Abstract
The envelope glycoprotein, GP64, of the baculovirus Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is a class III viral fusion protein that mediates pH-triggered membrane fusion during virus entry. Viral fusion glycoproteins from many viruses contain a short region in the ectodomain and near the transmembrane domain, referred to as the pre-transmembrane (PTM) domain. In some cases, the PTM domain is rich in aromatic amino acids and plays an important role in membrane fusion. Although the 23-amino-acid (aa) PTM domain of AcMNPV GP64 lacks aromatic amino acids, we asked whether this region might also play a significant role in membrane fusion. We generated alanine scanning and single and multiple amino acid substitutions in the GP64 PTM domain. We specifically focused on amino acid positions conserved between baculovirus GP64 and thogotovirus GP75 proteins, as well as hydrophobic and charged amino acids. For each PTM-modified construct, we examined trimerization, cell surface localization, and membrane fusion activity. Membrane merger and pore formation were also examined. We identified eight aa positions that are important for membrane fusion activity. Critical positions were not clustered in the linear sequence but were distributed throughout the PTM domain. While charged residues were not critical or essential, three hydrophobic amino acids (L465, L476, and L480) played an important role in membrane fusion activity and appear to be involved in formation of the fusion pore. We also asked whether selected GP64 constructs were capable of rescuing a gp64null AcMNPV virus. These studies suggested that several conserved residues (T463, G460, G462, and G474) were not required for membrane fusion but were important for budding and viral infectivity.
Baculoviruses are enveloped viruses with large double-stranded DNA genomes ranging from approximately 80 to 180 kbp. They infect only invertebrates, and the majority of baculoviruses described are from insects in the order Lepidoptera. The type species of the family Baculoviridae is the Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) (11, 34). Budded virions (BV) of AcMNPV enter cells via a low-pH-dependent endocytic pathway (16). During entry by endocytosis, the major envelope glycoprotein GP64 mediates low-pH-triggered membrane fusion (3). Baculovirus GP64 proteins are highly conserved among the group I alphabaculoviruses, and the only known proteins with amino acid sequence similarities to baculovirus GP64 proteins are the GP75 envelope glycoproteins from thogotoviruses, a subgroup of the Orthomyxoviridae. GP64 is a type I integral membrane protein that is present on the infected cell surface and on the virion as a homotrimer (22). Monomers of GP64 are associated in the trimer by a single intermolecular disulfide bond (12, 37), and GP64 does not appear to require protease cleavage for activation or function. GP64 has host cell receptor-binding activity (6), and a region important for receptor-binding was recently mapped to the N-terminal portion of the ectodomain (39). GP64 is necessary and sufficient for pH-dependent membrane fusion during viral entry (3, 13, 40), and in addition to its essential role in virus entry, GP64 is also necessary for efficient budding and production of infectious virions (20, 21). Recently, the crystal structure of the low-pH (postfusion) conformation of the AcMNPV GP64 ectodomain was reported (12). The GP64 postfusion structure is an elongated trimer comprised largely of beta sheets and a central core consisting of three extended coiled coils. Each GP64 monomer can be subdivided into five domains, and the overall structures of the monomer and trimer, as well as the linear arrangement of the domains, bear remarkable similarities to the postfusion structures of herpesvirus (herpes simplex virus type 1 and Epstein-Barr virus) gB glycoproteins and also to the structures of the vesicular stomatitis virus (VSV) G protein (1, 12). While these three proteins (GP64, VSV G, and herpesvirus gB) differ in size, have no apparent amino acid sequence similarity, and are from unrelated virus groups, they share common structural features, including the long central helix that forms a triple-stranded coiled coil at the heart of the trimer, and internal fusion loops that are likely associated with target membranes during membrane fusion. These similarities suggested that these three proteins belong to a new class of viral fusion proteins, now referred to as class III fusion proteins (1, 12). Although viral fusion proteins from different classes (I, II, and III) show a variety of molecular architectures, all are thought to catalyze fusion in similar manners (5). Following an initial triggering event, the fusion protein extends and interacts with the adjacent cellular membrane via the fusion peptide or fusion loop(s). This is followed by subsequent refolding to bring the transmembrane domain and the fusion peptide or loop(s) into close proximity. Mixing of outer leaflets of the two bilayers results in the formation of a hemifusion stalk, and this is followed by mixing of the inner leaflets to form a fusion pore. In some cases (such as the influenza hemagglutinin [HA] protein), the fusion pore first appears after some period (approximately 30 s after low-pH exposure) and begins to “flicker,” rapidly opening and closing. This is followed by a final open state of the fusion pore (31, 32). In contrast to the flickering pore of HA, previous studies suggest that the GP64 fusion pore opens almost immediately (∼0.6 s after being triggered by low pH) and expands rapidly with no flickering (27, 28).
Many viral fusion proteins contain a short region in the ectodomain near the transmembrane domain, referred to as the membrane-proximal region, stem region, or pre-transmembrane (PTM) domain. This PTM domain is often rich in aromatic amino acids (17) and in prior studies of class I viral fusion proteins, the PTM domains and particularly the conserved aromatic residues within the PTM domain, were shown to play important role(s) in membrane fusion. Examples include PTM domains from the fusion proteins of human immunodeficiency virus type 1 (29), feline immunodeficiency virus (4), Ebola virus (26), human parainfluenza virus type 2 (35), and severe acute respiratory syndrome coronavirus (8, 27, 28). Various mechanistic roles have been proposed for these PTM domains, including (i) contributions to the stabilization of a six-helix bundle structure, (ii) interactions with the membrane(s), and (iii) induction of membrane destabilization. The PTM domain of the VSV G protein plays roles in both membrane fusion and virion budding (10, 25). Interestingly, while deletions or substitutions of residues from the VSV-G PTM domain had dramatic negative effects on membrane fusion, only modest effects on virus infectivity were observed. In addition, single or multiple substitutions of the conserved aromatic residues (W457, F458, and W461) have only modest effects on cell-cell fusion activity (9).
The PTM domain of the baculovirus GP64 protein was not specifically examined for function in prior studies. However, it was recently demonstrated that 22 amino acids (aa) (residues 461 to 482) from the membrane-proximal PTM region of GP64, together with the predicted transmembrane domain (residues 483 to 505), cytoplasmic tail, and 38 aa from the mature N terminus of the GP64 ectodomain, were sufficient for rescuing the budding defect of a gp64null virus and targeting the protein to BV (38). Whether this region played a role in membrane fusion, however, was unknown. To examine the functional role(s) of the GP64 PTM domain in detail, we generated a bank of mutations in the 23-aa PTM domain and analyzed the effects of single and multiple alanine substitution mutations in this region. Our results indicate that the PTM domain of GP64 is not essential for stable expression, trimerization, or transport and localization of GP64 on the cell surface. However, the PTM domain was essential for GP64-mediated membrane fusion, and three PTM leucine residues were critical for that activity. Using recombinant baculoviruses encoding GP64 proteins with various PTM domain mutations, we also examined and characterized the effects of PTM domain mutations on virion budding, GP64 incorporation into virions, and virus infectivity.
MATERIALS AND METHODS
Cells, transfections, and infections.
Spodoptera frugiperda (Sf9) cells and GP64-expressing cell line Sf9Op1D (23) were cultured at 27°C in TNMFH medium (7) containing 10% fetal bovine serum. Cells were transfected using CaPO4 precipitation (2). For viral infections, budded virus was added to cells at a multiplicity of infection (MOI) of 5 and then incubated for 1 h, and cells were washed once in TNMFH. The zero time postinfection (p.i.) indicates the time of viral inoculum removal.
