Abstract
Serine protease inhibitors (serpins) are a superfamily of proteins, most of which control protease-mediated processes by inhibiting their cognate enzymes. Sequencing of the silkworm genome provides an opportunity to investigate serpin structure, function, and evolution at the genome level. There are thirty-four serpin genes in Bombyx mori. Six are highly similar to their Manduca sexta orthologs that regulate innate immunity. Three alternative exons in serpin1 gene and four in serpin28 encode a variable region including the reactive site loop. Splicing of serpin2 pre-mRNA yields variations in serpin2A, 2A′ and 2B. Sequence similarity and intron positions reveal the evolutionary pathway of seven serpin genes in group C. RT-PCR indicates an increase in the mRNA levels of serpin1, 3, 5, 6, 9, 12, 13, 25, 27, 32 and 34 in fat body and hemocytes of larvae injected with bacteria. These results suggest that the silkworm serpins play regulatory roles in defense responses.
Keywords: Bombyx mori, insect immunity, serine protease inhibitor, hemolymph protein, melanization, spätzle processing
Introduction
Serine proteases (SPs) mediate crucial physiological processes such as blood coagulation, complement activation, melanotic encapsulation, and spätzle processing in vertebrates and invertebrates [1, 2]. After accomplishing their missions, active SPs need to be promptly defunctionalized and removed from circulation. Serine protease inhibitors (serpins) have evolved to regulate SPs and maintain homeostasis [3]. To date, over 800 serpins have been identified in animals, plants, bacteria, and viruses. Typical mature serpins contain 350~400 amino acid residues and share a conserved three-dimensional structure composed of nine a-helices and three β-sheets [4]. An exposed reactive site loop (RSL), located near the carboxyl-terminus, connects β-sheets A and C and acts as bait for protease binding and cleavage. The serpin RSL, upon formation of a covalent bond with its target enzyme, undergoes a major conformational change and inserts into β-sheet A as the fourth strand (s4A). In this thermodynamically favored process, the protease molecule is dragged to the other end of serpin and becomes inactive due to the distortion of its catalytic machinery [5].
Serpins have been purified and cloned from insects including Bombyx mori [6, 7], Manduca sexta [8–13], Mythimna unipuncta [14], and Aedes aegypti [15]. Biochemical investigations suggest that most serpins in hemolymph can regulate plasma proteases. For instance, Manduca serpins inhibit hemolymph proteases (HPs) involved in the prophenoloxidase (proPO) activation pathway, a SP-mediated defense response against pathogen infection and tissue damage [16, 17]. Hemolymph proteins responsible for proPO activation and melanization include HP14, HP21, PAPs (for proPO-activating proteases), HP1, HP6, and SPHs (for SP homologs). High Mr complexes of SPH1 and SPH2, which lack the active site Ser residue or catalytic activity, are required for generating active PO by PAPs. We have identified the following physiological pairs of serpin-protease: serpin1I-HP14, serpin3/1J-PAPs, serpin4-HP1/HP6/HP21, serpin5-HP1/HP6, and serpin6-HP8/PAP1/PAP3 [9, 11, 12, 18, 19].
Genetic analyses have revealed physiological functions of several serpins in D. melanogaster [20, 21] and A. gambiae [22–24]. Drosophila Serpin43Ac regulates the Toll pathway in response to fungal infection in the adults. Serpin27A, an ortholog of M. sexta serpin3, controls melanization by inhibiting a proPO activating enzyme. It also affects embryonic development by inhibiting easter, a member of the SP pathway that establishes the dorsoventral axis [25]. Drosophila serpin4A is an intracellular serpin that regulates protein processing in the secretory pathway of cells [26]. In Anopheles, the relationships between serpins and malaria pathogens have been explored. The accumulation of serpin10 in the midgut epithelial cell correlates with the malaria parasite transmission [27]. Serpin6 expression affects Plasmodium berghei clearance by inhibiting proPO activation or promoting parasite lysis [23]. Serpin2, the A. gambiae ortholog of M. sexta serpin3, has a drastic effect on survival of P. berghei but its underexpression does not impact P. falciparum [24, 28].
Insect genome projects have uncovered many serpin genes in the fly [29], mosquitoes [30, 31], honeybee [32], and beetle [33], although their physiological roles remain mostly unknown. To facilitate the research on these important regulatory molecules, we have explored serpin genes in the silkworm genome [34, 35] and compared them with serpins from M. sexta and other insects. It is our hope that such a comparison will stimulate biochemical studies on B. mori serpins. In this paper, we describe the initial sequence analysis of silkworm serpins, which provides a perspective for their functional investigation and a landmark for comparative genomic analysis of insect serpins.
Materials and methods
Insects and collection of hemocytes, and fat body
B. mori eggs and diet were purchased from Carolina Biological Supply, and the larvae were reared at ambient temperature. Day 2, 5th instar larvae were injected with a mixture of 5×107 Escherichia coli cells, 15 μg Micrococcus luteus, and 15 μg curdlan suspended in 50 μl phosphate buffered saline (10 mM phosphate, 138 mM NaCl, 2.7 mM KCl, pH 7.4). Hemocyte and fat body samples were collected from the injected and uninjected (control) larvae 24 h later as previously described [18].
Database searching and sequence retrieving
M. sexta serpin1~6 protein sequences were used as queries for BLAST search of the local B. mori database (http://darwin.okstate.edu/blast/blast.html) at a cutoff E-value of 0.1. This standalone service was established using National Center for Biotechnology Information (NCBI) BLAST Tool Kit. B. mori genome sequences and protein prediction files were downloaded from Silkworm Genome Database, SilkDB (http://silkworm.genomics.org.cn/). Based on the ESTs downloaded from NCBI (http://www.ncbi.nlm.nih.gov/), a B. mori UniGene database was established using TGI Clustering Tools (http://www.tigr.org/software/other.shtml). Protein sequences resulting from the initial search were used as queries for another round of BLAST search, and this step was repeated until no new sequence was found. According to the combined list of accession numbers, corresponding nucleotide sequences of putative serpins were retrieved from SilkDB for BLAST searches of the EST database to validate the gene predictions. In case of discrepancies, cDNA sequences were used as references to correct prediction errors. Respective EST clones were either completely sequenced or assembled using CAP3 [36]. Exon-intron organization and alternative splicing was confirmed by comparing cDNA with the genome sequence using Est2genome (http://bioweb.pasteur.fr/docs/EMBOSS/est2genome.html). D. melanogaster, A. gambiae, A. aegypti, A. mellifera and T. castaneum serpin sequences were retrieved from FlyBase (http://flybase.bio.indiana.edu/), Ensembl (http://www.ensembl.org/index.html for the mosquitoes), NCBI (for the honeybee), and BeetleBase (http://www.hgsc.bcm.tmc.edu/projects/tribolium/). Other insect serpin sequences were retrieved from NCBI.
