Abstract
The Epstein-Barr virus BGLF4 and BGLF5 genes encode a protein kinase and an alkaline exonuclease, respectively. Both proteins were previously found to regulate multiple steps of virus replication, including lytic DNA replication and primary egress. However, while inactivation of BGLF4 led to the downregulation of several viral proteins, the absence of BGLF5 had the opposite effect. Using recombinant viruses that lack both viral enzymes, we confirm and extend these initial observations, e.g., by showing that both BGLF4 and BGLF5 are required for proper phosphorylation of the DNA polymerase processivity factor BMRF1. We further found that neither BGLF4 nor BGLF5 is required for baseline viral protein production. Complementation with BGLF5 downregulated mRNA levels and translation of numerous viral genes, though to various degrees, whereas BGLF4 had the opposite effect. BGLF4 and BGLF5 influences on viral expression were most pronounced for BFRF1 and BFLF2, two proteins essential for nuclear egress. For most viral genes studied, cotransfection of BGLF4 and BGLF5 had only a marginal influence on their expression patterns, showing that BGLF4 antagonizes BGLF5-mediated viral gene shutoff. To be able to exert its functions on viral gene expression, BGLF4 must be able to escape BGLF5's shutoff activities. Indeed, we found that BGLF5 stimulated the BGLF4 gene's transcription through an as yet uncharacterized molecular mechanism. The BGLF4/BGLF5 enzyme pair builds a regulatory loop that allows fine-tuning of virus protein production, which is required for efficient viral replication.
The Epstein-Barr virus (EBV) is a large double-stranded DNA virus that has more than 100 genes, the large majority of which are required for virus replication (29). Lytic genes are sequentially expressed and therefore classified as immediate-early, early, or late genes. Expression of EBV lytic genes is positively regulated by transactivators such as the immediate-early BZLF1 and BRLF1 (5, 9, 20), but the alkaline exonuclease BGLF5 (AE) has also been identified as a negative regulator (8). Through its ability to reduce expression of major histocompatibility complex class I and class II genes, BGLF5 was initially identified as a mediator of immune evasion (41, 54). BGLF5 host shutoff properties were subsequently found to extend to viral genes; expression of the BFLF2 gene, an early gene required for primary egress, was found to be enhanced in a ΔBGLF5 recombinant EBV (8, 17). Conversely, reintroduction of BGLF5 reverted this effect. The study of this mutant revealed multiple functions for AE during virus replication; the viral exonuclease was required for optimal total DNA synthesis and efficient processing of linear viral genomes. In the absence of BGLF5, viral encapsidation, nuclear egress, and virus production were substantially reduced (8).
These phenotypic traits are strongly reminiscent of those previously reported for an EBV producer cell line in which the BGLF4 gene, the only hitherto-identified EBV protein kinase (PK) gene, had been knocked down using a specific small interfering RNA; partial inhibition of viral DNA replication and retention of capsids in the nucleus were observed as a result (14). Despite these similarities, BGLF4 and BGLF5 were found to have opposite effects on viral gene expression; BGLF4 knockdown inhibited expression of some late viral genes such as the gp350 gene and of the early protein BFLF2, whereas deletion of BGLF5 resulted in an upregulation of the BFLF2 gene (and possibly of the BFRF1 gene, another early EBV gene), suggesting that both proteins might have opposite influences on viral protein expression (8, 14).
BGLF4 is a nuclear serine/threonine PK that displays structural and functional homologies with herpes simplex virus type 1 (HSV-1) UL13 and cytomegalovirus (CMV) UL97 (12, 26, 30, 34, 48). It has been identified as a virion tegument component, suggesting that it plays a role after virus penetration as already described for other viral tegument proteins (25, 35, 40, 47). In addition, BGLF4 has been shown to promote disassembly of the nuclear lamina, which is thought to be required for nuclear egress (32). Several viral targets for the BGLF4 PK have been identified and include the lytic BMRF1 (4, 13) and BZLF1 (1) genes, as well as the EBV latent EBNA-LP (27) and EBNA2 (51) genes. BGLF4-induced phosphorylation of EBNA2 was found to reduce its ability to transactivate LMP-1 expression (51). BGLF4 colocalizes during lytic replication with BMRF1 (also called early protein [EA-D] or DNA polymerase processivity factor) in replication compartments. These and other viral proteins such as BGLF5, but not BZLF1, were recently confirmed as being BGLF4 targets using an extensive EBV protein array (53). In addition, BGLF4 was found to interact with EBNA1 (53). BGLF4's targets also include cellular proteins such as the translation elongation factor 1δ (EF-1δ) (28) and members of the interferon regulatory factor 3 signaling pathway (46).
We have taken these issues further by constructing a BGLF4 mutant that lacks most of the BGLF4 gene's open reading frame. Unexpectedly, this mutant was found to be also deficient for BGLF5 expression, thereby providing an opportunity to study the interplay between the enzymes in their effects on viral gene expression and viral protein synthesis. We validated our findings with a mutant that lacks both BGLF4 and BGLF5 gene open reading frames.
MATERIALS AND METHODS
Primary cells and cell lines.
HEK293 is a neuroendocrine cell line obtained by transformation of embryonic epithelial kidney cells with adenovirus (16, 43). Raji is a human EBV-positive Burkitt's lymphoma cell line (39). WI38 is a line of primary human lung embryonic fibroblasts (21). Mononuclear cells were purified from fresh blood buffy coats by density gradient centrifugation. CD19-positive B cells were isolated from total lymphocytes using M-450 CD19 (PanB) magnetic beads (Dynal). All cell lines were routinely grown in RPMI 1640 medium supplemented with 10% fetal calf serum (Biochrom).
Recombinant plasmids.
A BZLF1 expression plasmid (p509) was used for initiation of the lytic cycle (19). The entire BGLF4 gene (B95.8 coordinates 122328 to 123692) was PCR amplified and cloned into an expression plasmid carrying the p38 parvovirus promoter to yield plasmid B300. The BGLF5 gene was PCR amplified (B95.8 coordinates 120932 to 122341) and placed under the control of a CMV promoter in the pCDNA3.1(+) expression plasmid (B332). Sequencing confirmed the integrity of the BGLF4 and BGLF5 sequences.
Recombinant EBV genomes.
The wild-type EBV recombinant plasmid (p2089) used in this study (6) was cloned onto the prokaryotic F factor origin of replication and carries the green fluorescent protein (GFP) gene, the chloramphenicol (cam) resistance gene, and the hygromycin (hyg) resistance gene. The EBV BGLF4-negative mutant was constructed by replacing the BGLF4 gene (B95.8 coordinates 122478 to 123692; GenBank accession number V01555) with the kanamycin (kan) resistance gene using homologous recombination (37). To this end, composite primers whose internal parts (24 bp; underlined) are specific for the kan resistance gene and whose external parts (40 bp) are specific for the BGLF4 gene were used. These primers (5′-ACCAAGACTCAATTTCCAAAAATCAACTACAAGCAGCTAGCCAGTCACGACGTTGTAAAACGAC-3′ and 5′-TTGAA CCTCTTTTTAAGGCTCCGGCACCACTGCAAGAATTGAACAGCTATG ACCATGATTACGCC-3′) allowed PCR-mediated amplification of the kan resistance gene through their internal sequences and then homologous recombination of the amplified PCR product with the EBV wild-type genome via their external sequences. The BGLF4-BGLF5 double mutant was constructed using the same strategy as that outlined above. EBV B95.8 coordinates 121348 to 123692 were replaced with the kan resistance gene using the following primers: 5′-ACCAAGACTCAATTTCCAAAAATCAACTACAAGCAGCTAGCCAGTCACGACGTTGTAAAACGAC-3′ and 5′-TTGACTGGGACCCGGTCTTTAATACCAATGCGCCCCGCATTTACTCAGCTAAACAGCTATGACCATGATTACGCC-3′ (underlined, kan-specific parts; italics, introduced stop codons). PCR amplification products were incubated with the restriction enzyme DpnI to remove traces of the parental plasmid and introduced by electroporation (1,000 V, 25 μF, 100 Ω) into Escherichia coli DH10B cells carrying the recombinant virus p2089 and the temperature-sensitive pKD46 helper plasmid encoding the phage lambda red recombinase to foster homologous recombination. Cells were grown in Luria broth (LB) with cam (15 μg/ml) at 37°C for an hour and then plated onto LB agar plates containing cam (15 μg/ml) and kan (10 μg/ml). Incubation at 42°C led to a progressive loss of the helper plasmid. After double selection, DNA of positive clones was purified and analyzed with the BamHI restriction enzyme to confirm correct recombination.
A revertant virus was generated by chromosomal building (37). To this end, the BGLF4 gene and its flanking left and right regions (B95.8 coordinates 121151 to 124342) were cloned onto a targeting vector (pSTAZ2) that consisted of a temperature-sensitive bacterial origin of replication, the ampicillin resistance gene, and the RecA and LacZ gene operons to yield plasmid B293. In addition, we introduced a deletion of the BglII restriction site in the noncoding region of the targeting vector located between the BGLF3 and BGLF4 genes that allowed quick and unequivocal distinction between the revertant clone and wild-type genomes. This allowed exclusion of potential contamination at all steps of the mutant construction. Homologous recombination was performed in two steps, with initial building of a cointegrate between B293 and ΔBGLF4 plasmids that contained both the mutant and wild-type BGLF4 genes, followed by resolution of these cointegrates to generate resolvants, some of which had successfully restored the BGLF4 gene locus.
Stable clone selection.
Recombinant EBV plasmid DNA was transfected into HEK293 cells using Lipofectamine reagent (Invitrogen) as described previously (24). One day posttransfection, the cells were transferred to a cell culture dish (150 mm in diameter) and hyg (100 μg/ml) was added to the culture medium for selection of stable HEK293 clones carrying the EBV recombinant plasmid. Outgrowing GFP-positive colonies were expanded for further investigation. The cell clones used in this study are referred to as 293/ΔBGLF4, 293/ΔBGLF4-Rev, and 293/ΔΔBG4/5 to denote clones carrying the BGLF4 knockout mutant, its revertant, and the BGLF4-BGLF5 double mutant, respectively.
Plasmid rescue in E. coli.