Mutagenesis and construction of plasmids and bacmids.
Modified GP64 constructs were generated using an overlap PCR method with the plasmid pGEM3ZGP64 (14) as the template as described previously (15). Primer sequences are available upon request. Briefly, the PCR products were purified and digested with unique restriction enzymes NotI and EcoRI and used for subcloning into plasmid pBiepA previously digested with the same enzymes. Recombinant baculoviruses expressing the modified GP64 proteins were generated by first subcloning overlap PCR products into plasmid pGEM3ZGP64 (NotI and HindIII sites), excising the promoter and modified GP64 open reading frame with KpnI and EcoRI, and subcloning them into the same sites of pFastBac1 (Invitrogen). Note that the polyhedrin promoter was removed. The construct containing a modified gp64 gene was inserted into the polyhedrin locus of an AcMNPV gp64null bacmid (vAc64−) by Tn7-mediated transposition (18) and confirmed by restriction enzyme analysis and DNA sequencing.
cELISA, syncytium formation, and fusion assays.
Relative levels of cell surface-localized GP64 protein were analyzed by a cell surface enzyme-linked immunosorbent assay (cELISA) (14, 15). Briefly, after transfection, Sf9 cells were incubated for 36 h and then fixed in 0.5% glutaraldehyde so that cells were not permeabilized. Relative levels of cell surface-localized GP64 were measured by cELISA using monoclonal antibody (MAb) AcV5 as described previously in detail (15). For analysis of membrane fusion by syncytium formation assays, Sf9 cells were plated in 12-well plates, transfected with plasmids encoding wild-type (WT) or modified forms of GP64, incubated 36 h at 27°C, and then exposed to phosphate-buffered saline (PBS) at pH 5.0 for 3 min as described previously (14, 15). After a 4-h incubation at 27°C, cells were fixed with methanol and stained with a HEMA3 stain kit (Fisher Scientific LLC), and the number of nuclei found in syncytia was scored. Syncytial masses were defined as fused cells containing ≥5 nuclei. Five representative fields were analyzed for each construct, and relative levels of fusion activity were determined by dividing the number of nuclei in syncytia by the total number of nuclei in a field and then normalizing the results to parallel data from cells expressing WT GP64 and localized to the cell surface at equivalent levels.
Immunofluorescence analysis of cell surface GP64.
Cell surface GP64 localization was confirmed by immunofluorescence analysis of transfected cells. Sf9 cells (4 × 105 cells/well) were transfected with plasmids expressing either WT or modified GP64 proteins. At 36-h posttransfection (p.t.), cells were fixed with 4% paraformaldehyde in PBS (pH 7.4), washed with PBS, and then immunostained for GP64 with primary anti-GP64 monoclonal antibody AcV1 (1:10 dilution in PBS) and a secondary goat anti-mouse monoclonal antibody conjugated to Alexa Fluor 488 (Molecular Probes, Invitrogen), as described previously (14, 15). Florescence was observed and documented with an Olympus IX70 epifluorescence microscope.
Analysis of hemifusion and pore formation.
To examine initial membrane merger (hemifusion) and pore formation, sheep red blood cells (RBCs) (HemoState Laboratories) were dual labeled with octadecyl rhodamine B chloride (R18) and calcein-acetoxymethyl (AM) (Molecular Probes, Invitrogen) (14, 15). Sf9 cells (4 × 105 cells/well) were transfected with plasmids expressing GP64 constructs and then incubated with R18- and calcein-labeled RBCs in the following manner. At 36-h p.t., dual-labeled RBCs were added to Sf9 cells and incubated for 20 min at room temperature, and then unbound RBCs were removed by washing them three times with PBS (pH 7.4). Sf9 cells were then exposed to low pH by incubation in PBS (pH 5.0) for 3 min at room temperature. After rinsing cells in PBS (pH 7.4), cells were placed in TNMFH medium and incubated for 20 min at 27°C. Analysis of hemadsorption and the transfer of fluorescence were determined by phase-contrast and epifluorescence microscopy. For each construct, five randomly selected fields were scored for dye transfer.
Transfection-infection assays.
To determine whether GP64 constructs with modified PTM domains were capable of supporting viral infection and rescuing a gp64null virus, bacmid DNAs encoding PTM-modified GP64 gene constructs were used to transfect Sf9 cells (or control Sf9Op1D cells) using calcium phosphate precipitation. At 96 h p.t., the supernatant was removed, clarified by centrifugation (10 min at 2,200 × g), and then used to infect Sf9 cells, which were subsequently examined for β-glucuronidase (GUS) activity at 96 h p.i. as described previously (19). Both transfected and infected cells were stained for GUS activity. To confirm the viability of each bacmid genome and preparation, the same AcMNPV bacmids were used to transfect Sf9Op1D cells (which express a wild-type GP64 protein), and supernatants were used to infect Sf9 cells.
Viral growth curves.
To generate viral growth curves, Sf9 cells (1 × 106) were infected in triplicate with each virus (vAcGP64WT, vAcGP64G460A, vAcGP64S464A, vAcGP64S466A, vAcGP64D467A, vAcGP64D467E, and vAcGP64G474A) in six-well plates, at an MOI of 5. After a 1-h incubation period, the inoculum was removed and exchanged with TNMFH. Supernatants were collected at the indicated times p.i., and the titers of all supernatants were determined by 50% tissue culture infective dose assays on Sf9 cells.
Virion budding and GP64 incorporation into virions.
The effects of GP64 substitutions on virion budding efficiency and incorporation into BV were examined in the following manner: viruses expressing the PTM-modified GP64 constructs were amplified, and titers of the virus in Sf9OP1D cells, which constitutively express OpMNPV GP64 (23), were determined. The resulting virus stocks were used to infect Sf9 cells (5 × 106 cells; MOI of 5), and virion budding and incorporation were examined as described previously (14, 15, 21). Briefly, at 15 h p.i., cells were starved for 1 h in 2 ml methionine-free Grace's medium (Grace−met; Invitrogen), and then the medium was replaced with 2.2 ml of Graces−met containing 200 μCi protein labeling mix (35S EasyTag Express, 1175.0 Ci/mmol; Perkin-Elmer Life Sciences). At 30 h p.i., the medium was supplemented with methionine by adding 0.8 ml of TNMFH. Supernatants were harvested at 40 h p.i. After clearing cell debris by centrifugation for 10 min at 3,000 × g at 4°C, the virus-containing supernatant was loaded onto a 25% sucrose cushion and centrifuged at 80,000 × g for 90 min at 4°C (SW60 rotor). Virus pellets were resuspended in 200 μl Laemmli buffer (4% sodium dodecyl sulfate [SDS], 20% glycerol, 10% 2-mercaptoethanol, 0.04% bromophenol blue, 0.125 M Tris, pH 6.8) containing protease inhibitors (Complete; Roche Applied Science), and proteins were separated on SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE) gels. To identify and quantify labeled virion proteins, dried gels were exposed on phosphorimager screens, scanned on a phosphorimager (Molecular Dynamics), and individual protein bands were quantified using ImageQuant TL software (Amersham, GE).
Western blot analysis.
Analysis of GP64 proteins by reducing and nonreducing SDS-PAGE (6% or 10% polyacrylamide gels) was described previously (21). GP64 was detected on Western blots using MAb AcV5 at a dilution of 1:1,000 (14, 15).