Identification and characterization of B. mori serpins
To confirm classification as serpins, the protein sequences were scanned for domain features using CDART (http://www.ncbi.nlm.nih.gov/structure/lexington/lexington.cgi?cmd=rps), PROSITE (http://us.expasy.org/prosite/), and SMART (http://smart.embl-heidelberg.de/smart). Signal peptides were predicted by SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/). Cleavage sites were predicted according to the conserved features of serpin reactive site loop [3, 37].
Multiple sequence alignment and phylogenetic analysis
Complete serpin domains were aligned using ClustalX 1.83 (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/). Phylograms were displayed by neighbor-joining analysis through Treeview (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). A Blosum 30 matrix, with a gap penalty of 10 and an extension gap penalty of 0.05 were selected for the multiple sequence alignment.
RNA extraction and reverse transcription (RT)-PCR analysis
Total RNA samples were extracted from fat body or hemocytes of naïve and induced B. mori larvae using Micro-to-Midi Total RNA Purification System (Invitrogen Life Technologies). First-strand cDNA synthesis was performed using 2–4 μg total RNA, 10 pmol oligo(dT)17, and 200 U MMLV reverse transcriptase (Invitrogen Life Technologies) at 37°C for 1 h. B. mori actin cDNA was used as an internal standard to normalize the templates in a preliminary PCR experiment. After template adjustment, PCRs were performed to detect relative levels of serpin cDNAs using the specific primers (Table S1). The thermal cycling conditions were: 94°C, 30s; 50°C, 40s; 72°C, 90s. PCR cycle numbers were empirically chosen to show comparable band intensity and avoid saturation. After separation by 1.5% agarose gel electrophoresis, intensities of the PCR products were quantified and compared using Kodak Digital Science 1D Gel Analysis Software and then categorized into different clusters based on their tissue specificity and inducibility.
Results
Overview of the silkworm serpin genes
Determination of the silkworm genome sequence allowed us to examine serpin genes in the entire genome. A search of Silkworm Genome Database (SilkDB) yielded 34 serpin gene sequences (Table 1). We sequenced the complete cDNA clones for serpin1 through 13 (Fig. S1). The remaining serpins (except for 21, 22, 25~27, 29~34) were partially or completely confirmed by expressed sequence tags (ESTs) deposited at NCBI. The cDNA/EST sequences improved the initial gene predictions and indicated that at least 32 of these genes are transcriptionally active – expression of serpin14~25, 27~29, 31~34 was later confirmed by RT-PCR and sequencing (see below). The total gene count is comparable to that of D. melanogaster (28), T. castaneum (31) or A. aegypti (23), but much higher than that of A. gambiae (17) or A. mellifera (7).
Table 1.
Structural features of Bombyx mori serpins
| Serpin |
Drosophila homolog |
Gene # BGIBMGA |
Lengtha (residues) |
Signal peptide |
Predicted cleavage site |
Target proteaseb |
EST c | Exon #d | Scaffold and contig #e |
Ch. #f |
|---|---|---|---|---|---|---|---|---|---|---|
| 1g | spn4/42Da | 009953 | 16+376/375/375 | MA-AV | TR-SS/VF-MS/LL-SA | T/C/C | + | 9+1 | 003014, 001788, 008229, c264931, c449174 | ? |
| 2g | spn4/42Da | 007720 | 374/375/378 | no | MR-CA/ML-CP/AF-CS | T/C/C | + | 8+2 | 013721, 018575, 000026 | 15 |
| 3 | spn27A | 010212 | 22+436 | TA-NI | NK-FG | T | + | 8+0 | 011782, 011630, c601391 | 7 |
| 4 | spn77Ba | 013852 | 17+393 | FA-QN | NR-IG | T | + | 1+0 | 012696 | 28 |
| 5 | spn77Ba | 013849 | 16+379 | YC-DV | DR-IS | T | + | 1+0 | 002283 | 28 |
| 6 | spn5/88Ea | 007729 | 17+396 | TS-QC | FR-SS | T | + | 7+1 | 003042, 015788 | 15 |
| 7 | spn77Ba | 013932 | 19+380 | YG-QC | AR-ML | T | + | 1+0 | 016063 | 28 |
| 8 | spn77Ba | 013904 | 23+408 | EA-QS | VA-TGh | E | + | 2+0 | 000113, 010384 | 28 |
| 9 | spn6/55B | 001983 | 16+379 | QA-FY | SL-SR | C | + | 9+0 | 008353 | 19 |
| 10 | spn100A | 010214 | 17+496 | SA-RW | PL-SEh | C | + | 7+1 | 006039, 005642B | 7 |
| 11 | spn6/55B | 010213 | 19+393 | DC-LD | PL-IN | C | + | 7+0 | 016390, 017062, c23733 | 7 |
| 12 | spn6/55B | 010216 | 18+484 | DS-QS | AL-SF | C | + | 7+0 | 005642A, 021829, c191638 | 7 |
| 13 | spn28D | 004955 | 19+415 | WC-NT | AR-SA | T | + | 8+1 | 015853, 004674 | 25 |
| 14 | spn77Ba | 013958 | 17+382 | TA-LK | II-SG | C | + | 1+0 | 011155 | 28 |
| 15 | spn85F | 008831 | 18+379 | EA-NR | SK-CSh | T | + | 9+0 | 001243B | 3 |
| 16 | spn85F | 003292 | 20+372 | CT-QN | AE-SVh | ? | + | 8+0 | 005540, 020795, c318038 | ?i |
| 17 | spn85F | 008830 | 18+379 | EA-NR | SK-CSh | T | + | 9+0 | 001243A, c43137 | 3 |
| 18 | spn85F | 003292 | 21+371 | CA-QN | AA-SVh | E | + | 8+0 | 007756 | ?i |
| 19 | spn85F | 004735A | 18+396 | AA-GF | EV-CPh | E | + | 9+0 | 000577 | 22 |
| 20 | spn85F | 008827 | 17+382 | LA-TD | KA-SPh | E | + | 9+0 | c008305 | 3 |
| 21 | spn4/42Da | no | 374 | no | RR-AH | T | + | 8+0 | 010908, 000965 | ? |
| 22 | spn85F | 003292 | 21+371 | CT-KN | AA-SVh | E | + | 8+? | 003823, 021354 | ?i |
