription region, which is involved in fixing the site of transcription initiation precisely (9). Further upstream is the “promoter” region, which contains two 21-bp tandem repeats and a 22-bp element that has a very similar sequence. The promoter region contains six GC-rich motifs that are binding sites for Sp1 and are indispensable for gene expression (9, 27, 35). Next, there is an “enhancer” region that contains two 72-bp perfect repeats. During SV40 infection, there is a shift in the initiation sites used for early transcription. During the early phase, transcription is initiated from sites downstream of the origin of DNA replication, while transcripts produced later are initiated from upstream sites on the upstream side; this shift is mediated by T antigen (15). The synthesis of T antigen is autoregulated, i.e., T antigen downregulates the overall level of early transcription by binding to two sites within the NCCR (96). T antigen also acts to upregulate late viral gene expression, independent of its function in amplifying templates through DNA replication (14, 52).
The BKV NCCR is characterized by the highest degree of variation between strains due to the occurrence of multiple rearrangements in the late proximal enhancer element observed for different isolates. The archetypal or unrearranged BKV NCCR (WW strain; GenBank accession no. M15987), which is predominant in the urine and is the transmissible form of the virus (99, 129), is arbitrarily divided into five regions, named the O, P, Q, R, and S elements (42, 71, 77). The O element is the early proximal element between the T-antigen start codon and the 5′ end of the enhancer element and includes the origin of DNA replication, the start site for early transcription, and the early 5′ untranslated region, followed by the enhancer elements P, Q, R, and S. S is the leader region of the late transcript leading up to the agnoprotein translation start codon. The prototypical Dunlop strain of BKV has a rearranged NCCR with the configuration OPP′P"S, i.e., the late proximal region contains an imperfect triple-repeat enhancer element (77). These triple tandem repeats are shown in Fig. 1. Deletion analysis experiments have demonstrated that for BKV, early promoter activity is dependent upon elements that lie both upstream and downstream of the transcription start site. In contrast, for SV40, downstream elements are not significantly involved in regulating early promoter transcription (77).
Like the BKV NCCR, the JCV NCCR is variable in nature due to rearrangements and yet largely confers the tissue-specific expression of the viral early and late genes (32, 121). A comparison of NCCR sequences among a number of JCV isolates revealed that most of the variability is confined to the 98-bp tandem repeat region (Fig. 1). Based on the occurrences of deletions and duplications, JCV isolates are assigned to two classes (32, 121). The class I viruses are characterized by the presence of the 98-bp tandem repeat within the NCCR, e.g., Mad-1 (Fig. 1), which is the prototypical strain of JCV. The class II viruses contain strains that exhibit variations from the NCCR of class I with deletions and insertions. Thus, rearrangements of the NCCR are common in polyomaviral diseases, but their exact role in pathogenesis and the mechanism for their generation remain poorly understood.
EARLY PROXIMAL ELEMENTS OF THE NCCR
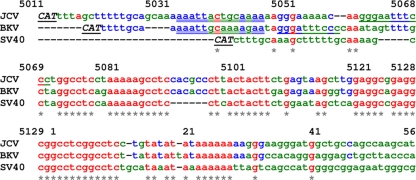
As discussed above, the early proximal region is highly conserved between different strains of the same virus, while differences between strains are found in the late proximal enhancer element. This conservation suggests that the early proximal region contains sites that are important for replication and/or transcription. The early proximal region contains several sequence elements with symmetrical arrangements as shown in Fig. 1, including the core element of the origin of replication. This region also contains initiation sites for transcription of the early region (reviewed in reference 91). The region is bounded on the early side by the initiation codon for T-antigen translation and on the late side by the poly(A/T) tract, which in JCV is part of the 98-bp repeat (Fig. 1). We compared the sequences of the NCCRs of the three viruses using the CLUSTAL multiple sequence alignment program (44). As shown in Fig. 2, there are multiple regions of sequence identity in the early proximal part of the NCCR but not in the sequences distal to the poly(A) tract, i.e., the nucleotides shown in green that are 3′ to nucleotide 31. There is a marked area of sequence identity between nucleotides 5096 and 29, which contain the core element of DNA replication (nucleotides 5119 to 11), and in the adjacent poly(A) tract (nucleotides 20 to 29). The core element and the poly(A) tract represent the minimum essential region for DNA replication (22, 25). The core element is an area of dyad symmetry (also shown in Fig. 1) and contains multiple pentamer GAGGC (or the reverse complement GCCTC) elements that bind T antigen (23, 25, 33). The palindromic sequence between nucleotides 5069 to 5089 is also highly conserved, contains multiple pentamer elements, and is also a binding site for T antigen (23, 25, 33). The region that is 5′ to nucleotide 5069 varies in length. That for SV40 lies only 24 nucleotides from the CAT trinucleotide that marks the T-antigen start codon, while the corresponding early leader regions for JCV and BKV are considerably larger, being 55 and 46 nucleotides in size, respectively. Interestingly, while this early leader region contains few nucleotides that are identical for all three viruses (Fig. 2, red), there are significant areas where nucleotides are conserved between JCV and BKV (Fig. 2, blue). These include an NF-κB site, which has been verified experimentally and is present in both JCV (72) and BKV (40) (Fig. 2, single underlining). NF-κB, an inducible transcription factor, binds to this NF-κB site and modulates transcription from the JCV early and late promoters (93). NF-κB is a family of transcription factors which are inducible in response to a wide variety of extracellular stimuli, including phorbol esters and cytokines. While constitutively expressed subunits p50 and p52 activate transcription from the D domain (92), subunit p65 activates transcription from this early proximal NF-κB motif (93). NF-κB family members p50 and p65 were also shown to indirectly influence JCV gene transcription, through a 23-bp element present within the regulatory regions of many JCV variants (101). Since this NF-κB site is stimulated by extracellular cytokines, such as tumor necrosis factor alpha (TNF-α), it may be crucial in determining the balance between JCV latency and reactivation in proinflammatory situations (72, 93). In BKV, cytokine signaling through this NF-κB site has also been implicated in BKV early promoter activation (40). Adjacent to this site is a binding site for another transcription factor, CCAAT/enhancer binding factor beta (C/EBPβ), that itself can be regulated by cytokines (Fig. 2, double underlining). The p65 subunit of NF-κB stimulates transcription of the BKV early promoter in CV-1 cells, and C/EBPβ showed a powerful synergistic effect on this activation, suggesting a functional cooperativity between these two transcription factors at this site (40). Another interesting feature of the NF-κB site in the case of BKV was revealed by experiments with the HIV type 1 (HIV-1) transactivator protein Tat. Tat was found to be a powerful transactivator of the BKV early promoter, and mutagenesis studies demonstrated that Tat activation was mediated through this NF-κB site and a second element, called BK virus transactivation-responsive element (41). HIV-1 Tat has the ability to transactivate a number of different viruses (125), and activation of BKV has been reported in a few case reports of AIDS patients with nephropathy, e.g., that by Crum-Cianflone et al. (18).
FIG. 2.
Multiple sequence alignment for early proximal part of the NCCR. CLUSTAL multiple sequence alignment was performed for JCV, BKV, and SV40 using the GenBank entries specified in the legend to Fig. 1. Sequences in red are shared for all three viruses and are also indicated with an asterisk. Sequences shared between JCV and BKV are in blue. Nonmatching sequences are in green. The underlined CAT corresponds to the opposite-strand ATG start codon for the early genes. Experimentally verified binding sites are shown for NF-κB (single underline) and C/EBPβ (double underline). Numbering is relative to the Mad-1 strain of JCV (GenBank accession no. NC_001699).
