Abstract
Most heritable behavioral traits have a complex genetic basis, but few multigenic traits are understood at a molecular level. Here we show that the C. elegans strains N2 and CB4856 have opposite behavioral responses to simultaneous changes in environmental O2 and CO2. We identify two quantitative trait loci (QTL) that affect this trait, and map each QTL to a single-gene polymorphism. One gene, npr-1, encodes a previously described neuropeptide receptor whose high activity in N2 promotes CO2 avoidance. The second gene, glb-5, encodes a neuronal globin domain protein whose high activity in CB4856 modifies behavioral responses to O2 and combined O2/CO2 stimuli. glb-5 acts in O2–sensing neurons to increase O2-evoked calcium signals, implicating globins in sensory signaling. An analysis of wild C. elegans strains indicates that the N2 alleles of npr-1 and glb-5 arose recently in the same strain background, possibly as an adaptation to laboratory conditions.
Introduction
Genetic variation contributes to individual differences in many behaviors, including psychiatric conditions in humans and behavioral traits in animals (Kendler, 2001; Kendler and Greenspan, 2006; Flint, 2003). Despite substantial evidence for heritable effects, only a few behavioral traits have been traced to discrete molecular changes (de Bono and Bargmann, 1998; Osborne et al., 1997; Yalcin et al., 2004). The complex genetic structure of natural variation poses challenges for gene identification: most traits are thought to be affected by a few polymorphic genes with moderate effects, and many genes with small effects (Flint, 2003; Mackay, 2004; Kendler and Greenspan, 2006). A molecular understanding of multigenic traits is essential to determine how genetic changes arise, what genes they affect, how these genes interact, and how they influence behavior.
A common approach used to dissect complex genetic traits is QTL analysis, in which two strains are intercross, F2 progeny or inbred lines of progeny are characterized by genotype and phenotype, and quantitative genetics is used to find linkage to traits of interest. Classical QTL analysis has associated chromosomal regions with traits like anxiety, aggression, drug preference, and learning, but since a well-defined QTL in Drosophila or mouse typically covers 300–500 genes, moving from a QTL to the causative mutation is very difficult (Mackay, 2004; Mott and Flint, 2008; Flint, 2003). Indeed, the association of the G protein regulator Rgs2 with anxiety may be the only established single-gene behavioral QTL in mice (Yalcin et al., 2004). Developing methods to address the QTL-to-genetic-alteration problem is a major goal of the field. For example, new methods of interest combine QTL analysis with complementary approaches such as gene expression analysis or association studies of outbred strains (Mackay, 2004; Wang et al., 2008; Toma et al., 2002).
The resolution of QTL mapping has been greatly improved by whole-genome sequences and inexpensive resequencing, which allow strains to be genotyped at thousands of polymorphic loci instead of the dozens that were typical in the past. To take advantage of these high-resolution DNA maps, it is optimal to have chromosomes with many recombination breakpoints for fine genetic mapping. Recombinant inbred advanced intercrossed lines, or RIAILs, are modified inbred lines with many crossover points per chromosome that should in principle allow rapid mapping of QTLs to individual genes (Mott and Flint, 2008; Rockman and Kruglyak, 2008). A set of high-resolution C. elegans RIAILs has been generated by intercrossing the standard laboratory strain (N2) with a strain isolated in a Hawaiian pineapple field (CB4856, henceforth “HW”) for ten generations, inbred by selfing for ten generations, and genotyped at 1455 loci (Rockman and Kruglyak, 2009). The resulting RIAILs have been used to map several single-gene QTLs for reproductive traits and pathogen resistance (Hodgkin and Doniach, 1997; Seidel et al., 2008; Palopoli et al., 2008; Reddy et al., 2009). Here we use the RIAILs to identify two QTLs that affect a behavioral trait, and map both QTLs to single genetic changes.
C. elegans has strong behavioral responses to the gases O2 and CO 2, which are highly variable in its natural soil and compost environments due to metabolic activity (Sylvia et al., 1998; Greenway et al., 2006). Previous studies have identified the neuropeptide receptor gene npr-1 as a regulator of O2 and CO2 responses that differs between N2 and HW C. elegans strains (de Bono and Bargmann, 1998). N2 has a high activity npr-1 allele (215 valine), and as a result has weak responses to O2 on food, and strongly avoids CO2 (Gray et al., 2004; Bretscher et al., 2008; Hallem and Sternberg, 2008). HW has a low-activity npr-1 allele (215 phenylalanine), and as a result strongly avoids high O2 levels on food, and is not repelled by CO2. Two other behaviors observed in HW strains, aggregation into feeding groups and accumulation at the border of a bacterial lawn, are in part caused by avoidance of high O2 conditions (Gray et al., 2004; Cheung et al., 2005). Using QTL mapping, we find that npr-1 cooperates with another variable gene, the globin homolog glb-5, to affect behavioral responses to O2 and CO2. Using genetic studies of 203 wild strains, we trace the history of these mutations in C. elegans.