| 23 | spn85F | 004735B | 18+393 | TA-AF | EV-CSh | E | + | 9+0 | 008594A, c477626 | 22 |
| 24 | spn85F | 008828–9 | 20+381 | ES-VR | CY-SIh | C | + | 9+0 | 001581A, 004108 | 3 |
| 25 | spn85F | 008826 | 16+383 | TG-KE | EK-CSh | T | + | 9+? | 006592 | 3 |
| 26 | spn100A | no | >164 | ? | RR-AGh | T | − | >5+? | 007109 | ? |
| 27 | spn75F | 011587 | >438 | ? | TR-STh | T | + | >2+? | 001075 | 23 |
| 28g | spn4/42Da | 004726 | 349/353/355/354 | no | KR-SG/VN-SD/TL-SS/RA-TS | T/?/C/E | + | 7+1 | 022161, 011144, 017742, c515738 | 22 |
| 29 | spn100A | 007721 | 18+343 | TA-EN | TY-SMh | C | + | 6+? | c016235, c043739 | 15 |
| 30 | spn85F | 004736 | 408 | no | SI-CAh | C | − | 8+? | 008594B, c33981, c020793 | 22 |
| 31 | spn77Ba | 013903 | 347 | no | GF-IQh | C | + | 2+? | c435360, c529910 | 28 |
| 32 | spn77Ba | 013848 | 20+375 | NA-DY | DR-IL | T | + | 1+? | c457704 | 28 |
| 33 | spn28D | 001066 | 16+407 | YC-QQ | DR-VMh | T | + | 9+? | 007282, c015844 | 13 |
| 34 | spn75F | 003349 | 29+519 | ES-TD | ?h | ? | + | 3+? | c010889 | 15 |
signal peptide + mature protein;
trypsin (T), chymotrypsin (C), or elastase (E)-like specificity;
serpin21, 22, 25, 27, 29, 31~34 mRNA (not identified in the EST project) were detected by RT-PCR (Fig. 4 and Table S2) and confirmed by sequencing;
coding exons + noncoding ones;
serpin 26 gene incomplete;
chromosome number;
alternative splicing;
sequence features significantly different from a typical inhibitory serpin in the P20-P20′ region (see Fig. 2);
member of a gene cluster whose chromosomal location is unclear.
Twenty-seven of these genes encode serpins containing a signal peptide for secretion (Table 1). Serpin2, 21, 28, 30 and 31 lack signal peptides and are predicted to be intracellular, whereas serpin26 and 27 gene predictions are incomplete at the amino terminus. Most of the mature proteins are 350~400 residues long with an average size of 377. Due to the amino-terminal extension, serpin3, 8, 12, and 27 are composed of 436, 408, 484, and >438 residues, respectively. Serpin10, 13, and 33 contain an insert in the serpin domain, which increases their sizes to 496, 415, and 407 residues, respectively. The additional sequences may not affect the overall folding because, based on molecular modeling (data not shown), the additional sequences are rich in hydrophilic residues and exist in an external loop. In contrast, serpin34 which lacks a conserved carboxyl-terminus (including RSL) is not anticipated to fold properly with a long insert separating the partial serpin domain. This insert (>220 residues) contains five low complexity regions.
Multiple sequence alignment of the B. mori serpins (Fig. 1) revealed that their RSL near the carboxyl-terminus (Fig. 2) is hypervariable in length and sequence. We have made predictions of their proteolytic cleavage site (Table 1). Serpin1A, 2A, 3~7, 13, 15, 17, 21, 25~28A, 32, and 33 (with Arg or Lys located at the predicted P1 position) may inhibit trypsin-like enzymes. Serpin15, 17 and 21 has R/K at the P1 and P4 positions but none of them contain an endoplasmic reticulum (ER)-retention signal, R/K/H-E/D-E-L/F [38], at the carboxyl terminus to suggest that they may regulate processing enzymes in the secretory pathway. In fact, serpin21 does not even have a signal peptide to lead it into ER. B. mori serpin1B, 1C, 2A′, 2B, 9~12, 14, 24, 28C, and 29~31 (with Phe, Tyr, Leu, or Ile located at the P1 site) are anticipated to inhibit chymotrypsin-like SPs; serpin8, 18~20, 22, 23, and 28D (with Ala or Val located at the P1 position) could control elastase-like enzymes.
Fig. 1. Multiple sequence alignment of B. mori serpins.
The serpin regions, including the partial ones (serpin25 and 26), are compared as described in Materials and Methods. For simplicity, only one of the splicing variants of serpin1, 2, and 28 is included in the alignment. To indicate the intron positions, the first amino acid residue encoded by each succeeding exon is shaded yellow. Introns in the 5′ and 3′ untranslated regions are marked before and after the protein sequences, respectively. The putative signal peptides are underlined. The predicted P1 residues are shown in bold and red. Secondary structural elements of B. mori serpin1A are predicted based on its high similarity with M. sexta serpin1K (1SEK) [49]. a-helices A through I are labeled as hA~hI, and strand n (n = 1~6) in β-sheet X (X = A, B or C) are labeled snX (e.g. s5B). The conserved Ser residues (Ser52 and Ser54 in B. mori serpin1) are shown in bold, with the ones using TCN codon in blue. The only exception was found in serpin11, whose Ser54 (in red) uses AGY codon. The codon usage of the second Ser is used as a marker for serpin evolution analysis [50]. Positions of those introns, which correspond to similar sites in most of the serpin amino acid sequences, are numbered 1 through 7 (shaded yellow). The inserts in serpin10 and 13 (but not 33 or 34) are corroborated by cDNA sequences (Fig. S1). Note that the short insert in serpin13 pushes the RSL toward the carboxyl-terminus. The group names in front of individual sequences, A~F in bold, reveal the basis of phylogenetic relationships shown in Fig. 3.