In the case of JCV, another important control element occurs at the origin of replication and has been referred to as upTAR and GGA/C-rich sequence (GRS) due to its ability to respond to HIV-1 Tat (17, 116) and to GRS binding protein (GBPi) (90), respectively. It has been suggested that this sequence element (GGAGGCGGAGGC), which contains two pentameric GAGGC T-antigen binding sites, might also bind Sp1 (91). However, it was recently demonstrated in a series of gel shift experiments that Sp1 does not bind to GRS (98), and indeed, Mad-1 JCV lacks any Sp1 sites in the NCCR, although another strain of JCV, MH-1, does contain an Sp1 site (43). In the case of HIV-1 Tat, the activity of the JCV late promoter is increased by HIV-1 Tat, and a deletion mutational analysis implicated the GRS/upTAR element (17). Further analysis revealed that Tat formed a complex with the cellular transcription factor Purα to activate transcription through the GRS/upTAR element (60). This interaction of Purα/Tat at the GRS/upTAR element can also affect T-antigen binding and enhance viral replication (20). Since the majority of cases of PML occur in individuals who are HIV-1 positive, Tat-mediated upregulation of viral transcription and replication may be an important mechanism of JCV reactivation.
Another feature of the GRS is its ability to activate transcription in response to cytokines and phorbol myristate acetate. This activity is conferred by the rapid de novo synthesis of the cellular transcription factor designated GBPi, which binds to GRS (90). Subsequent studies revealed the identity of GBPi to be early growth response protein 1 (Egr-1) (98). Egr-1 is rapidly synthesized in cells treated with cytokines or phorbol myristate acetate, binds to the GRS element, and activates JCV late transcription. Interestingly, Egr-1 is upregulated upon JCV infection of cultured astrocytes and in immunohistochemistry of the oligodendrocyte inclusion bodies and bizarre astrocytes of patients with PML, suggesting that Egr-1 induction may be important in JCV and PML pathogenesis (98).
Finally, a possible function of the early proximal portion of the NCCR may be to act in trans via the expression of proteins that are encoded in this early leader region. For SV40, an early leader protein (SELP) has been detected during late infection by in vivo pulse labeling as a 2.7-kDa peptide (54).
Thus, much of the strong sequence conservation of this region in the three viruses likely reflects the shared requirements for fundamental processes, such as DNA replication and transcription initiation. The extreme 5′ end, which is shared by JCV and BKV but not SV40, may reflect a regulatory site for governing latency and reactivation that is restricted to the human viruses and may be important in viral pathogenesis.
LATE PROXIMAL ELEMENTS OF THE NCCR
The late side of SV40, JCV, and BKV comprises a tandem repeat element that functions as an enhancer element for transcription. In the case of SV40, there is a 72-bp repeat element, which acts as an enhancer for early transcription. These repeats are found immediately upstream of the SV40 21-bp repeats, and deletion studies have shown that these sequences are essential for optimal transcription in vivo from the early promoter, since their removal decreased promoter activity more than 10-fold. Important transcription factors that were discovered in early works on SV40 and that bind to the SV40 72-bp repeat enhancer include Sp1 (26), AP-1 (64, 74), AP-2 (76), and AP-4 (74).
The inducible transcription factor NF-κB also binds to the SV40 enhancer (68). Binding to mutated enhancer correlated with the effect of these mutations in vivo, suggesting that this binding is important for SV40 enhancer activity (68). Recently, it was shown that nuclear factor of activated T cells 3 (NFAT3) and NFAT4 are important regulators of the SV40 enhancer (70). Inhibition of NFAT activity reduced SV40 infection, and this block occurred at the level of viral transcription. Both NFAT3 and NFAT4 were found to bind to the κB sites located within the 72-bp repeated enhancer region (70). Interaction of transcription factors with their cognate motifs in the SV40 enhancer is complex and can involve cooperativity and hierarchical levels of functional organization of cell-specific activity (34, 82).
In the case of BKV, the prototypical Dunlop strain has a triplicate enhancer element based on imperfect duplications of an archetypal P element, i.e., the NCCR has the structure O,P,P′,P",S, where O is the early proximal origin region discussed in the previous section and S is the late leader region abutting the agnoprotein start codon (Fig. 1). The NF-1 and Sp1 sites within the BKV enhancer are required for efficient early gene transcription, while the Dunlop strain of BKV has an AP-1 site at each P-block junction. The AP-1 site is not present in the BKV archetype, and it strengthens early transcription (77). However, the high degree of rearrangements seen within the late proximal enhancer element of BKV makes it very difficult to draw conclusions about the importance of individual transcription factor binding sites in regulating BKV gene expression. It should be noted that nearly all of the different isolates of BKV that have been sequenced from urine as well as from kidney tissue show a high degree of conservation and lack of rearrangement of the O region, as discussed in the previous section, which mediates the activation of the BKV early promoter by the transcription factors NF-κB and C/EBPβ (40). The late leader region (S region) of the BKV NCCR also tends to be conserved between different BKV strains and, interestingly, contains a steroid response unit (78). Corticosteroids are often used in maintenance immunosuppressive therapy, and a prospective study of BK replication and nephropathy in renal transplant recipients found that antirejection treatment, particularly with corticosteroids, was associated with BKV replication and nephropathy (46). Studies with BKV showed that dexamethasone stimulated BKV late transcription and viral replication but had only a small effect on the early promoter (78). The late leader region also contains a fully consensus estrogen response element and progesterone, and the estrogen β-estradiol was also found to stimulate BKV late transcription and viral replication (78).
For JCV, the late proximal NCCR of the Mad-1 strain contains a 98-bp tandem repeat, which functions as an enhancer for transcription. This element is also responsible, at least in part, for the glial cell tropism of JCV, i.e., the restriction of JCV replication to astrocytes and oligodendrocytes. Thus, reporter constructs containing the JCV 98-bp repeat are expressed well in primary glial cells but not in HeLa cells or CV-1 cells, whereas reporter constructs containing the SV40 72-bp repeat are expressed well in all three cell lines (53). Numerous transcription factors, both general and tissue specific, have been found to bind to the JCV enhancer and are described in several comprehensive reviews (32, 57, 58, 90).
JCV transcription assays performed both in vivo and in vitro as well as cell fusion experiments have shown that there exist both positively acting transcription factors in glial cells and negatively acting transcription factors in nonglial cells (7, 8). For example, Beggs et al. (8) showed that suppression of the JCV early promoter occurred in heterokaryons formed by cell fusion of JCV-transformed hamster glial cells with fibroblasts. The cis-acting region that mediated the extinction of early expression was shown to reside within the viral NCCR containing the two 98-bp repeats (8). Studies of tissue culture and promoter-swapping experiments using transgenic mice models have also indicated that it is the 98-bp repeat that is largely responsible for the cell type-specific expression of JCV (29, 67). With regard to the late transcription, it has been found that viral DNA replication and late gene expression can occur only in the presence of large T antigen (61).
Both of the JCV Mad-1 98-bp tandem repeats contain a TATA box (Fig. 1), and the first of these is involved in the positioning of transcription start sites for viral early gene expression (65, 66). A number of ubiquitous and cell-specific factors as well as viral proteins interact with the cis-acting elements in the 98-bp repeat to regulate JCV transcription (32, 57, 58, 91). For example, the glial cell-specific transcription factor Tst-1, a POU family member, promotes JCV transcription and contributes to the glial tropism of JCV (123). With the onset of the late phase, T antigen initiates DNA replication (65, 117) and also autoregulates its own transcription and executes the transcriptional switch from early to late gene expression (61).
In addition to being regulated by cellular proteins and its own regulatory proteins (T antigen and agnoprotein), JCV can also be cross-regulated by the regulatory proteins of other viruses, including the immediate-early transactivator 2 (IE2) from cytomegalovirus (126) and the Tat protein of HIV-1, as discussed above (116).