Circular plasmid DNA from 293/ΔBGLF4, 293/ΔΔBG4/5, and 293/ΔBGLF4-Rev cells was extracted using a denaturation-renaturation method as described previously (18). E. coli strain DH10B was transformed with the viral recombinant DNA by electroporation as described before (37), and clones were selected on LB plates containing cam (15 μg/ml). Single bacterial colonies were expanded, and the plasmid DNA preparation was subjected to digestion with restriction enzymes BamHI and BglII.
Virus induction and infection of target cells.
Producer cell clones 293/EBV-wt (carrying p2089) (EBV-wt is wild-type EBV), 293/ΔBGLF4, and 293/ΔΔBG4/5 were transfected with a BZLF1 expression plasmid (0.5 μg/well) to induce the lytic cycle using lipid miscelles (Metafectene; Biontex). In transcomplementation assays, 293/ΔBGLF4 or 293/ΔΔBG4/5 cells were cotransfected with a BGLF4 (0.1 μg/well) and/or BGLF5 expression plasmid (0.7 μg/well). Empty expression plasmids were cotransfected in noncomplemented inductions. One, 2, or 4 days posttransfection, cells were harvested for analysis of viral gene expression. Virus supernatants were harvested 4 days posttransfection, filtered through a 0.8-μm filter, and stored at 4°C. Viral titers were determined by infecting 104 Raji cells with increasing dilutions of virus supernatants. Three days after infection, GFP-positive Raji cells were counted using a fluorescence microscope. For immortalization assays, primary B cells were admixed with infectious supernatants and seeded into U-bottom 96-well microtiter plates coated with gamma-irradiated WI38 feeder cells at a concentration of 103 cells per well. Wells containing outgrowing lymphoblastoid cell line (LCL) clones were counted, and a few LCL clones were expanded for further analysis.
Immunostaining.
Mouse monoclonal antibodies (MAb) against the following viral proteins were used for staining: BZLF1 (clone BZ.1) (50), BRLF1 (clone 8C12) (33), gp350/220 (clone 72A1) (45), BALF4 (anti-VCA-gp125, MAb 8184; Chemicon), EA-D encoded by the BMRF1 gene (MAb 818; Chemicon), BFRF1 (clone E10) (7), and BFLF2 (clone C1) (15). We further used rabbit polyclonal antibodies against BGLF4 (see below) and BGLF5 (3). Cells were washed three times in phosphate-buffered saline (PBS) and air dried onto glass slides. Fixation protocols were carried out for 20 min at room temperature in pure acetone (for BMRF1, gp110, and gp350) or 20 min in 4% paraformaldehyde followed by a 2-min treatment with 0.1% Triton X-100 for cell permeabilization (for BZLF1, BRLF1, BFRF1, BFLF2, BGLF4, and BGLF5). The slides were incubated with the antibody for 30 min, washed three times in PBS, and incubated with a secondary antibody conjugated with Cy-3 fluorochrome (Dianova). The slides were washed three times in PBS and embedded with 90% glycerol. Counterstaining of the nuclei was obtained by incubation with Hoechst 33258 (1:10,000 dilution) before embedding. Immunofluorescence was evaluated and recorded using a fluorescence microscope (Leica) or a confocal fluorescence microscope (Nikon).
Polyclonal antiserum against BGLF4.
A 1,372-bp BGLF4 gene fragment (B95.8 coordinates 122331 to 123691) was PCR amplified and cloned into the BamHI-HindIII sites of the pET21b vector (Novagen) to enable synthesis of a His-tagged BGLF4 fusion protein. The plasmid was transformed into E. coli strain BL21(DE3), and protein extracts were obtained by lysing the cells in 20 mM Tris-Hcl, pH 7.9, 0.5 M NaCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 5 mM imidazole. The BGLF4 fusion protein was purified from a 10% sodium dodecyl sulfate-polyacrylamide gel after elution in 4 M sodium acetate for 48 h at 4°C. One female Chinchilla bastard rabbit was immunized subcutaneously with 150 μg BGLF4 recombinant protein in the presence of incomplete Freund's adjuvant, followed by three subsequent boosts.
Electron microscopy.
Lytically induced producer cells or a virus pellet obtained after centrifugation of 5 ml virus supernatant for 2 h at 30,000 × g was fixed with 2.5% glutaraldehyde, and further preparation was carried out as described previously (17). Ultrathin sections were examined by electron microscopy (Zeiss).
qPCR.
Detection of viral DNA and calculation of viral titers were carried out by quantitative real-time PCR (qPCR) using EBV-specific primers and a probe as described previously (8). The DNA content was calculated by using serial dilution of DNA from the Namalwa cell line, a human Burkitt's lymphoma cell line that contains two EBV genome copies per cell, as a standard curve.
Binding assays.
The capacity for the binding of wild-type EBV- or ΔBGLF4 virus-containing supernatants to target cells was analyzed by incubating purified primary B cells at a multiplicity of infection (MOI) of 10 genome equivalents per cell. After incubation for 3 h on ice, the cells were washed four times with PBS and used for qPCR analysis.
Gardella gel electrophoresis and Southern blot analysis.
Genomic DNA extraction, DNA digestion, conventional or Gardella gel electrophoresis from induced producer cells, membrane blotting, and hybridization were performed as described previously (6, 24). DNA from cells carrying different viral genomes was digested with the BamHI restriction enzyme, blotted onto a Hybond XL membrane (Amersham), and hybridized with a 32P-labeled DNA fragment specific to the EBV BGLF3 gene, the gp350 gene, or an EBV terminal repeat.
Northern blot analysis and 5′RACE.
Total RNA was purified from lytically induced cells using an RNA purification kit (Nucleospin; Machery & Nagel). Five micrograms of RNA was separated on a 1% formaldehyde gel, blotted onto a Hybond XL membrane (Amersham), and hybridized with 32P-radiolabeled EBV-specific double-stranded DNA fragments. The BGLF5 gene transcriptional start site was determined using a 5′/3′ rapid amplification of cDNA ends (RACE) kit (Roche Applied Science). Briefly, reverse transcription was performed with 450 ng RNA from lytically induced 293/EBV-wt cells using a gene-specific primer (5′-CGTCAACAGATAGTCACCCT-3′; B95.8 coordinates 121481 to 121500). cDNA was amplified by PCR using another gene-specific primer (5′-ATCAGGTTCTCAGACTGCC-3′; B95.8 coordinates 121989 to 122007) and an oligo(dT) anchor primer. The PCR product was further amplified using a nested gene-specific primer (5′-GTCACAGAAGCCAGTTTCAC-3′; B95.8 coordinates 122141 to 122160) and a PCR anchor primer. The resulting PCR product was purified on an 8% polyacrylamide gel and cloned into plasmid pGEM-T Easy (Promega). DNA from seven different clones was extracted and sequenced.
Western blot analysis.
Cells were resuspended in PBS and lysed by sonication. Twenty micrograms of proteins was denatured in Laemmli buffer for 5 min at 95°C, separated on a 10% or 12.5% (for BGLF4 detection) sodium dodecyl sulfate-polyacrylamide gel, and electroblotted onto a Hybond ECL membrane (Amersham). After preincubation for 30 min in 5% milk powder in PBS, blots were incubated either with a rabbit polyclonal serum against BGLF4 (this work) or BGLF5 (3) or a mouse MAb against BFLF2 (clone C1) (15), BFRF1 (clone E10) (7), EA-D encoded by the BMRF1 gene (Chemicon; MAb 8182), BZLF1 (clone BZ.1) (50), BRLF1 (clone 8C12) (33), BNRF1 (10), or actin (clone ACTN05; Dianova) for 1 h at room temperature. After several washings in 0.1% Tween in PBS, blots were incubated for 1 h with goat anti-mouse antibody coupled with horseradish peroxidase (Promega) or protein A coupled with horseradish peroxidase (Sigma). Bound antibody was revealed using an ECL detection reagent (Amersham).
RESULTS
Construction of a BGLF4-negative EBV mutant strain and its revertant.
The BGLF4 and BGLF5 genes are located in tandem within a complex viral locus where most genes partly overlap and frequently share promoters or polyadenylation sites (2, 22, 23, 52). A 3.4-kb mRNA that spans both the BGLF4 and BGLF5 genes and a 1.7-kb BGLF5 gene-specific transcript can be detected in induced cells. However, the existence of a promoter or of an internal ribosomal entry site located directly upstream of the BGLF5 gene has not been reported.
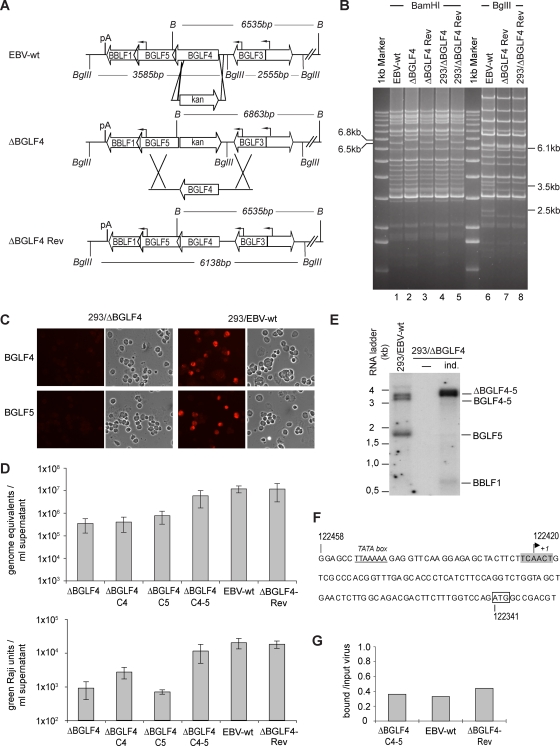
The BGLF4 gene knockout was obtained by deletion of a large part of the BGLF4 gene open reading frame. To this end, as depicted in Fig. 1A, we exchanged the entire BGLF4 gene open reading frame with the exception of its last fifty codons for the kan resistance gene by homologous recombination in E. coli. The DNA of the resulting recombinant EBV genomes was subjected to BamHI restriction enzyme analysis. The wild-type and mutant genomes differed in that the BGLF4 mutant carries a larger BamHI G fragment (6,863 bp compared to 6,535 bp in wild-type viruses) (Fig. 1A and B, lanes 1 and 2). The DNA preparation from one mutant clone was then transfected into HEK293 cells and subjected to hyg selection. Ten hyg-resistant and GFP-positive outgrowing cell clones were expanded and tested for permissivity to lytic replication. Induction of lytic replication was achieved through transient transfection of the immediate-early protein BZLF1 into the different cell clones (19). 293/EBV-wt cells, which contain the wild-type virus, were used as positive controls. Three days after transfection, cells were stained for glycoprotein gp350, a late marker of lytic replication, and the clone showing the highest percentage of positive cells (about 10%) was selected for further experiments. These findings already showed that BGLF4 is not required for gp350 expression at levels akin to those observed in permissive HEK293 clones that carry EBV wild-type genomes. The mutant viral genome present in this clone was then rescued in E. coli cells to determine its restriction profile anew. The results of this analysis, presented in Fig. 1B, showed that the mutant genome had retained an intact structure upon transfer into HEK293 cells (compare lanes 2 and 4). Apart from the expected size shift of the restriction fragments spanning the BamHI G fragment, no alterations of the mutant genome were visible. The HEK293 clone containing the ΔBGLF4 virus is referred to as 293/ΔBGLF4. We then performed an immunostaining to assess BGLF4 expression levels in lytically induced 293/ΔBGLF4 cells, which confirmed the absence of this protein in the mutant clone (Fig. 1C, top). In contrast, 293/EBV-wt cells stained positive for BGLF4 and served as a positive control in further experiments.
