Abstract
Host-encoded factors play an important role in virus multiplication, acting in concert with virus-encoded factors. However, information regarding the host factors involved in this process is limited. Here we report the map-based cloning of an Arabidopsis thaliana gene, TOM1, which is necessary for the efficient multiplication of tobamoviruses, positive-strand RNA viruses infecting a wide variety of plants. The TOM1 mRNA is suggested to encode a 291-aa polypeptide that is predicted to be a multipass transmembrane protein. The Sos recruitment assay supported the hypothesis that TOM1 is associated with membranes, and in addition, that TOM1 interacts with the helicase domain of tobamovirus-encoded replication proteins. Taken into account that the tobamovirus replication complex is associated with membranes, we propose that TOM1 participates in the in vivo formation of the replication complex by serving as a membrane anchor.
Multiplication of viruses involves host-encoded factors functioning in concert with virus-encoded factors (1, 2). For positive-strand RNA viruses of higher eukaryotes, however, specific information about such host factors is limited to a few examples including host proteins necessary for the activity of viral RNA-dependent RNA polymerases in vitro and those specifically interacting with virus-related RNAs or virus-encoded proteins (3–7). Alternatively, recessive host mutations affecting virus multiplication have been identified, and their corresponding wild-type gene products are suggested to be involved in virus multiplication (8–11). At present, the lack of abundant knowledge about host factors is one of the most serious bottlenecks in elucidating the replication mechanisms of RNA viruses.
The tobamovirus group includes tobacco mosaic virus (TMV) along with other viruses that infect a wide variety of plants. This group belongs to the alpha-like virus supergroup, a major class of positive-strand RNA viruses including agronomically and clinically important viruses of plants and animals (1). The genome of tobamovirus consists of a capped, single-stranded RNA of messenger sense. After invasion into a host cell, the genomic RNA is translated to produce a 130-kDa protein and its read-through product, a 180-kDa protein. These two proteins function to replicate the genomic RNA via the complementary RNA (1). They copurify with membrane-bound tobamoviral RNA-dependent RNA polymerase activity from infected plants and have been localized by indirect immunofluorescence microscopy to the endoplasmic reticulum (ER)-derived structures in infected cells (1, 12). The 130-kDa protein harbors two domains implicated in the capping and helicase functions, whereas the 180-kDa protein contains, in addition, a domain implicated in the polymerase function. These three domains are conserved among the members of the alpha-like virus supergroup (1), suggesting that their replication involves similar molecular mechanisms.
A mutation in Arabidopsis thaliana, termed tom1, suppresses efficient multiplication of tobamoviruses TMV-Cg and TMV-L at the single-cell level, whereas the mutation does not affect the multiplication of either cucumber mosaic virus (CMV, a cucumovirus), turnip yellow mosaic virus (a tymovirus), both of which belong to the alpha-like virus supergroup but are distinct from tobamoviruses, or turnip crinkle virus (a carmovirus), which is not an alpha-like virus (10, 13). The recessive nature of the tom1 mutation suggested that the wild-type TOM1 gene product is required for efficient intracellular multiplication of tobamoviruses. Here, we report the map-based cloning of TOM1.
Materials and Methods
Virus and Plants.
A crucifer strain of tobamovirus named TMV-Cg (14), which is closely related to Chinese rape mosaic virus (15), was used for the inoculation of A. thaliana (L.) Heynh. A. thaliana ecotype Columbia wild-type (Col-0) and Wassilewskija wild-type (WS) seeds were purchased from Lehle Seeds (Round Rock, TX). The A. thaliana tom1–1 and tom1–2 mutants were isolated from Col-0 and characterized as described (10). The tom1–3 mutant was isolated from WS by screening a T-DNA insertion library (16) for mutants with low-level accumulation of the coat protein (CP) of TMV-Cg as described (10). The tom1–3 plants showed similar accumulation patterns of TMV-Cg CP to those in A. thaliana tom1–1 and tom1–2 mutant plants. Genetic analysis suggested the causal mutation was within the TOM1 locus. The tom1–3 mutation was not linked with the T-DNA insertion. The conditions for plant growth, inoculation with TMV-Cg, and determination of the levels of TMV-Cg CP accumulation by SDS/PAGE were as described (10).
Mapping and Cloning of the TOM1 Gene.
