Summary
Hexameric helicases couple ATP hydrolysis to processive separation of nucleic acid duplexes, a process critical for gene expression, DNA replication and repair. All hexameric helicases fall into two families with opposing translocation polarities: the 3’→5’ AAA+ and 5’→3’ RecA-like enzymes. To understand how a RecA-like hexameric helicase engages and translocates along substrate, we determined the structure of the E. coli Rho transcription termination factor bound to RNA and nucleotide. Interior nucleic-acid binding elements spiral around six bases of RNA in a manner unexpectedly reminiscent of an AAA+ helicase, the papillomavirus E1 protein. Four distinct ATP-binding states, representing potential catalytic intermediates, are coupled to RNA positioning through a complex allosteric network. Comparative studies with E1 suggest that RecA and AAA+ hexameric helicases use different portions of their chemomechanical cycle for translocating nucleic acid, and track in opposite directions by reversing the firing order of ATPase sites around the hexameric ring.
Introduction
Oligomeric, ring-shaped ATPases are responsible for driving vital processes ranging from protein and nucleic acid metabolism to cellular homeostasis and energy production. A subset of these motors, the hexameric single-stranded nucleic acid helicases and translocases, are responsible for essential DNA and RNA rearrangements in all cells and many viruses. Using ATP to move along nucleic acid strands, these enzymes help direct numerous cellular events, including genome packaging, DNA replication and repair, and transcriptional regulation (Delagoutte and von Hippel, 2003; Patel and Picha, 2000; Singleton et al., 2007). To date, two major families of hexameric helicases have been identified (Berger, 2008; Iyer et al., 2004). Eukaryotes, eukaryotic viruses and archaea rely predominantly on one group, the ATPases Associated with various cellular Activities (AAA+) enzymes, which includes superfamily 3 (SF3) helicases and MCM proteins that translocate 3’→5’ along substrates. By contrast, bacteria and many of their phages typically utilize RecA-type helicases such as DnaB and T7gp4, which move in the 5’→3’ direction. Although the quaternary assemblies of RecA and AAA+ hexameric motors display distinct subunit orientations (Wang, 2004) (Figure S1), members of both families belong to the Additional Strand Conserved glutamatE (ASCE), P-loop ATPase super-group, and are linked by a common structural core (Leipe et al., 2003). The hexameric assembly of ASCE folds typically places the ATP binding motifs of one subunit near γ-phosphate sensor elements of an adjacent subunit, resulting in a radial array of bipartite active sites that cooperatively catalyze ATP hydrolysis.
The bacterial Rho factor is a conserved, RecA-family hexameric helicase that selectively terminates transcription at discrete genomic loci to control gene expression (Richardson, 2002). Rho loads onto nascent RNA strands at rut (Rho utilization) sites (Chen and Richardson, 1987), using a primary, cytosine-specific RNA-binding activity (Dolan et al., 1990; Dombroski and Platt, 1988). Loading permits RNA to interact with a secondary binding site in the central channel of the hexamer (Wei and Richardson, 2001), stimulating an RNA-dependent ATPase activity (Lowery-Goldhammer and Richardson, 1974). Coupling of ATP turnover to secondary-site binding is thought to translocate Rho 5’→3’ along the RNA (Brennan et al., 1987), toward a transcribing polymerase. Upon reaching the transcription complex, Rho may forcibly dissociate a paused RNA polymerase (Lau et al., 1983; Morgan et al., 1983) either by using its motor activity to separate the RNA-DNA heteroduplex (Brennan et al., 1987), or by pushing the polymerase forward to collapse the transcription bubble (Park and Roberts, 2006).
How Rho, and indeed hexameric motor proteins in general, coordinate ATP turnover between six catalytic centers to the processive translocation of a polymeric substrate remains an outstanding question. Crystallographic and biochemical studies have variously supported or weighed against three primary chemomechanical coupling models for these enzymes (Singleton et al., 2007), including: 1) a rotary mechanism in which individual ATP hydrolysis events proceed sequentially around the hexameric ring, 2) a concerted mechanism in which all active sites hydrolyze ATP simultaneously, and 3) a stochastic model whereby any ATPase site can hydrolyze nucleotide at random. Thus far, a paucity of structural data for fully-bound enzyme/substrate complexes has left the debate surrounding these mechanistic frameworks unresolved. However, an SF3 helicase, the papillomavirus E1 protein, has been imaged in a configuration akin to a catalytic-like state with both nucleotide and a single-stranded DNA (ssDNA) substrate (Enemark and Joshua-Tor, 2006). The structure revealed that E1, an AAA+ enzyme, binds DNA in the interior of its hexameric ring through a helical arrangement of hairpin-loops, and suggested that a sequential ATPase mechanism is used to propel the motor 3’→5’ along substrate with a net step size of one ATP/base. Whether this mechanism is preserved in other hexameric engines, such as RecA-type enzymes, has remained unclear. The molecular basis for differing substrate specificities (e.g. DNA or RNA) and translocation polarities between different motor subfamilies is similarly unknown.
To examine these issues, we determined the structure of a Rho hexamer bound to the ATP mimic ADP•BeF3 and a centrally-bound RNA oligonucleotide. Six bases of RNA are coordinated by a spiral staircase of loops that form contacts with the nucleic acid backbone. RNA binding coincides with the formation of an asymmetric particle that contains four distinct classes of ATP-binding sites, which together appear to recapitulate different catalytic states consistent with a sequential ATP hydrolysis mechanism. Comparison of nucleic-acid bound Rho and E1 hexamers reveals that the two motors bind nucleic acid substrates in a similar conformation and with the same relative polarity (Enemark and Joshua-Tor, 2006), but that different ATPase states are responsible for DNA or RNA binding. These features suggest that Rho and E1 translocate in opposite directions because the sequential firing orders of their respective ATPase sites may be reversed.
Results and Discussion
Structure solution
Initial crystals of full-length E. coli Rho dependent on RNA, ADP•BeF3, and Mg2+ ions were obtained in the spacegroup P6. Molecular replacement solutions revealed that the unit cell contained a six-fold and three-fold symmetric hexamer, both of which obscured the RNA bound in the center of the ring, a situation seen previously for this protein (Skordalakes and Berger, 2006). Inspection of some P6 crystals revealed twinning with a secondary pseudo-P6 lattice; optimization of both growth conditions and data collection strategies permitted the collection of datasets from P1 crystals with a single hexamer in the asymmetric unit (Supplemental Data) (Figure 1A–B). Initial phases obtained by molecular replacement revealed clear electron density for RNA and nucleotide bound to the protein (Figure 1C–D). Model building and refinement produced a final structure with good stereochemistry and an Rwork/Rfree of 27.0/29.5% at 2.8 Å resolution (Table 1).
Figure 1. Rho/RNA/ADP•BeF3•Mg2+ structure.
(A) Top down view of the Rho hexamer with differential coloring of the six subunits (see color key). Bound RNA is shown as orange sticks. ADP, BeF3 and Mg2+ are shown as magenta sticks, black sticks, and yellow-green spheres respectively.