REARRANGEMENTS OF THE NCCR
One of the most striking features of the primate polyomaviruses is the occurrence of rearrangement of the enhancer element of the NCCR. These include mutations, deletions, and duplications and, in the case of BKV and JCV, may be important because of their association with human disease. Indeed, the first strains of JCV and BKV that were isolated and characterized, including the prototypical Mad-1 JCV and Dunlop BKV strains, are each derived by rearrangement from an archetypal viral strain that lacks repeated enhancer elements and constitutes the transmissible form of the virus that is found in the kidney and shed in the urine. In the case of JCV, the archetype (CY) was isolated from the urine of nonimmunocompromised individuals by Yogo et al. (128). The archetype lacks a tandem repeat in the enhancer region but contains all the regulatory sequences necessary to generate the JCV isolates derived from PML patient samples through deletion and duplication (128). Thus, there are two forms of JCV, the transmissible archetype form of JCV (JCVCY), which is excreted in the urine (4, 128) and is found in sewage (11), and the pathological forms of JCV that have been isolated from the brain of patients with PML, which are characterized by rearrangement of the enhancer to give “PML-type” JCV, e.g., Mad-1 (32, 57, 58, 91). The archetype JCV is found in the kidneys of normal individuals and is thought to replicate episodically or at low levels, probably under slight immunodepressive conditions, to give rise to virus that is shed in the urine. The PML types of JCV are characterized by a tandem enhancer element generated by rearrangements of the archetype sequence, including deletions, duplications, and point mutations. A variety of rearrangements can occur in PML types, which can differ with respect to the site selection for DNA breakage and rejoining (reviewed in references 32 and 127). The molecular mechanism of rearrangement is unknown.
Similar to JCV, BKV also has a transmissible archetypal form that is shed into the urine and is the transmissible form of the virus, BKVWW (99, 129). The viral enhancer element in the NCCR can undergo an extremely diverse set of deletions, duplications, point mutations, and rearrangements in polyomavirus-associated nephropathy (42, 45, 77). The BKVWW NCCR is arbitrarily divided into regions O, P, Q, R, and S (71, 77). The O box represents the early proximal NCCR containing the origin of DNA replication, the start site for early transcription, and the early 5′ untranslated region. While the exact role of NCCR rearrangement in BKV pathogenesis is not established, it is possible that these changes in the NCCR confer alterations in the transcription and/or replication of BKV that confer a growth advantage to the virus and contribute to pathogenesis. Gosert et al. (42) recently reported the cloning of 10 rearranged NCCRs revealing diverse duplications and deletions; however, all displayed increased early promoter activity, replication, and cytopathology in vitro. This result suggests that the emergence of rearranged NCCRs is linked to increased replication capacity and hence disease (42). Rearrangements of the BKV NCCR can also occur when the archetype BKVWW is passaged in tissue culture (100).
Finally, rearrangements of the SV40 NCCR have been found to occur and may be of significance to SV40 regulation. While BKV and JCV can replicate in humans, causing diseases that involve rearrangements of the NCCR, no human disease has been ascribed to SV40 lytic replication. However SV40 can cause PML in Macaca mulatta, the rhesus monkey, which is the natural host for SV40. Newman et al. (80) analyzed SV40 DNA from brain, kidney, and urine samples from healthy and SIV-infected rhesus monkeys. In all animals, the NCCR had an archetype structure containing a single 72-bp enhancer element. Also, the NCCR from two animals lacked one of the three copies of the GC-rich 21-bp repeat. Thus, the laboratory reference strain 776 is not the predominant type of SV40 circulating in its natural host and likely evolved during the propagation of SV40 in tissue culture. In this regard, it has been reported that the archetype SV40 single 72-bp enhancer can be duplicated during virus growth in human cells and rhesus monkey kidney cells but not in green monkey kidney cells (83). Analyses of archival material of rhesus monkey kidney cells from the 1950s and early 1960s and early low-passage-number stocks of laboratory strain SV40 have provided strong evidence that the enhancer element duplicates during tissue culture (62, 87).
Thus, primate polyomaviruses are characterized by rearrangements of the enhancer elements which are, at least in the cases of JCV and BKV, likely to be important in pathogenesis. Understanding the molecular origins and the consequences of these rearrangements remains an important challenge in the field.
SIMPLE STRATEGY BY WHICH PRIMATE POLYOMAVIRUSES AVOID TRANSCRIPTIONAL SILENCING BY HOST-MEDIATED DNA METHYLATION
DNA methylation is a well-established mechanism of silencing eukaryotic gene expression (79, 97). Methylation of DNA is a postreplication process whereby cytosine residues in the dinucleotide sequence 5′-CG-3′ (CpG) are methylated at the cyclic carbon-5 position of the cytosine nucleotide. To examine the potential of polyomaviruses to be methylated, the sequences of the primate polyomaviruses were downloaded from GenBank and analyzed for their CpG content. Remarkably, the primate polyomaviruses appear to avoid methylation by the simple strategy of the near absence of the dinucleotide CpG. SV40 (GenBank accession no. NC_001669) possesses only 27 CpGs in a sequence with a length of 5,243 bp, whereas 218 CpGs would be expected based on the GC content of SV40 DNA if there was no bias. Similarly, JCV (NC_001699) and BKV (NC_001538) possess only 16 and 12 CpGs, respectively. Analyses of the sequences of retroviruses, which replicate via a DNA intermediate, indicate that a similar mechanism may be at play, e.g., HIV-1 (NC_001802) has only 82 CpGs in 9,181 bp, which is only 20% of the number expected based on GC content. This pronounced lack of CpG in small DNA and RNA viruses may represent a simple but powerful evolutionary mechanism to escape gene silencing by DNA methylation. While the statistical chances of this lack of CpG occurring by chance are extremely small (P < 10−15 for SV40), its biological function has not yet been explored experimentally. Note that a similar analysis was previously reported by Karlin et al. (51), who assessed the dinucleotide bias of DNA and RNA viral genomes of vertebrate species by odds ratio measurements and found that the dinucleotide CpG is statistically underrepresented in most small viruses (length < 30 kb) but has a normal relative abundance in most large viruses (length ≥ 30 kb).
RECENT STUDIES OF PRIMATE POLYOMAVIRUSES
Although most of the studies of the regulation of primate polyomaviruses were done in the 1970s through the 1990s, this is still an active area of research. For example, the study of the regulation of SV40 transcription continues to be investigated as a model system of basic transcriptional mechanisms. Experiments with the SV40 NCCR have revealed that the Sp1 and AP-1 elements are involved in directing chromatin remodeling in SV40 chromosomes during early transcription (75). In another study designed to identify promoter sequence elements that affect pre-mRNA splicing patterns, a motif positioned within the core promoter, comprised of eight T residues directly upstream of the SV40 early TATA box that affected splice site selection during pre-mRNA splicing, was identified (39).
Recent studies have also indicated a role for microRNAs (miRNAs) in the regulation of SV40 transcription (112). SV40-encoded miRNAs (SVmiRNAs) produced from a pre-miRNA precursor expressed in a late polarity (Fig. 1) were found to accumulate at late times in the infection cycle, and these SVmiRNAs were perfectly complementary to early viral mRNAs, thus targeting those mRNAs for cleavage. By creating a mutant virus lacking SVmiRNAs, it was shown that SVmiRNAs are not necessary for generation of infectious virus but do reduce the sensitivity of cells to cytotoxic T cells. Thus, SVmiRNAs that reduce T-antigen expression late in infection may represent a mechanism to reduce the immune response to SV40-infected cells (112). JCV and BKV also encode miRNAs which appear to have the same function as the SVmiRNAs (107). These miRNAs could be detected in JCV-infected brain tissue from PML patients (107).