FIG. 1.
Design, construction, and initial characterization of a ΔBGLF4 null mutant and its revertant. (A) Schematic representation of the strategy followed to construct the ΔBGLF4 mutant. The figure also depicts some of the viral genes present on the BamHI G fragment. A PCR-amplified DNA fragment containing the kan restriction gene flanked by oligonucleotides homologous to the BGLF4 locus was subjected to homologous recombination with the EBV wild-type genome. Recombination led to a BamHI G fragment size shift from 6,535 bp to 6,863 bp and resulted in the disruption of the BGLF4 gene open reading frame with the exception of its last 50 codons. Exactly the reverse strategy was followed to construct the ΔBGLF4-Rev revertant virus. The targeting vector consisted of the wild-type BGLF4 gene sequence recombined with the mutant BGLF4 virus genome. The BglII restriction site located between the BGLF3 and BGLF4 genes was excised from this targeting vector; digestion with BglII can therefore distinguish between DNA from the wild-type and revertant viruses. B, BamHI; pA, poly(A) site. (B) ΔBGLF4 and ΔBGLF4-Rev genome restriction fragment analysis. The ΔBGLF4 and the ΔBGLF4-Rev genome restriction patterns were determined at different steps of mutant construction to ensure integrity and exactness of the viral recombinants. Bacterial DNAs of these EBV genomes cloned into E. coli were digested with BamHI and separated on an agarose gel. EBV-wt DNA provided a positive control. The ΔBGLF4 mutant showed an altered 6.8-kb BamHI G fragment that replaced the wild-type 6.5-kb BamHI G fragment (lane 2), whereas the ΔBGLF4-Rev revertant had regained the wild-type allele (lane 3). Recombinant genome structure was reassessed after stable transfection into 293 cells and rescue in E. coli (293/ΔBGLF4 and 293/ΔBGLF4-Rev). The ΔBGLF4-Rev DNA was further digested with BglII restriction enzymes. This analysis confirmed the integrity and the absence of the BglII restriction site in the ΔBGLF4-Rev genome (lanes 7 and 8). (C) Induced 293/ΔBGLF4 cells are negative for both BGLF4 and BGLF5. Induced 293/ΔBGLF4 cells were immunostained with polyclonal antibodies specific for BGLF4 or BGLF5 (top). Induced 293/EBV-wt cells were used as positive controls (bottom). The cell morphology under phase-contrast illumination is also shown. (D) Viral titers in various induced cell lines. Viral genome DNA equivalents per ml of supernatant (physical titers) were quantified by qPCR amplification of the viral BALF5 gene (top) or by infection of Raji cells at limiting dilutions (functional titers that represent the number of infectious viruses per ml of supernatant) (bottom). Shown are mean values from five independent experiments. (E) Northern blot analysis of BamHI G transcripts. mRNA from noninduced (−) and induced (ind.) 293/ΔBGLF4 cells and induced 293/EBV-wt cells was hybridized with a BGLF5-specific probe. Induced 293/ΔBGLF4 cells produce a 328-bp-larger transcript as a consequence of the exchange between the BGLF4 gene and the kan gene. (F) Sequence of the BGLF5 gene upstream region. The transcription start site (+1) within a putative initiator element (highlighted in gray) was determined by 5′ RACE using 293/EBV-wt RNA. The predicted TATA box (underlined) and the BGLF5 ATG (box) are indicated. Numbers refer to B95.8 coordinates. (G) Complemented viruses display normal binding properties. B cells were exposed to supernatants from induced 293/EBV-wt, 293/ΔBGLF4-Rev, or 293/ΔBGLF4-C4-5 cells at an MOI of 10 genome equivalents per cell. Unbound viruses were washed off, and the number of viruses bound per cell was determined by qPCR. The ratios between bound viruses and input virus load are indicated.
We then constructed a revertant virus by recombining the ΔBGLF4 mutant genome with a targeting vector that carried the wild-type BGLF4 gene and its left and right flanking regions (Fig. 1A and B, lanes 3). A BglII restriction site located in the noncoding region of the wild-type EBV genome was removed from the targeting vector to allow unequivocal identification of the revertant virus (Fig. 1B, compare lanes 6 and 7). A producer cell line carrying the revertant genome was then selected on the basis of its ability to support the virus lytic cycle as described for the 293/ΔBGLF4 mutant. This clone, designated 293/ΔBGLF4-Rev, had an intact genome structure after plasmid rescue in E. coli cells and BamHI and BglII restriction enzyme analysis (Fig. 1B, lanes 5 and 8).
The 293/ΔBGLF4 phenotype can be complemented with BGLF4 in combination with BGLF5.
Defective mutants can usually be complemented by introduction of an expression plasmid carrying the missing gene into the mutant or the virus target cell line. Successful complementation guarantees with a high probability that the defects introduced into the mutant were restricted to the altered gene, i.e., that the remainder of the viral genome is intact. We first tested several BGLF4 expression plasmids for their ability to produce BGLF4 at levels similar to those observed in induced wild-type cells; expression plasmids based on CMV-derived promoters (pRK5 and pCDNA3.1) produced very high levels of protein. We therefore screened several expression systems until we identified the parvovirus p38 promoter (p38-BGLF4) as being able to reproduce BGLF4 expression levels on a par with those seen in induced 293/EBV-wt cells (data not shown). 293/ΔBGLF4, 293/ΔBGLF4-Rev, 293/ΕΒV-wt, and p38-BGLF4-transcomplemented 293/ΔBGLF4 cells (293/ΔBGLF4-C4) were then lytically induced. Virus production was monitored by quantifying cell-free encapsidated viral DNA in the supernatants from induced cell lines (expressed as genome equivalents) and by Raji cell infection using limiting dilutions of the infectious supernatants (expressed as green Raji units). This analysis revealed that viral titers in 293/ΕΒV-wt and 293/ΔBGLF4-Rev supernatants were very similar (mean values from five infection experiments yielded titers around 1.2 × 107 genome equivalents/ml supernatant and 2 × 104 infectious particles/ml supernatant for both cell lines) (Fig. 1D). From the observation that the revertant virus displayed a normal phenotype it can be inferred that the viral sequence outside the modified BGLF4 locus was intact in the 293/ΔBGLF4 genome and that the reconstruction process had been successful. Supernatants from multiple induction experiments with 293/ΔBGLF4 cells showed 34- and 22-fold-lower physical and functional titers (3.5 × 105 genome equivalents/ml and 9 × 102 infectious particles/ml), respectively, than those obtained with wild-type viruses (Fig. 1D). The introduced mutation therefore substantially curbs virus replication. Surprisingly, complementation with p38-driven BGLF4 had only limited effects on viral titers (4 × 105 genome equivalents per ml and 2.7 × 103 infectious particles per ml) (Fig. 1D). This suggested that the ΔBGLF4 mutant was defective for more functions than those fulfilled by BGLF4. We therefore sequenced the regions that flank the mutated BGLF4 gene. This analysis revealed an absolutely intact sequence (data not shown). We further assessed BGLF5 expression in the 293/ΔBGLF4 cell line by immunostaining with a rabbit polyclonal antiserum. 293/EBV-wt cells were included as positive control. This assay revealed an unexpected absence of BGLF5 expression in induced 293/ΔBGLF4 cells (Fig. 1C, bottom). Furthermore, a Northern blot analysis showed complete abolition of the 1.7-kb BGLF5 gene mRNA transcript (Fig. 1E). These findings suggested the existence of a thus far unidentified promoter located directly upstream of BGLF5. Analysis of the BGLF5 gene 5′ sequence identified a putative TATA box (B95.8 coordinates 122452 to 122446) and a transcription start site downstream thereof (Fig. 1F). We then tested this prediction by using a 5′ RACE-based strategy on RNA isolated from lytically induced 293/EBV-wt cells. Reverse transcription and amplification of cDNAs led to the identification of short BGLF5 transcripts. Cloning and sequencing of six of these revealed that they were all initiated 25 nucleotides (nt) downstream of the predicted TATA box at the adenosine +1 of a putative initiator element (Fig. 1F) (44) and 79 nt upstream of the BGLF5 gene ATG start codon (Fig. 1F). A seventh clone included additional upstream sequences and was probably derived from the previously identified 3.4-kb mRNA that spans the BGLF4 and BGLF5 genes.