RESULTS
Conservation in the PTM domain of GP64 proteins.
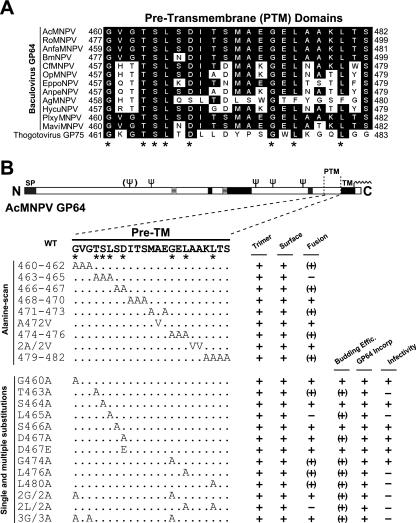
To examine conservation in the PTM domain of GP64, we performed a sequence alignment of the PTM domains from baculovirus GP64 proteins and thogotovirus GP75. The PTM domain of AcMNPV GP64 examined in this study was defined as the 23-aa region, comprising aa 460 to 482 immediately upstream of the transmembrane domain. Although aromatic amino acids in the PTM domain are important for the function of many class I viral fusion proteins, aromatic amino acids are not conserved in the PTM domain of baculovirus GP64 proteins. While some GP64 proteins contain one to four aromatic residues (F, Y, W, or H), others, such as the AcMNPV GP64 protein, contain no aromatic amino acids in the PTM domain (Fig. 1A). However, by amino acid sequence alignment of the PTM domains of GP64 proteins and thogotovirus GP75 (which shares approximately 28% overall amino acid sequence similarity with baculovirus GP64 proteins), we identified several highly conserved amino acid positions in the PTM domain. These conserved positions include G460, T463, S464, L465, D467, G474, L476, and L480 (Fig. 1A).
FIG. 1.
Sequence alignment and mutations of the baculovirus GP64 PTM domain. (A) Sequence alignment of the PTM domains from baculovirus GP64s and thogotovirus GP75 was performed using the Clustal W program. Asterisks indicate the conserved amino acids. Virus abbreviations and GenBank accession numbers are as follows: AcMNPV, Autographa californica multicapsid nucleopolyhedrovirus (GenBank accession no. L22858); RoMNPV, Rachiplusia ou (accession no. AY145471); BmNPV Bombyx mori (accession no.L33180); CfMNPV, Choristoneura fumiferana (accession no. AF512031); OpMNPV, Orgyia pseudotsugata (accession no. U75930); HycuNPV Hyphantria cunea (accession no. AAF09194); AnpeNPV, Antheraea pernyi (accession no. NC_008035); PlxyMNPV, Plutella xylostella (accession no. ABE68511); MaviMNPV, Maruca vitrata (accession no. ABL76049); EppoNPV, Epiphyas postvittana (accession no. AY043265); AgMNPV, Anticarsia gemmatalis (accession no. AAM82816); AnfaMNPV, Anagrapha falcifera (accession no. AAB53360); and thogotovirus GP75, accession no. P28977. (B) Amino acid sequence of the AcMNPV GP64 PTM (Pre-TM) domain and locations of mutations. N, N terminus; C, C terminus; SP, signal peptide; ψ, N-glycosylation sites; TM, transmembrane. Dots indicate amino acids identical to WT AcMNPV GP64. The name of each GP64 construct is shown on the left of the sequence of the PTM domain. Results from trimerization, surface localization, membrane fusion, virion budding efficiency, GP64 incorporation, and viral infectivity assays are summarized on the right. Symbols represent the following: +, moderate or high-level activity; (+), low level activity; −, no detectable activity. Asterisks indicate the conserved amino acid positions identified in panel A.
Substitution mutations in the GP64 PTM domain.
We used two strategies to examine the requirements of the GP64 PTM domain. First, we performed an alanine scan of the GP64 PTM domain, substituting two, three, or four alanines for amino acids of the PTM domain sequence (Fig. 1B). Alanines in the wild-type PTM domain (positions A472, A477, and A478) were substituted with valine residues. Next, we generated single and multiple amino acid substitutions in various PTM positions, based primarily on sequence conservation among GP64 and GP75 proteins (Fig. 1A and B). Amino acid positions G460, T463, S464, L465, D467, S466, G474, L476, and L480 were substituted individually and in some cases, in combinations (Fig. 1A and B).
Expression and cell surface localization of PTM-modified GP64 proteins.
GP64 proteins containing substitutions in the PTM domain were transiently expressed from plasmids in Sf9 cells, and cell lysates were examined at 36 h p.t., by Western blot analysis using either reducing or nonreducing conditions for SDS-PAGE. As GP64 trimers contain a disulfide bond between monomers in the trimer, nonreducing conditions permit the assessment of GP64 oligomerization (6, 22). Oligomers representing GP64 trimers (14, 22) were detected from all GP64 constructs with substitutions in the PTM domain (see Fig. S1A and B in the supplemental material). These data suggested that substitutions within the PTM domain of GP64 did not significantly affect the expression or oligomerization of the resulting proteins.
Because cell surface levels of GP64 can affect the assessment of function, we asked whether GP64 constructs containing substitutions in the GP64 PTM domain were transported to the cell surface and displayed on the plasma membrane. Using a cELISA protocol, we measured the level of each GP64 construct at the cell surface relative to that detected from WT GP64. Of the nine alanine scanning constructs examined, four resulted in cell surface GP64 levels of >20% of that from wild-type GP64, and five resulted in surface levels below 10% (Fig. 2A). In all cases, the measured cell surface levels were sufficient for detection of membrane fusion activity from wild-type GP64. Alanine residues found in the wild-type GP64 PTM domain were substituted with valine residues. Substitution of alanines with valines resulted in either minor or moderately reduced surface levels of GP64 (Fig. 2A, constructs A472V and 2A/2V). Similar to the results from alanine-scanning mutations, surface levels of GP64 constructs with single or multiple amino acid substitutions in the PTM domain were frequently lower than surface levels of wild-type GP64 (Fig. 2B), although some constructs (S464A, G460A, 2G/2A, and 3G/3A) had only minor or moderate reductions in cell surface levels. All other constructs were displayed at the cell surface at levels of ≤10% of that from WT GP64. However, the detected cell surface levels were sufficient for detection of membrane fusion activity from wild-type GP64. To confirm the presence and native conformation of GP64 constructs at the cell surface, we also used a conformation-specific MAb (MAb AcV1) to detect each GP64 construct at the cell surface by indirect immunofluorescence (see Fig. S1C and D in the supplemental material). MAb AcV1 recognizes only the prefusion conformation of GP64, and the epitope is lost upon exposure of GP64 to low pH (40). All GP64 constructs containing substitution mutations in the GP64 PTM domain were detected at the cell surface by MAb AcV1, confirming cell surface localization and indicating that the GP64 constructs were in the native prefusion conformation.
FIG. 2.