(B) Side view of Rho rotated 90° with respect to (A), and shown with a transparent surface. Subunits A and B have been removed to highlight the RNA.
(C) Representative Fo−Fc electron density in the ATPase active site calculated prior to including nucleotide in the model (3 σ contouring). The refined ADP•BeF3•Mg2+ model is shown to highlight the starting map quality.
(D) Representative Fo−Fc electron density (green) for RNA calculated prior to including the nucleic acid substrate in the model (left, 2.5 σ contouring), and refined 2Fo−Fc electron density (blue) for the final model (right, 1.25 σ contouring). The refined RNA model is shown in orange with oxygen and nitrogen atoms colored red and blue, respectively.
Table 1.
Data Collection and Refinement
| Data | ||
|---|---|---|
| Space Group | P1 | |
| Unit Cell | a=69.23 b=127.0 c=127.2 α=60.48 β=90.26 γ=89.77 |
|
| Wavelength (Å) | 1.11 | |
| Resolution (Å) | 42 – 2.8 | |
| Unique Reflections | 87,859 | |
| Redundancy | 3.8 (3.6) | |
| Completeness (%) | 94.6 (66.6)a | |
| I/σ | 15.5 (3.8) | |
| Rmerge | 0.079 (0.342) | |
| Refinement | ||
| Rwork (%) | 27.0b | |
| Rfree (%) | 29.5c | |
| RMSD bonds (Å) | 0.013 | |
| RMSD angles (°) | 0.970 | |
| Ramachandrand | Preferred (%) | 94.4 |
| Allowed (%) | 5.6 | |
| Outliers (%) | 0 | |
| Number of Atoms | 19,558 | |
| Protein | 19,141 | |
| Ligands | 333 | |
| Water | 84 | |
Values in parenthesis correspond to the high resolution bin of 2.9–2.8 Å
Data is > 90% complete in all resolution bins to 2.9 Å
Rwork =Σ||Fo|-|Fc||/ |Fo|
Rfree is calculated using 5% of the data omitted from refinement
As reported by Molprobity (Davis et al., 2007)
RNA is absent from Rho’s primary binding site
In contrast to previous structures (Skordalakes and Berger, 2003, 2006), there was no evidence of RNA associated with Rho’s primary binding site located in the N-terminal oligonucleotide-binding (OB) folds of the protein. This absence may result from the sub-stoichiometric ratio of RNA to Rho monomer used during crystallization. Although this observation does not contradict the “tethered tracking” model (Steinmetz and Platt, 1994), a scheme for Rho function in which a portion of RNA is thought to remain associated with the primary RNA binding site during translocation, it does suggest that Rho can attain a closed-ring conformation in the absence of RNA/primary-site interactions.
Protein-RNA contacts in the secondary binding site
The secondary RNA-binding site in Rho is composed of two elements, the Q and R loops (Miwa et al., 1995), which are structurally analogous to the L1 and L2 loops of other RecA-family members (Story et al., 1992). In the Rho structure, both motifs line the channel that passes through the center of the ring, associating with six bases of single-stranded RNA. The polarity of bound nucleic acid places the 5’-end proximal to the N-terminal domain of Rho, an orientation consistent with data for Rho and other RecA-family hexameric helicases (Egelman et al., 1995; Jezewska et al., 1998a; Jezewska et al., 1998b; Lo et al., 2008; Skordalakes and Berger, 2003). In contrast to the non-hexameric superfamily 1A (SF1A) helicases (Velankar et al., 1999), but more similar to SF1B and SF2 enzymes (Singleton et al., 2007) (Saikrishnan et al., 2009), Rho appears to almost exclusively bind the phosphodiester backbone of nucleic acid, making only minimal contacts with the bases.
The Q/R loops of subunits A–E form a spiral staircase that tracks along the RNA backbone (Figure 2A–B), while subunit F, which transitions from the top to the bottom of the staircase to close the ring, makes only limited contact with a single RNA base. All nucleic acid-binding loops are visible in the electron density, although the Q loop from chain F was poorly resolved and hence modeled as poly-alanine. The RNA makes a complete turn in six bases to interact with these binding loops, giving rise to a stoichiometry of one base/subunit. Compared to idealized A-form RNA, this configuration sharply increases the average helical twist of the RNA by ~20°/base, while decreasing the average helical rise by ~0.6 Å/base (Figure S2). This manner of binding RNA is reminiscent of the DNA binding configuration seen in E1 (Enemark and Joshua-Tor, 2006) (Figure 2A), suggesting that hexameric RecA and AAA+ helicases may have converged upon a common means for engaging nucleic acid substrates, despite pronounced structural differences between the two enzymes (Figure S1) (Wang, 2004).
Figure 2. Translocation loop/RNA interactions.
(A) The Rho Q loops (left) and E1 β-hairpin loops (right) bind the nucleic acid backbone in a spiral staircase configuration. Both sets of nucleic acid binding loops are structured in a manner suggesting that they pull (arrows), rather than push, on the substrate to achieve a particular translocation polarity (5’→3’ for Rho vs. 3’→5’ for E1). The Rho Q loops for subunits A–E are shown (see key color key, panel B); subunit F, which does not contact the RNA, is removed for clarity. The E1 loops are colored similarly to those in Rho based on their position in the staircase. Nucleic acids are colored orange and illustrated as sticks on a cartoon backbone. The orientation of the Rho hexamer is similar to that shown in Figure 1B.
(B) The Rho R loops form a spiral staircase that engages the RNA phosphates. The R loops for all six subunit are colored (see key). The 5’ end of the RNA (orange and red sticks) projects out, toward the viewer. The side-chains of K326 (sticks), project into the center of the ring and interact with the RNA phosphate groups.
(C) Specific RNA contacts with the Q and R loops of subunits B and C (see color key, panel B). Backbone atoms and side-chains for the RNA-binding residues are shown as sticks. Dashed lines indicate hydrogen-bonds or salt bridges.