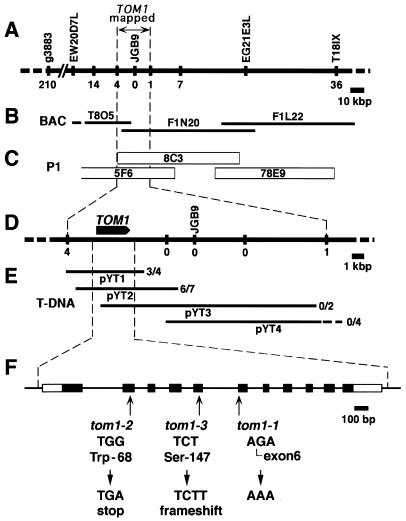
A contig encompassing the TOM1 locus (Fig. 1C) was constructed by screening of the A. thaliana P1 genomic library (obtained from Mitsui Plant Biotechnology Research Institute, Tsukuba, Japan) for clones hybridizing with the JGB9 DNA that is tightly linked to the TOM1 locus (Fig. 1A), and subsequent chromosome walking (17). Fine mapping of the TOM1 locus was performed by using the tom1–3 mutant as follows: DNA was prepared from one of the leaves of each F2 plant derived from a cross between tom1–3 (WS background) and Col-0 wild-type plants by using the NaOH extraction method (18). The genotype for markers g3883 and T18ISX (Fig. 1A) was examined for each F2 plant, and individuals having a chromosomal recombination event between these two markers were selected. DNA then was extracted from the leaves of each selected F2 plant by using the cetyltrimethylammonium bromide method (19), and purified by chloroform extraction, isopropanol precipitation, and MgCl2 precipitation (20). By using this DNA, genotypes for the cleaved amplified polymorphic sequence marker JGB9 and restriction fragment length polymorphism (RFLP) markers as indicated in Fig. 1 A and D were determined. The RFLP markers were prepared from the P1 genomic clones by restriction digestion. Information on the RFLP markers is available on request. Preparation of probes and Southern hybridization were performed by using the Gene Images labeling and detection system (Amersham Pharmacia). The cleaved amplified polymorphic sequence marker T18ISX (Fig. 1A), which gives an XbaI RFLP between ecotypes Col-0 and WS, was created based on the finding that the end probe of the P1 clone 78E9 (Fig. 1C) showed an XbaI RFLP between Col-0 and WS, and on the sequence information of bacterial artificial chromosome contig (ref. 21; Fig. 1B). Primers for T18ISX were T18ISXF 5′-CTGAGAATGTTTATCCCAGCTG-3′ and T18ISXR 5′-GTAATGCTTGAATCTCTTGATATC-3′. The PCR markers g3883 and JGB9 are described in the Arabidopsis Information Resources (22).
Figure 1.
Genetic mapping and positional cloning of TOM1. (A) Genetic map around the TOM1 locus on chromosome 4. Vertical bars represent DNA markers. Each number represents recombination events between the marker and the TOM1 locus among 3,103 F2 plants. (B and C) Contigs of ESSA II bacterial artificial chromosome clones and Mitsui P1 clones encompassing the TOM1 locus, respectively. (D) Fine map around the TOM1 locus. Vertical bars and numbers are the same as in A. The position and orientation of the TOM1 gene is shown by the arrow. (E) T-DNA contigs. T-DNA clones derived from the P1 clones 8C3 or 5F6 were organized into an overlapping set that spanned the TOM1 locus. These T-DNA clones were used to genetically transform tom1–1 mutant plants, which subsequently were tested for complementation of the TMV-Cg multiplication phenotype. The ratio of numbers of transformed lines that showed wild-type level TMV-Cg multiplication vs. total lines tested for TMV-Cg multiplication is indicated at the right of each T-DNA clone. (F) Intron-exon organization of the TOM1 gene and the tom1 mutations. Exons are indicated by boxes. Open boxes indicate noncoding regions, and filled boxes indicate coding regions. Mutations in the three tom1 alleles are shown below the intron/exon structure.
Complementation Analysis of the tom1 Mutation.
The T-DNA clones pYT1 and pYT2 were constructed by digesting the P1 genomic clones 8C3 with SalI + ClaI, and 5F6 with XhoI, respectively, and by inserting appropriate DNA fragments into the T-DNA vector pCLD04541 (22). T-DNA clones pYT3 and pYT4 were constructed by partial digestion of the P1 genomic clones 8C3 and 5F6, respectively, with EcoRI, and by inserting the generated DNA fragments into pCLD04541. T-DNA clones were electroporated into Agrobacterium tumefaciens C58C1 (pGV2260) and used to transform A. thaliana plants by the vacuum infiltration method (23). T2 (5–8) plants were used to determine the TMV-Cg multiplication phenotype for each transformed line. The presence of the neomycin phosphotransferase II gene (NPTII) in T2 plants was examined in two and four of the complemented lines transformed by pYT1 and pYT2, respectively. The primers 5′-CTATGACTGGGCACAACAGACAATC-3′ and 5′-GCGATAGAAGGCGATGCGCT-3′ were used to amplify the NPTII DNA sequence by PCR. Preparation of protoplasts, introduction of viral RNAs into the protoplasts by electroporation, and subsequent RNA analysis were performed as described (13). To check the quality of protoplasts, we inoculated protoplasts with CMV RNA by electroporation and confirmed that CMV RNA multiplied normally.