These results prompted us to cotransfect 293/ΔBGLF4 cells with BGLF4 and BGLF5 upon lytic induction (the resulting cells are referred to as 293/ΔBGLF4-C4-5). This treatment increased the genome equivalent concentration in supernatants by a factor of 17 (6 × 106/ml genome equivalents, mean value from five experiments), reaching values close to the ones observed for the 293/EBV-wt or 293/ΔBGLF4-Rev clones (Fig. 1D). Similarly, functional titers in the complemented null mutant reached 55% of those for their wild-type counterparts (1.1 × 104 infectious viruses/ml supernatant, mean value from five experiments) (Fig. 1D). We further assessed the binding properties of virions in the supernatants from complemented 293/ΔBGLF4-C4-5 cells by performing a binding assay followed by a qPCR analysis (Fig. 1F). The results of this analysis revealed similar binding efficiencies for 293/ΔBGLF4-C4-5, 293/ΔBGLF4-Rev, and 293/EBV-wt, ranging from 33 to 44% bound viruses per input virus. We concluded from the above findings that 293/ΔBGLF4 cells are in fact defective for both BGLF4 and BGLF5. In the sequel, we therefore systematically assessed the effect of BGLF4, BGLF5, or a combination of BGLF5 and BGLF4 in 293/ΔBGLF4 cells (referred to as 293/ΔBGLF4-C4, 293/ΔBGLF4-C5, or 293/ΔBGLF4-C4-5 cells, respectively) on the various viral functions.
293/ΔBGLF4 cells complemented with BGLF4 reproduce the phenotypic traits of a BGLF5-null mutant.
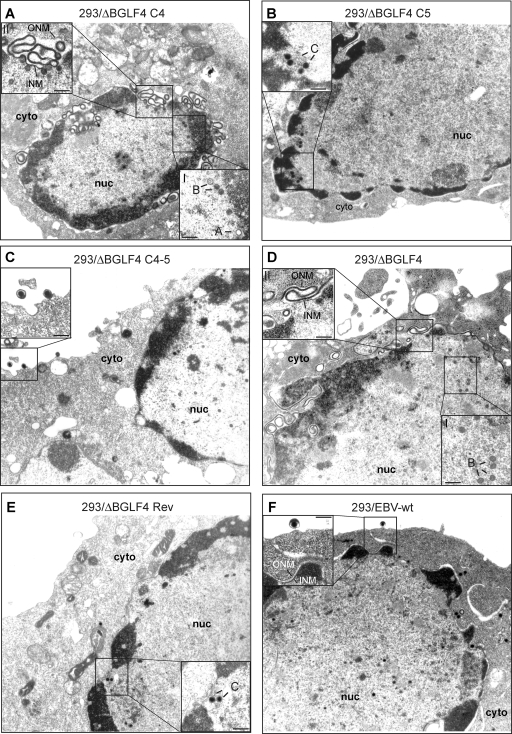
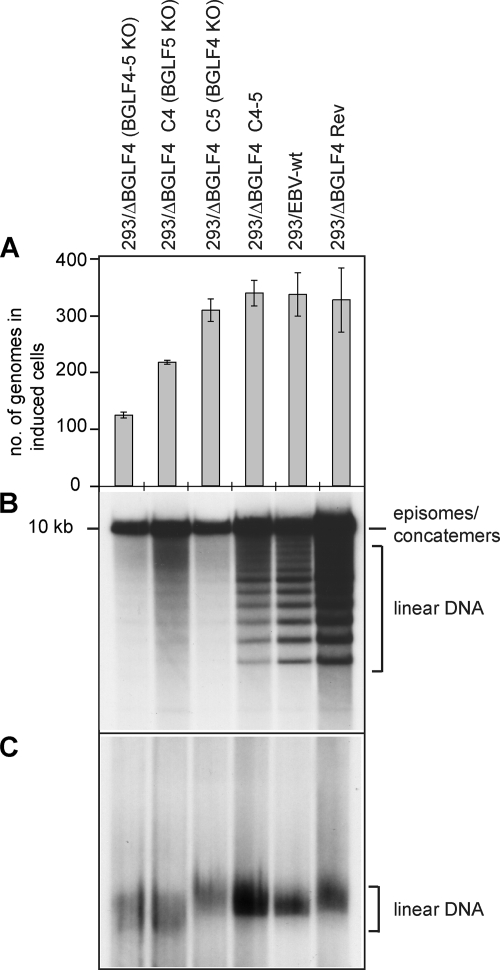
Complementation of 293/ΔBGLF4 with BGLF4 should reproduce the phenotype of the 293/ΔBGLF5 mutant, whose phenotype has been described previously (8). As previously mentioned, induced 293/ΔBGLF4-C4 displayed reduced physical and functional titers relative to positive controls (Fig. 1D). Approximately 11% of 293/ΔBGLF4-C4 cells observed by electron microscopy contained viral structures, compared to 15% in 293/ΔBGLF4-C4-5 cells (Table 1). Replicating cells exhibited a maturation block that resulted in an abnormally high proportion of immature A- and B-type nucleocapsid forms at the expense of DNA-filled mature C-type capsids, which were rarely observed (96% A and B capsids, 4% C capsids) (Fig. 2A and Table 1). Intracytoplasmic or extracellular virions were not visible. The nuclear membrane showed striking abnormalities, with images of reduplication and of complex projections (Fig. 2A, inset II). The block in capsid maturation can result from deficient packaging or from inefficient viral DNA replication. We investigated the latter hypothesis by several methods and found that DNA replication was qualitatively and quantitatively altered; total viral DNA replication was reduced by 1.6-fold, as measured by qPCR with EBV-specific primers (Fig. 3A); Southern blot analysis with a probe specific to the EBV terminal repeats showed reduced synthesis of linear genomes (Fig. 3B); and linear viral DNAs displayed an aberrant electrophoretic migration pattern when assessed in a Gardella gel analysis (Fig. 3C). These phenotypic traits could be reverted by complementation with BGLF4 and BGLF5. These data are fully congruent with those gathered from the study of the 293/ΔBGLF5 mutant (8), thereby indirectly confirming that 293/ΔBGLF4 is deficient for BGLF5 and that complementation was effective.
TABLE 1.
Quantitative analysis of viral structures in induced cells by electron microscopy
| Cell linea | % of cells containing viral structuresb | % of all nucleocapsids (no.) that were: |
No. in cytoplasm/ extracellularc | ||
|---|---|---|---|---|---|
| A form | B form | C form | |||
| 293/ΔBGLF4 | 7 (15/207) | 7 (14) | 89 (178) | 4 (7) | 2/1 |
| 293/ΔBGLF4-C4 | 11 (26/244) | 18 (46) | 78 (202) | 4 (11) | 0/0 |
| 293/ΔBGLF4-C5 | 3.6 (7/194) | 13 (11) | 39 (33) | 48 (41) | 0/0 |
| 293/ΔBGLF4-C4-5 | 15 (15/97) | 12 (37) | 58 (183) | 30 (88) | 6/3 |
| 293/ΔBGLF4-Rev | 14 (14/99) | 21 (35) | 40 (66) | 39 (64) | 31/3 |
| 293/EBV-wt | 10 (7/69) | 13 (24) | 57 (105) | 30 (56) | 51/6 |
Analyzed lytically induced cells.
Numbers of cells containing viral structures among the total numbers of cells analyzed are in parentheses.
Total numbers of viruses observed in cytoplasm or extracellular space in the examined samples.
FIG. 2.
Electron micrographs of induced producer cells that carry various recombinants. (A) Induced 293/ΔBGLF4-C4 cells contained immature nucleocapsids (A and B capsids) lacking an electron-dense DNA core (inset I). The inner nuclear membrane was markedly thickened and showed accumulation of electron-dense material between outer and inner nuclear membranes. Images of extensive membrane folding in the perinuclear space are shown in inset II. (B) Induced 293/ΔBGLF4-C4 cells showed intranuclear capsid maturation. The nuclear membrane displayed normal morphological features. Intracytoplasmic or extracellular virions were not visible. (C) Induced and complemented 293/ΔBGLF4-C4-5 cells showed evidence of intranuclear maturation, with numerous DNA-filled nucleocapsids displaying an electron-dense core. Further, intracytoplasmic and extracellular virions that had successfully undergone primary and secondary egress were identified (inset). (D) Induced 293/ΔBGLF4 cells that lack BGLF4 and BGLF5 displayed morphological features that were very similar to those observed in 293/ΔBGLF4-C4 cells. Micrographs revealed an obvious capsid maturation block (inset I) and morphological nuclear membrane abnormalities (inset II). (E and F) 293/ΔBGLF4-Rev and 293/EBV-wt cells showed normal virus maturation, as described for panel C. cyto, cytoplasm; nuc, nucleus; ONM, outer nuclear membrane; INM, inner nuclear membrane; A, B, and C, A-, B-, and C-type capsids. Inset bars, 200 nm.
FIG. 3.
Role of BGLF4 and BGLF5 in viral lytic DNA replication. (A) Viral DNA replication in induced cells was quantitated by qPCR amplification. Mean values and standard deviations from three independent experiments are presented. (B) Southern blot analysis of BamHI-cleaved DNA fragments hybridized with a terminal repeat-specific probe. The 10-kb fragment results from restriction of complete BamHI Nhet fragments that are present only in nonlinear genomes, i.e., circular genomes or genome concatemers. In contrast, the smaller fragments are generated by restriction of single unit length linear genomes. (C) Gardella gel electrophoresis coupled to Southern blot analysis using a nonrepetitive gp350-specific probe. This assay allows distinction between circular and unit length linear DNA molecules. Note the faster electrophoresis migration pattern in induced 293/ΔBGLF4 and 293/ΔBGLF4-C4 cells.