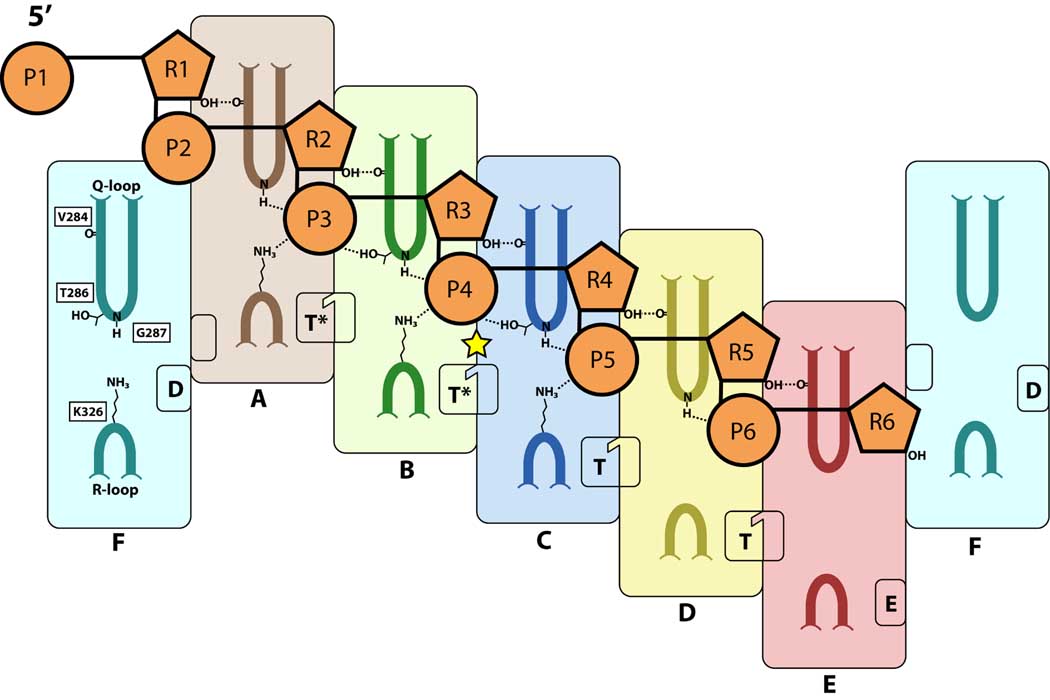
Except for chain F, each Q loop makes up to three contacts with RNA through a highly conserved set of amino acids (Figure 2C). A backbone carbonyl from V284A–E (where the subscript indicates the hexamer subunit) hydrogen bonds with the 2’-OH of RNA ribose1–5 (where the subscript indicates the RNA nucleotide), likely accounting for the specificity of Rho for RNA substrates (Lowery-Goldhammer and Richardson, 1974). A tight turn in the Q loop occurs at the location of two adjacent glycines (G287 and G288), allowing the backbone amide of G287A–D to hydrogen bond with RNA phosphates3–6. A final set of contacts arises between phosphates3–4 and the hydroxyl of T286B–C. Overall, these contacts cause the Q loops to drape down and hook around the 3’-side of the RNA phosphodiester backbone, where they engage the 2’-OH of the ribose and the O1P atom of the phosphate group. Interestingly, a similar “loop and hook” feature is used by the E1 helicase, albeit with translocation loops wrapping over the 5’-edge of the phosphodiester backbone to make polar contacts with the O1P atom of the phosphate group and van der Waals contacts with the deoxyribose (Enemark and Joshua-Tor, 2006). Both substrate-binding mechanisms are consistent with a need to pull on the nucleic acid substrate in a particular direction, 5’→3’ for Rho and 3’→5’ for E1 (Figure 2A). Given that the nucleic acid binding loops of Rho and E1 are unrelated in sequence and project from distinct portions of the core ASCE fold, this arrangement may represent yet another convergent nucleic-acid binding approach between hexameric RecA-like and AAA+ enzymes.
The R loops make a single contact to the nucleic acid substrate through a nearly invariant basic residue, K326A–C, which bonds with RNA phosphates3–5 (Figure 2C). K326D,F also projects into the center of the ring and appears poised to interact with nearby phosphates. Together, these lysines form a set of positively-charged steps in the protein staircase. The only R-loop lysine that is not positioned to interact with RNA, K326E, lies at the bottom of the staircase and points down and away from the pore, creating a groove that allows the incoming 3’ base of RNA to enter Rho’s central channel (Figure 2B).
Structural asymmetry generates four different ATP binding states
Just as the central RNA-binding site is composed of a unique set of asymmetric contacts within the hexamer, the ATPase sites of Rho also are non-uniform (Figure 3). Although ADP and BeF3 are bound to all six catalytic centers, the active sites partition into four distinct states. Several criteria distinguish these states, such as: 1) the spatial separation between subunits at the active site interface, 2) relative nucleotide temperature-factors (which are consistent with the observation of three tight and three weak ATP binding sites in Rho (Geiselmann and von Hippel, 1992; Xu et al., 2003)) (Figure S3), 3) the placement of catalytic groups and water molecules with respect to ADP•BeF3•Mg2+ (Figure S4 and Figure S5), and 4) the conformation of the ADP•BeF3 moiety. Using these characteristics as a metric, the sites appear to reflect a set of prospective catalytic intermediates, including a nucleotide exchange (E) state, an ATP-bound (T) state, a hydrolysis-competent (T*) state, and a product (D) state (see below). Structural comparisons with the F1-ATPase (Abrahams et al., 1994), a well-studied hexameric ATPase with ~40% sequence similarity to Rho (Dombroski and Platt, 1988), further support these active site assignments.
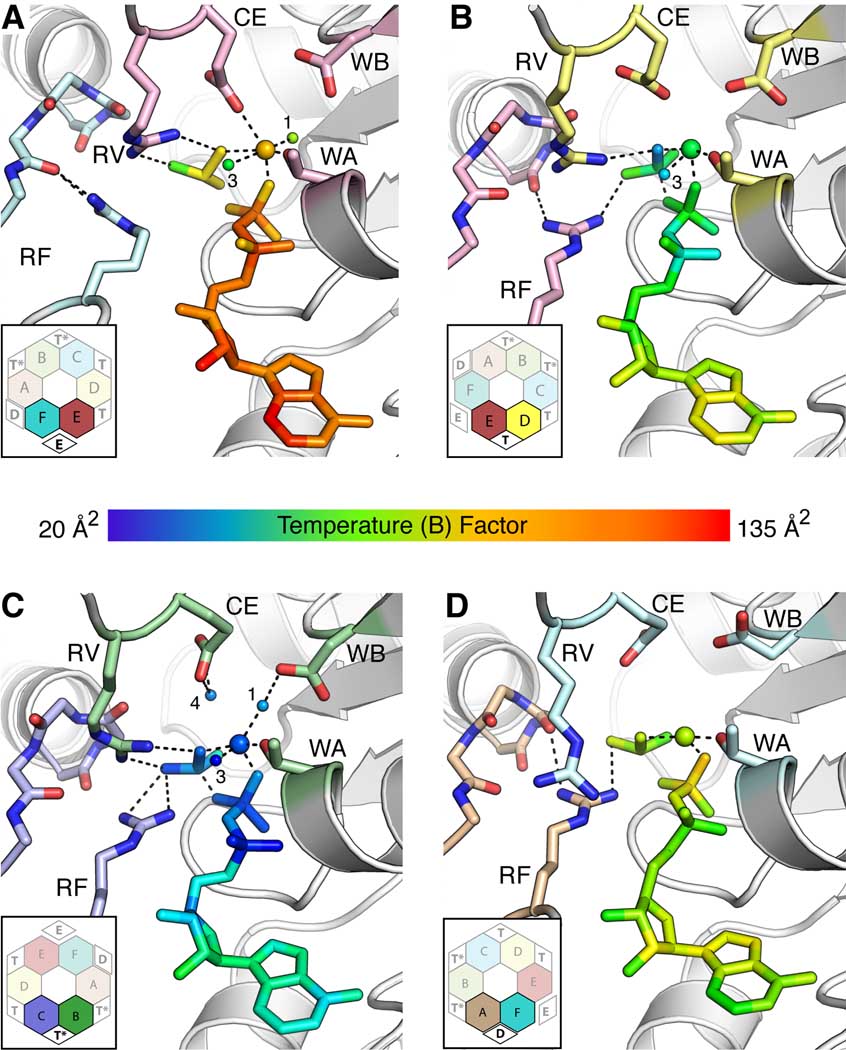
Figure 3. Structural asymmetry generates four ATP binding states.