Fig. 2. Multiple sequence alignment of RSL in B. mori serpins.
Based on the comparison of serpin domains (Fig. 1), the P18-P15′ region of serpin4 and corresponding sequences from the other proteins are shown. Gap positions in the adopted sequences are manually adjusted for proper alignment with Gly360-Thr361 and Pro385-Phe386 of serpin4. These and other conserved residues at P15, P14, P11′, and P12′ sites are shaded yellow. Prediction of P1 residue (in bold and red) is mainly based on the distance between P14 and P1 including gap(s). Minor adjustments are made by considering that most inhibitory serpins have Ser/Thr as well as small hydrophobic residues at P1′ site [4].
A closer examination of the RSL region suggests that some residues may affect the inhibitory activity of these serpins (Fig. 2). The conserved Gly (or Ala) residue at P15 is substituted by Asn, Asp, His, Gln, or Pro in serpin8, 16, 18~20, 22, 23, 25~27, 30, and 33, which may interfere with the insertion of RSL into β-sheet A. The small hydrophobic residues between P11 and P8 are replaced by bulky residues in serpin8, 10, 14~20, 22~27, 29~31, and 33. Therefore, the predicted inhibitory activity and selectivity (Table 1) of these serpins needs to be tested experimentally.
Evolutionary relationships among the insect serpins
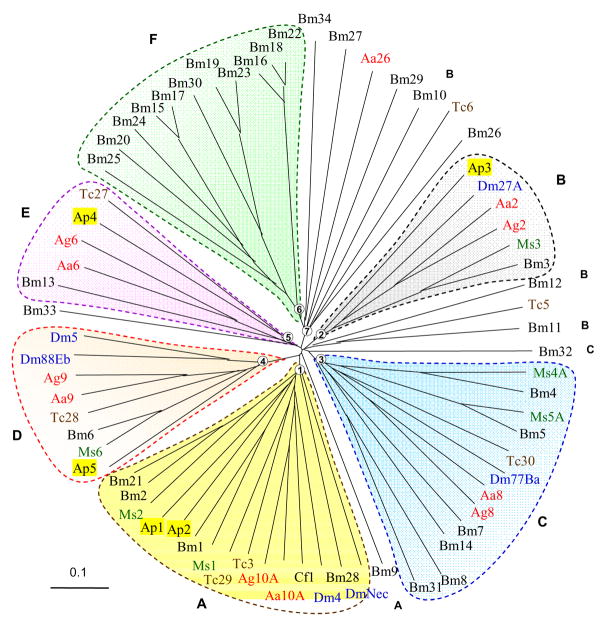
Based on our phylogenetic analysis, six groups of serpin genes have been distinguished, each with a bootstrap support of 660 or higher (Fig. 3):
Fig. 3. Phylogenetic relationships among the serpins from B. mori, M. sexta, D. melanogaster, and other insects.
Based on a multiple sequence alignment similar to the one shown in Fig. 1, an unrooted neighbor-joining tree is constructed as described in Materials and Methods. Bootstrap values for nodes 1 through 7 are 854, 973, 784, 1000, 660, 993, and 771, respectively (1000 trials). The corresponding clades form six distinct phylogenetic groups named A through F (shaded yellow, grey, blue, orange, pink, and green). Based on the similarity in intron locations, B. mori serpin9, 10, 11, 12, and 32 are assigned to groups A, B, B, B, and C, respectively. All group F members (serpin15~20, 22~25, and 30), some group C (serpin8, 14, and 31) and B (serpin10) members, as well as serpins 26, 27, 29, 33, and 34 are predicted to be noninhibitory.
Group A consists of serpin1, 2, 21, 28 and serpins from the other insect species, including M. sexta serpin1 and 2, A. mellifera serpin1 and 2, T. castaneum serpin3 and 29, Ctenocephalides felis serpin1, A. gambiae and A. aegypti serpin10A, and D. melanogaster serpin4 (i.e. spn42Da) and Necrotic (i.e. spn43Ac). One characteristics of this group is the existence of alternative exons for RSL in some of its members. These proteins are also related to a group of fourteen Drosophila genes (spn28Da, 28Db, 28B, 28F, 31A, 38F, 42Db, 42Dc, 42Dd, 42De, 43Aa, 43Ab, 47C, and 55B) that arose from a lineage-specific family expansion (data not shown).
Group B is composed of B. mori, M. sexta, A. mellifera serpin3, D. melanogaster spn27A, A. gambiae and A. aegypti serpin2. B. mori serpin3, 10, 11, 12 genes are syntenic and similar in exon-intron organization, even though their sequence identity is low. Genetic and biochemical analyses indicated that M. sexta serpin3 and D. melanogaster spn27A regulate proPO and spätzle activation [11, 23, 25].
Group C includes B. mori serpin4, 5, 7, 8, 14, and 31, M. sexta serpin4 and 5, A. gambiae and A. aegypti serpin8, T. castaneum serpin30, and D. melanogaster spn77Ba, 77Bb and 77bc (data not shown). B. mori serpin4, 5 (and 32) genes, serpin7 and 14 genes, serpin8 and 31 genes form three clusters on the same chromosome (Table 1). Serpin4 is most similar to serpin5, and so are serpin7 and 8 to serpin14 and 31, respectively. Biochemical study has demonstrated that the M. sexta serpin4 and 5 inhibit multiple HPs involved in immune responses [12].