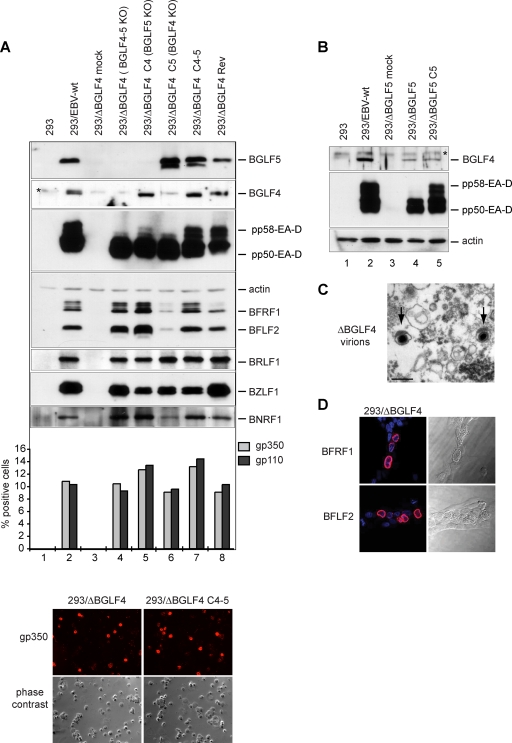
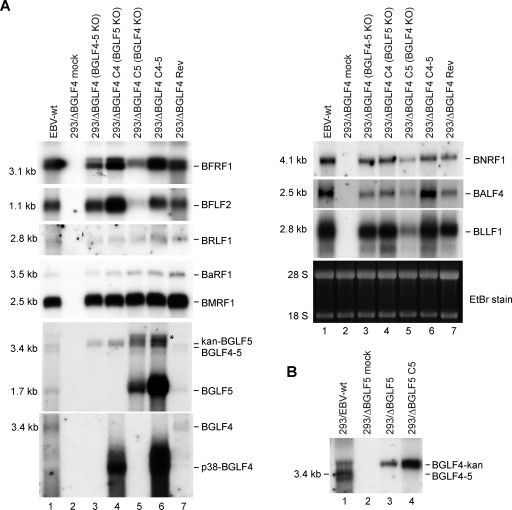
We then extended the characterization of the 293/ΔBGLF4-C4 phenotype by assessing its viral protein expression pattern by Western blot analysis (Fig. 4A, lane 5). Positive controls were provided by the wild-type virus (lane 2), the revertant virus (lane 8), and complemented 293/ΔBGLF4-C4-5 (lane 7) cells. Interpretation of the signals must take into account the percentage of gp350-positive cells (11, 9, and 13% in these controls, respectively) (Fig. 4A). Some early proteins such as BFLF2 and to a certain extent BFRF1 (UL31 and UL34 are their respective positional homologs in HSV-1) were significantly upregulated relative to levels in both 293/ΔBGLF4-C4-5 and 293/ΔBGLF4 (Fig. 4A, compare lane 5 with lanes 4 and 7). This indicates that the BGLF4 protein stimulates expression of BFLF2 and BFRF1 but also suggests that this effect is abolished when BGLF5 is present in wild-type cells. The immediate early BRLF1 protein expression was not altered in the absence of BGLF5 (Fig. 4A). The late proteins gp110 and gp350 showed minimal variations; the percentage of gp350-positive cells went from 10% to 12% after BGLF4 transfection, which compares to 13% in the BGLF4-BGLF5 double-complemented virus, and the percentage of gp110-positive cells increased from 9 to 13% after complementation with BGLF4 and to 14% after complementation with BGLF4 and BGLF5 (Fig. 4A, compare lanes 4, 5, and 7). The early protein EA-D (encoded by the BMRF1 gene), which has been shown to be a BGLF4 substrate (13), showed more-subtle alterations; while total expression of this protein was not substantially altered in the absence of the viral exonuclease, the phosphorylation pattern appeared modified by the absence of the viral exonuclease. Mono- and hyperphosphorylated forms of EA-D can be detected in replicating cells as discrete signals of increasing size. While hyperphosphorylated forms (pp58) were visible in positive controls (Fig. 4A, lanes 2, 7, and 8), they were clearly underrepresented in induced 293/ΔBGLF4-C4 cells (lane 5). Further support for this claim came from EA-D-specific immunoblotting carried out with induced 293/ΔBGLF5 cells, which showed unequivocal downregulation of the hyperphosphorylated EA-D forms (Fig. 4B, lane 4).
FIG. 4.
Viral protein expression patterns in the various mutants. (A) Western blot analysis and immunostains of viral proteins in mutant producer cell lines and their wild-type parent or revertant. The expression of members from all three classes of viral proteins in cells induced by transfection of BZLF1 is shown. Induced cells were analyzed 1 day (immediate early BZLF1 and BRLF1 genes), 2 days (early EA-D/BMRF1, BGLF4, BGLF5, BFRF1, and BFLF2 genes), or 3 days (late BNRF1, gp110, and gp350 genes) after induction. Actin was stained as a uniform loading control. The BZLF1 staining identifies a stronger signal in cells cotransfected with BZLF1 and an empty pCDNA vector than in cells cotransfected with BZLF1 and BGLF4 and/or BGLF5. This, however, did not impede activation of the lytic program, as shown by the very similar BRLF1 levels in all transfected-cell populations. The immunoblot against EA-D/BMRF1 recognizes the variably phosphorylated forms of this protein. Expression of gp110 and gp350 is difficult to evaluate by Western blotting and was instead assessed by immunofluorescence. The percentages of positive cells are given. Also shown is an example of an immunostain against gp350 as well as a phase-contrast picture. The BGLF4 antibody shows a nonspecific signal (indicated by an asterisk) that migrates directly above the BGLF4-specific band. This cross-reactivity was observed with several different BGLF4-specific antibodies (11, 31), suggesting that BGLF4 shares immunogenic epitopes with a cellular protein. Cross-reactivity was not visible in immunostains. (B) Western blot analysis of BGLF4 and EA-D/BMRF1 in 293/ΔBGLF5 cells. (C) Supernatant from induced 293/ΔBGLF4 cells was pelleted by ultracentrifugation and after fixation was examined by electron microscopy. The picture shows rare examples of mature virions with electron-dense cores (arrows). Bar, 200 nm. (D) Immunostains of induced 293/ΔBGLF4 cells with BFLF2- and BFRF1-specific antibodies. Nuclei were visualized by chromatin staining with Hoechst 33258 (blue), and fluorescent signals were recorded with a confocal microscope. Nomarski pictures revealing fine morphological features of induced cells are shown at the right. Note the obvious thickening of the nuclear membranes in the induced cells.
We wished to learn whether the observed modifications of the protein expression pattern were due to enhanced viral transcription. A Northern blot analysis, presented in Fig. 5, showed that BFLF2 and BFRF1 gene mRNAs were more abundant in 293/ΔBGLF4-C4 cells than in their wild-type counterparts (compare lane 4 with lanes 1, 6, and 7). The expression levels of all other tested lytic genes were either unaltered or only marginally stronger in 293/ΔBGLF4-C4 cells than in 293/ΔBGLF4 cells.
FIG. 5.
Total mRNA expression of a panel of viral genes in the various mutants. The mRNA expression profile was assessed by Northern blot analysis using specific probes. (A) Induced 293/ΔBGLF4 cells transfected with the indicated plasmids were analyzed 1 day (immediate early BZLF1 and BRLF1 genes; left), 2 days (early EA-D/BMRF1, BGLF4, BGLF5, BFRF1, and BFLF2 genes; left), or 3 days (late BNRF1, gp110, and gp350 genes; right) after induction. Ethidium bromide (EtBr) staining of the denaturing gel was used as a loading control. Hybridization with a BGLF5-specific probe identified a 1.7-kb mRNA (BGLF5), a 3.4-kb mRNA (BGLF4-5), and a 3.7-kb mRNA in cells where the BGLF4 gene was replaced by the kan resistance gene (kan-BGLF5) (see Fig. 1A). An unspecific band (*) is observed in cells transfected with the BGLF5 expression plasmid. The BGLF4-specific probe detects only the 3.4-kb mRNA or the gene expressed from the BGLF4 expression plasmid (p38-BGLF4). (B) BGLF4 mRNA expression in induced 293/ΔBGLF5 cells. A BGLF4-specific probe detects the 3.4-kb BGLF4 mRNA in EBV-wt cells and a 3.7-kb transcript in ΔBGLF5 cells carrying the kan resistance gene (BGLF4-kan).
293/ΔBGLF4-C5 cells show impaired primary egress coupled to impaired viral DNA and viral protein synthesis.
We followed the same strategy to analyze BGLF4's functions during virus replication. 293/ΔBGLF4-C5 cells, which are equivalent to a BGLF4-null mutant, were induced, and virus titers were assessed. These assays revealed an important role for BGLF4 in virus replication: concentration in virus DNA equivalents dropped by 15-fold, and functional titers, assessed by infection of Raji cells, were 28-fold lower (7.7 × 105 genome equivalents/ml and 7 × 102 infectious particles/ml) than those for positive controls (293/EBV-wt and 293/ΔBGLF4-Rev) (Fig. 1D). The analysis of induced 293/ΔBGLF4-C5 cells by electron microscopy is shown in Fig. 2B; the percentage of cells containing viral structures was reduced compared to the percentage of 293/ΔBGLF4-C4-5 cells (Table 1) (3.6% versus 15%). Transfection of BGLF5 in the absence of BGLF4 therefore reduces the percentage of cells producing virus. This, however, did not reflect a generally impaired viral induction, as the total number of cells that expressed the late marker gp350 was only marginally changed by transfection of BGLF5 (293/ΔBGLF4, 10%; 293/ΔBGLF4-C5, 9%; 293/ΔBGLF4-C4-5, 13%) (Fig. 4A). In cells undergoing viral replication, we observed viral capsid maturation, as attested by a normal distribution between the mature and immature nucleocapsid forms (52% A and B capsids and 48% C capsids) (Table 1). In contrast, primary egress appeared reduced; virions were absent both from the cytoplasm and the extracellular space. The nuclear membrane was unremarkable. Systematic analysis of viral DNA replication identified a 10% reduction in total viral DNA synthesis, as assessed by qPCR after normalization on the basis of the percentage of gp350-positive cells (Fig. 3A). Southern blot hybridization with a probe specific to the EBV terminal repeats further revealed a decreased production of linear genomes (Fig. 3B). Gardella gel analysis of free linear virions produced by induced 293/ΔBGLF4-C5 cells showed mildly reduced linear DNA production but no abnormal electrophoretic migration pattern (Fig. 3C). Western blot analysis and immunostains revealed multiple abnormalities in the viral protein expression pattern of induced 293/ΔBGLF4-C5 cells (Fig. 4A, lane 6). Some proteins such as BFLF2 or BFRF1 were markedly downregulated in the absence of BGLF4 relative to the wild-type controls and the double mutant (compare lane 6 with lanes 2, 4, and 7). A similar though less pronounced effect was visible for BNRF1. Others proteins such as EA-D and BRLF1 were produced at nearly the same level in 293/ΔBGLF4-C5 cells, 293/ΔBGLF4 cells, and the positive controls. Immunostains against gp350 and gp110 yielded similar results regardless of the presence or absence of BGLF5. In particular, staining intensity was not influenced by the AE (Fig. 4A and data not shown). The alterations in the BMRF1 phosphorylation pattern observed in 293/ΔBGLF4-C5 cells were even more pronounced than in 293/ΔBGLF4-C4 cells, with only the monophosphorylated form visible (Fig. 4A, compare lanes 5 and 6).