The field of JCV gene regulation is also still an active one. In addition to the recent identification of Egr-1 as an important regulator as described above (98), other studies have implicated NFAT4 as being important for JCV infection of glial cells (69). In contrast, the transcription factor NF-1A was found to negatively regulate JCV (94). Thus, downregulation of NF-1A expression in JCV-nonsusceptible cells, such as HeLa cells, resulted in a susceptibility for JCV multiplication. Recently, the DEAD box protein 1 (DDX1), an RNA helicase, and the cleavage stimulation factor (CstF) were shown to form a complex that binds to the JCV NCCR (113, 114). DDX1 is expressed at much higher levels in the JCV-susceptible cell line IMR-32 than in nonsusceptible cell lines. DDX1 significantly increased transactivation of the JCV promoter and enhanced the expression of JCV proteins in JCV-infected cells, while knockdown of DDX1 using small interfering RNA suppressed the expression of JCV proteins (113, 114). The small JCV regulatory protein agnoprotein has also been shown to have a critical role in JCV regulation (55). Treating JCV-infected cells with small interfering RNA to agnoprotein results in a marked inhibition both of viral protein expression and of virus production (84, 89). Agnoprotein can bind to the cellular transcription factor YB-1, which together with its interaction with T antigen, regulates JCV transcription (102). Further investigation of this interaction by functional assays demonstrated that agnoprotein negatively regulated YB-1-mediated gene transcription (103). JCV agnoprotein contains several potential phosphorylation sites, including Ser7, Ser11, and Thr21, that are predicted to be potential targets for the serine/threonine-specific protein kinase C (PKC). In vitro and in vivo kinase assays demonstrated that agnoprotein is a target for phosphorylation by PKC. When each of the PKC phosphorylation sites was mutated to Ala singly and in combination, virus containing each of these mutations failed to propagate (104). Thus, phosphorylated forms of agnoprotein may have essential functions in the viral life cycle. Further investigation revealed that agnoprotein phosphorylation is regulated by protein phosphatase 2A (PP2A), a serine/threonine-specific protein phosphatase, and that JCV small t antigen is involved in this regulation (105). PP2A associates with agnoprotein and dephosphorylates it at the PKC-specific sites. Small t antigen also interacts with PP2A, and this interaction inhibits the dephosphorylation of agnoprotein by PP2A. Downregulation of PP2A caused a significant reduction in the level of JCV replication. These results suggest that there is interplay between agnoprotein, small t antigen, and PP2A with respect to the regulation of the JCV life cycle (105).
Agnoprotein is an essential trans-acting factor for the JCV infectious cycle, and a recent study has also investigated the cis-acting potential of the agnogene region of the genome, which encodes agnoprotein. It was found that agnogene contained DNA elements that play cis-acting regulatory roles in the JCV lytic cycle (5). Thus, transcription factor binding sites that regulate the activity of JCV gene expression are not restricted exclusively to the NCCR.
More-recent studies continue to explore the regulation of BKV. Like that of JCV, phosphorylation of BKV agnoprotein at Ser-11 by PKC has also been recently reported to have an important regulatory function (50). Additional recent studies have continued to explore the regulation of BKV at the level of transcriptional control of the NCCR, in particular the notion that cytokines, and the transcription factors that lie downstream of them, may be involved in the regulation of the balance between BKV latency and reactivation. A study of the role of gamma interferon (IFN-γ) in regulating lytic infection of primary human kidney proximal tubule epithelial cells by BKV (TU, Dunlop, and Proto-2 strains) showed that IFN-γ inhibited the early expression of T antigen and of the late protein VP1 in a dose-dependent manner. IFN-γ-mediated inhibition was shown to occur at the level of transcription and reduced the level of virus production as much as 50- to 80-fold (2). Kidney proximal tubule epithelial cells (the host cells for BKV infection) are thought to interact with neighboring cells and immune cells via production of cytokines such as interleukin-6 (IL-6), IL-8, IL-15, TNF-α, monocyte chemoattractant protein 1 (MCP-1), and transforming growth factor β (TGF-β) (19), and the role of cytokines in mediating the regulation of BKV replication has been investigated. In the case of TGF-β, a cytokine that can be upregulated in immunosuppression, the activity of the BKV early promoter (TU strain of BKV) was stimulated by this cytokine. Site-directed mutagenesis mapped the site of stimulation by TGF-β to a single predicted Smad3 site in the TU-strain BKV NCCR (1). It is thus possible that reactivation of BKV in immunocompromised patients may involve signals, such as stimulation by TGF-β and the absence of IFN-γ (1). Proinflammatory cytokines such as TNF-α and IL-6 can also activate NF-κB, and the role of NF-κB and C/EBPβ (a transcription factor whose activity is also controlled by cytokines) in the activation of BKV early transcription via an early proximal binding site in the BKV NCCR was discussed above (40).
OTHER PRIMATE POLYOMAVIRUSES
At least two other primate polyomaviruses are known to exist. Complete genome sequence information is available for simian agent 12 (SA12) and African green monkey lymphotropic polyomavirus (LPV). SA12, whose natural host is thought to be the chacma baboon, has a 5,230-bp genome, is very closely related to BKV, and shares many of the structural features of BKV, including the same viral proteins and miRNAs and a similar regulatory region (16). LPV has a 5,270-bp genome, contains a 63-bp tandem repeat putative enhancer element at the late proximal side of the regulatory region, infects only cells of B-lymphocyte origin, and is more phylogenetically distant from the other primate polyomaviruses; it is more closely related to the rodent polyomaviruses, although it does not contain a middle T antigen (86). Thus, as far as can be judged from sequence data, many of the mechanisms that we have explored in this review may also operate in these viruses.
NEWLY DISCOVERED HUMAN POLYOMAVIRUSES KIV, WUV, AND MCV
New high-throughput methods of DNA analysis have recently led to the discovery of three new human polyomaviruses which are only distantly related to JCV, BKV, and SV40. Much less is known about these novel viruses. At the Karolinska Institute (KI), large-scale molecular screening of human diagnostic samples of libraries prepared from nasopharyngeal aspirates led to the identification of a third human polyomavirus, Karolinska Institute Virus (KIV) (6). The genome is 5,040 bp in size and is organized in the same way as the other primate polyomaviruses except that it lacks agnoprotein, as judged by the lack of an open reading frame in the late leader sequence. The KIV NCCR contains three potential large T-antigen binding sites, compared to the usual four for other polyomaviruses. The sequence shows putative binding sites for c-Ets-1, Oct-1, and NF-1 but not Sp1. At Washington University (WU), a similar approach was used to identify a novel polyomavirus, WUV (38). The WUV genome is 5,229 bp in size and also lacks an agnogene. Unusual features of the Washington University Virus (WUV) NCCR region include two partially overlapping binding sites for large T antigen and slightly variant spacing between these binding sites compared to those for SV40, BKV, and JCV. Phylogenetically, WUV is more closely related to KIV than to SV40, BKV, and JCV. Geographically, WUV and KIV have been reported to be found around the world (10). It is not clear that these viruses are associated with any respiratory disease (81).
Merkel cell carcinoma (MCC) is a rare, aggressive, primary skin cancer exhibiting neuroendocrine differentiation. A third novel polyomavirus was detected using digital transcriptome subtraction analysis, in which cDNAs from MCC samples were exhaustively sequenced and then known cellular sequences subtracted to reveal foreign DNA. This led to the discovery of a novel polyomavirus, MCC virus (MCV) (30). MCV has a genome size of 5,387 bp and lacks agnoprotein. Phylogenetically, it is remote from both the KIV/WUV and SV40/BKV/JCV groups of polyomaviruses. The MCV NCCR is highly conserved with other polyomaviruses including features such as a poly(T) tract, inverted repeats, and conserved pentameric large T-antigen binding boxes. The clonal integration of the MCV genome in MCC strongly suggests a role in tumor initiation. A common feature of such integration events involves inactivation of the replicative ability of T antigen (110). Similar to SV40, BKV, and JCV discussed above, MCV has miRNAs encoded in the late polarity, which may function to downregulate early transcripts (108).