Northern Hybridization Analysis of TOM1 mRNA.
Total RNA was extracted from frozen plant tissues and purified by using ISOGEN LS (Nippon Gene, Toyama, Japan) according to the manufacturer's instructions. Northern blotting and hybridization was performed as described (13). 32P-labeled probes were prepared by using a Multiprime DNA labeling system (Amersham Pharmacia).
Isolation of TOM1 cDNA and Sequence Analysis.
We screened an A. thaliana 5′-STRETCH cDNA library (CLONTECH) containing 1 × 105 plaques for clones hybridizing with the overlapping region of the inserts of T-DNA clones pYT1 and pYT2 and obtained two such cDNA clones. Inserts of the cDNA clones were amplified by PCR and sequenced. Primers used to amplify and sequence the cDNA inserts were 5′-CGCCTCCATCAACAAACTTTCTTG-3′ and 5′-GTTCTGGTAAAAAGCGTGGTC-3′. The mRNA sequence carrying the entire coding sequence (GenBank accession no. AB016925) was identified by the 5′ and 3′ rapid amplification of cDNA ends method by using a SMART PCR cDNA Synthesis kit (CLONTECH). A 377 DNA sequencer and BigDye Terminator Sequencing kit (Perkin–Elmer) were used for sequencing.
The Sos Recruitment System.
Saccharomyces cerevisiae cdc25–2 strain and plasmids to express p110–5′Sos and 5′Sos-F are as described (24). TOM1–5′Sos is a fusion of full-length TOM1 (amino acids 1–291) to the N terminus of 5′Sos (amino acids 1–1066). 5′Sos-Hel and 5′Sos-HelΔ are fusions of C-terminal parts of the TMV-Cg 130-kDa protein (amino acids 609-1103 and 713-1103, respectively) (14) to the C terminus of 5′Sos. TOM1fs is a TOM1 derivative with a four-base insertion at the BamHI site in TOM1 ORF, resulting in a frameshift after amino acid residue 60. These constructs were expressed from the S. cerevisiae ADH1 promoter (25) on 2-μm plasmid vectors (26).
Results
Positional Cloning of the TOM1 Gene.
In our previous report, the tom1 mutation was mapped to the long arm of chromosome 4 between the RFLP markers m600 and m557 (27) with F2 lines generated by a cross between tom1–1 (Col-0 background) and the wild-type ecotype of Landsberg (La-0) (11). The position of the tom1 mutation was further confined with these F2 lines to the region between the yeast artificial chromosome ends EW20D7L and EW21E3L (28) (Fig. 1A). For more precise mapping, we used the tom1–3 mutant, which was derived from WS. Among 3,103 F2 plants obtained from a cross between Col-0 and tom1–3 plants, 246 individuals had chromosomal recombination in the region between the DNA markers g3883 and T18ISX (Fig. 1A), where TOM1 is present. Together with determination of the TMV-Cg-multiplication phenotype of these recombinants, further examination with other DNA markers as shown in Fig. 1 A and D (vertical bars with numbers) revealed that TOM1 is located in an ≈22-kbp region containing the JGB9 marker (Fig. 1 A and D).
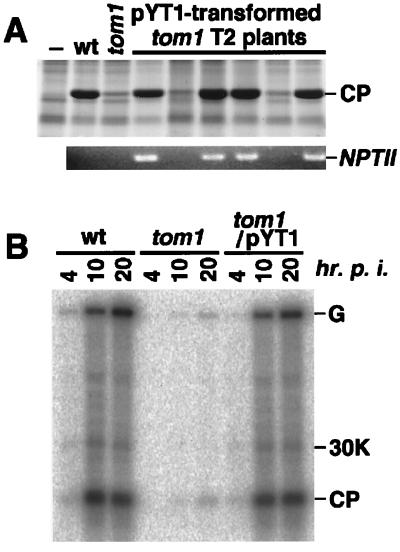
This 22-kb region was covered by two overlapping P1 genomic clones, 8C3 and 5F6 (Fig. 1C). To further localize the TOM1 gene within this region, DNA fragments of the P1 clones were subcloned into a T-DNA vector (pYT1 to 4; Fig. 1E) and used to stably transform tom1–1 plants. Each transformant (T1 generation) was inoculated with TMV-Cg, and the accumulation of the CP was examined. The results showed that the transformants obtained with pYT1 or pYT2 supported multiplication of TMV-Cg at a level comparable to that observed in the wild-type plants, whereas those obtained with pYT3 or pYT4 did not (Fig. 1E). As far as we examined, all of the T2 plants in which TMV-Cg multiplication was recovered carried the NPTII sequence that was present within the T-DNAs (Fig. 2A).