BGLF5 was further found to have more general effects at the RNA level than expected by the results of the protein analysis. Northern blots with probes specific to BFLF2, BFRF1, or BLLF1 clearly showed a reduced RNA synthesis following transfection of BGLF5 (Fig. 5A, lane 5), compared to that in 293/ΔBGLF4 cells (Fig. 5A, lane 3) or the positive controls (Fig. 5A, lanes 1, 6, and 7). Milder effects were visible with BNRF1 and BALF4. BMRF1 and BRLF1 mRNA expression remained unchanged in the presence of the exonuclease.
BGLF4 and BGLF5 are not required for viral protein synthesis and reciprocally activate their own expression.
The findings gathered so far indicated that BGLF4 and BGLF5 both target viral DNA replication and primary egress, resulting in decreased viral replication. However, the influences of both proteins on viral gene expression were found to be opposite: BGLF4 stimulates expression of BFLF2 and BFRF1 genes, resulting in increased production of both proteins, whereas BGLF5 markedly reduces expression of these genes. This observation suggested that both proteins, AE and PK, may act in concert to modulate gene expression of some viral genes, but it was unclear whether they were at all required for viral protein production. These questions prompted us to analyze the phenotype of the BGLF4-BGLF5 double null mutant (293/ΔBGLF4). As already mentioned, viruses from induced 293/ΔBGLF4 cells produced low virus titers both in qPCR and Raji assays (Fig. 1D). As expected, both phenotypic traits seen in 293/ΔBGLF4-C5 and 293/ΔBGLF4-C4 added together in the double mutant; a reduced proportion of cells displayed images of viral replication (7% versus 15% for 293/ΔBGLF4-C4-5), and those that did exhibited impaired primary egress, blockage in viral maturation, and abnormal nuclear membranes (Fig. 2D and Table 1). Direct examination of pelleted supernatants from 293/ΔBGLF4 cells confirmed the marked reduction in mature virions released into the extracellular milieu; intact mature virions with normal morphology could nevertheless be identified (Fig. 4C). Lytic DNA replication and linear DNA synthesis were reduced to one-third of wild-type levels, and 293/ΔBGLF4 linear genomes shared with those of 293/ΔBGLF4-C4 an abnormal electrophoretic mobility (Fig. 3C, lane 1). The percentages of gp350- and gp110-positive cells went from 10% to 13% and from 9 to 14% after complementation, respectively, suggesting that more cells completed the lytic cycle in the presence of both BGLF4 and BGLF5 (Fig. 4A). Interestingly, the viral protein production pattern in 293/ΔBGLF4 cells was in general similar to the one observed in the control cells, 293/EBV-wt, 293/ΔBGLF4-Rev, and 293/ΔBGLF4-C4/5, after normalization by the number of gp350-positive cells, indicating that PK and AE are dispensable for baseline viral protein production (Fig. 4A, compare lane 4 with lanes 2, 7, and 8). In addition, BFLF2- and BFRF1-specific immunostains revealed unaltered distribution of both proteins, with a typical accumulation at the nuclear membrane (Fig. 4D). This shows that proper location of these proteins is independent of PK and AE.
We then extended our characterization of the viral protein expression pattern by asking whether BGLF4's upregulating effect on viral transcription extended to BGLF5; coexpression of BGLF4 and BGLF5 from two expression plasmids indeed led to increased BGLF5 gene mRNA expression (Fig. 5A, lane 6). This enhancing effect on BGLF5 gene transcription was not, however, followed by a substantial increase in BGLF5 protein synthesis (Fig. 4A, lane 7). Interestingly, and in contrast with its effects on other viral genes, BGLF5 increased BGLF4 gene transcription (Fig. 5A, lane 6). Here again, modulation of mRNA levels did not result in increased protein synthesis (Fig. 4A, lane 7). Both BGLF4 and BGLF5 were expressed at high levels in the transfected cells (Fig. 4A); this reflects the fact that both plasmid transfections reached more than 60% of cells, though a minority of these actually underwent lytic replication. BGLF5's enhancing effect was not limited to a BGLF4 expression plasmid; transfection of BGLF5 into induced 293/ΔBGLF5 cells produced the same effect on BGLF4 expressed from the complete virus under the control of its natural promoter (Fig. 5B, lanes 3 and 4).
Phenotypic traits of a BGLF4-BGLF5 double knockout.
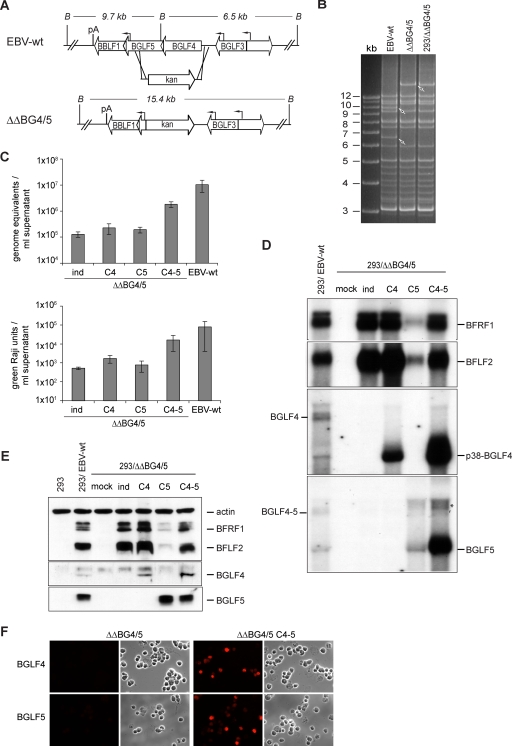
We wished to confirm the central results regarding the ability of BGLF4 and BGLF5 to modulate viral gene expression obtained with the BGLF4 knockout virus. To this end, we constructed a mutant that lacked both the BGLF4 and the BGLF5 gene open reading frames, designated here as 293/ΔΔBG4/5 (Fig. 6A and B). Furthermore we addressed potential clonal effects by studying viral replication in seven 293/ΔΔBG4/5 clones. The results of this analysis are summarized in Fig. 6C and show a low level of replication in induced 293/ΔΔBG4/5 cells (average value, 1.3 × 105 genome equivalents/ml supernatant). Complementation with BGLF4 and BGLF5 increased titers by 14-fold. However, as previously observed with the BGLF4-null mutant, titers remained lower (by fivefold) than in a panel of four wild-type clones. Similar results were obtained after assessment of the functional titers using infected Raji cells (Fig. 6C, bottom). We further extended these observations by inducing one 293/ΔΔBG4/5 clone complemented with either BGLF4 or BGLF5 (Fig. 6C). Here again, complementation with only one of those enzymes led to only a modest increase in virus titers.
FIG. 6.
Construction and analysis of a BGLF4-BGLF5 double mutant. (A) Schematic map of the BGLF4-BGLF5 genes in EBV-wt and in the double-mutant (ΔΔBG4/5) virus after homologous recombination with the kan resistance gene targeting vector. The BamHI cleavage sites and the expected fragment sizes are given. B, BamHI; pA, poly(A) site. (B) BamHI restriction fragment analysis of EBV-wt and ΔΔBG4/5 genomes after construction in E. coli or after rescue from stably transfected 293 cells (293/ΔΔBG4/5). (C) Viral titers in various induced cell lines. Viral genome DNA equivalents per ml of supernatant as measured by qPCR (top) or the numbers of infectious viruses per ml of supernatant as measured by Raji cell infection (bottom) are presented. Values are means from seven different mutant clones and four different EBV-wt clones. Complementation with either BGLF4 or BGLF5 alone was performed with one 293/ΔΔBG4/5 cell clone. All data were obtained from three independent experiments. (D) Northern blot analysis of BFRF1, BFLF2, BGLF4, and BGLF5 mRNA expression in induced and complemented 293/ΔΔBG4/5 cells and 293/EBV-wt cells. *, unspecific band observed in cells transfected with the BGLF5 expression plasmid (see Fig. 5A). (E) Western blot analysis of BFRF1, BFLF2, BGLF4, and BGLF5 protein expression in induced and complemented 293/ΔΔBG4/5 cells. Protein extracts from HEK293 cells and 293/EBV-wt cells served as negative and positive controls, respectively. (F) Immunostaining of induced or complemented 293/ΔΔBG4/5 cells with BGFL4- and BGLF5-specific antibodies confirms the absence of both proteins in 293/ΔΔBG4/5 cells.
We went on to assay BFLF2 and BFRF1 expression at the protein and the mRNA levels (Fig. 6D and E) in 293/ΔΔBG4/5 cells in the presence or absence of BGLF4 and/or BGLF5. These investigations confirmed our previous observations; BGLF4 induces expression of both BFRF1 and BFLF2, whereas BGLF5 has the opposite effect. Coexpression of both enzymes restored protein and mRNA expression to levels similar to those observed in knockout cells.
The BGLF4 mutant transforms primary B cells.