Finally, it has recently been reported that KIV, WUV, and MCV can be reactivated in immunosuppressed individuals (109), as suggested by an analysis of autopsy samples of lymphoid tissue from AIDS-immunosuppressed subjects. Interestingly, the NCCR sequences from KIV and WUV showed a number of point mutations and insertions that were absent in viruses recovered from respiratory tract specimens. This result suggests that NCCR changes can occur that potentially lead to transcriptional deregulation, which may have pathogenic consequences as seen for JCV and BKV.
CONCLUSIONS
We have mainly considered the role of transcription factors with respect to the replicative life cycle of the virus. When polyomaviruses infect cells that are nonpermissive for viral replication, oncogenic transformation can occur. For example, JCV produces tumors in experimental animals, such as rodents and monkeys. Early transcription occurs in these tumors, resulting in the expression of early T antigen, but there is no late transcription, no capsid protein production, nor any viral DNA replication. The reason for this failure of late transcription is not known, but possible causes have been discussed in a recent review (24).
The existence of a transmissible archetype with a highly conserved NCCR for each polyomavirus suggests that the exact physical arrangement of NCCR elements is important for viral function and transmission and that the polyomaviruses evolve along with the host. This is consistent with the extremely narrow host range of the polyomaviruses. On the other hand, the rearrangement of the NCCR, including enhancer duplications, during disease, suggests that the polyomaviruses can adapt for periods of rapid lytic replication. While a great deal has been learned about polyomaviruses, there are still many aspects that remain unknown. These include the mode of transmission between individuals, trafficking of virus within the body, the events involved in viral entry into the cell and exit from the cell, the molecular mechanism and consequences of rearrangement of the NCCR, the nature of the virus in the latent state, and the details of the transcriptional switching events involved in the transition from latency to active viral lytic infection. Understanding these events is important not just to advance our biological understanding of polyomaviruses but also because the diseases associated with BKV (polyomavirus-associated nephropathy in kidney allograft recipients) and JCV (PML in HIV-1/AIDS patients) are both significant public health issues for which effective therapies are sorely lacking.
Acknowledgments
We thank past and present members of the Department of Neuroscience and Center for Neurovirology for their continued support, insightful discussions, and sharing of reagents and ideas. We also wish to thank C. Schriver for editorial assistance.
This work was made possible by grants awarded by the NIH to M.K.W., M.S., and K.K.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Abend, J. R., and M. J. Imperiale. 2008. Transforming growth factor-beta-mediated regulation of BK virus gene expression. Virology 378:6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abend, J. R., J. A. Low, and M. J. Imperiale. 2007. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J. Virol. 81:272-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abend, J. R., A. E. Joseph, D. Das, D. B. Campbell-Cecen, and M. J. Imperiale. 2009. A truncated T antigen expressed from an alternatively spliced BK virus early mRNA. J. Gen. Virol. 90:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agostini, H. T., C. F. Ryschkewitsch, and G. L. Stoner. 1998. Rearrangements of archetypal regulatory regions in JC virus genomes from urine. Res. Virol. 149:163-170. [DOI] [PubMed] [Google Scholar]
- 5.Akan, I., I. K. Sariyer, R. Biffi, V. Palermo, S. Woolridge, M. K. White, S. Amini, K. Khalili, and M. Safak. 2006. Human polyomavirus JCV late leader peptide region contains important regulatory elements. Virology 349:66-78. [DOI] [PubMed] [Google Scholar]
- 6.Allander, T., K. Andreasson, S. Gupta, A. Bjerkner, G. Bogdanovic, M. A. Persson, T. Dalianis, T. Ramqvist, and B. Andersson. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amemiya, K., R. Traub, L. Durham, and E. O. Major. 1992. Adjacent nuclear factor-1 and activator protein binding sites in the enhancer of the neurotropic JC virus: a common characteristic of many brain-specific genes. J. Biol. Chem. 267:14204-14211. [PubMed] [Google Scholar]
- 8.Beggs, A. H., R. J. Frisque, and G. A. Scangos. 1988. Extinction of JC virus tumor-antigen expression in glial cell-fibroblast hybrids. Proc. Natl. Acad. Sci. USA 85:7632-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benoist, C., and P. Chambon. 1981. In vivo sequence requirements of the SV40 early promotor region. Nature 290:304-310. [DOI] [PubMed] [Google Scholar]
- 10.Bialasiewicz, S., D. M. Whiley, S. B. Lambert, K. Jacob, C. Bletchly, D. Wang, M. D. Nissen, and T. P. Sloots. 2008. Presence of the newly discovered human polyomaviruses KI and WU in Australian patients with acute respiratory tract infection. J. Clin. Virol. 41:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bofill-Mas, S., P. Clemente-Casares, E. O. Major, B. Curfman, and R. Girones. 2003. Analysis of the excreted JC virus strains and their potential oral transmission. J. Neurovirol. 9:498-507. [DOI] [PubMed] [Google Scholar]
- 12.Bollag, B., C. Prins, E. L. Snyder, and R. J. Frisque. 2000. Purified JC virus T and T′ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology 274:165-178. [DOI] [PubMed] [Google Scholar]
- 13.Bollag, B., L. H. Kilpatrick, S. K. Tyagarajan, M. J. Tevethia, and R. J. Frisque. 2006. JC virus T′135, T′136 and T′165 proteins interact with cellular p107 and p130 in vivo and influence viral transformation potential. J. Neurovirol. 12:428-442. [DOI] [PubMed] [Google Scholar]
- 14.Brady, J., J. B. Bolen, M. Radonovich, N. Salzman, and G. Khoury. 1984. Stimulation of simian virus 40 late gene expression by simian virus 40 tumor antigen. Proc. Natl. Acad. Sci. USA 81:2040-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchman, A. R., M. Fromm, and P. Berg. 1984. Complex regulation of simian virus 40 early-region transcription from different overlapping promoters. Mol. Cell. Biol. 4:1900-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantalupo, P., A. Doering, C. S. Sullivan, A. Pal, K. W. Peden, A. M. Lewis, and J. M. Pipas. 2005. Complete nucleotide sequence of polyomavirus SA12. J. Virol. 79:13094-13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdhury, M., M. Kundu, and K. Khalili. 1993. GA/GC-rich sequence confers Tat responsiveness to human neurotropic virus promoter, JCVL, in cells derived from central nervous system. Oncogene 8:887-892. [PubMed] [Google Scholar]
- 18.Crum-Cianflone, N., M. Quigley, G. Utz, and B. Hale. 2007. BK virus-associated renal failure among HIV patients. AIDS 21:1501-1502. [DOI] [PubMed] [Google Scholar]
- 19.Daha, M. R., and C. Van Kooten. 2000. Is the proximal tubular cell a proinflammatory cell? Nephrol. Dial. Transplant. 15:(Suppl. 6):41-43. [DOI] [PubMed] [Google Scholar]
- 20.Daniel, D. C., M. J. Wortman, R. J. Schiller, H. Liu, L. Gan, J. S. Mellen, C.-F. Chang, G. L. Gallia, J. Rappaport, K. Khalili, and E. M. Johnson. 2001. Coordinate effects of human immunodeficiency virus type 1 protein Tat and cellular protein Pur α on DNA replication initiated at the JC virus origin. J. Gen. Virol. 82:1543-1553. [DOI] [PubMed] [Google Scholar]
- 21.Daniels, R., D. Sadowicz, and D. N. Hebert. 2007. A very late viral protein triggers the lytic release of SV40. PLoS Pathog. 3:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deb, S., A. L. DeLucia, C. P. Baur, A. Koff, and P. Tegtmeyer. 1986. Domain structure of the simian virus 40 core origin of replication. Mol. Cell. Biol. 6:1663-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeLucia, A. L., B. A. Lewton, R. Tjian, and P. Tegtmeyer. 1983. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J. Virol. 46:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Valle, L., M. K. White, and K. Khalili. 2008. Potential mechanisms of the human polyomavirus JC in neural oncogenesis. J. Neuropathol. Exp. Neurol. 67:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deyerle, K. L., F. G. Sajjadi, and S. Subramani. 1989. Analysis of origin of DNA replication of human papovavirus BK. J. Virol. 63:356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dynan, W. S., and R. Tjian. 1983. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell 32:669-680. [DOI] [PubMed] [Google Scholar]
- 27.Everett, R. D., D. Baty, and P. Chambon. 1983. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 11:2447-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feigenbaum, L., K. Khalili, E. Major, and G. Khoury. 1987. Regulation of the host range of human papovavirus JCV. Proc. Natl. Acad. Sci. USA 84:3695-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feigenbaum, L., S. H. Hinrichs, and G. Jay. 1992. JC virus and simian virus 40 enhancers and transforming proteins: role in determining tissue specificity and pathogenicity in transgenic mice. J. Virol. 66:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng, H., M. Shuda, Y. Chang, and P. S. Moore. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiers, W., R. Contreras, G. Haegemann, R. Rogiers, A. Van de Voorde, H. Van Heuverswyn, J. Van Herreweghe, G. Volckaert, and M. Ysebaert. 1978. Complete nucleotide sequence of SV40 DNA. Nature 273:113-120. [DOI] [PubMed] [Google Scholar]
- 32.Frisque, R. J., and F. A. White. 1992. The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy, p. 25-160. In R. P. Roos (ed.), Molecular neurovirology. Humana Press, Inc., Totowa, NJ.