Group D contains B. mori and M. sexta serpin6, A. mellifera and D. melanogaster serpin5 (i.e. spn88Ea), Drosophila 88Eb, T. castaneum serpin28, A. gambiae and A. aegypti serpin9 (Fig. 3). M. sexta serpin6 forms covalent complexes with HP8 and PAP1/PAP3 [13, 19]. Group E comprises B. mori serpin13, A. mellifera serpin4, A. aegypti and A. gambiae serpin6, and T. castaneum serpin27. There was a family expansion in the mosquito, which gave rise to A. gambiae serpin4, 5, 6, and 16 (data not shown). No biochemical analysis has been carried out for any of the group members so far.
Group F encompasses eleven B. mori serpins (15~20, 22~25, 30), which evolved from a major lineage-specific family expansion. B. mori serpin15, 17, 20, 24 and 25 genes form a cluster on chromosome 3 (Table 1); serpin19-23-30 gene cluster resides on chromosome 22; serpin16, 18, 22 genes are highly similar and syntenic. The close genomic locations and high sequence identity indicate recent gene duplications. However, since little functional data is available for these serpins, we are not certain whether there is any functional gain from the group expansion.
The remaining serpins (9~12, 26, 27, 29, 32~34), radiating from the phylogenetic tree center (Fig. 3), seem to be evolutionarily ancient. Unlike members of groups A~D, these proteins are not orthologous to serpins from other species with known functions. Therefore, physiological roles of these serpins are to be elucidated experimentally.
RSL variation and alternative exon usage
B. mori serpin1 consists of three RSL variants, each encoded by exons 1~8 and exon 9A/B/C (Fig. S1–1). Due to a stop codon in the end of the alternative exons, exon 10 is noncoding and B. mori serpin1A, 1B and 1C contain an amino-terminal constant region and a carboxyl-terminal variable part. The exon 9-coding region is less variable in regions that form the secondary structures (s1C, s4B, and s5B) and more variable in the loop region that directly interacts with a target protease. With the RSL sequences of TMTR*SSKV, AVVF*MSAA and VELL*SAVI, serpin1A, 1B, and 1C are predicted to have different inhibitory selectivity (Table 1).
We have also detected sequence variations in B. mori serpin2 (Fig. S1–2). Serpin2A and 2A′ are nearly identical in most of the sequence except for the region near the carboxyl-terminus. Since the serpin2 gene contains a gap, we are unable to conclude whether alternative exon usage or allelic variation has led to the sequence difference. In another cDNA clone, the exon 9A region is absent and exon 10 codes for a sequence similar to that encoded by exon 9A. The predicted P1 residue suggests that serpin2A inhibits a protease with trypsin specificity and that serpin2A′ and 2B may regulate chymotrypsin-like enzymes. Comparison of the cDNA and partial gene sequence revealed alternative splicing sites in the 5′ and 3′ untranslated regions.
Another member of group A, B. mori serpin28, may use the same mechanism to generate sequence diversity in the RSL (Fig. S1–28). Exons 7A~7D correspond in size and position to the exon 9 variants in the serpin1 gene. Although four alternative exons are identified in the gene, cDNA sequences only confirm the use of exon 7D. No EST support is available for B. mori serpin21 (Fig. S1–21). Based on the phylogenetic analysis (Fig. 3), the common ancestor of serpin1, 2, 21, and 28 genes may contain 7 or 8 exons that encode an intracellular serpin, and there was probably an increase in exon number when serpin1 and 2 evolved.
Expression analysis
We carried out RT-PCR analysis to test whether the silkworm serpin genes are actively transcribed in fat body or hemocytes and if the mRNA levels change after an immune challenge. The serpin1, 3, 5, 6, 9, 12, 13, 25, 27, 32 and 34 transcript levels increased in both fat body and hemocytes after bacteria injection (Fig. 4 and Table S2, cluster I). Serpin23 mRNA concentration increased in fat body. Similar to serpin15, 17 and 19, serpin23 transcripts were mainly (if not solely) found in fat body (cluster II). Tissue-specific expression of serpin31 gene occurred in hemocytes, and the mRNA was detected at a high level in the induced sample (cluster III). While transcriptional up-regulation in hemocytes was observed for serpin4 and 28, their transcripts remained stable in fat body before and after the immune challenge (Table S2, cluster IV). The mRNA levels of serpin7, 11, 14, 21 and 29 did not significantly change in either tissue (Fig. 4, cluster V). In comparison, a decrease in transcript abundance (cluster VI) was observed for serpin2. The mRNA levels were below the detection limit for the other ten serpins (8, 10, 16, 18, 20, 22, 24–26, 30, cluster VII). Some of these genes may be transcribed in a different tissue or developmental stage. For instance, B. mori serpin16 and 18 are strongly expressed in the silk gland [39].
Fig. 4. RT-PCR analyses of B. mori serpin transcripts.
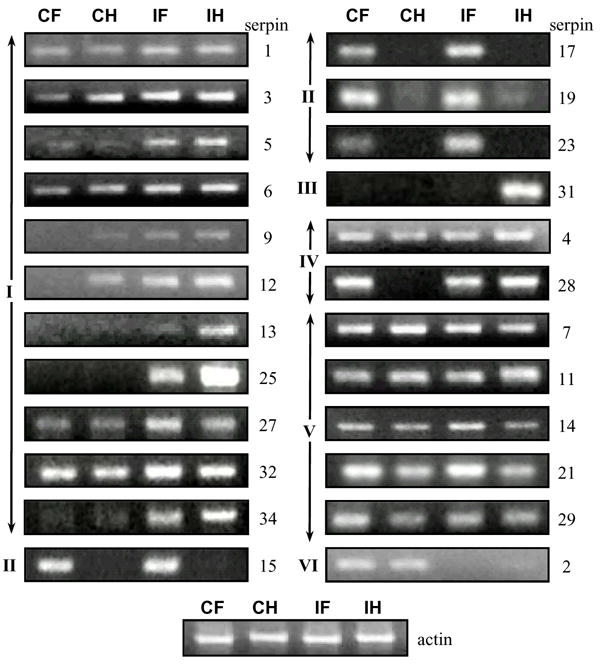
RNA samples from naïve or bacteria-injected larvae were prepared for cDNA synthesis and PCR amplification using gene-specific oligonucleotide primers (Table S1). Except for serpin25 (50 cycles) and serpin29 (45 cycles), reaction mixtures from 35 cycles of amplification were analyzed by agarose gel electrophoresis. The diluted cDNA samples were normalized with B. mori actin as shown at the bottom. No product was detected for serpin8, 11, 16, 18, 20, 22, 24, 25, 26, or 30. CH and IH, control and induced hemocytes; CF and IF, control and induced fat body.