Views of (A) Exchange - E; (B) ATP - T; (C) ATP hydrolysis - T*; (D) ADP - D active sites. Each inset shows the interface location (bold and black text) with respect to the rest of the hexamer (faded and grey text). For nucleotide and associated Mg2+/water molecules, coloring is by B-factor, with blue indicating low values (20–35 Å2) and red high values (120–135 Å2). By contrast, selected catalytic motifs are shown in stick representation and colored by subunit in accordance with Figure 1A. ADP and BeF3 are shown as sticks. Bound Mg2+ and associated water molecules are shown as large and small spheres, respectively; the water molecules are numbered according to their position in the active site. Bonding interactions are shown as dashed lines. Abbreviations: WA – Walker A, WB – Walker B, CE – Catalytic Glutamate, RV – Arginine Valve, RF – Arginine Finger.
The widest opening between adjacent subunits occurs at the E/F protomer interface, which buries substantially less surface area compared to the other five interfaces (~2500 Å2 vs. ~3200–3600 Å2). As a result of this conformation, a number of catalytic residues exhibit atypical conformations compared to other subunits (Figure 3A). The arginine finger (RF), R366F, is flipped away from the nucleotide bound to the P-loop of chain E, and instead hydrogen bonds to a backbone carbonyl group in its own polypeptide chain (G339F). The catalytic glutamate (CE), E211E, which typically coordinates a nucleophilic water molecule, instead directly ligands the Mg2+ ion bound in the active site. Walker-B residue D265E, which generally interacts with Mg2+ via a water molecule, does not appear to bind the metal-nucleotide complex. Although ADP•BeF3 is present, it exhibits atomic B-factors that are significantly higher than those seen for nucleotides in the other five ATP-binding sites, and the BeF3 group does not bond optimally with ADP. Together, these factors suggest that the E/F interface is in an exchange-like (E) state that is either preparing to release product or has just bound an incoming nucleotide.
Proceeding counterclockwise around the ring, the intersubunit distance in successive ATPase sites decreases significantly, the B-factors of the bound ADP•BeF3 drop, and the ADP•BeF3 complex becomes optimally coordinated through the additive binding of catalytic residues (Figure 3). In the D/E subunit interface, the NH2 group of the arginine finger moves into the active site to interact with the F1 atom of bound BeF3, and the catalytic glutamate begins to pull away from the Mg2+ ion and move into a cavity behind the BeF3 (Figure 3B). In the C/D interface, the catalytic glutamate moves further into the cavity while the Walker-B residue shifts to coordinate Mg2+ through a water molecule. In the B/C and A/B subunit interfaces, the NH1 group of the arginine finger forms an additional bond to the F1 atom of BeF3 (with the NH1 group simultaneously breaking a bond with an intra-chain carbonyl as it moves), and a water molecule becomes hydrogen bonded to the catalytic glutamate (Figure 3C). Although not perfectly positioned for an in-line attack on the γ-phosphate mimic (BeF3), this water occupies a position similar to that of the prospective catalytic water in the structure of F1-ATPase bound to ADP•BeF3 (Kagawa et al., 2004) (the arginine finger of F1 shifts similarly between active site states as seen here for Rho). As an ensemble, these structural changes suggest that the D/E and C/D subunit active sites reside in a pre-hydrolysis, ATP-bound (T) state, while the B/C and A/B sites adopt a more hydrolysis competent (T*) conformation.
The fourth state appears at the F/A interface, where several nucleotide-binding residues withdraw from the active site (Figure 3D). The most critical is R212F, referred to hereafter as the “arginine valve” (RV), which breaks its otherwise persistent contact with BeF3. In the catalytic β-subunit of bovine F1, the corresponding arginine (R189) is one of the primary structural determinants between the ATP and ADP states (Abrahams et al., 1994; Enemark and Joshua-Tor, 2008). The Walker-B aspartate also moves out of the F/A subunit active site, disengaging from the Mg2+ ion. These observations, along with the intermediate B-factors of the nucleotide compared to the E and T/T* states, and the sub-optimal ADP•BeF3 geometry, suggest that this site reflects a type of post-hydrolysis (D) state that has not yet released product.
We note that some active sites in the structure exhibit strained or incomplete Mg2+-coordination spheres. Though we cannot rule out the possibility that apparent non-octahedral geometries may be linked to the modest resolution of our structure (2.8Å), the changes are consistent with observed active site rearrangements such as movement of Walker B residue (D265) and the catalytic glutamate (E211). The partial coordination observed in some active sites also may explain why Rho can utilize metals that prefer coordination spheres less than six such as Cd2+, Co2+, Ni2+ and Zn2+ (Weber et al., 2003); however, we note that differential coordination and strain on the scissile phosphate linkage likewise could play a role in catalysis. Such a mechanism has been postulated to occur in other metal-dependent enzymes such as RNase H (Nowotny and Yang, 2006).
RNA binding is coupled to ATPase state
The hexameric ring serves to structurally link RNA binding and ATP turnover by associating each protein subunit with a unique ATP-binding state and RNA coordination geometry (Figure 4). Subunit E, which sits at the bottom of the staircase, and whose active site resides in an exchange-like (E) state, makes only one RNA contact. Moving up the staircase, the T- and T*-state active sites progressively bind ATP more tightly, concurrent with repositioning of the Q and R loops to form additional RNA contacts. Enzyme/nucleic-acid interactions reach a maximum in subunits B and C (each of which makes four hydrogen bonds or salt bridges to the RNA), and then relax slightly to three protein-RNA bonds in subunit A. As subunit F transitions between the top and bottom staircase positions, it disengages from the phosphodiester backbone. The active site of subunit F also appears to occupy a type of post-hydrolysis product (D) state, indicating that the stable formation of ADP+Pi may be linked to RNA release.
Figure 4. Correlation between ATP and RNA binding.
Schematized view of RNA-binding contacts and ATP-binding states. Protein subunits are illustrated as large, rounded rectangles and colored as per Figure 1A. Q and R loops are drawn with darker, colored lines to highlight their positions. The perspective is similar to Figure 1B, except that the subunits are pulled open and spread flat on the page. Subunit F is shown twice to highlight its orientation with respect to subunits A and E. The two halves of the bipartite ATP-binding site are illustrated as small rounded rectangles; linked, notched rectangles represent insertion of the arginine finger into the active site of T and T* states. Ribose (R) and phosphate (P) moieties of the RNA backbone are colored orange and numbered according to the structure. Protein residues contacting RNA are labeled, and chemical groups that bond with the RNA are shown (dashed lines). The yellow star indicates the B/C interface in which adjacent subunits have maximized their protein-RNA contacts.