- 33.Frisque, R. J., G. L. Bream, and M. T. Cannella. 1984. Human polyomavirus JC virus genome. J. Virol. 51:458-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fromental, C., M. Kanno, H. Nomiyama, and P. Chambon. 1988. Cooperativity and hierarchical levels of functional organization in the SV40 enhancer. Cell 54:943-953. [DOI] [PubMed] [Google Scholar]
- 35.Fromm, M., and P. Berg. 1982. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J. Mol. Appl. Genet. 1:457-481. [PubMed] [Google Scholar]
- 36.Garcea, R. L., and M. J. Imperiale. 2003. Simian virus 40 infection of humans. J. Virol. 77:5039-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner, S. D., A. M. Field, D. V. Coleman, and B. Hulme. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253-1257. [DOI] [PubMed] [Google Scholar]
- 38.Gaynor, A. M., M. D. Nissen, D. M. Whiley, I. M. Mackay, S. B. Lambert, G. Wu, D. C. Brennan, G. A. Storch, T. P. Sloots, and D. Wang. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gendra, E., D. F. Colgan, B. Meany, and M. M. Konarska. 2007. A sequence motif in the simian virus 40 (SV40) early core promoter affects alternative splicing of transcribed mRNA. J. Biol. Chem. 282:11648-11657. [DOI] [PubMed] [Google Scholar]
- 40.Gorrill, T. S., and K. Khalili. 2005. Cooperative interaction of p65 and C/EBPbeta modulates transcription of BKV early promoter. Virology 335:1-9. [DOI] [PubMed] [Google Scholar]
- 41.Gorrill, T., M. Feliciano, R. Mukerjee, B. E. Sawaya, K. Khalili, and M. K. White. 2006. Activation of early gene transcription in polyomavirus BK by human immunodeficiency virus type 1 Tat. J. Gen. Virol. 87:1557-1566. [DOI] [PubMed] [Google Scholar]
- 42.Gosert, R., C. H. Rinaldo, G. A. Funk, A. Egli, E. Ramos, C. B. Drachenberg, and H. H. Hirsch. 2008. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J. Exp. Med. 205:841-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henson, J. W. 1994. Regulation of the glial-specific JC virus early promoter by the transcription factor Sp1. J. Biol. Chem. 269:1046-1050. [PubMed] [Google Scholar]
- 44.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 45.Hirsch, H. H. 2005. BK virus: opportunity makes a pathogen. Clin. Infect. Dis. 41:354-360. [DOI] [PubMed] [Google Scholar]
- 46.Hirsch, H. H., W. Knowles, M. Dickenmann, J. Passweg, T. Klimkait, M. J. Mihatsch, and J. Steiger. 2002. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N. Engl. J. Med. 347:488-496. [DOI] [PubMed] [Google Scholar]
- 47.Hou, J., and E. O. Major. 2000. Progressive multifocal leukoencephalopathy: JC virus induced demyelination in the immune compromised host. J. Neurovirol. 6(Suppl. 2):S98-S100. [PubMed] [Google Scholar]
- 48.Imperiale, M. J., and E. O. Major. 2007. Polyomaviruses, p. 2263-2298. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 49.Javier, R. T., and J. S. Butel. 2008. The history of tumor virology. Cancer Res. 68:7693-7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johannessen, M., M. R. Myhre, M. Dragset, C. Tümmler, and U. Moens. 2008. Phosphorylation of human polyomavirus BK agnoprotein at Ser-11 is mediated by PKC and has an important regulative function. Virology 379:97-109. [DOI] [PubMed] [Google Scholar]
- 51.Karlin, S., W. Doerfler, and L. R. Cardon. 1994. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 68:2889-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keller, J. M., and J. C. Alwine. 1984. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell 36:381-389. [DOI] [PubMed] [Google Scholar]
- 53.Kenney, S., V. Natarajan, D. Strike, G. Khoury, and N. P. Salzman. 1984. JC virus enhancer-promoter active in human brain cells. Science 226:1337-1339. [DOI] [PubMed] [Google Scholar]
- 54.Khalili, K., J. Brady, and G. Khoury. 1987. Translational regulation of SV40 early mRNA defines a new viral protein. Cell 48:639-645. [DOI] [PubMed] [Google Scholar]
- 55.Khalili, K., M. K. White, H. Sawa, K. Nagashima, and M. Safak. 2005. The agnoprotein of polyomaviruses: a multifunctional auxiliary protein. J. Cell. Physiol. 204:1-7. [DOI] [PubMed] [Google Scholar]
- 56.Khalili, K., J. Gordon, and M. K. White. 2006. The polyomavirus, JCV, and its involvement in human disease. Adv. Exp. Med. Biol. 577:274-287. [DOI] [PubMed] [Google Scholar]
- 57.Khalili, K., M. Safak, L. Del Valle, and M. K. White. 2008. JC virus molecular biology and the human demyelinating disease, progressive multifocal leukoencephalopathy, p. 190-211. In C. S. Reiss (ed.), Neurotropic virus infections, Chapter 10. Cambridge University Press, Cambridge, United Kingdom.
- 58.Kim, H.-S., J. W. Henson, and R. J. Frisque. 2001. Transcription and replication in the human polyomaviruses, p. 73-126. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. John Wiley & Sons, Inc., New York, NY.
- 59.Knowles, W. A. 2001. The epidemiology of BK virus and the occurrence of antigenic and genomic subtypes, p. 527-584. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. Wiley-Liss, Inc., New York, NY.