Results
O2- and CO2-evoked responses of two C. elegans isolates
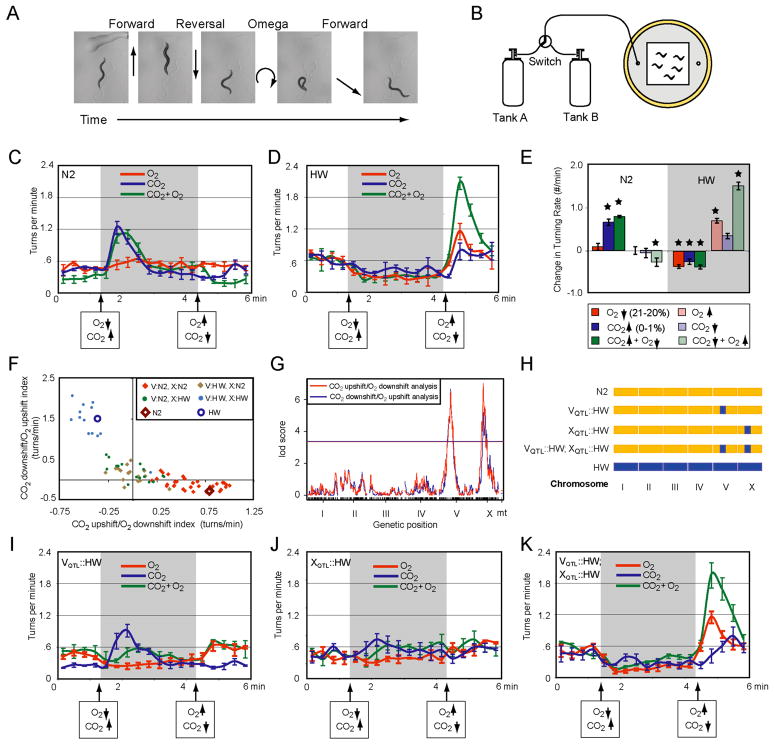
C. elegans increases its frequency of spontaneous reversals and high-amplitude turns when exposed to nociceptive and repulsive stimuli, and suppresses reversals and turns when exposed to attractants (Chalasani et al., 2007; Ryu and Samuel, 2002). These behavioral responses contribute to a biased random walk strategy for navigating to a favorable environment (Pierce-Shimomura et al., 1999). To learn more about O2 and CO2 responses of C. elegans, we scored turning behaviors (Fig. 1A) in freely-moving adult animals on a thin lawn of OP50 bacteria, while switching the chamber between two different gas mixtures every three minutes for sixty minutes (Fig. 1B). Video recordings were analyzed using automated tracking software that recorded instantaneous speed, reversals, and turns (Ramot et al., 2008). We focused on omega turns, which are high-amplitude forward turns that reorient movement by >90°.
Fig. 1. Quantitative genetic analysis of O2 and CO2 responses in two C. elegans strains.
A. Forward movement interrupted by a reversal and omega turn.
B. Behavioral arena for turning assay. 20–30 animals were recorded in the presence of OP50 bacteria as O2 and CO2 concentrations were controlled by regulated gas flow.
C,D. Average turn rates of N2 and CB4856 (HW) strains across nine six-minute pulse sequences (see Methods). In all Figures, O2 changes were 21% O2->20% O2->21% O2, CO2 changes were 21% O2->21% O2/1% CO2->21% O2, and simultaneous O2 and CO2 changes were 21% O2->20% O2/1% CO2->21% O2. The balance in all mixtures was N2.
E. Average change in turning rate before and after the indicated step change (data in C and D). Stars denote significant effects of the step change (P<0.05, t-test).
F. Behavioral responses of 78 RIAILs to simultaneous O2 and CO2 step changes. Y-axis, difference in turning frequency after CO2 downshift/O2 upshift; X-axis, difference in turning frequency after CO2 upshift/O2 downshift. N2 and HW reference strains are included; each RIAIL is color-coded by genotype at the QTLs on chromosome V and X.