Due to the close resemblance of several members in group F (i.e. serpin15-17 cDNAs: 92% identical, serpin16-18-22 cDNAs: 92–95% identical, and serpin19-23 cDNAs: 89% identical), we tried to amplify the PCR products for serpin14 through 32 by nested PCR and sequenced the products from the second PCR (Table S2). Detection of serpin14~25, 27~29, 31~34 sequences confirmed that the predicted genes are actively transcribed, even though the expression levels were very low for serpin16, 18, 20, 22, 24, 25, 29 and 33. ESTs for serpin 21, 22, 25~27, 29, 31~34 sequences have not previously been identified by the EST project.
Discussion
Serpins are a superfamily of proteins evolving at unusually high rates, especially in the RSL region [40, 41]. Although sequence identities among some of its members are lower than 20%, the domain size and overall folding remain largely unchanged in the long history of evolution [4, 42]. With its members spread over each domain of living organisms as well as some viruses, many ways of structural diversification have been selected to fulfill specific biological functions. Consequently, several studies have explored serpin evolution at the level of major taxonomic groups including prokaryotes, archaea, plants, arthropods, and mammals [16, 42–44]. In this study, we have focused on the serpins in a lepidopteran insect, B. mori. Through data mining, sequence confirmation, expression profiling, and phylogenetic analysis, we have gained insights into the gene architectures, protein structures, and potential biochemical functions of the silkworm serpins. Such fundamental information is useful for understanding the evolutionary dynamics of insect serpins and will guide our functional analyses in this and other closely related species.
We have found that serpin gene duplication has occurred quite extensively in the silkworm and other insects to generate new functions. The rapid expansion of group F is a good example for this mechanism (Fig. 3), where successive gene duplications occurred at multiple genomic locations and resulted in large gene clusters. The group F members account for nearly one third of the entire gene family in the silkworm genome. Similarly, a major family expansion gave rise to eighteen serpin genes in Drosophila [29]. An extreme case was uncovered in the red flour beetle genome: as many as sixteen serpin genes constitute a large cluster in a 50 kb region [33]. While the reactive site loops in group F serpins lack the characteristic features of inhibitory serpins [37], it is worth testing their biological roles by genetic and biochemical studies. Some serpins do not function as protease inhibitors, and others do inhibit proteases even though the conserved features are missing [15, 40]. Functional assays may provide justifications for the existence of a large number of closely related serpin genes in the insects.
Apart from gene duplication, diversity of serpins can also be generated through alternative exon splicing [45, 46]. This mechanism, first discovered in M. sexta [47], represents a parsimonious way of producing variations in RSL sequence and inhibitory selectivity. This phenomenon appears to be quite prevalent in invertebrates. In this work, we provide evidence that this mechanism is not limited to serpin-1 and its orthologs in other insect species. Other group A members (e.g. serpin2 and 28) also have the potential to generate multiple transcripts from the same pre-mRNA. Alternative splicing gives rise to differences in RSL, signal peptide, and untranslated regions.
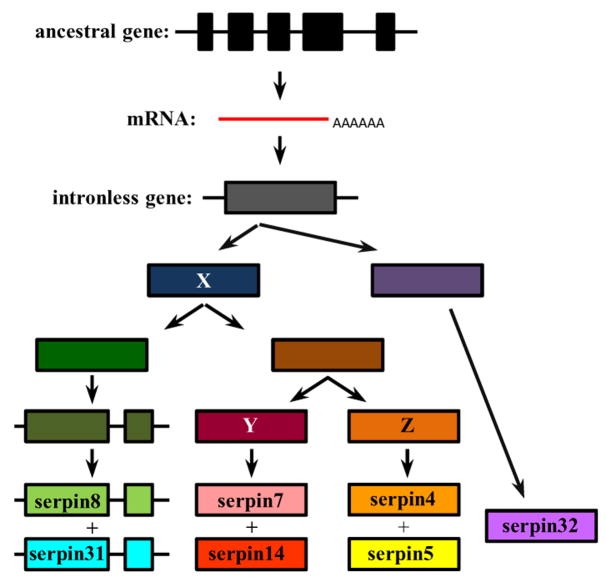
The intron positions can be highly informative about the process of serpin gene family evolution [48]. In group C, serpin4, 5, 7, 14, and 32 genes do not contain any intron, whereas serpin8 and 31 have one located at the same position (Fig. 1 and Fig. 3). One possible evolutionary pathway is that there was an intron loss during the evolution of these genes: the ancestor of serpin8 and 31 genes contain a single intron and, after duplication, one copy gave rise to serpin8 and 31 genes while the other copy lost the intron and was the precursor of the intron-less genes of serpin4, 5, 7 and 14 through two rounds of duplication. An alternative explanation is that the parental gene of serpin8 and 31 gained an intron after it diverged from the intron-less ancestor of serpin4, 5, 7 and 14 genes. The second process is more plausible since: 1) other serpin genes in the genome (except for serpin32) contain at least six exons (and, therefore, it is much more straightforward to produce a common ancestor of group C by losing all the introns in a single step through reverse transcription and transposition of processed mRNA, perhaps); 2) the intron position in serpin8 and 31 is at least 40 bp away from the conserved intron positions 6 and 7 in other serpins (Fig. 1) and, therefore, the single intron in serpin8 and 31 genes did not originate from ancestors of those genes; 3) the gene of B. mori serpin32, which is the closest outgroup for group C, does not contain any intron. In light of this phylogenetic analysis, we can now reconstruct the evolutionary steps of these seven genes (Fig. 5): 1) a complete loss of introns by retro-transposition generated the common ancestor of group C; 2) duplication of the common ancestor produced serpin32 and an intermediate gene X on chromosome 28; 3) after duplication of X, one copy acquired a novel intron and the other duplicated again to yield intermediate genes Y and Z; 4) the two-exon gene gave rise to single-intron genes of serpin8 and 31 next to each other on nscaf3099; 5) gene Y duplicated to form two adjacent genes (serpin7 and 14) on a different region of nscaf3099 whereas gene Z produced the serpin4-5 pair on nscaf3098. The serpin4 gene is next to serpin32 and is two genes away from serpin5 on nscaf3098.