As a whole, the most tightly-bound ATP states of Rho make the greatest number of contacts with the RNA, whereas the product release and exchange states exhibit no or few interactions. This coupling between ATPase state and RNA binding contrasts with the AAA+ helicase E1, which binds DNA via a combination of ATP (T) and product-like (D) states (Enemark and Joshua-Tor, 2006). Consistent with these observations, the DNA-bound state of T7gp4 (a RecA-type enzyme) is stabilized by ATP binding (Hingorani and Patel, 1993), whereas the DNA-bound state of M. thermoautotrophicus MCM (an AAA+ enzyme) is stabilized by ATP hydrolysis (Sakakibara et al., 2009). Thus, despite general similarities in how nucleic acid segments are bound, the chemomechanical coupling mechanisms of AAA+ and RecA-like hexameric helicases appear to be fundamentally different. The functional consequences of these differences are not known, but may reflect an ability of RecA and AAA+ enzymes to utilize distinct phases of the ATPase cycle, such as ATP binding or Pi release, to power substrate movement (Enemark and Joshua-Tor, 2006).
A dynamic allosteric network positions the catalytic glutamate
Our structure suggests that the linkage between ATP turnover and RNA movement requires significant inter-subunit flexibility (Figure 5A and Movie S1). The structural transition between these states results from inter-subunit clamping motions, which alter the relative positions of adjacent Q/R loops and their associated helices Qα1, Qα2 and Rα1. The extent of the conformational change is highlighted by residues E333 and R347, which together form an inter-subunit salt bridge in the T and T* states, but are separated by ~11 Å in the E state. The structure also reveals conformational changes that take place within each Rho subunit, the most significant of which occur in the Q and R loops (Figure 5B).
Figure 5. Inter- and intra-subunit conformational changes position the catalytic glutamate via a conserved allosteric network.
(A) Inter-subunit conformational changes in Rho. Three different interfaces (F/A, E/F, B/C) are shown. The inset identifies the subunits and active site states represented by each interface. The product-release (D) and exchange (E) states are relatively open (i.e. more accessible to solvent), whereas the ATP bound (T and T*) states (illustrated here by the B/C interface) are closed (sequestered from solvent). These changes alter the relative positions of adjacent Q and R loops and their associated secondary structural elements, Rα1, Qα1 and Qα2, which are labeled and colored by subunit (see key, panel B). Selected residues involved in inter-subunit contacts are labeled and shown as sticks, and bonding interactions as dashed lines.
(B) Intra-subunit conformational changes illustrated by structural superposition of the RecA-folds from all six subunits. The core ASCE fold is colored white, while the Q and R loops, and the Rα1, Qα1 and Qα2 structural elements are colored by subunit (see key). While most of the motor domain shows minimal intra-subunit conformational changes, the RNA binding elements exhibit significant variations.
(C) The position of the allosteric network with respect to RNA (orange cartoon) and the presence of a prospective catalytic water molecule (large red spheres) in the ATPase active site. The Rα1, Qα1 and Qα2 structural elements encircle the RNA binding site and form a large portion of the subunit interface within the hexamer. Charged residues (sticks) project from Rα1, Qα1 and Qα2, forming a network between subunits. Bonding interactions (dashed lines) within the network vary around the hexamer depending on nucleotide state. Protein subunits are differentially colored (see key, panel B).
(D) The completely bonded allosteric network located at the T* interface between subunits B and C. The inter-subunit conformational changes shown in panel A are linked to an interaction between R347 and E333. All members of the network except R347 project from the Rα1, Qα1 and Qα2 structural elements of adjacent subunits and form a complex network of salt bridges that affects the position of the catalytic glutamate (E211B) responsible for activating the nucleophilic water molecule. ADP and BeF3 are colored as magenta and black sticks. Protein side-chains are illustrated as sticks and colored by subunit (see key, panel B). The Mg2+ ion and catalytic water molecule are shown as yellow-green and red spheres, respectively. See also supplemental movie S1 and movie S2.
These intra- and inter-subunit conformational changes not only reposition the RNA binding loops, but also alter the interactions between a set of invariant, charged amino acids projecting primarily from helices Qα1, Qα2 and Rα1. Inter-subunit conformational changes appear to be sensed through the aforementioned R347/E333 salt bridge (Figure 5A). Residues D328, R272, E334, K298 and R269 generate additional inter-subunit contacts that vary between the six protomer interfaces in the hexamer (Figure 5C and Movie S2). This ion-pair network ultimately plugs into the active site through an R269/E211 bond that orients the catalytic glutamate for binding a prospective nucleophilic water molecule. The only active site that contains a completely bonded network lies at the B/C interface (Figure 5D), between the two subunits that together enclose a T* state and that display the largest number of protein-RNA contacts. Thus, the network appears to function as a communication relay that links RNA binding to the proper positioning of catalytic amino acids to support ATP hydrolysis. A wealth of biochemical data supports this premise, showing that a variety of substitutions in the network can indirectly perturb substrate binding and ATP turnover, and further explaining why mutations in residues that directly bind RNA often disrupt ATPase activity and vice-versa (Table S1).
Chemomechanical coupling in a RecA-like hexameric helicase
The question of how ATP turnover is linked to substrate translocation has been one of the most central issues surrounding RecA-family hexameric helicases, and oligomeric motor proteins in general (Delagoutte and von Hippel, 2002; Enemark and Joshua-Tor, 2008; Patel and Picha, 2000; Singleton et al., 2007). Analysis of our present structure leads to a straightforward mechanistic model describing how RNA binding stimulates the ATPase activity of Rho, and how this activity is in turn coupled to RNA movement (Figure 6A).
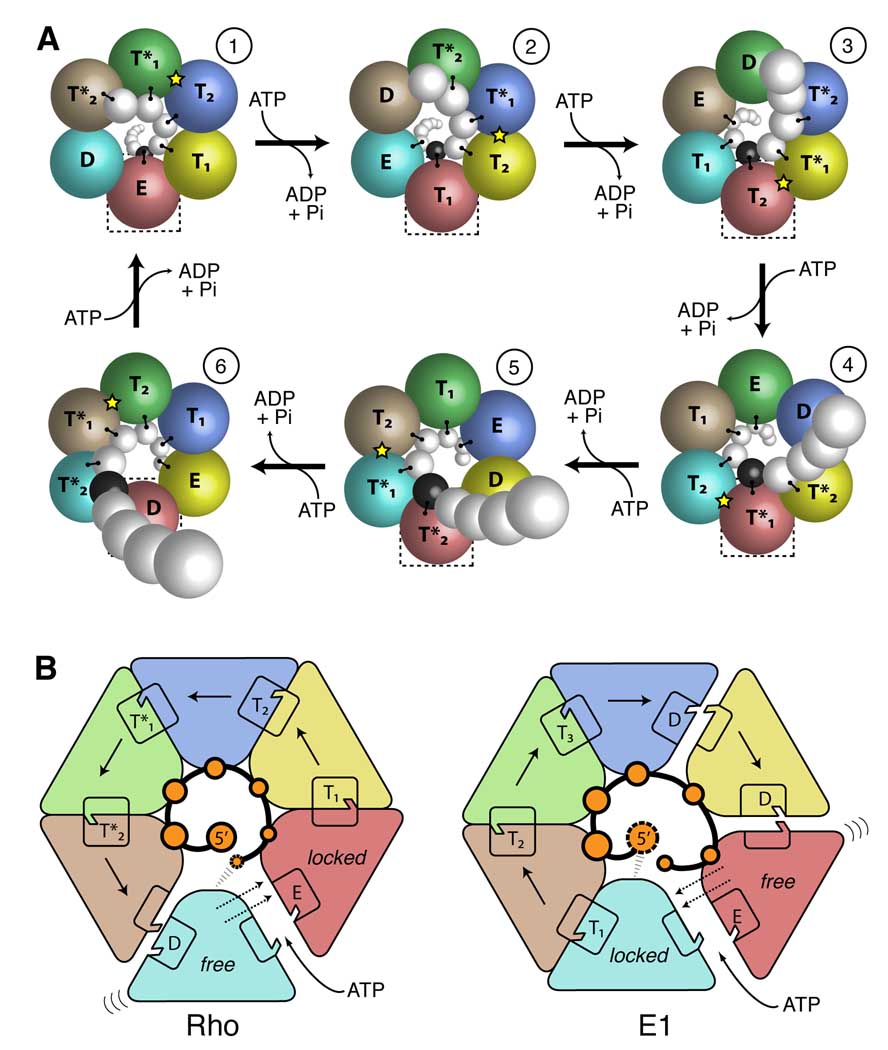
Figure 6. Translocation mechanism and directional polarity.