Figure 2.
Complementation of tom1 mutation with T-DNA clone pYT1. (A) Cosegregation of T-DNA and complementation. T2 progenies derived from a tom1–1 mutant transformed with T-DNA clone pYT1 were inoculated with TMV-Cg. Two weeks after inoculation, total protein was prepared from the inoculated plants, separated by SDS/PAGE, and stained with Coomassie brilliant blue (Upper). For each T2 plant, the NPTII sequence was amplified from purified genomic DNA by PCR with specific primer sets and analyzed by agarose gel electrophoresis followed by ethidium bromide staining (Lower). The positions of TMV-Cg CP and the PCR-amplified NPTII DNA fragment are indicated. (B) Complementation at protoplast level. Direct descendants of a T2 progeny derived from a tom1–1 mutant transformed with T-DNA clone pYT1 and carrying the transgene homozygously were used to prepare protoplasts. Protoplasts were inoculated with TMV-Cg RNA by electroporation, cultured for 4, 10, or 20 h, and the accumulation of TMV-Cg-related RNAs was examined by Northern blot hybridization. The position of genomic RNA (G), subgenomic mRNAs for 30-kDa protein (30K), and CP are indicated at the right. As controls, analysis with nontransformed Col-0 (wt) and tom1–1 (tom1) protoplasts were performed in parallel.
To further confirm the complementation, protoplasts were isolated from tom1 plant lines that had been transformed with pYT1 or pYT2 and that carried the transgenes homozygously. TMV-Cg RNA was introduced into these protoplasts by electroporation, and amplification of the viral RNAs was examined by Northern blot hybridization. The pattern of TMV-Cg-related RNA accumulation in pYT1- or pYT2-transformed tom1 protoplasts was similar to that in wild-type protoplasts (Fig. 2B and data not shown), indicating that the cloned fragments complemented the tom1 mutation at the protoplast level. These results suggested that the TOM1 gene is located within the 6-kbp region shared by the clones pYT1 and pYT2 (Fig. 1 D and E).
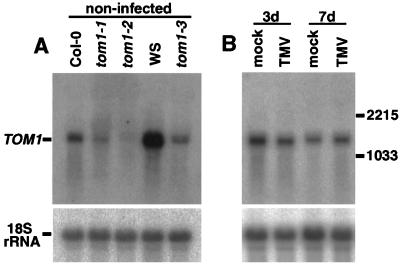
Northern blot hybridization with this 6-kbp region as a probe detected a single RNA species of ≈1.4 kb in length within the total RNA extracted from wild-type plants. In tom1–1, -2, and -3 plants, the accumulation levels of the 1.4-kb RNA were lower than those in the corresponding wild-type plants (Fig. 3A), suggesting that the mutations affected this transcription unit. The accumulation level of the mRNA species in wild-type plants was not significantly influenced on TMV-Cg infection (Fig. 3B). The presence of the RNA species in noninfected plants is consistent with our results, suggesting that TOM1 function is necessary for the establishment of tobamovirus infection from an early stage after the invasion of viral RNA (13).
Figure 3.
Northern blot hybridization analysis of the TOM1 mRNA. (A) TOM1 mRNA accumulation in wild-type and mutant plants. Total RNA was extracted from aerial tissues of 25-day-old noninoculated wild-type (Col-0 and WS) and mutant (tom1–1, tom1–2, and tom1–3) plants. Note that tom1–1 and tom1–2 are derived from Col-0, whereas tom1–3 is from WS (see Materials and Methods). RNA samples (10 μg) were denatured by glyoxal, separated by 1% agarose gel electrophoresis, and blotted onto a nylon membrane. Duplicate blots were prepared and probed with 32P-labeled DNAs hybridizing with either TOM1 or 18S rRNA (43) sequences. To prepare the TOM1-specific probe, a DNA fragment corresponding to the predicted TOM1 ORF was amplified by PCR from a cDNA clone and gel purified. (B) TOM1 mRNA levels are not altered by TMV-Cg infection. Total RNA was extracted from aerial tissues of mock-inoculated (mock) and TMV-Cg-inoculated (TMV) Col-0 plants. Twenty-day-old plants were inoculated and samples were harvested 3 or 7 days after inoculation. Northern blot hybridization was performed as in A. The positions of TOM1 mRNA and 18S rRNA are indicated (A Left). The positions of CMV (Y-strain) RNA3 (2,215 nt) and RNA4 (1,033 nt) used as size markers are shown (B Right).