Analysis of cell surface levels and membrane fusion activity of GP64 PTM constructs. (A and B) Relative cell surface levels of GP64 constructs were measured by cELISA, using MAb AcV5. Each GP64 construct was expressed by transfecting Sf9 cells with 2 μg of the appropriate plasmid DNA. Values represent the means from triplicate transfections and are normalized to those for cells transfected with 2 μg of the plasmid DNA expressing WT GP64. A standard curve generated from WT GP64 expression is shown on the left, and results from GP64 constructs containing PTM domain mutations are shown on the right. Error bars represent the standard deviation from the mean, and the amount of DNA per transfection for WT GP64 is indicated by numbers below the graph (μg). (C and D) Analysis of membrane fusion by syncytium formation assays. Sf9 cells were transfected with plasmids expressing WT GP64 or GP64 PTM constructs. At 36 h posttransfection, cells were exposed to pH 5.0 for 3 min, incubated for 4 h, and observed and photographed under phase-contrast microscopy (magnification, ×200). (E and F) Analysis of syncytium formation efficiency. Relative fusion activity was determined in the following manner: for each field, the number of nuclei in syncytia was divided by the total number of nuclei in a field. Those percentages were normalized to parallel syncytium formation data from WT GP64 that was localized to the cell surface at equivalent levels. For each sample, five fields were examined. The means and standard deviations of triplicate transfections are shown. Alanine scanning substitutions (panels A, C, and E) and single and multiple amino acid substitutions (panels B, D, and F).
Fusion activity of mutant GP64s.
We evaluated the membrane fusion activity of each GP64 PTM construct by measuring fusion efficiency in a semiquantitative syncytium formation assay. Because surface levels of different GP64 constructs varied, the fusion activity of each modified GP64 construct was normalized by comparison with the activity from WT GP64 that was localized to the cell surface at the same level. Analysis of the alanine scanning substitutions showed that substitutions in all regions except one (468 to 470) substantially reduced membrane fusion activity of the GP64 constructs (Fig. 2C and E). The normalized fusion activities of constructs 460 to 462, 466 to 467, 471 to 473, 474 to 476, and 479 to 482 were very low, only 1 to 5% compared with that of WT GP64, and substitutions at positions 463 to 465 (TSL changed to AAA) resulted in a complete loss of detectable fusion activity. Replacement of PTM alanines with valines (constructs A472V and A477 to 478V) had no substantial effect on the fusion activity of GP64 (Fig. 2C and E). Thus, substitutions in most regions of the PTM domain had a dramatic negative effect on membrane fusion activity (Fig. 2E). However, fusion activity was substantially unaffected by substitutions in six aa positions (468 to 470, 472, 477, and 478) in this analysis.
Next, we examined conserved amino acid positions in the PTM domain by introducing single or multiple amino acid substitutions (Fig. 1B, lower panel). Many of these substitution mutations had dramatic effects on GP64 fusion activity. The most substantial effects were observed with alanine substitutions L465A and 2L/2A (2L/2A represents alanine substitutions in positions L476 and L480), which resulted in GP64 proteins with no detectable fusion activity (Fig. 2D and F). In addition, GP64 PTM substitutions T463A, G474A, L476A, and L480A mediated only very low levels of membrane fusion, with normalized fusion activities of approximately 1.4% (L476A and L480A), 3.1% (G474A), and 4.2% (T463A), compared with WT GP64 (Fig. 2F). Substitution of charged residue D467 with alanine or glutamic acid, resulted in a 90% decrease in fusion activity. In general, analysis of alanine substitutions in conserved glycine residues of the PTM domain revealed no dramatic negative effects. Construct G460A retained fusion activity at levels near wild-type levels (∼90% of wild-type activity), and double and triple glycine substitutions (2G/2A, positions G460 and G474; 3G/3A, positions G460, G462, and G474) resulted in only modestly decreased fusion activities (∼70% and 60%, respectively), compared with wild-type GP64 (Fig. 2D). It is of interest that the single G474A substitution resulted in dramatically reduced GP64 fusion activity, yet when the same substitution was included with one or two additional glycine substitutions (in constructs 2G/2A and 3G/3A), there was only a minor negative effect of the G474A substitution on membrane fusion activity. Substitutions in conserved and semiconserved serine residues resulted in no detectable effect on fusion activity of GP64 (Fig. 1B and 2F, S464A and S466A). In total, our analysis of alanine scanning substitutions in combination with single and multiple amino acid substitutions indicated that the specific amino acids at certain positions (G460, S464, S466, I468, T469, S470, A472, A477, and A478) were not critical for membrane fusion. In contrast, the loss or dramatic reduction of fusion activity from constructs containing single or multiple substitution mutations at other positions (T463, L465, D467, and L476, and L480) identified these residues as important for membrane fusion activity.
Infectivity, budding efficiency, and GP64 incorporation into virions.
Because GP64 plays multiple roles in the infection cycle, we also examined the overall effects of the PTM on viral infection and propagation. Each gp64 gene construct containing single or multiple substitutions in the PTM domain was cloned under the transcriptional control of the WT GP64 promoter and inserted into the polyhedrin locus of a gp64null bacmid using standard Tn7-based transposition (18, 19). The donor plasmid used for transposition also encoded a GUS reporter gene under the control of the AcMNPV p6.9 promoter. As a positive control, the wild-type AcMNPV gp64 gene was inserted into the same donor plasmid and was used to rescue the gp64null bacmid. A similar donor plasmid containing no gp64 gene was used to generate a negative control bacmid. Each bacmid DNA construct was then used to transfect Sf9 cells in a transfection-infection assay (see Materials and Methods) (19).
Transfected and infected cells were scored for GUS activity as an indicator of viral replication (when detected after transfection) and an indicator of the generation of infectious virions (when detected after infection). Transfection of Sf9 cells with bacmids expressing gp64 PTM constructs S464A, D467A, D467E, G460A, and G474A resulted in rescue of gp64null AcMNPV and production of infectious virions. In contrast, transfection with bacmids expressing constructs T463A, L465A, 2G/2A, 3G/3A, L476A, L480A, and 2L/2A did not result in rescue of viral infectivity (Fig. 1B, infectivity; Fig. 3A, left). Interestingly our previous analysis showed that gp64 protein constructs T463A, 2G/2A, 3G/3A, L476A, and L480A were capable of mediating membrane fusion (Fig. 2B and D), but those constructs were unable to rescue viral infectivity when inserted into and expressed from the gp64null bacmid genome. To ensure that all gp64 protein constructs were expressed in Sf9 cells after transfection of the bacmids, lysates of the transfected Sf9 cells were analyzed by Western blot analysis. We detected expression of all of the gp64 protein constructs in the bacmid-transfected cells (data not shown). In addition, to confirm that any defects detected were not due to other lethal mutations in the bacmid, all the mutant bacmids were transfected into Sf9Op1D cells, which constitutively express the OpMNPV GP64 protein. In each case, infectious virions were produced (data not shown), indicating that the lack of infectious virion production in Sf9 cells was due to the lack of a functional GP64 protein and did not result from a second-site mutation.
FIG. 3.