G. QTL analysis of data from F, showing significant QTL peaks on the left arms of V and X. Horizontal line represents the genome-wide significance threshold (P=0.05).
H. Schematic of three introgression strains with small regions of HW DNA around the QTLs on V and X introduced into an N2 background.
I–K. Turning responses of introgression strains, as in C and D.
In all figures, error bars indicate standard error of mean (SEM), and Supplementary Tables 1 and 2 have full datasets and statistics.
Strikingly, N2 and HW hermaphrodites, both nominally wild-type, had opposite turning responses to simultaneous changes in CO2 and O2 concentrations (Fig. 1C, D). When N2 animals were shifted from a mixture of 21% O2/79% N2 to 1% CO2/20% O2/79% N2, they generated a burst of turns that peaked after ~30s and fell to baseline ~60s later (Fig. 1C, E). The reciprocal switch back to 21% O2/79% N2 suppressed turning. By contrast, HW animals shifted from 21% O2/79% N2 to 1% CO2/20% O2/79% N2 suppressed turning, and the reciprocal switch back to 21% O2/79% N2 caused a burst of turning that peaked after ~30s and fell to baseline ~60s later (Fig. 1D, E). Since transient bursts of turning accompany the appearance of a repellent, these results suggest that HW and N2 have opposite preferences for the two gas mixtures.
To determine which gas was most important for turning behavior, CO2 and O2 levels were changed separately. Upon a shift from 0% to 1% CO2, N2 responded with a burst of turning but HW did not respond; a reciprocal shift from 1% to 0% CO2 had no effect on N2, but stimulated turning slightly in HW (Fig. 1C–E). These results suggest that N2 avoids CO2, and HW is weakly attracted to CO2. A shift from 21% to 20% O2, or from 20% to 21% O2, had little effect on turning in N2 (Fig. 1C, E). By contrast, HW showed a transient 2-fold increase in turning upon a 20% to 21% O2 upshift and reduced turning upon the reciprocal O2 downshift, suggesting a preference for 20% O2 (Fig. 1D, E).
Quantitative analysis of the behaviors revealed significant interactions between the gas responses (Supplementary Tables 1, 2). The N2 turning response in the mixture was dominated by CO2, and was consistent with the known avoidance of CO2 by N2 (Bretscher et al., 2008; Hallem and Sternberg, 2008). The turning response in HW was dominated by a preference for 20% over 21% O2, and was significantly stronger when both CO2 and O2 were changed simultaneously, suggesting a slight attraction to CO2 and an interaction between the responses.
Two genetic loci control the difference in CO2/O2 behavior between HW and N2
The genetic basis of the behavioral response to simultaneous small changes in O2 and CO2 was determined by characterizing 78 RIAILs (Fig. 1F). Based on their turning behaviors, the RIAILs fell into a nearly continuous distribution with three general groups: (1) HW-like lines that turned upon simultaneous O2 increases/CO2 decreases and suppressed turning upon simultaneous O2 decreases/CO2 increases; (2) N2-like lines that turned upon O2 decreases/CO2 increases and suppressed turning upon O2 increases/CO2 decreases; (3) intermediate lines that did not turn much in response to either change. The continuous distribution of phenotypes and the existence of a novel intermediate behavioral class indicate that this is a complex genetic trait, i.e. that multiple loci influence the behavior.
To identify loci that contribute to the behaviors of the RIAILs, quantitative trait locus (QTL) mapping was performed on the O2 increase/CO2 decrease response and on the O2 decrease/CO2 increase response using genotypes of the RIAILs at 1455 SNP markers. Both analyses identified two significant QTLs (Fig. 1G), one QTL on chromosome V (lod scores 6.5 and 6.6 for the O2 decrease/CO2 increase and O2 increase/CO2 decrease, respectively, genome-wide corrected P < 0.0001 each) and one QTL on chromosome X (lod scores 7.0 and 5.2, P < 0.0001 and P = 0.0003 for the O2 decrease/CO2 increase and O2 increase/CO2 decrease, respectively). For the O2 decrease/CO2 increase response, the two QTLs account for 79% of the among-line variance, explaining 33% (V) and 45% (X) of the variance with no significant interaction between them (F1 = 3.87, P = 0.053). For the O2 increase/CO2 decrease response, the main effects of the two loci explain 34% (V) and 35% (X) of the variance, and a significant interaction effect between the two loci accounts for 15% of the among-line variance (F1 = 63, P < 10−10; see Methods for further discussion).