Structure of the TOM1 Gene.
We identified a single 1,363-nt mRNA sequence (GenBank accession no. AB016925) containing an ORF of 876 nt (291 aa; Fig. 4A) within the 6-kbp region shared by the T-DNA clones pYT1 and pYT2, when the first ATG codon was taken to be the initiation codon. Comparison with the genomic sequence (bacterial artificial chromosomes T8O5 and F1N20; ref. 21; Fig. 1B) suggested that this mRNA sequence is composed of 11 exons (Fig. 1F). All three tom1 alleles (ref. 10; also Materials and Methods) carried mutations destructive to this ORF: a point mutation at the splicing acceptor site for intron 5 (tom1–1), a nonsense mutation in exon 2 (tom1–2), and a one-base insertion in exon 5 (tom1–3) (Fig. 1F), further suggesting that this ORF encodes TOM1.
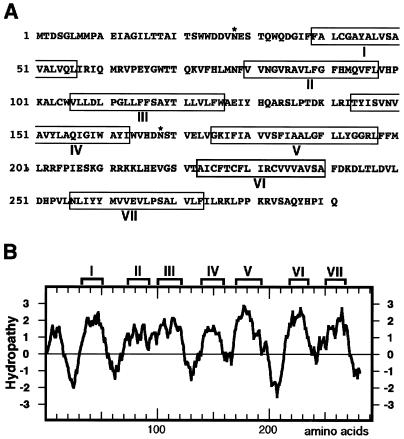
Figure 4.
Structure of TOM1. (A) Deduced amino acid sequence of TOM1. Boxed amino acid residues represent those belonging to putative membrane-spanning regions, as predicted by the sosui program (29). The program phdthtm (30) did not list the region VII, and psort (31) did not list the region II as a transmembrane region. Asn residues marked by * are putative glycosylation sites. (B) Hydropathy plot for the deduced amino acid sequence of TOM1. The hydropathy plot was created by the method described by Kyte and Doolittle (44). The regions predicted to be membrane spanning in A are indicated above the plot.
The deduced amino acid sequence of the TOM1 protein (Fig. 4A) contains several highly hydrophobic regions (Fig. 4B). Computer programs predicted that the protein is a seven-pass (sosui; ref. 29) or six-pass (phdthtm, ref. 30; psort, ref. 31) transmembrane protein with its N terminus exoplasmic (phdthtm, ref. 30; psort, ref. 31). Two possible N-glycosylation sites were found at amino acids 28–31 and 168–171 (Fig. 4A). No apparent N-terminal or other signal sequences necessary for targeting to specific organelles were found (psort; ref. 31). Database searches for proteins with similar amino acid sequences to that of TOM1 listed only integral membrane proteins with weak similarity: blast 2.0 (32) detected a weak similarity to a human putative seven-pass transmembrane protein TM7SF1 whose function is not known (ref. 33; 60 identical amino acid residues in 279-aa overlap) whereas fasta 3 (34) detected a weak similarity to cytochrome b from many eukaryotic organisms (e.g., 33 identical amino acid residues in 160-aa overlap with cytochrome b from maize). We speculate that these hits do not necessarily represent functional similarity, but rather represent structural similarity among integral membrane proteins.
Interaction Between TOM1 and the TMV-Cg-Encoded Replication Proteins.
To determine whether TOM1 interacts with tobamovirus-encoded replication proteins, the Sos-recruitment system was used (24). This system is based on the observation that the N-terminal fragment (5′Sos) of the Ras guanine nucleotide exchange factor, the human homolog of Drosophila melanogaster Son of Sevenless protein, suppresses temperature-sensitive growth caused by the cdc25–2 mutation in S. cerevisiae if 5′Sos is targeted to the plasma membrane in the vicinity of Ras.