Analysis of virus infectivity, virion budding, and efficiency of GP64 incorporation into virions by viruses expressing GP64 constructs with PTM domain substitutions. (A) The results of a transfection-infection assay (left) are indicated by + or − for each gp64null bacmid expressing a PTM-modified GP64 construct. Transfection-infection assays were performed as described in Materials and Methods. Virus one-step growth curves are shown at right. BV preparations generated from bacmids containing WT GP64 or GP64 constructs with the PTM domain substitutions (G460A, S464A, S466A, D467A, D467E, and G474A) were used to infect Sf9 cells (MOI of 5). Infectious BV yields were determined at the indicated time points by the 50% tissue culture infective dose assays (TCID50). Data points indicate averages for infections performed in triplicate, and error bars represent standard deviations. (B) Analysis of virion budding efficiency by analysis of labeled progeny virions. Infected cells were pulse labeled with [35S]methionine, and progeny BV purified as described in Materials and Methods. Labeled viral proteins were analyzed by SDS-PAGE and phosphorimager analysis. Control viruses and viruses expressing GP64 constructs with PTM domain substitutions are indicated above the lanes, and the positions of GP64 and VP39 proteins are indicated between the panels. Controls included an AcMNPV virus expressing WT GP64 and an AcMNPV virus containing a GP64 knockout (KO). Each lane represents virus purified from an equivalent volume of the cell culture supernatant. (C) Comparison of virion budding efficiencies. Relative budding efficiencies were determined by comparing the quantity of [35S]methionine in the major capsid protein (VP39) band from each virion preparation with that from a virus expressing WT GP64. (D) Comparison of efficiencies of GP64 incorporation into BV. For each virion preparation (shown in panel B), the [35S]methionine incorporated into the GP64 construct was divided by the [35S]methionine incorporated into the VP39 capsid protein, and each bar in the graph represents that ratio. Virions containing GP64 constructs with PTM domain substitutions were compared with virions expressing WT GP64, and construct names are indicated below each bar in the graph.
To further examine the GP64 PTM constructs that rescued infectivity (G460A, S464A, S466A, D467A, D467E, and G474A), we performed one-step growth curve studies of Sf9 cells. Infectious virus production from viruses vAcGP64G460A, vAcGP64S464A, and vAcGP64S466A was similar to that from a control virus expressing wild-type GP64, with no substantial differences at most times examined (Fig. 3A, right). In contrast, GP64 constructs D467A, D467E, and G474A resulted in infectious virus titers that were approximately 10-fold lower than those of the control virus expressing WT GP64 at 48 to 120 h p.i. We noted that these reduced titers correlated somewhat with reduced fusion activity for these same constructs (Fig. 2F, D467A, D467E, and G474A).
In addition to its role in membrane fusion during entry, GP64 also plays an important role in virion budding. Budding is severely reduced in the absence of GP64 (21). To examine possible effects of the PTM domain on virion budding, we examined virion budding from viruses expressing GP64 PTM mutations. Each of the above viruses expressing a PTM-modified GP64 construct was used to infect Sf9 cells. Cells were pulse labeled with [35S]methionine, and labeled progeny virions were purified. The relative quantities of progeny virions from cell culture supernatants were estimated by quantifying the [35S]methionine label from the major virion capsid protein VP39 as described previously (21) (see Materials and Methods). Comparison of each virus with a control virus expressing WT GP64 revealed that nine of the PTM domain GP64 constructs had substantial or dramatic negative effects on virion budding efficiency (Fig. 3B and C). These included constructs T463A, L465A, D467A, G474A, L476A, L480A, 2L/2A, 2G/2A, and 3G/3A, in which budding efficiencies were reduced to approximately 3 to 14% relative to that mediated by WT GP64. In some cases, PTM substitutions had very moderate or no effect on virion budding efficiency. Viruses expressing constructs D467E, G460A, S464A, and S466A had measured budding efficiencies of approximately 48 to 92% relative to that mediated by WT GP64. Analysis of these single and multiple point mutations confirmed the importance of the GP64 PTM domain in virion budding.
We also examined the effects of PTM domain substitutions on the incorporation of GP64 into virions. As a measure of the relative quantity of GP64 incorporation into virions, we measured the ratio of GP64 to the major capsid protein VP39 in each virion preparation. As shown in Fig. 3B and D, all of the mutant GP64s were incorporated into BV as efficiently as WT GP64. Thus, although the PTM domain was important for virion budding, we did not identify any residues necessary or important for GP64 incorporation into BV.
Conserved leucine residues.
In models of some fusion proteins, the PTM domain is proposed to interact directly with membranes, perturbing local membrane stability. In such models, aromatic or hydrophobic residues of the PTM domain would play important roles in membrane fusion. We therefore examined in more detail, the most highly conserved hydrophobic PTM domain residues of GP64. For this analysis we examined residues L465, L476, and L480 (Fig. 4A). In our initial studies, substitutions in these positions (constructs L465A and 2L/2A) resulted in no detectable membrane fusion (Fig. 2D), suggesting an important functional role for these residues. To determine if hydrophobicity at these positions was critical or whether leucine residues specifically were required, we generated both conservative and nonconservative substitutions at these positions. Each leucine residue was substituted with isoleucine, lysine, or asparagine (Fig. 4A). GP64 constructs containing these PTM modifications were transiently expressed in Sf9 cells and examined by Western blot analysis of cell lysates by the use of reducing and nonreducing conditions. The profiles of the PTM domain-substituted GP64 constructs were similar to that of WT GP64, indicating that expression and trimerization were not substantially affected by any of these substitutions (see Fig. S1E in the supplemental material). Analysis of cell surface localization showed that substitution of the hydrophobic amino acid leucine by isoleucine (Fig. 4A, constructs L465I and 2L/2I) had the least effect on GP64 localization to the cell surface. Surface levels for construct L465I were similar to those for wild-type GP64, and levels for construct 2L/2I were reduced to 13% of that for wild-type GP64. Substitutions of polar and charged residues for leucines at 465, 476, and 480 (Fig. 4A, constructs L465N, 2L/2N, L465K, and 2L/2K) had more substantial negative effects on surface localization, reducing surface levels of the latter constructs to less than 3% of that from wild-type GP64. In addition, all of these GP64 constructs were detected at the cell surface by the monoclonal antibody AcV1, suggesting that these constructs were in the prefusion conformation (data not shown). Analysis of membrane fusion activity by GP64 constructs containing substitutions in conserved leucines showed that fusion activity was similar to that of the wild type when the hydrophobic amino acid isoleucine was substituted for L465, and fusion was completely abolished when the same amino acid (L465) was substituted with polar or charged amino acids (Fig. 4C, L465I, L465N, and L465K). These results were not caused by reduced cell surface levels since expression of WT GP64 at similar levels resulted in readily detectable membrane fusion. Substitution of both leucines L476 and L480 with isoleucine had a negative effect on the fusion activity of GP64, but remaining fusion activity was estimated at approximately 10% of that from wild-type GP64. As with the L465 position, substitution of L476 and L480 by polar or charged amino acids resulted in no detectable fusion activity (Fig. 4C, 2L/2I versus 2L/2N and 2L/2K). Overall, the results of substitutions in the conserved leucine residues suggest that the PTM domain of GP64 is essential for membrane fusion activity and that hydrophobicity at positions L465, L476, and L480 is critical.
FIG. 4.
Analysis of conserved leucine residues in the GP64 PTM domain. (A) A sequence alignment of PTM domain sequences from a variety of baculovirus GP64 proteins and from the thogotovirus GP75 protein is shown (top), and conserved amino acids are indicated by asterisks below the alignment. The conserved leucine residues are boxed. Single- or double-substitution mutations (hydrophobic [I], polar [N], or charged [K]) generated in the conserved leucine positions of GP64 constructs (bottom) are shown. The construct names are indicated on the left. (B) Analysis of relative cell surface levels of GP64 constructs. Relative cell surface levels of GP64 constructs containing PTM domain substitutions were measured by cELISA. Each mutant GP64 construct was expressed by transfecting 2 μg plasmid DNA encoding the modified GP64 construct. A standard curve was generated by transfecting cells with various quantities of plasmid DNA expressing WT GP64. Quantities are indicated below the graph (μg). (C) Analysis of relative fusion activity of GP64 constructs. Relative fusion activity was evaluated using efficiency of syncytium formation calculated from representative random fields and normalized to parallel syncytium formation data from WT GP64 that was localized to the cell surface at equivalent levels, as described in the legend for Fig. 2E and F. The means and standard deviations of triplicate transfections are shown. (D) Analysis of hemifusion and fusion-pore formation by GP64 protein constructs. RBCs that were dual labeled with membrane-restricted (R18) and cytosolic (calcein-AM) dyes were bound to Sf9 cells transiently expressing GP64 protein constructs. The bound cells were exposed to acidic PBS (pH 5.0) for 3 min to induce fusion and examined after a 20 min incubation period. Membrane dye transfer (R18, left) or cytosolic dye transfer (calcein-AM, right) were monitored by fluorescence microscopy. Each GP64 construct (or control) expressed after transfection is indicated on the left.