The genotypes of the two loci on V and X in the 78 RIAILs correlated with the three behavioral classes noted above (Fig. 1F). Although the behavioral scores appeared to fall in a continuous distribution, most RIAILs that had N2 alleles of both QTLs behaved like N2 (n=33), and most RIAILs with HW alleles of both QTLs behaved like HW (n=13). RIAILs with N2 alleles at one locus and HW alleles at the other had weak responses to either O2/CO2 step change (n=32) (Fig. 1F).
To further analyze the mixed allele strains, HW alleles of both QTLs were individually introgressed into N2 by backcrossing based on linked SNPs for at least 10 generations, and then the two introgressed strains were crossed together (Fig. 1H). The crosses yielded one strain with nearly pure N2 DNA except for the QTL region on chromosome V (referred to as VQTL:HW), one strain with nearly pure N2 DNA except for the QTL region on chromosome X (referred to as XQTL:HW), and one strain with nearly pure N2 DNA with both V and X QTLs from HW (referred to as VQTL:HW; XQTL:HW). The three strains were then tested for turning responses to O2 step changes, CO2 step changes, and combined O2/CO2 step changes (Fig. 1I–K, Supplementary Fig. 1). In all three conditions, VQTL:HW; XQTL:HW were indistinguishable from HW, confirming that two discrete loci account for most of HW behavior (Fig. 1K). The VQTL:HW and XQTL:HW strains with one HW locus were distinguishable from each other and from both starting strains. XQTL:HW had minimal responses in all six step changes (Fig. 1J). VQTL:HW increased turns in response to individual CO2 increases, but did not respond when these were paired with O2 decreases (Fig. 1I).
The QTL on X is caused by a point mutation in npr-1
The QTL on chromosome X spans the region from 4–525 Mb, centered on the npr-1 gene. The high activity npr-1(215V) allele is required for CO2 avoidance by N2 (Bretscher et al., 2008; Hallem and Sternberg, 2008); since HW animals have the low activity npr-1(215F) allele, an npr-1 contribution was a plausible explanation for the QTL on X. To test this possibility, a plasmid containing the high-activity N2 npr-1(215V) allele was injected into XQTL:HW animals. The resulting transgenic animals behaved like N2 (Fig. 2A and Supplementary Fig. 1), indicating that expression of the N2 allele of npr-1 is sufficient for N2-like turning behavior in XQTL:HW animals. Injecting a comparable low-activity HW npr-1(215F) plasmid did not affect the behavior of XQTL:HW animals (Fig. 2A and Supplementary Fig. 1). Animals bearing the EMS-induced npr-1(ad609) loss-of-function mutant in an N2 background behaved like XQTL::HW animals (Fig. 2A), confirming the importance of npr-1 in the turning assay. These results indicate that variation in the npr-1 coding sequence can explain the QTL for CO2 and O2 behavior on the X chromosome.
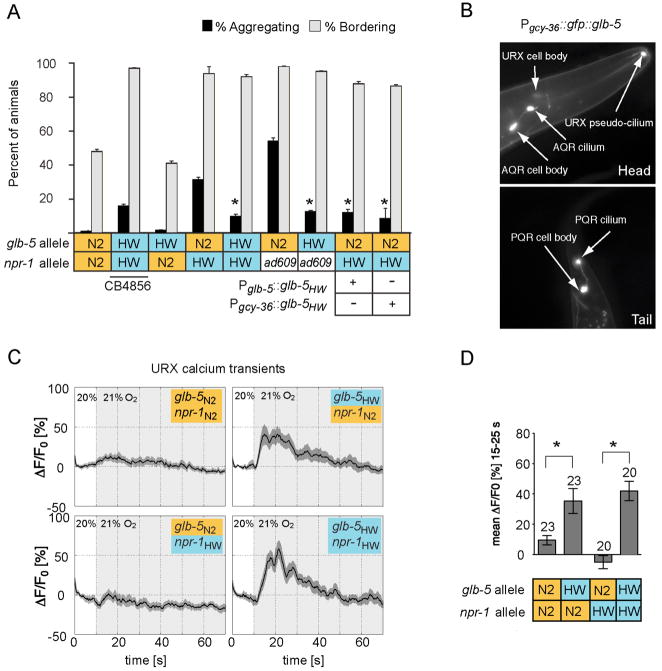
Fig. 2. npr-1 and glb-5 polymorphisms account for the two behavioral QTLs.