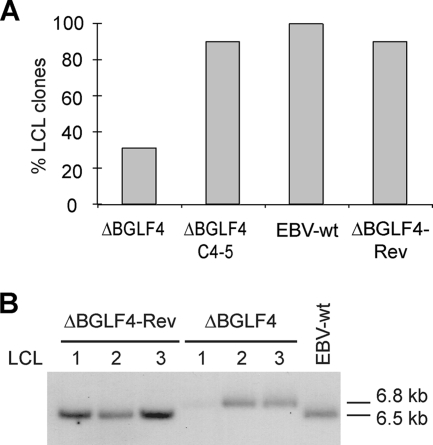
Experiments performed thus far were indicative of reduced but not abolished virus production in 293/ΔBGLF4 cells. We therefore assessed the ability of these infectious particles to infect and transform target cells by exposing primary B cells to 293/ΔBGLF4, 293/ΔBGLF4-C4-5, 293/ΔBGLF4-Rev, and 293/EBV-wt supernatants at an MOI of 10 genome equivalents per cell. One representative experiment, depicted in Fig. 7A, showed that all positive controls yielded cell colonies at similar transformation efficiencies. However, only 31% of wells containing B cells admixed with 293/ΔBGLF4 supernatants contained outgrowing B-cell lines. To confirm that these LCL clones carried the viruses they had been putatively exposed to, DNA was extracted from some of them and subjected to Southern blot analysis using a BGLF3-specific probe. This probe allows distinction between wild-type and mutant virus as it hybridizes with a 6.8-kb fragment in the ΔBGLF4 mutant genome and a 6.5-kb fragment in the wild-type EBV genome. The result of this experiment, shown in Fig. 7B, confirmed the genotype of the viruses present in these LCLs. Altogether, we concluded that BGLF4 and BGLF5 are not absolutely required for maintenance of B-cell transformation.
FIG. 7.
The BGLF4 and BGLF5 double null mutant retains EBV B-cell-transforming properties. (A) BGLF4 and BGLF5 are not required for maintenance of the transformed phenotype. Resting B cells were exposed to supernatants from 293/ΔBGLF4, 293/ΔBGLF4-C4-5, 293/ΔBGLF4-Rev, and 293/EBV-wt cells at an MOI of 10 genome equivalents per cell. B-cell transformation efficiency was assessed by counting the wells containing outgrown cell clones. (B) Southern blot analysis of LCLs obtained by infection of resting B cells with supernatants from induced 293/ΔBGLF4, 293/ΔBGLF4-Rev, and 293/EBV-wt cells. Genomic DNA was cleaved with BamHI, separated by gel electrophoresis, and hybridized with a BGLF3-specific probe that allows distinction between wild-type and recombinant genomes (the wild-type BamHI G fragment is 6.5 kb, whereas the recombinant fragment is 6.8 kb).
DISCUSSION
The construction of mutant viruses that lack one gene requires either partial or total deletion of its open reading frame or the introduction of stop codons shortly after the initiation codon. The former strategy runs the risk of inadvertently destroying unknown genes running antiparallel to the gene of interest or important regulatory sequences such as uncharacterized promoters. Efforts to interrupt an open reading frame through insertion of stop codons can be marred by the presence of cryptic methionine start codons that efficiently initiate translation of the gene of interest, resulting in the production of a truncated gene product. The 293/ΔBGLF4 mutant described in this paper illustrates these difficulties. The existence of several successive methionines in the 5′ part of the BGLF4 gene open reading frame that could act as alternative translation initiation codons prompted us to use the first method. However, deletion of part of the BGLF4 gene coding sequence also completely inhibited BGLF5's expression at the protein level (Fig. 1C). This also correlated with the total disappearance of the 1.7-kb BGLF5 mRNA signal in Northern blotting (Fig. 1E), suggesting that this transcript is initiated from a promoter located directly upstream of the BGLF5 gene that had been inadvertently destroyed during the mutant's construction. Indeed, two observations support this hypothesis. First, we identified, 74 nt upstream of the BGLF5 gene's translation start site, a degenerate TATA box and a putative initiator element. Second, induced 293/EBV-wt cells produced a transcript that started at the adenosine +1 of this initiator element (Fig. 1F).
Complementation of ΔBGLF4 with BGLF4 and BGLF5 led to a 17-fold increase in virus titers. However, this represents only 50% of those measured in the revertant and wild-type viruses. To exclude the possibility that the observed imperfect complementation resulted from an incomplete deregulation of BGLF5 expression, we repeated inductions with multiple clones of a genuine BGLF4/BGLF5 mutant that lacks both open reading frames. Unexpectedly, the difference in viral titers between complemented 293/ΔΔBG4/5 cell clones and their wild-type counterparts was even larger. The reason for this effect is unclear but might result from the intricacies of the BGLF gene locus, in which multiple genetic elements overlap; larger deletions are more likely to exert adverse effects on gene expression. Nevertheless, we are confident that the imperfect complementation does not result from introduction of adventitious mutations. Indeed, similar results were obtained with two independent mutants (ΔBGLF4 and ΔΔBG4/5 mutants) and multiple clones thereof. Furthermore, we have constructed a revertant virus from 293/ΔBGLF4 that perfectly replicated and have sequenced the viral region in the ΔBGLF4 mutant that was replaced by the revertant targeting vector. We rather suspect that complementation with two enzymes is difficult to perform with expression plasmids. In this vein, complementation of CMV UL97 PK and HSV UL12 alkaline nuclease mutants could not be fully achieved (38, 42). Similarly, our own complementation attempts with BGLF4 proved difficult, as high levels of protein production appear to be detrimental to efficient virus replication; only small amounts of a plasmid driven by a very weak parvovirus promoter were tolerated. This might be explained by BGLF5's potentiating effects on BGLF4 gene transcription. Cotransfection of BGLF4 and BGLF5 upon induction of the lytic cycle is probably unable to reproduce the molecular events observed in cells infected with a wild-type virus. In particular, it is likely that BGLF5 is, at least in part, temporally expressed before BGLF4, as otherwise BGLF4 would block BGLF5's beneficial effects on viral replication. Perfect complementation would therefore require successive expression of these two enzymes. Yet another possibility is that an additional undefined viral sequence has been destroyed during mutant construction. However, we can exclude the possibility that this sequence is the putative BGLF3.5 gene, as complementation with BGLF4, BGLF5, and BGLF3.5 was not more effective than that with BGLF4 and BGLF5 alone (data not shown).
Complementation of the ΔBGLF4 mutant with BGLF5 confirmed BGLF4's functions initially identified by the study of a BGLF4 knockdown virus (14) and from BGLF4 transient expression studies (32). Analysis of a panel of lytic genes at the mRNA level revealed a negative effect of BGLF5 on transcription of several viral genes, compared to noncomplemented ΔBGLF4 cells (Fig. 5A, lanes 5). This shows that BGLF5 host shutoff properties extend to viral genes. However, the transcription of other genes such as the immediate early BRLF1 gene and the early BMRF1 gene was not substantially influenced by the exonuclease; BGLF5's viral shutoff properties are therefore not universal. This mRNA downregulation correlated with a reduced level of protein synthesis that was particularly evident for the early proteins BFLF2 and BFRF1 in both 293/ΔBGLF4 and 293/ΔΔBG4/5 cells (Fig. 4, lane 6, and 6D and F). As expected from earlier data (13, 36), in the absence of BGLF4, hyperphosphorylated forms of EA-D could not be detected. This could at least partly explain the reduced lytic DNA replication, as it has been suggested that the hyperphosphorylated EA-D form enhances viral DNA replication (49).
Transfection of BGLF4 in 293/ΔBGLF4 cells reproduced the phenotypic traits of a ΔBGLF5 mutant and confirmed that BGLF5, like BGLF4, serves important functions during DNA replication and primary egress (8). However, our findings also evidenced clear differences between BGLF4 and BGLF5. Indeed, in contrast to induced 293/ΔBGLF4-C4 cells, induced 293/ΔBGLF4-C5 cells displayed normal capsid maturation and nuclear membrane morphology (Fig. 2) and produced linear genomes whose electrophoretic migration patterns did not differ from that of the wild type (Fig. 3). This suggests that the capsid maturation block observed in the absence of BGLF5 is related to an abnormal configuration of the linear genomes rather than to the relative reduction in linear forms observed both with BGLF4 and BGLF5. Transfection of BGLF4 in the absence of BGLF5 did not enhance viral transcription or translation relative to levels seen in 293/ΔBGLF4 cells except for transcription of the BFLF2 and BFRF1 genes. However, cotransfection of BGLF4 and BGLF5 did not result in the viral gene shutoff seen when BGLF5 was transfected without BGLF4. BGLF5 was further found to affect EA-D posttranslational maturation, as 293/ΔBGLF5 and 293/ΔBGLF4-C4 cells exhibited reduced amounts of the hyperphosphorylated forms compared to wild-type controls (Fig. 4A and B). We conclude from these observations that BGLF4 enhances the expression of a minority of viral genes but that it more generally neutralizes the BGLF5-induced viral shutoff.
In the absence of both BGLF4 and BGLF5, viral titers were reduced (Fig. 1D and 6C). However, induced 293/ΔBGLF4 cells contained viral structures, albeit in a reduced percentage of the cells, showing that both enzymes are not required for the initial stages of capsid assembly. 293/ΔBGLF4 cells still released infectious viruses with transforming capacities. Whether the observed threefold reduction in immortalization efficiency in 293/ΔBGLF4 compared to 293/ΔBGLF4-C4-5 can be attributed to a role of one of these enzymes in the initiation of transformation requires more-detailed investigations.
As expected, 293/ΔBGLF4 cells displayed phenotypic traits of both 293/ΔBGLF4-C5 and 293/ΔBGLF4-C4 cells; DNA replication, packaging, and primary egress were impaired (Fig. 1, 2, and 3). However, viral lytic gene transcription and protein expression levels were generally close to those observed in wild-type controls, although BALF4 and BLLF1 were expressed at lower levels in 293/ΔBGLF4 cells than in 293/ΔBGLF4-C4-5 cells (Fig. 4A and 5A). This could mean that BGLF4 and BGLF5 actions globally result in enhanced synthesis of these late proteins. Alternatively, it could simply be the consequence of a more efficient early phase of viral replication, in particular at the level of viral DNA replication.
We infer from this collective set of data that neither AE nor PK is required for basal viral gene expression. It is mainly when BGLF5 is produced that BGLF4 becomes required to regain normal levels of viral gene expression. BGLF4 and BGLF5 therefore provide an additional layer of viral gene control. The precise mechanisms through which BGLF4 counteracts BGLF5's repressing effects on transcription are currently unknown. Interestingly, BGLF5 has recently been shown to be a substrate for phosphorylation by BGLF4 (53). Whether this is the case in replicating cells is still unknown, but the 293/ΔBGLF4 mutant or 293/ΔΔBG4/5 double mutant should help in clarifying this issue.