(A) Schematic of a Rho translocation cycle in which six ATP molecules are hydrolyzed to move six nucleotides of RNA. Helicase subunits are illustrated as colored spheres. RNA is shown as a chain of white spheres spiraling out of the plane of the paper. Protein-RNA contacts are indicated by lines connecting the protein and RNA spheres; the black RNA sphere serves as a reference point, and moves toward the viewer as the boxed red subunit transitions through six steps in the translocation cycle. A yellow star represents activation of the allosteric network that likely promotes hydrolysis. See also supplemental movie S3–movie S6.
(B) Schematics of Rho and E1 (chains A–F) illustrating their respective sequential ATP hydrolysis directions. Protein subunits are colored as in Figure 1. Nucleic acid phosphates observed in the structures are illustrated as bold orange circles, with the incoming phosphate shown as a dashed orange circle. Rectangles represent the two halves of the bipartite active site. Interlocked rectangles show insertion of the arginine finger in ATP-bound states. Solid arrows outline the progression toward subsequent steps in the ATPase cycle. Dotted arrows show the movement of the mobile “transition” subunit, upon binding ATP, toward a partner subunit locked in place by ATP-dependent (T-state) contacts within the ring.
During translocation, each Rho subunit is thought to transition through a round of ATP binding, hydrolysis and product release. Our structure suggests that Rho uses an [E →T1 →T2 →T*1→T*2 →D] ATPase cycle, where a full circuit involves the release of ADP and binding of a new ATP at the lone E state, progressively tighter binding of ATP in the T states, ATP hydrolysis in one of the T* states, and stable product formation in the D state (Movie S3). Although the precise timing of Pi release is not evident from our model, hydrolysis likely occurs in the allosterically activated (Figure 5D) T*1 state (B/C), with Pi release delayed until the partially-open product (D), or fully-open E (E/F) interface, is reached. This model is consistent with pre-steady state kinetic analyses illustrating that T7-gp4 and Rho release Pi only after ~2 or ~3.5 ATP molecules have been hydrolyzed respectively (Adelman et al., 2006; Liao et al., 2005).
Concomitant with these events, the subunits change their relative conformation with respect to each other in a closed, cyclic wave that pulls RNA through the central channel of the helicase (Figure 6A and Movie S4). As ATP is bound by an E-state subunit, the protomer latches onto an incoming 3’-RNA nucleotide (stage 1). As the subunit transitions into T and then T* states, it chaperones that same nucleotide up through the ring (stages 2–5). As RNA prepares to exit from Rho, structural changes in the Q loop reduce the number of protein-RNA contacts (Figure 4, subunit A), priming the nucleic acid for release; the structure suggests that a recoil of the compressed RNA constrained by subunits B and C could participate in this release event, similar to a concept proposed for the bacteriophage φ8 RNA packaging motor (Lisal et al., 2004). To complete a cycle, the subunit enters into a D state, transitioning from the top to the bottom of the staircase, and disengaging from the RNA (stage 6). In this scheme, the structure appears to strongly support a strictly sequential, rotary ATP-hydrolysis mechanism, in which each ATP turnover event translocates the motor by one RNA base (Movie S5 and Movie S6).
What is the coupling that links ATPase activity to RNA movement and vice-versa? In Rho, these events are tied together by the global structure of the hexamer, as well as by the intricate allosteric network that links the ATP and RNA binding sites (Movie S2). The network is activated by the tight binding of RNA to a pair of adjacent subunits (Figure 4, subunits B and C in our structure); less optimal RNA contacts are seen in T-state active sites that do not appear competent to hydrolyze ATP. This linkage explains how Rho can bind multiple ATP molecules tightly, yet hydrolyze them in an ordered, sequential manner rather than a concerted fashion (Adelman et al., 2006). Moreover, although the presence of an arginine finger is required for tight ATP binding by Rho (Miwa et al., 1995), it is not the sole determinant of hydrolysis. Rather, it is the synergistic effect of both the arginine finger and the positioning of a catalytic glutamate by the allosteric network that precisely times ATP hydrolysis.
We note that the mechanism described here espouses some concepts similar to previous structure-based models proposed for RecA-family hexameric helicases (Mancini et al., 2004; Singleton et al., 2000), but is also distinct. For example, the RNA binding configuration in our structure indicates that Rho does not act as a symmetric particle, such as a trimer of dimers (Seifried et al., 1992; Skordalakes and Berger, 2006). Instead the hexamer appears to function as an asymmetric particle in which five subunits simultaneously contact nucleic acid, each in a unique manner. The structure also indicates that there is no handoff of the RNA from one subunit to the next; rather, a single subunit stays associated with one RNA nucleotide throughout the catalytic cycle, and only engages every seventh (n+6) nucleotide in a chain (Movie S7). It seems likely that the absence of certain substrates in previous structures of RecA-like hexameric helicases may have precluded capture of a fully configured and asymmetric state, allowing only some elements of a complete catalytic cycle to be observed. This proposed sequential mechanism is generally consistent with a number of studies on Rho and other hexameric RecA-family motors, including the F1-ATPase (Adelman et al., 2006; Crampton et al., 2006; Noji et al., 1997; Notarnicola and Richardson, 1993; Richardson and Ruteshouser, 1986).
RecA and AAA+ hexameric helicases: the structural basis for translocation polarity
A major distinguishing feature of nucleic acid helicases and translocases is the polarity of their movement along DNA or RNA (Singleton et al., 2007). Among hexameric helicases, RecA-family enzymes move 5’→3’, while AAA+ proteins track 3’→5’. What accounts for this difference? A priori, one could envision that each helicase’s ATPase active sites fire sequentially in the same direction around the ring (i.e. clockwise or counterclockwise), but that nucleic acid strands bind to the two enzymes in opposite orientations. Alternatively, RecA and AAA+ helicases might bind substrate equivalently, but invert their sequential order of active site turnover.