- 60.Krachmarov, C. P., L. G. Chepenik, S. Barr-Vagell, K. Khalili, and E. M. Johnson. 1996. Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Pur alpha. Proc. Natl. Acad. Sci. USA 93:14112-14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lashgari, M. S., H. Tada, S. Amini, and K. Khalili. 1989. Regulation of JCVL promoter function: transactivation of JCVL promoter by JCV and SV40 early proteins. Virology 170:292-295. [DOI] [PubMed] [Google Scholar]
- 62.Lednicky, J. A., and J. S. Butel. 1997. Tissue culture adaptation of natural isolates of simian virus 40: changes occur in viral regulatory region but not in carboxy-terminal domain of large T-antigen. J. Gen. Virol. 78:1697-1705. [DOI] [PubMed] [Google Scholar]
- 63.Reference deleted.
- 64.Lee, W., P. Mitchell, and R. Tjian. 1987. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell 49:741-752. [DOI] [PubMed] [Google Scholar]
- 65.Lynch, K. J., and R. J. Frisque. 1990. Identification of critical elements within the JC virus DNA replication origin. J. Virol. 64:5812-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lynch, K. J., and R. J. Frisque. 1991. Factors contributing to the restricted DNA replicating activity of JC virus. Virology 180:306-317. [DOI] [PubMed] [Google Scholar]
- 67.Lynch, K. J., S. Haggerty, and R. J. Frisque. 1994. DNA replication of chimeric JC virus-simian virus 40 genomes. Virology 204:819-822. [DOI] [PubMed] [Google Scholar]
- 68.Macchi, M., J. M. Bornert, I. Davidson, M. Kanno, R. Rosales, M. Vigneron, J. H. Xiao, C. Fromental, and P. Chambon. 1989. The SV40 TC-II(κ B) enhanson binds ubiquitous and cell type specifically inducible nuclear proteins from lymphoid and non-lymphoid cell lines. EMBO J. 8:4215-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manley, K., B. A. O'Hara, G. V. Gee, C. P. Simkevich, J. M. Sedivy, and W. J. Atwood. 2006. NFAT4 is required for JC virus infection of glial cells. J. Virol. 80:12079-12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manley, K., B. A. O'Hara, and W. J. Atwood. 2008. Nuclear factor of activated T-cells (NFAT) plays a role in SV40 infection. Virology 372:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Markowitz, R. B., and W. S. Dynan. 1988. Binding of cellular proteins to the regulatory region of BK virus DNA. J. Virol. 62:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayreddy, R. P., M. Safak, M. Razmara, P. Zoltick, and K. Khalili. 1996. Transcription of the JC virus archetype late genome: importance of the κB and the 23-base-pair motifs in late promoter activity in glial cells. J. Virol. 70:2387-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mázló, M., H. G. Ressetar, and G. L. Stoner. 2001. The neuropathology and pathogenesis of progressive multifocal leukoencephalopathy, p. 257-335. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. Wiley-Liss, Inc., New York, NY.
- 74.Mermod, N., T. J. Williams, and R. Tjian. 1988. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature 332:557-561. [DOI] [PubMed] [Google Scholar]
- 75.Milavetz, B. I. 2002. SP1 and AP-1 elements direct chromatin remodeling in SV40 chromosomes during the first 6 hours of infection. Virology 294:170-179. [DOI] [PubMed] [Google Scholar]
- 76.Mitchell, P. J., C. Wang, and R. Tjian. 1987. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell 50:847-861. [DOI] [PubMed] [Google Scholar]
- 77.Moens, U., and O. P. Rekvig. 2001. Molecular biology of BK virus and clinical and basic aspects of BK virus renal infection, p. 359-408. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. Wiley-Liss, Inc., New York, NY.
- 78.Moens, U., N. Subramaniam, B. Johansen, T. Johansen, and T. Traavik. 1994. A steroid hormone response unit in the late leader of the noncoding control region of the human polyomavirus BK confers enhanced host cell permissivity. J. Virol. 68:2398-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakao, M. 2001. Epigenetics: interaction of DNA methylation and chromatin. Gene 278:25-31. [DOI] [PubMed] [Google Scholar]
- 80.Newman, J. S., G. B. Baskin, and R. J. Frisque. 1998. Identification of SV40 in brain, kidney and urine of healthy and SIV-infected rhesus monkeys. J. Neurovirol. 4:394-406. [DOI] [PubMed] [Google Scholar]
- 81.Norja, P., I. Ubillos, K. Templeton, and P. Simmonds. 2007. No evidence for an association between infections with WU and KI polyomaviruses and respiratory disease. J. Clin. Virol. 40:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ondek, B., L. Gloss, and W. Herr. 1988. The SV40 enhancer contains two distinct levels of organization. Nature 333:40-45. [DOI] [PubMed] [Google Scholar]
- 83.O'Neill, F. J., J. E. Greenlee, and H. Carney. 2003. The archetype enhancer of simian virus 40 DNA is duplicated during virus growth in human cells and rhesus monkey kidney cells but not in green monkey kidney cells. Virology 310:173-182. [DOI] [PubMed] [Google Scholar]
- 84.Orba, Y., H. Sawa, H. Iwata, S. Tanaka, and K. Nagashima. 2004. Inhibition of virus production in JC virus-infected cells by postinfection RNA interference. J. Virol. 78:7270-7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Padgett, B. L., G. M. Zu Rhein, D. L. Walker, R. Echroade, and B. Dessel. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet i:1257-1260. [DOI] [PubMed] [Google Scholar]
- 86.Pawlita, M., A. Clad, and H. zur Hausen. 1985. Complete DNA sequence of lymphotropic papovavirus: prototype of a new species of the polyomavirus genus. Virology 143:196-211. [DOI] [PubMed] [Google Scholar]
- 87.Peden, K., L. Sheng, R. Omeir, M. Yacobucci, M. Klutch, M. Laassri, K. Chumakov, A. Pal, H. Murata, and A. M. Lewis, Jr. 2008. Recovery of strains of the polyomavirus SV40 from rhesus monkey kidney cells dating from the 1950s to the early 1960s. Virology 370:63-76. [DOI] [PubMed] [Google Scholar]
- 88.Prins, C., and R. J. Frisque. 2001. JC virus T′ proteins encoded by alternatively spliced early mRNAs enhance T antigen-mediated viral DNA replication in human cells. J. Neurovirol. 7:250-264. [DOI] [PubMed] [Google Scholar]
- 89.Radhakrishnan, S., J. Gordon, L. Del Valle, J. Cui, and K. Khalili. 2004. Intracellular approach for blocking JC virus gene expression by using RNA interference during viral infection. J. Virol. 78:7264-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Raj, G. V., and K. Khalili. 1994. Identification and characterization of a novel GGA/C-binding protein, GBP-i, that is rapidly inducible by cytokines. Mol. Cell. Biol. 14:7770-7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raj, G. V., and K. Khalili. 1995. Transcriptional regulation: lessons from the human neurotropic polyomavirus, JCV. Virology 213:283-291. [DOI] [PubMed] [Google Scholar]
- 92.Raj, G. V., M. Safak, G. H. MacDonald, and K. Khalili. 1996. Transcriptional regulation of human polyomavirus JC: evidence for a functional interaction between RelA (p65) and the Y-box-binding protein, YB-1. J. Virol. 70:5944-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ranganathan, P. N., and K. Khalili. 1993. The transcriptional enhancer element, kappa B, regulates promoter activity of the human neurotropic virus, JCV, in cells derived from the CNS. Nucleic Acids Res. 21:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ravichandran, V., and E. O. Major. 2008. DNA-binding transcription factor NF-1A negatively regulates JC virus multiplication. J. Gen. Virol. 89:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reddy, V. B., B. Thimmappaya, R. Dhar, K. N. Subramanian, B. S. Zain, J. Pan, P. K. Ghosh, M. L. Celma, and S. M. Weissman. 1978. The genome of simian virus 40. Science 200:494-502. [DOI] [PubMed] [Google Scholar]