To be able to counteract BGLF5, the BGLF4 mRNA must be able to withstand BGLF5's shutoff properties. Crucially, BGLF4 was found to be insensitive to BGLF5's negative effects on viral transcription. AE was even required to reach normal BGLF4 gene transcription (Fig. 5A and B and 6D). Here again, the molecular basis that underlies this process is not known, but it interesting to note that not only the BGLF4 gene open reading frame (p38-BGLF4) but also the larger mRNA initiated at the BGLF4 gene promoter (BGLF4-kan) were transactivated by BGLF5 (Fig. 5A and B).
What could be the function of the BGLF4-BGLF5 regulatory loop? BGLF5 has previously been shown to downregulate expression of HLA class I and class II molecules and thereby to contribute to immune evasion (41, 54). This essential viral function can potentially harm viral replication and needs to be counteracted. We have shown here that this role can be fulfilled by BGLF4. Furthermore, we also confirmed that BGLF5 is directly required for virus maturation; in the absence of BGLF5, BFLF2 and to a lesser extent BFRF1 were upregulated. Under these circumstances, the nuclear membrane becomes highly abnormal, a situation that could be deleterious for efficient primary egress and that can be corrected by BGLF5 (8, 15).
In conclusion, we have genetically identified a regulatory loop that is involved in the control of primary egress. Additional work will be required to understand the molecular functions of BFRF1 and BFLF2 and why these proteins need to be tightly regulated.
Acknowledgments
We are very grateful to F. Grässer and N. Müller-Lantsch for the BGLF5 antibody, to J. Rommelaere for the p38 expression plasmid, and to Martin Rowe and Jiamin Zuo for their help with the BGLF5 Western blot analysis. We also thank H. Lips and B. Hub for expert technical assistance.
Confocal microscope images were recorded, processed, and analyzed at the Nikon imaging center at the University of Heidelberg.
Footnotes
Published ahead of print on 26 August 2009.
REFERENCES
- 1.Asai, R., A. Kato, K. Kato, M. Kanamori-Koyama, K. Sugimoto, T. Sairenji, Y. Nishiyama, and Y. Kawaguchi. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 80:5125-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tufnell, and B. G. Barell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 3.Chen, H. F., M. Sauter, P. Haiss, and N. Muller-Lantzsch. 1991. Immunological characterization of the Epstein-Barr virus phosphoprotein PP58 and deoxyribonuclease expressed in the baculovirus expression system. Int. J. Cancer 48:879-888. [DOI] [PubMed] [Google Scholar]
- 4.Chen, M. R., S. J. Chang, H. Huang, and J. Y. Chen. 2000. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J. Virol. 74:3093-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Countryman, J., H. Jenson, R. Seibl, H. Wolf, and G. Miller. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farina, A., R. Santarelli, R. Gonnella, R. Bei, R. Muraro, G. Cardinali, S. Uccini, G. Ragona, L. Frati, A. Faggioni, and A. Angeloni. 2000. The BFRF1 gene of Epstein-Barr virus encodes a novel protein. J. Virol. 74:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feederle, R., H. Bannert, H. Lips, N. Müller-Lantzsch, and H. J. Delecluse. 2009. The Epstein-Barr virus alkaline exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 83:4952-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feederle, R., B. Neuhierl, G. Baldwin, H. Bannert, B. Hub, J. Mautner, U. Behrends, and H. J. Delecluse. 2006. Epstein-Barr virus BNRF1 protein allows efficient transfer from the endosomal compartment to the nucleus of primary B lymphocytes. J. Virol. 80:9435-9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gershburg, E., M. Marschall, K. Hong, and J. S. Pagano. 2004. Expression and localization of the Epstein-Barr virus-encoded protein kinase. J. Virol. 78:12140-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gershburg, E., and J. S. Pagano. 2008. Conserved herpesvirus protein kinases. Biochim. Biophys. Acta 1784:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gershburg, E., and J. S. Pagano. 2002. Phosphorylation of the Epstein-Barr virus (EBV) DNA polymerase processivity factor EA-D by the EBV-encoded protein kinase and effects of the l-riboside benzimidazole 1263W94. J. Virol. 76:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gershburg, E., S. Raffa, M. R. Torrisi, and J. S. Pagano. 2007. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 81:5407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonnella, R., A. Farina, R. Santarelli, S. Raffa, R. Feederle, R. Bei, M. Granato, A. Modesti, L. Frati, H. J. Delecluse, M. R. Torrisi, A. Angeloni, and A. Faggioni. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 79:3713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 17.Granato, M., R. Feederle, A. Farina, R. Gonnella, R. Santarelli, B. Hub, A. Faggioni, and H. J. Delecluse. 2008. Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 82:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin, B. E., E. Bjorck, G. Bjursell, and T. Lindahl. 1981. Sequence complexity of circular Epstein-Bar virus DNA in transformed cells. J. Virol. 40:11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 20.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayflick, L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614-636. [DOI] [PubMed] [Google Scholar]
- 22.Hummel, M., and E. Kieff. 1982. Epstein-Barr virus RNA. VIII. Viral RNA in permissively infected B95-8 cells. J. Virol. 43:262-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hummel, M., and E. Kieff. 1982. Mapping of polypeptides encoded by the Epstein-Barr virus genome in productive infection. Proc. Natl. Acad. Sci. USA 79:5698-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 74:10142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 80:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, K., A. Yokoyama, Y. Tohya, H. Akashi, Y. Nishiyama, and Y. Kawaguchi. 2003. Identification of protein kinases responsible for phosphorylation of Epstein-Barr virus nuclear antigen leader protein at serine-35, which regulates its coactivator function. J. Gen. Virol. 84:3381-3392. [DOI] [PubMed] [Google Scholar]
- 28.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1δ. J. Virol. 77:2359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieff, E., and A. B. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2603-2654. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 30.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, C. P., J. Y. Chen, J. T. Wang, K. Kimura, A. Takemoto, C. C. Lu, and M. R. Chen. 2007. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J. Virol. 81:5166-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, C. P., Y. H. Huang, S. F. Lin, Y. Chang, Y. H. Chang, K. Takada, and M. R. Chen. 2008. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 82:11913-11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Roux, F., A. Sergeant, and L. Corbo. 1996. Epstein-Barr virus (EBV) EB1/Zta protein provided in trans and competent for the activation of productive cycle genes does not activate the BZLF1 gene in the EBV genome. J. Gen. Virol. 77:501-509. [DOI] [PubMed] [Google Scholar]
- 34.Marschall, M., A. Marzi, P. aus dem Siepen, R. Jochmann, M. Kalmer, S. Auerochs, P. Lischka, M. Leis, and T. Stamminger. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357-33367. [DOI] [PubMed] [Google Scholar]
- 35.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
- 36.Murata, T., H. Isomura, Y. Yamashita, S. Toyama, Y. Sato, S. Nakayama, A. Kudoh, S. Iwahori, T. Kanda, and T. Tsurumi. 2009. Efficient production of infectious viruses requires enzymatic activity of Epstein-Barr virus protein kinase. Virology 389:75-81. [DOI] [PubMed] [Google Scholar]
- 37.Neuhierl, B., and H. J. Delecluse. 2005. Molecular genetics of DNA viruses: recombinant virus technology. Methods Mol. Biol. 292:353-370. [DOI] [PubMed] [Google Scholar]
- 38.Prichard, M. N., N. Gao, S. Jairath, G. Mulamba, P. Krosky, D. M. Coen, B. O. Parker, and G. S. Pari. 1999. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 73:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulvertaft, R. J. V. 1964. Cytology of Burkitt's lymphoma (African lymphoma). Lancet i:238-240. [DOI] [PubMed] [Google Scholar]
- 40.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses, p. 2501-2601. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 41.Rowe, M., B. Glaunsinger, D. van Leeuwen, J. Zuo, D. Sweetman, D. Ganem, J. Middeldorp, E. J. Wiertz, and M. E. Ressing. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. USA 104:3366-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao, L., L. M. Rapp, and S. K. Weller. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196:146-162. [DOI] [PubMed] [Google Scholar]
- 43.Shaw, G., S. Morse, M. Ararat, and F. L. Graham. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16:869-871. [DOI] [PubMed] [Google Scholar]
- 44.Smale, S. T., A. Jain, J. Kaufmann, K. H. Emami, K. Lo, and I. P. Garraway. 1998. The initiator element: a paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harbor Symp. Quant. Biol. 63:21-31. [DOI] [PubMed] [Google Scholar]
- 45.Tanner, J., Y. Whang, J. Sample, A. Sears, and E. Kieff. 1988. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J. Virol. 62:4452-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, J. T., S. L. Doong, S. C. Teng, C. P. Lee, C. H. Tsai, and M. R. Chen. 2009. Epstein-Barr virus BGLF4 kinase suppresses the interferon regulatory factor 3 signaling pathway. J. Virol. 83:1856-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, J. T., P. W. Yang, C. P. Lee, C. H. Han, C. H. Tsai, and M. R. Chen. 2005. Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 86:3215-3225. [DOI] [PubMed] [Google Scholar]
- 48.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 98:1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, P. W., S. S. Chang, C. H. Tsai, Y. H. Chao, and M. R. Chen. 2008. Effect of phosphorylation on the transactivation activity of Epstein-Barr virus BMRF1, a major target of the viral BGLF4 kinase. J. Gen. Virol. 89:884-895. [DOI] [PubMed] [Google Scholar]
- 50.Young, L. S., R. Lau, M. Rowe, G. Niedobitek, G. Packham, and F. Shanahan. 1991. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J. Virol. 65:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yue, W., E. Gershburg, and J. S. Pagano. 2005. Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase suppresses transactivation of the LMP1 promoter. J. Virol. 79:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, C. X., G. Decaussin, M. de Turenne Tessier, J. Daillie, and T. Ooka. 1987. Identification of an Epstein-Barr virus-specific desoxyribonuclease gene using complementary DNA. Nucleic Acids Res. 15:2707-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, J., G. Liao, L. Shan, J. Zhang, M. R. Chen, G. S. Hayward, S. D. Hayward, P. Desai, and H. Zhu. 2009. Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J. Virol. 83:5219-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuo, J., W. Thomas, D. van Leeuwen, J. M. Middeldorp, E. J. Wiertz, M. E. Ressing, and M. Rowe. 2008. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J. Virol. 82:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]