The structure of Rho presented here, compared to the DNA-bound structure of the AAA+ helicase E1 (Enemark and Joshua-Tor, 2006), reveals that these two enzymes share a common nucleic acid binding polarity. Viewed from the 5’ end of the DNA or RNA strand, each hexamer’s ATP-binding sites are formed with the Walker A/B motifs situated counterclockwise from the neighboring arginine finger (Figure 6B). This arrangement indicates that the opposing translocation movements of the two enzymes arise from a reversal in the direction of the rotary ATPase wave. Such a “rewiring” of step direction within a common motor framework is not without precedent; for example, the related RecB and RecD SF1 helicases move in opposing directions, even though ssDNA substrates bind to the two helicases with the same polarity (Saikrishnan et al., 2009; Singleton et al., 2004).
Why is the sequential order of ATP hydrolysis inverted between Rho and E1? Superficially, the two motors appear very similar – they share a common nucleic acid binding polarity and a common spiral configuration for engaging substrate. In addition, the fully-liganded structures show that both helicases contain an exchange/empty (E) ATP-binding site sequestered between a tightly-bound ATP (T) state and a weakly-bound product (D) state. However, the relative order of flanking active sites for Rho is flipped in comparison with E1 (Figure 6B). We suggest that this configuration is responsible for biasing the direction of subunit movement, with the protomer linked to the T state effectively being locked down, and the subunit between the E and D states free to move upon ATP binding. This inverted orientation would lead to a counterclockwise shift of the “free” subunit upon binding ATP in Rho, and a clockwise shift in E1. Thus, by reorienting a common motor domain within two different hexameric assemblies (Figure S1) (Wang, 2004), and by reversing the relative direction of movement between an ATP binding site and its adjoining arginine finger, nature has evolved two families of hexameric helicases with opposing translocation polarities.
Concluding remarks
Our structure of the E. coli Rho transcription termination factor bound to both RNA and an ATP mimetic provides new insights into the chemomechanical coupling mechanism of a RecA-family hexameric helicase. Comparisons with the E1 papillomavirus protein suggest that RecA- and AAA+-family hexameric helicases employ different ATPase states to drive substrate binding, and that the two motors sequentially hydrolyze ATP in opposite directions.
In light of the remarkable functional diversity of RecA- and AAA+-type ring-shaped motors, the generality of the Rho and E1 models for understanding other hexameric motor proteins is still unknown. For instance, studies on the phage φ12 packaging protein (RecA-like) and the archaeal MCM helicase (AAA+) suggest that these enzymes may bind nucleic acid segments with a polarity opposite that seen for Rho and E1, respectively (Huiskonen et al., 2007; Mancini et al., 2004; McGeoch et al., 2005). Recent single molecule data on the pentameric phage φ29 double-stranded DNA packaging motor suggests there may exist unique translocation mechanisms with non-integer step sizes (Moffitt et al., 2009). Finally, biochemical studies of MCMs and the ClpX protein translocase have shown that some ring systems have relaxed dependencies on ATPase firing order that may approach a “loosely-sequential” or even stochastic ATP turnover mechanism (Martin et al., 2005; Moreau et al., 2007). While it is likely that functional specialization has led to important differences between and even within motor families, the general principles revealed by fully liganded structures of F1-ATPase, E1 and now Rho, provide a set of powerful tools to inform further research on these remarkable molecular machines.
Experimental Procedures
Crystallization and data collection
Full-length, selenomethionine labeled Escherichia coli Rho protein was prepared as described previously (Skordalakes and Berger, 2003). Rho was then concentrated to 60 mg/mL by ultrafiltration (Millipore – Amicon Ultra) and dialyzed into 10 mM Tris-HCl pH 7.5, 300mM NaCl and 1mM TCEP. A solution of ADP•BeF3 (prepared at a 1:3:15 molar ratio of ADP:BeCl2:NaF) was mixed with a 1:1 molar equivalent of MgCl2:ADP. To form the complex, rU12 RNA (IDT) was mixed with protein at a 1.1:1 stoichiometry of rU12 :Rho hexamer, and incubated on ice for 15 minutes. The ADP•BeF3•Mg2+ solution was added to the protein-RNA complex, and the mixture equilibrated for another 15 minutes. The final protein solution contained 20 mg/mL Rho, 0.28 mg/mL of rU12 RNA, 2.5 mM ADP•BeF3, 2.5 mM MgCl2, 10mM Tris-HCl (pH 7.5), 300mM NaCl and 1mM TCEP.
Microbatch crystals were grown by mixing 2µL of protein solution with 2µL of a solution containing 5% MPD, 100mM HEPES (pH 7.9), 20mM NaCl, and 10mM spermidine-HCl, and incubating under paraffin oil at 18°C. Crystals (hexagonal rods) were cryoprotected by stepwise exchange of the mother liquor with an identical solution supplemented with 25% MPD over a period of 15 minutes. Crystals were looped, flash frozen in liquid nitrogen, and diffraction data were collected on Beamline 8.3.1 at the Advanced Light Source (MacDowell et al., 2004).
Structure solution and refinement
Despite the use of selenomethionine-derivatized protein, we were unable to obtain useful maps from SAD datasets, likely due to radiation damage accrued over the course of collecting a P1 dataset at the absorption edge. To minimize this problem, a single “native” dataset was collected at a low-energy remote wavelength away from the selenium edge. HKL-2000 was used to process the data (Otwinowski and Minor, 1997). Phases were obtained by molecular replacement (MR) using PHASER (McCoy et al., 2007). The most useful search model consisted of residues 1–126 of chain A and 155–415 of chain C from the open ring Rho-RNA structure (1PVO) (Skordalakes and Berger, 2003), combined with residues 127–154 of an unpublished, higher-resolution open-ring Rho structure.
Initial rigid body and grouped B-factor refinement were performed using PHENIX (Adams et al., 2002). At this stage, Fo−Fc difference electron density maps showed clear density for RNA and nucleotide (Figure 1C–D). Iterative rounds of multi-domain, NCS-restrained, simulated annealing in PHENIX (Adams et al., 2002), 6-fold multi-domain NCS averaging and density modification using DM (Cowtan, 1994), and manual model building in Coot (Emsley and Cowtan, 2004), generated a starting model. In subsequent refinements, NCS restraints were relaxed and un-averaged maps were used. Model bias was removed using prime-and-switch and composite-omit maps, both calculated by PHENIX (Adams et al., 2002). The RNA and ADP•BeF3•Mg2+ were added in the last stages of refinement, along with manually-placed waters and two spermidine molecules. The final model was refined in PHENIX using weak, multi-domain backbone NCS restraints for the OB and RecA folds, individual B-factor modeling, and TLS modeling of protein domains obtained from the TLSMD server (Painter and Merritt, 2006).