A. Genetic identification of QTLs on V and X. Turning responses of strains with transgenes bearing N2 or HW alleles of npr-1, or N2 or HW alleles of glb-5; transgenes were in bicistronic transcripts followed by SL2::GFP. ad609 is a loss of function allele of npr-1 in the N2 background; “Het” indicates F1 progeny of a XQTL::HW and VQTL::HW; XQTL::HW cross. Complete datasets are in Supplementary Figure 1.
B. Sequence polymorphisms in glb-5 between N2 and HW. In N2, a 765 bp duplication covering the 6th exon truncates the predicted GLB-5 protein. cc, coiled-coil domain; ct, alternate C-terminal domain caused by the duplicated exon.
C. Expression of bicistronic Pglb-5::glb-5::SL2::gfp transgene in L1 animal.
D. Rescue of XQTL:HW animals by expressing the HW allele of glb-5 in URX, AQR, and PQR neurons (Pgcy-36::glb-5HW in bicistronic SL2::GFP clone; Pgcy-36::gfp::glb-5HW, N-terminally GFP-tagged glb-5HW).
E. Killing URX, AQR, and PQR with qaIs2241 eliminates the effects of glb-5HW. In panels A, D and E, control strains from Fig. 1 are included for reference.
The QTL on chromosome V is caused by a duplication/insertion in glb-5
The QTL on chromosome V was bounded by markers at 5.46 and 5.62 Mb, a region of 160 kb. Nine RIAILs had breakpoints between these markers, and finer breakpoint mapping of these strains narrowed the region to 90 kb, or 19 genes. Two interesting candidates in this interval, glb-5 and glb-6, are predicted to encode proteins with globin domains that can potentially bind O2 and CO2 (Hoogewijs et al., 2008). The sequence of the coding region of glb-6 was identical between N2 and HW, but the genomic sequence of glb-5 contained a 765 bp duplication/insertion in N2 compared to HW (Fig. 2B). glb-5 cDNA analysis demonstrated that the DNA polymorphism resulted in substantially different mRNAs and predicted GLB-5 proteins in N2 and HW. The duplicated exon in N2 led to an in-frame stop codon in the glb-5 cDNA, resulting in a truncation of the last 179 amino acids of the protein compared to HW, and the inclusion of 40 different residues (Fig. 2B).
To ask whether glb-5 regulates O2/CO2 responses, we first tested heterozygous offspring of VQTL:HW; XQTL:HW and XQTL:HW animals to determine which allele of the QTL on V was dominant when npr-1 genotype was held constant. These offspring behaved like VQTL:HW; XQTL:HW (Fig. 2A), indicating that the HW variant on chromosome V was dominant. To ask whether the QTL corresponded to glb-5, a glb-5 cDNA encoding the HW allele of glb-5 driven by its own promoter was injected into XQTL:HW animals. The resulting transgenic animals behaved like HW animals, indicating that glb-5 transgene can mimic the presence of the HW locus on V (Fig. 2A and Supplementary Fig. 1). Injection of a similar transgene bearing the N2 allele of glb-5 did not affect XQTL:HW turning behavior (Fig. 2A and Supplementary Fig. 1). These results indicate that the 765 bp duplication/insertion in glb-5 is the likely cause of the QTL on chromosome V.
glb-5 acts in URX, AQR, and PQR O2–sensing neurons
The expression of glb-5 was characterized with a bicistronic transgene that encoded the active HW allele of glb-5 followed by the coding region of GFP. In first larval stage (L1) animals, GFP was expressed prominently in the URX and BAG sensory neurons, with weaker and inconsistent expression in the ASG and ADF sensory neurons, the pharynx, and a few intestinal cells (Fig. 2C). In older animals, GFP fluorescence was also observed in AQR and PQR sensory neurons. URX, AQR, and PQR are O2-sensing neurons that are required for npr-1 mutants to avoid high O2 on food (Chang et al., 2006; Cheung et al., 2005; Gray et al., 2004), suggesting a possible functional link between glb-5 and the O2 response. Indeed, expressing the HW allele of glb-5 in XQTL:HW animals under the gcy-36 promoter, which is selectively expressed in the URX, AQR, and PQR neurons, was sufficient for HW-like behavior (Fig. 2D and Supplementary Fig 1).