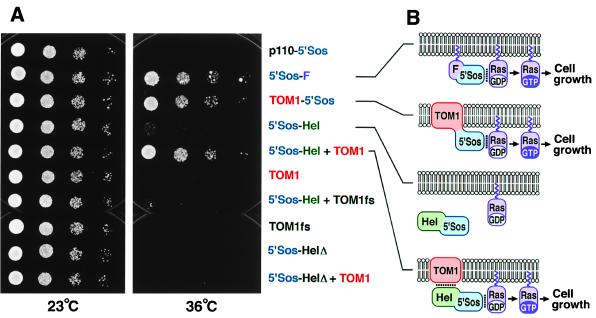
Expression of a fusion protein between TOM1 and 5′Sos (TOM1–5′Sos) but not TOM1 protein alone suppressed the temperature sensitivity (Fig. 5A), suggesting TOM1 is at least in part targeted to the plasma membrane and that the C terminus of the fusion protein is exposed to the cytoplasm when expressed in yeast. Coexpression of the TOM1 protein and a fusion protein between 5′Sos and a C-terminal half of the 130-kDa replication protein of TMV-Cg containing the helicase domain (5′Sos-Hel) also suppressed the temperature sensitivity, although the expression of 5′Sos-Hel alone suppressed it only very weakly (Fig. 5A). The weak suppression by 5′Sos-Hel alone may suggest a weak interaction of the helicase domain with the yeast plasma membrane. Similar results were obtained for the helicase domain of TMV-L (Kenji Kubota and T.M., unpublished results). Introduction of either an internal deletion in 5′Sos-Hel (5′Sos-HelΔ) or a frameshift mutation in TOM1 (TOM1fs) abolished this suppression (Fig. 5A). These results support the idea that 5′Sos-Hel fusion protein is recruited to the plasma membrane by the action of TOM1 protein, presumably through an interaction between the helicase domain and plasma membrane-bound TOM1 protein (Fig. 5B).
Figure 5.
Interaction between TOM1 and the TMV-Cg-encoded replication proteins. (A) cdc25–2 yeast strains harboring plasmids designed to constitutively express indicated proteins were diluted in sterile water to absorbance at 600 nm of 0.2, 0.025, 0.003, and 0.0004 (8-fold serial dilutions). Each dilution (2 μl) was spotted onto YAPD plates [1% (wt/vol) yeast extract/2% (wt/vol) peptone/2% (wt/vol) glucose/0.004% (wt/vol) adenine hemisulfate/2% (wt/vol) agar] and cultured at 23°C for 52 h or 36°C for 66 h. Part of p110 β, a subunit of phosphatidylinositol-3-phosphate kinase, fused to 5′Sos (p110–5′Sos), and 5′Sos fused to Ras farnesylation signal (5′Sos-F) are negative and positive controls, respectively, for suppression of cdc25–2 temperature sensitivity (24). TOM1fs and 5′Sos-HelΔ are frameshifting and deletion derivatives of TOM1 and 5′Sos-Hel, respectively. (B) Models explaining the results of A. Lipid bilayers indicate plasma membranes with the lower sides cytoplasmic. Noncovalent interactions are indicated by dotted lines. Covalent linkage of 5′Sos polypeptide with TOM1 or a noncovalent interaction between TOM1 and the helicase domain of TMV-Cg-encoded replication proteins in 5′Sos-Hel recruits 5′Sos to the plasma membrane to activate Ras signaling.
Discussion
We report here the identification of the A. thaliana TOM1 gene by positional cloning. Recessiveness of the tom1 mutation, resulting in reduced accumulation levels of tobamoviruses, suggested that that the wild-type TOM1 protein is necessary for the efficient multiplication of tobamoviruses in plants (10). Delayed accumulation of virus-related RNAs observed in tom1 protoplasts inoculated with tobamovirus RNA, together with a lower percentage of inoculated protoplasts accumulating detectable levels of viral CP, also indicated that TOM1 functions to support tobamovirus multiplication from an early stage of infection after the uncoating of virions and when the invasion of viral RNA is not yet destined either to abortive infection or productive virus multiplication (13). Such processes include the replication of tobamovirus RNA. The deduced amino acid sequence of TOM1 identified in this study contained several highly hydrophobic regions, and computer programs predicted that TOM1 is a multipass transmembrane protein. We have demonstrated the involvement of a host transmembrane protein in the intracellular multiplication of positive-strand RNA viruses.
The replication of most or all eukaryotic positive-strand RNA viruses including tobamoviruses is thought to occur in membrane-bound complexes (1). The docking to membranes therefore appears to be critical for the formation of active replication complexes of this class of viruses. For several positive-strand RNA viruses, virus-encoded proteins are suggested to serve as lipophilic anchors to retain the replication complexes on membranes (35–37). In contrast, neither transmembrane regions nor posttranslational modification facilitating membrane association have been predicted for tobamovirus-encoded replication proteins (29–31) and they are thought to form a replication complex in association with host proteins localized on membranes. In keeping with this possibility, the Sos-recruitment assay supported the hypothesis that TOM1 associates with membranes and interacts with the helicase domain of tobamovirus-encoded replication proteins. This may indicate that TOM1 participates in the in vivo formation of the tobamovirus replication complex by functioning to anchor the complex to membranes. The Sos recruitment assay results suggested that TOM1 localizes at least in part on the plasma membrane in yeast, whereas the tobamovirus replication complex localizes on ER membranes in plant cells (12). These two observations are not necessarily contradictory because the localization of TOM1 in yeast might be different from that in plant cells, and/or TOM1 might localize on both the plasma membrane and ER membranes.