Structural Analysis
RNA conformation was analyzed with 3DNA (Lu and Olson, 2003), structural superpositions and figures were prepared with PyMol (DeLano, 2002), and linear interpolations of the six structural states were prepared using PyMol in conjunction with CNS (Brunger et al., 1998) and software written by the Yale Morph Server (Krebs and Gerstein, 2000).
Supplementary Material
Acknowledgements
We thank James Keck, Andreas Martin, Allyn Schoeffler and Nancy Crisona for helpful discussions and editing of the manuscript, and Emmanuel Skordalakes and Berger lab members for assistance in various stages of the project. Special thanks go to the staff at SSRL beamlines 9.2 and 11.1; Scott Classen, George Meigs and James Holton at ALS beamlines 12.3.1 and 8.3.1; Steve Gamblin for assistance with data processing; and Nat Echols for assistance with programs from the Yale Morph Server. This work was supported by funding from the NIH (GM071747) and the G. Harold and Leila Y. Mathers Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Coordinates for the fully liganded Rho structure have been deposited in the RCSB Protein Data Bank under ID code 3ICE.
References
- Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Adelman JL, Jeong YJ, Liao JC, Patel G, Kim DE, Oster G, Patel SS. Mechanochemistry of transcription termination factor Rho. Mol Cell. 2006;22:611–621. doi: 10.1016/j.molcel.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Berger JM. SnapShot: nucleic acid helicases and translocases. Cell. 2008;134:888–888. doi: 10.1016/j.cell.2008.08.027. e881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, Dombroski AJ, Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chen CY, Richardson JP. Sequence elements essential for rho-dependent transcription termination at lambda tR1. J Biol Chem. 1987;262:11292–11299. [PubMed] [Google Scholar]
- Cowtan K. 'dm': An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- Crampton DJ, Mukherjee S, Richardson CC. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol Cell. 2006;21:165–174. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structures and properties of isolated helicases. Q Rev Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q Rev Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Dolan JW, Marshall NF, Richardson JP. Transcription termination factor rho has three distinct structural domains. J Biol Chem. 1990;265:5747–5754. [PubMed] [Google Scholar]
- Dombroski AJ, Platt T. Structure of rho factor: an RNA-binding domain and a separate region with strong similarity to proven ATP-binding domains. Proc Natl Acad Sci U S A. 1988;85:2538–2542. doi: 10.1073/pnas.85.8.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci U S A. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiselmann J, von Hippel PH. Functional interactions of ligand cofactors with Escherichia coli transcription termination factor rho. I. Binding of ATP. Protein Sci. 1992;1:850–860. doi: 10.1002/pro.5560010703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani MM, Patel SS. Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry. 1993;32:12478–12487. doi: 10.1021/bi00097a028. [DOI] [PubMed] [Google Scholar]
- Huiskonen JT, Jaalinoja HT, Briggs JA, Fuller SD, Butcher SJ. Structure of a hexameric RNA packaging motor in a viral polymerase complex. J Struct Biol. 2007;158:156–164. doi: 10.1016/j.jsb.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W. Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB Helicase? Fluorescence energy transfer studies. J Biol Chem. 1998a;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- Jezewska MJ, Rajendran S, Bujalowski W. Functional and structural heterogeneity of the DNA binding site of the Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1998b;273:9058–9069. doi: 10.1074/jbc.273.15.9058. [DOI] [PubMed] [Google Scholar]
- Kagawa R, Montgomery MG, Braig K, Leslie AG, Walker JE. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23:2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF, Roberts JW, Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J Biol Chem. 1983;258:9391–9397. [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J Mol Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- Liao JC, Jeong YJ, Kim DE, Patel SS, Oster G. Mechanochemistry of t7 DNA helicase. J Mol Biol. 2005;350:452–475. doi: 10.1016/j.jmb.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Lisal J, Kainov DE, Bamford DH, Thomas GJ, Jr., Tuma R. Enzymatic mechanism of RNA translocation in double-stranded RNA bacteriophages. J Biol Chem. 2004;279:1343–1350. doi: 10.1074/jbc.M309587200. [DOI] [PubMed] [Google Scholar]
- Lo YH, Tsai KL, Sun YJ, Chen WT, Huang CY, Hsiao CD. The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery-Goldhammer C, Richardson JP. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with rho termination factor. Proc Natl Acad Sci U S A. 1974;71:2003–2007. doi: 10.1073/pnas.71.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDowell AA, Celestre RS, Howells M, McKinney W, Krupnick J, Cambie D, Domning EE, Duarte RM, Kelez N, Plate DW, et al. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- Miwa Y, Horiguchi T, Shigesada K. Structural and functional dissections of transcription termination factor rho by random mutagenesis. J Mol Biol. 1995;254:815–837. doi: 10.1006/jmbi.1995.0658. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Morgan WD, Bear DG, von Hippel PH. Rho-dependent termination of transcription. II. Kinetics of mRNA elongation during transcription from the bacteriophage lambda PR promoter. J Biol Chem. 1983;258:9565–9574. [PubMed] [Google Scholar]
- Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- Notarnicola SM, Richardson CC. The nucleotide binding site of the helicase/primase of bacteriophage T7. Interaction of mutant and wild-type proteins. J Biol Chem. 1993;268:27198–27207. [PubMed] [Google Scholar]
- Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr., Sweet RM, editors. Methods in Enzymology. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. TLSMD web server for the gernation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci U S A. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- Richardson JP. Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- Richardson JP, Ruteshouser EC. rho factor-dependent transcription termination. Interference by a mutant rho. J Mol Biol. 1986;189:413–419. doi: 10.1016/0022-2836(86)90313-x. [DOI] [PubMed] [Google Scholar]
- Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5'-3' translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- Sakakibara N, Schwarz FP, Kelman Z. ATP Hydrolysis and DNA Binding Confer Thermostability on the MCM Helicase (dagger) Biochemistry. 2009 doi: 10.1021/bi801921j. [DOI] [PubMed] [Google Scholar]
- Seifried SE, Easton JB, von Hippel PH. ATPase activity of transcription-termination factor rho: functional dimer model. Proc Natl Acad Sci U S A. 1992;89:10454–10458. doi: 10.1073/pnas.89.21.10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Platt T. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc Natl Acad Sci U S A. 1994;91:1401–1405. doi: 10.1073/pnas.91.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Wang J. Nucleotide-dependent domain motions within rings of the RecA/AAA(+) superfamily. J Struct Biol. 2004;148:259–267. doi: 10.1016/j.jsb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Weber TP, Widger WR, Kohn H. Metal dependency for transcription factor rho activation. Biochemistry. 2003;42:1652–1659. doi: 10.1021/bi020601y. [DOI] [PubMed] [Google Scholar]
- Wei RR, Richardson JP. Identification of an RNA-binding Site in the ATP binding domain of Escherichia coli Rho by H2O2/Fe-EDTA cleavage protection studies. J Biol Chem. 2001;276:28380–28387. doi: 10.1074/jbc.M102444200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Johnson J, Kohn H, Widger WR. ATP binding to Rho transcription termination factor. Mutant F355W ATP-induced fluorescence quenching reveals dynamic ATP binding. J Biol Chem. 2003;278:13719–13727. doi: 10.1074/jbc.M212979200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.