The role of URX, AQR, and PQR in turning responses was confirmed using a strain in which the URX, AQR, and PQR neurons are selectively killed by the BH3 protein egl-1 (the transgene qaIs2241)(Chang et al., 2006). AQR, PQR, and URX were essential for the O2 and O2/CO2 responses in the VQTL::HW; XQTL::HW background, but they were not required for turning responses in N2 (Fig. 2E and Supplementary Fig. 1). An interesting result was observed in the VQTL:HW animals with glb-5HW and npr-1N2. Killing URX, AQR, and PQR in this strain resulted in an N2-like turning response, although the parent strain only responded to CO2 alone (Fig. 2E and Supplementary Fig. 1). Thus, the cell ablations uncovered a neuronal function that depends on the specific genetic background: URX, AQR, and PQR neurons with the glb-5HW allele inhibited N2-like O2/CO2 responses in VQTL:HW animals, but enhanced HW-like O2 and O2/CO2 responses in animals with HW alleles of both glb-5 and npr-1.
glb-5 modifies aggregation behavior
Animals from the HW strain aggregate into feeding groups and accumulate strongly at the border of a bacterial lawn; these behaviors require the URX, AQR, and PQR neurons, low npr-1 activity, and high O2 levels (Coates and de Bono, 2002; Gray et al., 2004). To ask whether glb-5 contributes to these more complex O2–dependent behaviors, aggregation and bordering were quantified in N2, HW, and introgressed strains. Indeed, aggregation in an npr-1HW genetic background was significantly modulated by the glb-5 allele: strains with glb-5N2 aggregated more strongly than strains with glb-5HW (Fig. 3A). glb-5HW also suppressed the aggregation of an npr-1(ad609) loss of function allele, but in the npr-1N2 genetic background, there was virtually no aggregation regardless of glb-5 genotype (Fig. 3A). As expected from the dominant nature of the glb-5HW allele, aggregation of the glb-5N2 strain was suppressed by expressing glb-5HW under its own promoter or under another promoter expressed in URX, AQR, and PQR neurons (Fig. 3A). The glb-5 allele had relatively little effect on bordering behavior, which was largely determined by npr-1 genotype (Fig. 3A).
Fig. 3. glb-5 affects aggregation behavior and URX sensory responses.
A. Aggregation and bordering behaviors in N2, HW (CB4856), introgression strains, and transgenic rescued lines. Asterisks indicate results significantly affected by glb-5HW genotype or transgene at P<0.01 compared to the appropriate glb-5N2 control.
B. GFP-tagged GLB-5HW protein (Fig. 2D) is enriched in AQR and PQR cilia and in the anterior tip of URX.
C. Calcium responses of URX neurons in strains with four combinations of npr-1 and glb-5 alleles, as indicated. Fluorescence increases in the G-CaMP indicator are caused by Ca++ increases, likely associated with depolarization. Light shading indicates time at 21% O2; dark shading indicates SEM; n= 20–23 animals for each genotype.
D. Average calcium increase at t=15–25s from (C). Asterisks, results different at P<0.01.
glb-5 sensitizes URX responses to small O2 changes
C. elegans encodes 33 distinct globins (Hoogewijs et al., 2007), most functionally uncharacterized (Hoogewijs et al., 2008). In principle, glb-5 could affect O2 sensation in URX, AQR, and PQR, or synaptic release onto other neurons, among other possibilities. A functional, GFP-tagged GLB-5 protein encoded by the HW allele of glb-5 was highly enriched in sensory endings of URX, AQR, and PQR, with less expression in cell bodies, axons, and dendrites (Fig. 3B). This localization suggests a role for GLB-5 in sensory transduction.
Previous studies with genetically-encoded calcium sensors have shown that the URX neurons are activated by O2 upshifts from 10→21% or 15→21% O2 (Zimmer et al., 2009). The glb-5-dependent behaviors in HW suggest that glb-5 might affect URX responses to smaller O2 changes. Therefore, calcium response of URX were monitored in response to upshifts from 20→21% O2, the conditions in which N2 and HW behaved differently. In the VQTL:HW animals with the glb-5HW allele, these small O2 20→21% upshifts induced rapid calcium transients in URX (Fig. 3C, D) but in the N2 strain, similar upshifts had no apparent effect. Similar URX responses to 20→21% O2 were observed in glb-5HW strains with either npr-1HW and npr-1N2 alleles, but not in any glb-5N2 strains (Fig. 3C, D). These results suggest that the HW allele of glb-5 sensitizes URX responses to small 20→21% O2 upshifts.