Fig. 5. Evolutionary pathway of B. mori serpin genes in group C.
Exons (colored bars) and introns (thin lines) indicate the intron loss or acquisition during evolution. Duplicated genes and their closest common ancestor are labeled with similar colors. Increase in color brightness represents the continuous change of group C from an ancient intron-rich gene to two single-intron genes (serpin8 and 31) and five single-exon genes (serpin4, 5, 7, 14 and 32).
Serpin1, 2, 21, 28 (group A members) and 9 genes contain seven introns located at all seven conserved positions in their coding regions (Fig. 1). Based on this evidence, we include serpin 9 in group A and consider the common ancestor of this group as an extracellular serpin. Interestingly, nearly all of the group F members contain a signal peptide for secretion and their genes have seven introns present at the same sites and phases in the coding sequences. This conserved exon-intron organization indicates that groups A and F may have a common origin. The architectures of serpin10, 26, 27, 29 and 34 genes are significantly different from the group F members (Fig. 1), even though the bootstrap value is 771 at node 7 (Fig. 3). Based on the resemblance in gene structure, we place serpin32 in group C and serpin10~12 in group B. Serpin3 and 12 genes have five introns located at the same positions, three of which are shared by serpin11 and 10 (Fig. 1).
From the alignment of RSL sequences (Fig. 2), we predict that thirteen of the silkworm serpins (1~7, 11~13, 21, 28 and 32) function as protease inhibitors. Combined with results from the sequence comparison and expression studies, we propose that some of these serpins regulate extracellular SPs that mediate innate immune responses (e.g. proPO activation and spätzle processing) and embryonic development (Toll pathway activation). The expansion of group F seems to suggest that these serpins play important physiological roles in the silkworm. However, since the conserved features for serpins to inhibit proteases are missing, such functions may not be related to protease inhibition.
The presence of multiple serpins provides possible ways of regulation in metazoan development and defense. While many studies have already yielded useful genetic and biochemical data on their biological functions in insects, limited information is available regarding serpin gene structure and evolution at the genome level. We have annotated the B. mori serpins and compared their sequences with orthologs from other insects. Multiple sequence alignment, in conjunction with exon-intron organizations, allows us to understand the molecular mechanisms underlying the family expansion and functional diversification. The information framework established in this work may facilitate biochemical investigations of insect serpins, which complement and stimulate the genetic analyses in the fruitfly and mosquitoes in a scenario of pathogen-host interaction.
Supplementary Material
Acknowledgments
We greatly appreciate our colleagues in the Institute of Sericulture and Systems Biology at Southwest University in China for the initial gene prediction, scaffold, contig and EST sequences, and other useful information provided at SilkDB. Dr. Jun Ishibashi at National Institute of Agrobiological Sciences in Japan kindly provided unpublished sequences of serpin20, 25, 33 and 34 which allowed us to improve our dataset. We would also like to thank Drs. Michael Kanost, Ulrich Melcher, and Jack Dillwith for their helpful comments on the manuscript. This work was supported by the National Institutes of Health Grants GM58634 (to H.J). The article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and supported in part under project OKLO2450.
Footnotes
The nucleotide sequences reported in this paper have been submitted to the GenBank™/EBI Data Bank with accession numbers AY566164~5, EU935610~34, FJ183804~5.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27:67–74. doi: 10.1016/s0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- 2.Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 3.Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;10:1845–1864. doi: 10.1101/gr.gr-1478r. [DOI] [PubMed] [Google Scholar]
- 4.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 5.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki T, Kobayashi K. Isolation of two novel proteinase inhibitors from hemolymph of silkworm larva, Bombyx mori. J Biochem. 1984;95:1009–1117. doi: 10.1093/oxfordjournals.jbchem.a134688. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki T. Patchwork-structure serpins from silkworm (Bombyx mori) larval hemolymph. Eur J Biochem. 1991;202:255–261. doi: 10.1111/j.1432-1033.1991.tb16370.x. [DOI] [PubMed] [Google Scholar]
- 8.Kanost MR, Prasad SV, Wells MA. Primary structure of a member of the serpin superfamily of proteinase inhibitors from an insect, Manduca sexta. J Biol Chem. 1989;264:965–972. [PubMed] [Google Scholar]
- 9.Jiang H, Kanost MR. Characterization and functional analysis of 12 naturally occurring reactive site variants of serpin-1 from Manduca sexta. J Biol Chem. 1997;272:1082–1087. doi: 10.1074/jbc.272.2.1082. [DOI] [PubMed] [Google Scholar]
- 10.Gan H, Wang Y, Jiang H, Mita K, Kanost MR. A bacteria-induced, intracellular serpin in granular hemocytes of Manduca sexta. Insect Biochem Mol Biol. 2001;31:887–898. doi: 10.1016/s0965-1748(01)00034-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y, Wang Y, Gorman MJ, Jiang H, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J Biol Chem. 2003;278:46556–46564. doi: 10.1074/jbc.M309682200. [DOI] [PubMed] [Google Scholar]
- 12.Tong Y, Jiang H, Kanost MR. Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activation pathway. J Biol Chem. 2005;280:14932–14942. doi: 10.1074/jbc.M500532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Jiang H. Purification and characterization of Manduca sexta serpin-6: a serine proteinase inhibitor that selectively inhibits prophenoloxidase-activating proteinase-3. Insect Biochem Mol Biol. 2004;34:387–395. doi: 10.1016/j.ibmb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Cherqui A, Cruz N, Simoes N. Purification and characterization of two serine protease inhibitors from the hemolymph of Mythimna unipuncta. Insect Biochem Mol Biol. 2001;31:761–769. doi: 10.1016/s0965-1748(00)00172-7. [DOI] [PubMed] [Google Scholar]