In brome mosaic virus-infected cells, virus-encoded replication proteins colocalize at the sites of viral RNA synthesis on ER membranes (38). In contrast, it is proposed that the site of synthesis of turnip yellow mosaic virus or alfalfa mosaic virus RNA is on the cytoplasmic surface of the chloroplast outer membranes (39, 40), whereas that of CMV in cytopathic structures associated with the tonoplast membranes (41). Although the sites of RNA synthesis have not been determined, it is also known that the replication proteins of tobacco mosaic virus localize to ER (12), and those of Semliki Forest virus and Sindbis virus both localize to endosome and lysosome membranes (42). Therefore, the intracellular sites of viral RNA synthesis seem to differ even among these alpha-like viruses that encode replication proteins with conserved domains. If this is the case, factors that anchor viral replication complexes to membranes also are expected to be different from one virus group to another. It is plausible that the replication complexes of CMV and turnip yellow mosaic virus are linked to membranes in a manner independent of TOM1, possibly explaining why the tom1 mutation does not affect the multiplication of these viruses (10).
Consistent with the recessiveness of the tom1 mutations, sequence analysis revealed that all three tom1 alleles contained mutations that would seriously affect TOM1 gene expression, probably resulting in almost complete loss of gene function. Nevertheless, tobamovirus multiplication was not completely inhibited in any of the tom1 mutants (10, 13). This suggests either that TOM1 function is not absolutely necessary for tobamovirus multiplication, or that there are redundant genes of TOM1 in the A. thaliana genome. Recently, we have identified a TOM1-like gene in A. thaliana that would have parallel functions to TOM1. In preliminary studies, simultaneous mutations in TOM1 and the TOM1-like gene resulted in undetectable levels of tobamovirus multiplication, suggesting their function is likely to be essential for tobamovirus multiplication (T.Y., Rena Satoh, S.N., and M.I., unpublished results).
Despite the anticipated serious damage to TOM1 gene expression by the tom1 mutations, the mutant plants appear to grow normally under our growth conditions. From the view of application, this is encouraging toward the utilization of the gene for antiviral strategies, because it may be possible to generate tobamovirus-resistant plants by artificially repressing the TOM1 gene expression without affecting the viability and growth of the plant itself. Expanding this possibility to other plant species, we recently have identified TOM1-like genes from tomato and tobacco (T.Y., Rena Satoh, S.N., and M.I., unpublished results). Identification of the TOM1 gene is an important step toward elucidation of the molecular mechanisms of tobamovirus replication in plants, as well as providing direction for the development of novel strategies for the artificial control of tobamovirus infection.
Acknowledgments
We thank P. Ahlquist for critical review, D. Bartlem for careful reading of the manuscript, T. Mizuno and K. Fujiwara for general assistance, and G. Coupland, D. Stevenson, A. Prescott, S. Doyle, J. Goodrich, C. Lister, D. Shibata, T. C. Osborn, C. Wetzel, A. Aronheim, Y. Komeda, and the Arabidopsis Biological Resource Center for materials. We used the facilities of the Biopolymer Analysis Laboratory in the Faculty of Agriculture, Hokkaido University and the Research Center for Molecular Genetics at Hokkaido University. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan to M.I. (No. 08680733) and S.N. (No. 06278102), and by a grant from the Japan Society for the Promotion of Science to M.I. (RFTF96L00603).