N2 alleles of npr-1 and glb-5 are rare in wild isolates of C. elegans
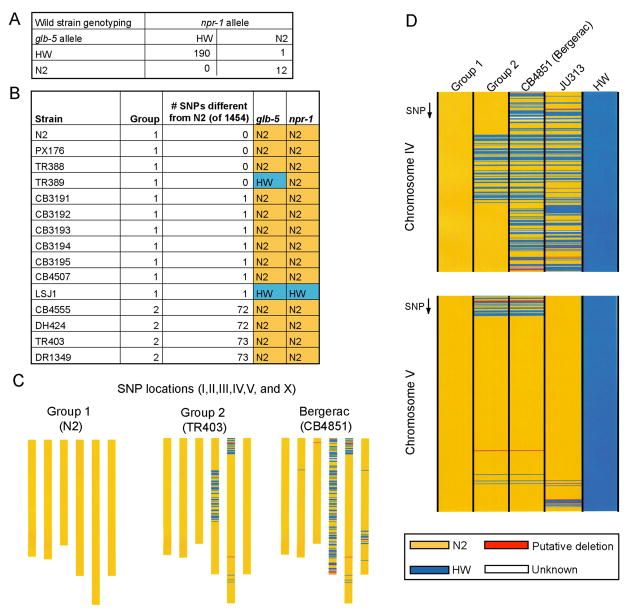
To assess the frequency and distribution of glb-5 and npr-1 alleles in the wild, each gene was characterized in 203 C. elegans isolates from Europe, North America, Africa, South America, Australia, and Japan (Supplementary Table 3). The genotypes were highly skewed: 190 strains had HW alleles of both genes, and 12 strains had N2 alleles of both npr-1 and glb-5, of which only one was isolated in the past 15 years (Fig. 4A). One strain (TR389) had the N2 allele of npr-1 and the HW allele of glb-5.
Fig. 4. Analysis of polymorphisms in wild strains.
A. glb-5 and npr-1 allele distributions in 203 wild strains.
B. Genotype analysis of 1454 SNPs of the strains containing N2 alleles of npr-1 or glb-5, and of LSJ1; data from (Rockman and Kruglyak, 2009). In 110 additional wild strains genotyped at these 1454 SNPs, there were on average 358 differences from N2, with a range of 112–1452 differences.
C. SNP genotypes on all chromosomes in Group 1 strains, Group 2 strains, and the Bergerac strain. All SNPs distinguishing Group 2 and N2 are on chromosome IV and V.
D. Comparison of SNPs in Group 2 strains with N2, HW and Bergerac on chromosomes IV and V. The most similar wild strain in the region shared between Group 2 animals and Bergerac on chromosome IV (JU313) is shown for reference.
Additional genotyping at 1454 loci revealed that all N2-like strains were genetically similar (Fig. 4B, generated from data taken from (Rockman and Kruglyak, 2009)). Nine of the N2-like strains were identical in at least 1453 out of 1454 HW/N2 single nucleotide polymorphisms (SNPs)(Group 1, Fig. 4B). The remaining four N2-like strains had blocks of HW SNPs on chromosome IV and V, but were otherwise N2-like (Group 2, Fig. 4B, C). The specific blocks of SNPs in these four strains, as well as the pattern of Tc1 transposon insertions (Egilmez et al., 1995), match blocks that are present in the genome of the Bergerac strain of C. elegans (Fig. 4C, D). A comparison across genotypes of these strains suggests that these four strains resulted from crosses between an N2-like strain and a Bergerac-like strain (Egilmez et al., 1995).
Since these results suggest that N2-like strains are rare in the wild, we specifically examined additional strains from the Pasadena area where nine of the thirteen N2-like strains were isolated. 55 more recently isolated strains all had HW-like alleles of npr-1 and glb-5 (Supplementary Table 3).
Discussion
N2 and HW strains of C. elegans have opposite behavioral responses to small changes in O2 and CO2. Despite the high genetic variability between these strains (Maydan et al., 2007; Wicks et al., 2001), two QTLs, one caused by a polymorphism in npr-1 and the other caused by a polymorphism in glb-5, account for most of the variation in these traits.
glb-5 is a member of the globin-domain superfamily, a widespread group of heme-binding proteins that bind O2 and typically act in O2-affiliated roles such as transport, storage, scavenging, or sensing (Weber and Vinogradov, 2001). There are 33 distinct globin domain-containing proteins encoded by the C. elegans genome (Hoogewijs et al., 2007); most are expressed exclusively in subsets of neurons, and one, GLB-10, is enriched at synapses (Hoogewijs et al., 2008; Sieburth et al., 2005). Like mammalian neuroglobins (Nienhaus and Nienhaus, 2007), the functions of neuronal C. elegans globins are largely unknown. Our results demonstrate a sensory role for GLB-5 that shapes behavioral responses to O2, a new insight into a poorly-understood chemosensory modality.