Hemifusion and pore formation.
To examine the step in membrane fusion that was affected by substitutions of hydrophobic amino acids of the PTM, we used membrane and cytosolic dyes to examine membrane merger and pore formation. Constructs with substitutions at positions L465, L476, and L480 were examined. Dual-labeled RBCs containing both the integral membrane dye (R18) and a cytosolic dye (calcein AM) were bound to Sf9 cells that were previously transfected with plasmids encoding wild-type or modified GP64 constructs. Membrane fusion was initiated by exposure of the RBC-bound Sf9 cells to low pH (pH 5.0) for 3 min. R18 redistribution from RBCs to Sf9 cell membranes in the absence of calcein transfer to the Sf9 cell cytosol indicates a merger of the outer membrane leaflets (hemifusion) and the absence of pore formation. All GP64 constructs with substitutions in the PTM domain were observed to mediate lipid dye (R18) transfer from RBCs to Sf9 cells (Fig. 4D and data not shown). In addition, those constructs positive for fusion also showed transfer of the cytosolic dye, calcein AM (Fig. 4D, L465I and 2L/2I; see also Fig. S2 in the supplemental material). However, we did not observe transfer of the cytosolic dye calcein-AM by constructs L465A, L465N, L465K, 2L/2A, 2L/2N, and 2L/2K (Fig. 4D). Thus, although the latter constructs are capable of mediating the initial membrane merger step (hemifusion), fusion pore formation did not appear to occur. Thus, these data suggest that reduction of the hydrophobic character of positions L465, L476, and L480 resulted in GP64 constructs capable of mediating membrane merger but unable to form fusion pores.
Analysis of charged amino acids in the PTM domain.
We observed that substitution of the conserved charged residue D467 caused dramatic decreases in fusion activity relative to wild-type GP64 (Fig. 2F). To examine the possible roles of other charged residues in the PTM domain of GP64, we generated GP64 substitution mutations of charged residues E473, E475, and K479. In each case we substituted either a neutral uncharged amino acid (alanine) or a similarly charged amino acid (aspartic acid or arginine) (Table 1). A GP64 construct containing a double alanine substitution for residues E473 and E475 (2E/2A) was also generated. All mutants were examined for trimerization, cell surface localization, membrane fusion activity, and virus infectivity. The GP64 proteins containing the above substitutions of charged amino acids were all expressed and trimerized (data not shown). While some constructs (E473A, E475A, and 2E/2A) resulted in reduced GP64 surface localization, all constructs exhibited relatively high levels of membrane fusion activity compared with wild-type GP64 (Table 1). In addition, all of the above substitutions of charged amino acids resulted in GP64 proteins that were capable of rescuing a gp64null AcMNPV bacmid in a transfection-infection assay (Table 1). Thus, unlike the dramatic effects observed from substitution of hydrophobic residues, we observed relatively minor effects from substitution mutations in charged residues E473, E475, and K479, suggesting that these charged residues do not play a critical or essential role in membrane fusion or other functions of GP64 in the infection cycle.
TABLE 1.
Mutational analysis of additional charged amino acids within the GP64 PTM domain
| Construct | PTM domain | Trimer | Surface (%) | Normalized fusion (%) | Infectivity |
|---|---|---|---|---|---|
| WT | GVGTSLSDITSMAEGELAAKLTS | + | 100 | 99.6 ± 0.3 | + |
| E473A | ·············A········· | + | 38.1 ± 5.6 | 97.2 ± 1.6 | + |
| E473D | ·············D········· | + | 59.9 ± 4.1 | 100 ± 0.2 | + |
| E475A | ···············A······· | + | 36.2 ± 4.8 | 87.3 ± 3.4 | + |
| E475D | ···············D······· | + | 61.8 ± 6.4 | 100 ± 0.6 | + |
| 2E/2A | ·············A·A······· | + | 3.7 ± 0.6 | 32.6 ± 4.2 | + |
| K479A | ···················A··· | + | 79.9 ± 7.9 | 100 ± 0.2 | + |
| K479R | ···················R··· | + | 97.9 ± 3.1 | 100 ± 0.3 | + |
DISCUSSION
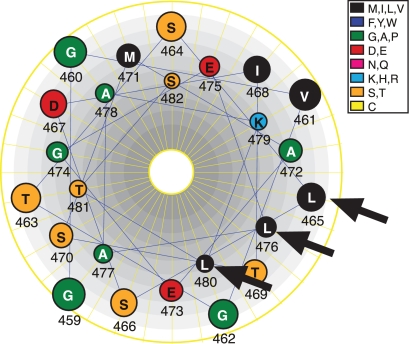
The PTM domains of many viral fusion proteins play an important role in membrane fusion. Little, however, was known about the role of the baculovirus GP64 PTM in membrane fusion. PTM domains of fusion proteins from many groups (including retroviruses, filoviruses, orthomyxoviruses, paramyxoviruses, rhabdoviruses, alphaviruses, coronaviruses, and flaviviruses) are enriched in aromatic amino acids (17, 33), particularly tyrosine and/or tryptophan. Mutational analysis of conserved PTM domain tryptophan residues in proteins such as VSV G (9, 10) and human immunodeficiency virus type 1 GP41 (29) demonstrates an important role for the aromatic residues in membrane fusion. It was previously suggested that the aromatic amino acid-rich PTM domain may play a role in destabilizing or perturbing the viral membrane during the fusion process. This model is supported by recent studies in which the gp41 PTM was functionally replaced by a tryptophan-rich antimicrobial peptide (indolicidin), a peptide that is known to disrupt membranes (36). Although the precise mechanism of membrane disruption by aromatic amino acids is unclear, it is thought that the indole ring of tryptophan may insert into the membrane-water interface and perhaps disrupt the phospholipid organization (30, 33). In contrast with many other membrane fusion proteins, the PTM domains of baculovirus GP64 or thogotovirus GP75 proteins do not contain highly conserved aromatic amino acid residues. Indeed, no aromatic residues are present in the AcMNPV GP64 PTM domain (Fig. 1). Although the PTM domain of GP64 is not visible in the crystal structure of postfusion GP64 (12), the PTM domain can be modeled as an alpha-helix. In this hypothetical model, the three conserved leucine residues that were identified as critical for GP64 function in membrane fusion, are found aligned along one face of the helix (Fig. 5). Analysis of fusion activity for GP64 constructs containing alanine substitutions in one (L465A) or two (L476A and L480A) of these positions resulted in loss of membrane fusion activity (Fig. 2F, L465A and 2L/2A). In addition, substitution of hydrophobic, polar, or charged residues for leucines at 465 or 476 and 480 (Fig. 4A) resulted in GP64 constructs with fusion activity that correlated with the degree of hydrophobicity at these positions. Thus, our results suggest that while hydrophobic amino acids at these positions are critical for membrane fusion activity, leucine was not specifically or absolutely required. Analysis of hemifusion and pore formation by duel dye-labeling experiments also suggested that these conserved leucine positions (L465, L476, and L480) are not required for the initial fusion of the outer membrane leaflets (and formation of the hemifusion intermediate), but hydrophobic residues in these positions are necessary for fusion pore formation.