- 15.Stark KR, James AA. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito, Aedes aegypti. J Biol Chem. 1998;273:20802–20809. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- 16.Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 17.Jiang H. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Sci. 2008;15:53–66. [Google Scholar]
- 18.Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost MR. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- 19.Zou Z, Jiang H. Manduca sexta serpin-6 regulates immune serine proteinases PAP-3 and HP8. cDNA cloning, protein expression, inhibition kinetics, and function elucidation. J Biol Chem. 2005;280:14341–14348. doi: 10.1074/jbc.M500570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levashina EA, et al. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- 21.De Gregorio E, et al. An immune-responsive serpin regulates the melanization cascade in Drosophila. Dev Cell. 2002;3:581–592. doi: 10.1016/s1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- 22.Danielli A, Kafatos FC, Loukeris TG. Cloning and characterization of four Anopheles gambiae serpin isoforms, differentially induced in the midgut by Plasmodium berghei invasion. J Biol Chem. 2003;278:4184–4193. doi: 10.1074/jbc.M208187200. [DOI] [PubMed] [Google Scholar]
- 23.Abraham EG, et al. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc Natl Acad Sci USA. 2005;102:16327–16332. doi: 10.1073/pnas.0508335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel K, et al. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc Natl Acad Sci USA. 2006;103:16858–16863. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto C, Kim DR, Weiss LA, Miller JW, Morisato D. Spatial regulation of developmental signaling by a serpin. Dev Cell. 2003;5:945–950. doi: 10.1016/s1534-5807(03)00338-1. [DOI] [PubMed] [Google Scholar]
- 26.Richer MJ, Keays CA, Waterhouse J, Minhas J, Hashimoto C, Jean F. The Spn4 gene of Drosophila encodes a potent furin-directed secretory pathway serpin. Proc Natl Acad Sci USA. 2004;101:10560–10565. doi: 10.1073/pnas.0401406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danielli A, Barillas-Mury C, Kumar S, Kafatos FC, Loukeris TG. Overexpression and altered nucleocytoplasmic distribution of Anopheles ovalbumin-like SRPN10 serpins in Plasmodium-infected midgut cells. Cell Microbiol. 2005;7:181–190. doi: 10.1111/j.1462-5822.2004.00445.x. [DOI] [PubMed] [Google Scholar]
- 28.Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6:891–897. doi: 10.1038/sj.embor.7400478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reichhart JM. Tip of another iceberg: Drosophila serpins. Trends Cell Biol. 2005;15:659–665. doi: 10.1016/j.tcb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Christophides GK, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 31.Waterhouse RM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou Z, Lopez DL, Kanost MR, Evans JD, Jiang H. Comparative analysis of serine protease-related genes in the honey bee genome: possible involvement in embryonic development and innate immunity. Insect Mol Biol. 2006;15:603–614. doi: 10.1111/j.1365-2583.2006.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou Z, et al. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia Q, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 35.Mita K, et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopkins PC, Carrell RW, Stone SR. Effects of mutations in the hinge region of serpins. Biochemistry. 1993;32:7650–7657. doi: 10.1021/bi00081a008. [DOI] [PubMed] [Google Scholar]
- 38.Ragg H. The role of serpins in the surveillance of the secretory pathway. Cell Mol Life Sci. 2007;64:2763–2770. doi: 10.1007/s00018-007-7157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou Y, et al. Studies on middle and posterior silk glands of silkworm (Bombyx mori) using two-dimensional electrophoresis and mass spectrometry. Insect Biochem Mol Biol. 2007;37:486–496. doi: 10.1016/j.ibmb.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 40.van Gent D, Sharp P, Morgan K, Kalsheker N. Serpins: structure, function and molecular evolution. Int J Biochem Cell Biol. 2003;35:1536–1547. doi: 10.1016/s1357-2725(03)00134-1. [DOI] [PubMed] [Google Scholar]
- 41.Irving JA, Askew DJ, Whisstock JC. Computational analysis of evolution and conservation in a protein superfamily. Nature Methods. 2004;32:73–92. doi: 10.1016/s1046-2023(03)00200-7. [DOI] [PubMed] [Google Scholar]
- 42.Law RH, et al. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts TH, Hejgaard J, Saunders NF, Cavicchioli R, Curmi PM. Serpins in unicellular eukarya, archaea, and bacteria: sequence analysis and evolution. J Mol Evol. 2004;59:437–447. doi: 10.1007/s00239-004-2635-6. [DOI] [PubMed] [Google Scholar]
- 44.Roberts TH, Hejgaard J. Serpins in plants and green algae. Funct Integr Genomics. 2008;8:1–27. doi: 10.1007/s10142-007-0059-2. [DOI] [PubMed] [Google Scholar]
- 45.Krüger O, Ladewig J, Köster K, Ragg H. Widespread occurrence of serpin genes with multiple reactive centre-containing exon cassettes in insects and nematodes. Gene. 2002;293:97–105. doi: 10.1016/s0378-1119(02)00697-2. [DOI] [PubMed] [Google Scholar]
- 46.Hegedus DD, Erlandson M, Baldwin D, Hou X, Chamankhah M. Differential expansion and evolution of the exon family encoding the serpin-1 reactive centre loop has resulted in divergent serpin repertoires among the Lepidoptera. Gene. 2008;418:15–21. doi: 10.1016/j.gene.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Jiang H, et al. Organization of serpin gene-1 from Manduca sexta. Evolution of a family of alternate exons encoding the reactive site loop. J Biol Chem. 1996;271:28017–28023. doi: 10.1074/jbc.271.45.28017. [DOI] [PubMed] [Google Scholar]
- 48.Kaiserman D, Bird PI. Analysis of vertebrate genomes suggests a new model for clade B serpin evolution. BMC Genomics. 2005;6:167. doi: 10.1186/1471-2164-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Wang Z, Canagarajah B, Jiang H, Kanost MR, Goldsmith EJ. The structure of active serpin 1K from Manduca sexta. Structure. 1999;7:103–109. doi: 10.1016/s0969-2126(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 50.Krem MM, Di Cera E. Conserved Ser residues, the shutter region, and speciation in serpin evolution. J Biol Chem. 2003;278:37810–37814. doi: 10.1074/jbc.M305088200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.