Abbreviations
- TMV
tobacco mosaic virus
- CP
coat protein
- RFLP
restriction fragment length polymorphism
- NPTII
neomycin phosphotransferase II gene
- CMV
cucumber mosaic virus
- Col-O
Columbia wild type
- WS
Wassilewskija wild type
- ER
endoplasmic reticulum
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB016924 and AB016925).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170295097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170295097
References
- 1.Buck K W. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai M M C. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 3.Quadt R, Kao C C, Browning K S, Hershberger R P, Ahlquist P. Proc Natl Acad Sci USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osman T A, Buck K W. J Virol. 1997;71:6075–6082. doi: 10.1128/jvi.71.8.6075-6082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardigon N, Strauss J H. J Virol. 1996;70:1173–1181. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBride A E, Schlegel A, Kirkegaard K. Proc Natl Acad Sci USA. 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waggoner S, Sarnow P. J Virol. 1998;72:6699–6709. doi: 10.1128/jvi.72.8.6699-6709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa M, Diez J, Restrepo-Hartwig M A, Ahlquist P. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diez J, Ishikawa M, Kaido M, Ahlquist P. Proc Natl Acad Sci USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa M, Obata F, Kumagai T, Ohno T. Mol Gen Genet. 1991;230:33–38. doi: 10.1007/BF00290647. [DOI] [PubMed] [Google Scholar]
- 11.Ohshima K, Taniyama T, Yamanaka T, Ishikawa M, Naito S. Virology. 1998;243:472–481. doi: 10.1006/viro.1998.9078. [DOI] [PubMed] [Google Scholar]
- 12.Heinlein M, Padgett H S, Gens J S, Pickard B G, Casper S J, Epel B L, Beachy R N. Plant Cell. 1998;10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa M, Naito S, Ohno T. J Virol. 1993;67:5328–5338. doi: 10.1128/jvi.67.9.5328-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamanaka T, Komatani H, Meshi T, Naito S, Ishikawa M, Ohno T. Virus Genes. 1998;16:173–176. doi: 10.1023/a:1007945723588. [DOI] [PubMed] [Google Scholar]
- 15.Aguilar I, Sanchez F, Martin M A, Martinez-Herrera D, Ponz F. Plant Mol Biol. 1996;30:191–197. doi: 10.1007/BF00017814. [DOI] [PubMed] [Google Scholar]
- 16.Feldmann K A, Marks M D. Mol Gen Genet. 1987;208:1–9. [Google Scholar]
- 17.Liu Y G, Mitsukawa N, Vazquez-Tello A, Whittier R F. Plant J. 1995;7:351–358. [Google Scholar]
- 18.Wang H, Qi M, Cutler A J. Nucleic Acids Res. 1993;21:4153–4154. doi: 10.1093/nar/21.17.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers S O, Bendich J. Plant Mol Biol. 1985;5:69–76. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto T, Horikoshi M. Nucleic Acids Res. 1995;23:3351–3352. doi: 10.1093/nar/23.16.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer K, Schüller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Düsterhöft A, Stiekema W, Entian K-D, Terryn N, et al. Nature (London) 1999;402:769–777. [Google Scholar]
- 22.Flanders D J, Weng S, Petel F X, Cherry J M. Nucleic Acids Res. 1998;26:80–84. doi: 10.1093/nar/26.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bechtold N, Pelletier G. Methods Mol Biol. 1993;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 24.Aronheim A, Zandi E, Hennemann H, Elledge S J, Karin M. Mol Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colicelli J, Birchmeier C, Michaeli T, O'Neill K, Riggs M, Wigler M. Proc Natl Acad Sci USA. 1989;86:3599–3603. doi: 10.1073/pnas.86.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz R D, Sigino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 27.Chang C, Bowman J L, DeJohn A W, Lander E S, Meyerowitz E M. Proc Natl Acad Sci USA. 1988;85:6856–6860. doi: 10.1073/pnas.85.18.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt R, West J, Cnops G, Love K, Balestrazzi A, Dean C. Plant J. 1996;9:755–765. doi: 10.1046/j.1365-313x.1996.9050755.x. [DOI] [PubMed] [Google Scholar]
- 29.Hirokawa T, Boon-Chieng S, Mitaku S. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 30.Rost B, Casadio R, Fariselli P, Sander C. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spangenberg C, Winterpacht A, Zabel B U, Löbbert R W. Genomics. 1998;48:178–185. doi: 10.1006/geno.1997.5170. [DOI] [PubMed] [Google Scholar]
- 34.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahola T, Lampio A, Auvinen P, Kaariainen L. EMBO J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Towner J S, Ho T V, Semler B L. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 37.Schaad M C, Jensen P, Carrington J C. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Restrepo-Hartwig M A, Ahlquist P. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.deGraaff M, Coscoy L, Jaspars E M J. Virology. 1993;194:878–881. doi: 10.1006/viro.1993.1335. [DOI] [PubMed] [Google Scholar]
- 40.Garnier M, Ramoun R, Bové J M. Virology. 1980;104:357–374. doi: 10.1016/0042-6822(80)90340-2. [DOI] [PubMed] [Google Scholar]
- 41.Hatta T, Francki R I B. J Gen Virol. 1981;53:343–346. [Google Scholar]
- 42.Froshauer S, Kartenbeck J, Helenius A. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi T, Komeda Y. Plant Cell Physiol. 1992;33:389–394. [Google Scholar]
- 44.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]