FIG. 5.
Helical wheel model of the PTM domain of AcMNPV GP64. The amino acid sequence of the PTM domain of AcMNPV GP64 (aa 459 to 482) is modeled as a helical wheel. Arrows indicate the positions of conserved leucine residues (L465, L476, and L480) on one face of the helix.
Interestingly, these positions are conserved in all GP64 proteins examined, except the AgMNPV GP64 protein, which differs considerably from other baculovirus GP64 proteins. While the AgMNPV protein contains a leucine at the homologous L465 position, the other two important positions identified in the current study (L476 and L480) are substituted in AgMNPV GP64 with aromatic (phenylalanine) residues (Fig. 1A, AgMNPV). In addition, the AgMNPV GP64 protein contains five aromatic amino acid residues in the PTM domain, whereas other GP64 proteins examined here contain a maximum of one aromatic residue. Taken together with the functional identification of leucines as critical residues for GP64-mediated fusion, these findings suggest that the proposed roles of aromatic residues in PTM domains of other viral membrane fusion proteins may be substituted by those of hydrophobic leucine residues in most baculovirus GP64 (and thogotovirus GP75) proteins.
We also asked whether charged residues in the PTM domain may play a role in membrane fusion activity. Of the single- and double-substitution mutations of residues E473, E475, and K479 (Table 1), all resulted in constructs capable of mediating membrane fusion and rescuing viral infectivity, suggesting that these charged residues do not play a critical or important role in GP64-mediated membrane fusion.
Because the PTM domain is not represented in the available postfusion structure of GP64 (14) and a prefusion structure is not available, it is not yet possible to accurately model the structure and mechanistic function of the PTM domain. However, several speculative models may be useful for exploring the data observed in the current study. These include the possible involvement of the PTM domain in protein-protein interactions within GP64 monomers or trimers. It is possible that the critical leucine residues (L465, L476, and L480) may be required for important hydrophobic interactions within the interior of the folded protein. Such interactions might stabilize the pre- or postfusion conformations of GP64 or mask the hydrophobic fusion loops. It is also possible that the PTM domain may facilitate the pH-triggered conformational change, possibly serving as a hinge-like region. Functional data provided in the current study may argue against the latter role since substitutions of the three glycines, which would presumably provide structural flexibility, did not abrogate GP64-mediated membrane fusion. Most attractive is the possibility that the PTM domain may serve as a membrane-disrupting domain that facilitates membrane merger, as has been suggested for PTM domains of other viral fusion proteins. The demonstrated importance of hydrophobic residues at positions L465, L476, and L480 is consistent with that role, and the evolutionary substitution of aromatic amino acids in two of those positions (positions 476 and 480) provides further support for this model.
Substitutions that disrupt membrane fusion may disrupt one or several of the steps involved in protein-mediated membrane fusion. These include but are not limited to triggering of the conformational change, interaction of a fusion peptide with the adjacent membrane, apposition to bring the two membranes into close contact, merger of the outer membrane leaflets to form a hemifusion intermediate, formation of a fusion pore by merger of the inner membrane leaflets, and expansion of the fusion pore. As described above, our results suggest that the PTM may not be involved in the initial stage of outer membrane leaflet merger but instead may be required for initiating or completing pore formation. Electrophysiological studies of triggering and pore formation by the influenza virus HA protein report a substantial delay (∼30 s) after triggering, which is followed by an initially low conductance (suggesting formation of a small fusion pore) and rapid flickering as the fusion pore opens and closes rapidly (31, 32). In contrast, GP64 fusion pores open rapidly (∼0.6 s) after low-pH triggering, with a high initial conductance, and do not flicker (23, 24). It is possible that such differences in the opening and stabilization of the fusion pore may be related to differences in PTM domain interactions within the membrane in the hemifusion intermediate.
In prior studies, it was shown that BV budding is dramatically reduced in the absence of GP64 (21). A GP64 construct containing only 38 aa from the mature N terminus and 52 aa from the C terminus (including only 22 aa of the PTM domain) of GP64 was capable of partially rescuing that budding defect (38). In the current study we extended the analysis of PTM domain requirements for budding by examining the effects of single and multiple amino acid substitutions in the PTM domain of a full-length GP64 protein. Virus budding efficiency was dramatically reduced (by 80 to 90%) when 6 of the 10 single amino acid substitutions were introduced into the GP64 PTM domain (Fig. 3C). The three constructs containing multiple amino acid substitutions in the GP64 PTM domain also showed dramatic reductions in budding efficiency. For many of these constructs, budding efficiency was near or only slightly above that of the GP64 knockout virus. Single amino acid substitutions that did not substantially affect budding included substitutions G640A, S464A, S466A, and D467E, which had only slight or modestly reduced budding efficiency (reduced by 10 to 50%) compared with that of WT GP64. In some cases, the effects of the PTM substitution on budding efficiency appeared to parallel cell surface expression levels, while in other cases, such as D467E, budding efficiency seemed to be independent of cell surface levels. We also examined the effect of the PTM on the incorporation of GP64 into virions by measuring the ratio of GP64 to VP39 in virion preparations purified from each construct. We observed no substantial effect on GP64 incorporation into virions in any of the single or multiple amino acid substitutions examined (Fig. 3D). Overall, these data indicate that the PTM domain is critically important for efficient virion budding. However, we did not identify any role for the PTM in regulating the incorporation of GP64 into virions.
Because GP64 is involved in multiple functions during entry and egress, we also examined the effects of PTM domain substitutions on overall infectivity. While membrane fusion assays showed that 11 of the 13 constructs retained some level of fusion activity, only 6 of the 13 constructs were able to rescue virus infectivity (Fig. 3A). Rescue of infectivity almost certainly depends on a combination of GP64 roles in entry (binding and membrane fusion) and egress (virion budding). Where fusion activity was absent in GP64 constructs, no rescue was observed (Fig. 1, fusion versus infectivity). Rescue of infectivity generally correlated with virion budding efficiency, although in two cases in which budding efficiency was low (Fig. 3C, D467A and G474A), the constructs were able to rescue infectivity. In those cases, however, virion production appeared to be somewhat reduced (Fig. 3A, rescued virus growth curves).
In summary, we found that the AcMNPV GP64 PTM domain serves a critical role in GP64-mediated membrane fusion. We identified three critical leucine residues (L465, L476, and L480) that abolished GP64 fusion activity when substituted either alone (L465) or in combination (L476 and L480). Mutations in the PTM domain did not substantially affect GP64 expression, trimerization, or the ability of GP64 to incorporate into BV. However, the PTM was important for GP64 trafficking to the plasma membrane, and conserved amino acids in the PTM domain were important for virion budding and virus infectivity. Because of its close proximity to the viral envelope, the importance of the PTM domain in membrane fusion is likely related to interactions of the PTM domain with the lipid bilayer of the viral envelope, although we cannot exclude other possibly important interactions of the PTM domain. Such interactions may stabilize the pre- or postfusion structure, and interactions within the GP64 protein or between GP64 and other proteins may also be important during virion budding. In future studies it will be important to examine the intra- and intermolecular interactions that mediate these fundamental processes. Such studies will enhance our understanding of the molecular mechanisms of membrane fusion during virus entry and will also contribute to an understanding of the complex nature of virion budding, the final step in virus assembly.