- 96.Rio, D. C., and R. Tjian. 1983. SV40 T antigen binding site mutations that affect autoregulation. Cell 32:1227-1240. [DOI] [PubMed] [Google Scholar]
- 97.Robertson, K. D. 2002. DNA methylation and chromatin—unraveling the tangled web. Oncogene 21:5361-5379. [DOI] [PubMed] [Google Scholar]
- 98.Romagnoli, L., I. K. Sariyer, J. Tung, M. Feliciano, B. E. Sawaya, L. Del Valle, P. Ferrante, K. Khalili, M. Safak, and M. K. White. 2008. Early growth response-1 protein is induced by JC virus infection and binds and regulates the JC virus promoter. Virology 375:331-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rubinstein, R., N. Pare, and E. H. Harley. 1987. Structure and function of the transcriptional control region of nonpassaged BK virus. J. Virol. 61:1747-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rubinstein, R., B. C. Schoonakker, and E. H. Harley. 1991. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J. Virol. 65:1600-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Safak, M., G. L. Gallia, and K. Khalili. 1999. A 23-bp sequence element from human neurotropic JC virus is responsive to NF-kappa B subunits. Virology 262:178-189. [DOI] [PubMed] [Google Scholar]
- 102.Safak, M., R. Barrucco, A. Darbinyan, Y. Okada, K. Nagashima, and K. Khalili. 2001. Interaction of JC virus Agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J. Virol. 75:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Safak, M., B. Sadowska, R. Barrucco, and K. Khalili. 2002. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. J. Virol. 76:3828-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sariyer, I. K., I. Akan, V. Palermo, J. Gordon, K. Khalili, and M. Safak. 2006. Phosphorylation mutants of JC virus agnoprotein are unable to sustain the viral infection cycle. J. Virol. 80:3893-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sariyer, I. K., K. Khalili, and M. Safak. 2008. Dephosphorylation of JC virus agnoprotein by protein phosphatase 2A: inhibition by small t antigen. Virology 375:464-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seif, I., G. Khoury, and R. Dhar. 1979. The genome of human papovavirus BKV. Cell 18:963-977. [DOI] [PubMed] [Google Scholar]
- 107.Seo, G. J., L. H. Fink, B. O'Hara, W. J. Atwood, and C. S. Sullivan. 2008. Evolutionarily conserved function of a viral microRNA. J. Virol. 82:9823-9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seo, G. J., C. J. Chen, and C. S. Sullivan. 2009. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology 383:183-187. [DOI] [PubMed] [Google Scholar]
- 109.Sharp, C. P., P. Norja, I. Anthony, J. E. Bell, and P. Simmonds. 2009. Reactivation and mutation of newly discovered WU, KI, and Merkel cell carcinoma polyomaviruses in immunosuppressed individuals. J. Infect. Dis. 199:398-404. [DOI] [PubMed] [Google Scholar]
- 110.Shuda, M., H. Feng, H. J. Kwun, S. T. Rosen, O. Gjoerup, P. S. Moore, and Y. Chang. 2008. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. USA 105:16272-16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stewart, S. E., B. E. Eddy, and N. G. Borgese. 1958. Neoplasms in mice inoculated with a tumor agent carried in tissue culture. J. Natl. Cancer Inst. 20:1223-1243. [DOI] [PubMed] [Google Scholar]
- 112.Sullivan, C. S., A. T. Grundhoff, S. Tevethia, J. M. Pipas, and D. Ganem. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682-686. [DOI] [PubMed] [Google Scholar]
- 113.Sunden, Y., S. Semba, T. Suzuki, Y. Okada, Y. Orba, K. Nagashima, T. Umemura, and H. Sawa. 2007. Identification of DDX1 as a JC virus transcriptional control region-binding protein. Microbiol. Immunol. 51:327-337. [DOI] [PubMed] [Google Scholar]
- 114.Sunden, Y., S. Semba, T. Suzuki, Y. Okada, Y. Orba, K. Nagashima, T. Umemura, and H. Sawa. 2007. DDX1 promotes proliferation of the JC virus through transactivation of its promoter. Microbiol. Immunol. 51:339-347. [DOI] [PubMed] [Google Scholar]
- 115.Sweet, B. H., and M. R. Hilleman. 1960. The vacuolating virus, S.V. 40. Proc. Soc. Exp. Biol. Med. 105:420-427. [DOI] [PubMed] [Google Scholar]
- 116.Tada, H., J. Rappaport, M. Lashgari, S. Amini, F. Wong-Staal, and K. Khalili. 1990. Trans-activation of the JC virus late promoter by the tat protein of type 1 human immunodeficiency virus in glial cells. Proc. Natl. Acad. Sci. USA 87:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tavis, J. E., and R. J. Frisque. 1991. Altered DNA binding and replication activities of JC virus T-antigen mutants. Virology 183:239-250. [DOI] [PubMed] [Google Scholar]
- 118.Reference deleted.
- 119.Trowbridge, P. W., and R. J. Frisque. 1995. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. J. Neurovirol. 1:195-206. [DOI] [PubMed] [Google Scholar]
- 120.Tyagarajan, S. K., and R. J. Frisque. 2006. Stability and function of JC virus large T antigen and T′ proteins are altered by mutation of their phosphorylated threonine 125 residues. J. Virol. 80:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vaz, B., P. Cinque, M. Pickhardt, and T. Weber. 2000. Analysis of the transcriptional control region in progressive multifocal leukoencephalopathy. J. Neurovirol. 6:398-409. [DOI] [PubMed] [Google Scholar]
- 122.Vilchez, R. A., and J. S. Butel. 2004. Emergent human pathogen simian virus 40 and its role in cancer. Clin. Microbiol. Rev. 17:495-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wegner, M., D. W. Drolet, and M. G. Rosenfeld. 1993. Regulation of JC virus by the POU-domain transcription factor Tst-1: implications for progressive multifocal leukoencephalopathy. Proc. Natl. Acad. Sci. USA 90:4743-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.White, M. K., and K. Khalili. 2004. Polyomaviruses and human cancer: molecular mechanisms underlying patterns of tumorigenesis. Virology 324:1-16. [DOI] [PubMed] [Google Scholar]
- 125.White, M. K., T. S. Gorrill, and K. Khalili. 2006. Reciprocal transactivation between HIV-1 and other human viruses. Virology 352:1-13. [DOI] [PubMed] [Google Scholar]
- 126.Winklhofer, K. F., I. Albrecht, M. Wegner, and R. Heilbronn. 2000. Human cytomegalovirus immediate-early gene 2 expression leads to JCV replication in nonpermissive cells via transcriptional activation of JCV T antigen. Virology 275:323-334. [DOI] [PubMed] [Google Scholar]
- 127.Yogo, Y., and C. Sugimoto. 2001. The archetype concept and regulatory region rearrangement, p. 127-148. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. John Wiley & Sons, Inc., New York, NY.
- 128.Yogo, Y., T. Kitamura, C. Sugimoto, T. Ueki, Y. Aso, K. Hara, and F. Taguchi. 1990. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J. Virol. 64:3139-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yogo, Y., S. Zhong, Y. Xu, M. Zhu, Y. Chao, C. Sugimoto, H. Ikegaya, A. Shibuya, and T. Kitamura. 2008. Conserved archetypal configuration of the transcriptional control region during the course of BK polyomavirus evolution. J. Gen. Virol. 89:1849-1856. [DOI] [PubMed] [Google Scholar]
- 130.Zerrahn, J., U. Knippschild, T. Winkler, and W. Deppert. 1993. Independent expression of the transforming amino-terminal domain of SV40 large T antigen from an alternatively spliced third SV40 early mRNA. EMBO J. 12:4739-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]