Supplementary Material
Acknowledgments
We thank Gerrit Heetderks for technical assistance.
This work was supported by NIH grant AI33657 and BTI project B00103-R06-1255.
Footnotes
Published ahead of print on 19 August 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Backovic, M., and T. S. Jardetzky. 2009. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blissard, G. W., and G. F. Rohrmann. 1991. Baculovirus gp64 gene expression: analysis of sequences modulating early transcription and transactivation by IE1. J. Virol. 65:5820-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg, H., F. J. Fuller, and W. A. Tompkins. 2004. Mechanism of feline immunodeficiency virus envelope glycoprotein-mediated fusion. Virology 321:274-286. [DOI] [PubMed] [Google Scholar]
- 5.Harrison, S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hefferon, K., A. Oomens, S. Monsma, C. Finnerty, and G. Blissard. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258:455-468. [DOI] [PubMed] [Google Scholar]
- 7.Hink, W. F. 1970. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature 226:466-467. [DOI] [PubMed] [Google Scholar]
- 8.Howard, M. W., E. A. Travanty, S. A. Jeffers, M. K. Smith, S. T. Wennier, L. B. Thackray, and K. V. Holmes. 2008. Aromatic amino acids in the juxtamembrane domain of severe acute respiratory syndrome coronavirus spike glycoprotein are important for receptor-dependent virus entry and cell-cell fusion. J. Virol. 82:2883-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeetendra, E., K. Ghosh, D. Odell, J. Li, H. P. Ghosh, and M. A. Whitt. 2003. The membrane-proximal region of vesicular stomatitis virus glycoprotein G ectodomain is critical for fusion and virus infectivity. J. Virol. 77:12807-12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeetendra, E., C. S. Robison, L. M. Albritton, and M. A. Whitt. 2002. The membrane-proximal domain of vesicular stomatitis virus G protein functions as a membrane fusion potentiator and can induce hemifusion. J. Virol. 76:12300-12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jehle, J. A., G. W. Blissard, B. C. Bonning, J. S. Cory, E. A. Herniou, G. F. Rohrmann, D. A. Theilmann, S. M. Thiem, and J. M. Vlak. 2006. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch. Virol. 151:1257-1266. [DOI] [PubMed] [Google Scholar]
- 12.Kadlec, J., S. Loureiro, N. G. Abrescia, D. I. Stuart, and I. M. Jones. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024-1030. [DOI] [PubMed] [Google Scholar]
- 13.Leikina, E., H. O. Onaran, and J. Zimmerberg. 1992. Acidic pH induces fusion of cells infected with baculovirus to form syncytia. FEBS Lett. 304:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Z., and G. W. Blissard. 2008. Functional analysis of the transmembrane (TM) domain of the Autographa californica multicapsid nucleopolyhedrovirus GP64 protein: substitution of heterologous TM domains. J. Virol. 82:3329-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Z., and G. W. Blissard. 2009. The Autographa californica multicapsid nucleopolyhedrovirus GP64 protein: analysis of transmembrane domain length and sequence requirements. J. Virol. 83:4447-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long, G., X. Pan, R. Kormelink, and J. M. Vlak. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 80:8830-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorizate, M., N. Huarte, A. Saez-Cirion, and J. L. Nieva. 2008. Interfacial pre-transmembrane domains in viral proteins promoting membrane fusion and fission. Biochim. Biophys. Acta 1778:1624-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lung, O., M. Westenberg, J. M. Vlak, D. Zuidema, and G. W. Blissard. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 76:5729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monsma, S. A., A. G. P. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297-314. [DOI] [PubMed] [Google Scholar]
- 22.Oomens, A. G. P., S. A. Monsma, and G. W. Blissard. 1995. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology 209:592-603. [DOI] [PubMed] [Google Scholar]
- 23.Plonsky, I., M. S. Cho, A. G. P. Oomens, G. W. Blissard, and J. Zimmerberg. 1999. An analysis of the role of the target membrane on the gp64-induced fusion pore. Virology 253:65-76. [DOI] [PubMed] [Google Scholar]
- 24.Plonsky, I., and J. Zimmerberg. 1996. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. J. Cell Biol. 135:1831-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saez-Cirion, A., M. J. Gomara, A. Agirre, and J. L. Nieva. 2003. Pre-transmembrane sequence of Ebola glycoprotein. Interfacial hydrophobicity distribution and interaction with membranes. FEBS Lett. 533:47-53. [DOI] [PubMed] [Google Scholar]
- 27.Sainz, B., Jr., E. C. Mossel, W. R. Gallaher, W. C. Wimley, C. J. Peters, R. B. Wilson, and R. F. Garry. 2006. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res. 120:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sainz, B., Jr., J. M. Rausch, W. R. Gallaher, R. F. Garry, and W. C. Wimley. 2005. The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: putative role during viral entry. Biochemistry 44:947-958. [DOI] [PubMed] [Google Scholar]
- 29.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schibli, D. J., R. C. Montelaro, and H. J. Vogel. 2001. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry 40:9570-9578. [DOI] [PubMed] [Google Scholar]
- 31.Spruce, A. E., A. Iwata, and W. Almers. 1991. The first milliseconds of the pore formed by a fusogenic viral envelope protein during membrane fusion. Proc. Natl. Acad. Sci. USA 88:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spruce, A. E., A. Iwata, J. M. White, and W. Almers. 1989. Patch clamp studies of single cell-fusion events mediated by a viral fusion protein. Nature 342:555-558. [DOI] [PubMed] [Google Scholar]
- 33.Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Theilmann, D. A., G. W. Blissard, B. Bonning, J. Jehle, D. R. O'Reilly, G. F. Rohrmann, S. Thiem, and J. M. Vlak. 2005. Baculoviridae, p. 177-185. In H. V. Van Regenmortel, D. H. L. Bishop, M. H. Van Regenmortel, and C. M. Fauquet (ed.), Virus taxonomy: eighth report of the international committee on taxonomy of viruses. Elsevier Academic Press, New York, NY.
- 35.Tong, S., F. Yi, A. Martin, Q. Yao, M. Li, and R. W. Compans. 2001. Three membrane-proximal amino acids in the human parainfluenza type 2 (HPIV 2) F protein are critical for fusogenic activity. Virology 280:52-61. [DOI] [PubMed] [Google Scholar]
- 36.Vishwanathan, S. A., and E. Hunter. 2008. Importance of the membrane-perturbing properties of the membrane-proximal external region of human immunodeficiency virus type 1 gp41 to viral fusion. J. Virol. 82:5118-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volkman, L. E. 1986. The 64K envelope protein of budded Autographa californica nuclear polyhedrosis virus. Curr. Top. Microbiol. Immunol. 131:103-118. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, J., and G. W. Blissard. 2008. Display of heterologous proteins on gp64null baculovirus virions and enhanced budding mediated by a vesicular stomatitis virus G-stem construct. J. Virol. 82:1368-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, J., and G. W. Blissard. 2008. Identification of a GP64 subdomain involved in receptor binding by budded virions of the baculovirus Autographica californica multicapsid nucleopolyhedrovirus. J. Virol. 82:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, J., and G. W. Blissard. 2006. Mapping the conformational epitope of a neutralizing antibody (AcV1) directed against the AcMNPV GP64 protein. Virology 352:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.