O2 sensation in glb-5-expressing neurons requires a cGMP second messenger and specific soluble guanylate cyclase homologs (sGCs) that detect O2 upshifts and downshifts (Zimmer et al., 2009). Members of this sGC subfamily can bind O2 through a heme group, and have been proposed to generate cGMP in an O2–regulated manner (Gray et al., 2004; Zimmer et al., 2009). sGCs are essential for all O2-sensitivity in URX, but the HW glb-5 polymorphism has a more subtle effect: it increases URX sensitivity to a small 20–21% O2 upshift, suggesting that it alters the most-preferred O2 level. This interpretation is consistent with glb-5 modulation of O2 dependent aggregation behavior, as aggregates have lower O2 levels than lawn borders or open lawns (Gray et al., 2004; Rogers et al., 2006). One appealing reason to study quantitative genetic variation is the ability to discover biologically important molecules with subtle phenotypes, like glb-5, that might not emerge from classical forward genetic screens.
Our identification of the causal mutations for two behavioral QTLs suggests that RIAIL strategies could be used to find causal mutations for other behavioral QTLs with similar effect magnitudes, and possibly QTLs with smaller effects (Mott and Flint, 2008). In mice, QTLs of comparable magnitude to those described here have been found for exploratory behaviors, open field activity, and drug preference (Flint, 2003). The most important technical advance for this work was the high mapping resolution of the RIAIL strains (Rockman and Kruglyak, 2008), which increased the information per strain compared to classical RIL analysis. The large number of breakpoints in each RIAIL between N2 and HW DNA and the high density of the SNP markers allowed us to localize the new QTL on chromosome V to 19 genes by phenotyping only 78 strains. RIAILs can also provide advantages over association studies in wild strains, because RIAILs are less subject to mapping difficulties caused by linkage disequilibrium or family structure (Rockman and Kruglyak, 2009).
When and where did the polymorphisms in npr-1 and glb-5 arise? The genotyping of wild strains showed that N2-like alleles are evolutionarily recent. Only 13 of 203 C. elegans strains had an N2 allele of either glb-5 or npr-1, and high-density analysis at 1454 SNP loci indicates that the npr-1 and glb-5 polymorphisms arose recently in a common, N2-like genetic background, which in one case mated with a Bergerac-like strain. In this context, it is possible that the strains with N2-like npr-1 and glb-5 alleles are not actually independent wild strains, but re-isolates of laboratory N2 or N2-derived strains. Many of the N2-like strains were isolated by procedures on exposed laboratory benches with a high potential for cross-contamination (P. Anderson, P. Phillips and C. Johnson, personal communication). Molecular results also raise suspicions about the N2-like strains CB4555 and DR1349, whose genotypes are inconsistent with their recorded history but consistent with N2 cross-contamination (Hodgkin and Doniach, 1997). The fact that the four “Group 2” strains can be explained as a cross between N2 and Bergerac is problematic, as Bergerac strains were maintained in the laboratory for over thirty years in association with N2; N2 and Bergerac were often intercrossed in the laboratory to generate strains with high transposon activity (Hodgkin and Doniach, 1997).
We suggest that the N2 alleles of npr-1 and glb-5 may have originated in the laboratory, where N2 was cultivated for nearly two decades before permanent frozen cultures were established in 1969. The N2 strain was originally isolated around 1951 by Warwick Nicholas (personal communication) from mushroom compost produced by L.N. Staniland; a sample was given to Ellsworth Dougherty at UC Berkeley, who sent a sample to Sydney Brenner, who isolated a single hermaphrodite whose progeny became N2 (Hodgkin and Doniach, 1997). From the outset, the Brenner strain had the non-aggregating behavior characteristic of the N2 npr-1 allele (S. Brenner, personal communication). A second strain derived from the Dougherty lab, LSJ1, is identical to N2 at 1453 of 1454 SNPs, but has HW alleles of glb-5 and npr-1 (Fig. 4B, Supplementary Table 3; N. Lu, personal communication). The existence of HW-like and N2-like alleles of npr-1 and glb-5 in one genetic background from the Dougherty lab, and only HW-like alleles in most wild-caught strains, suggests that the N2 alleles arose after N2 and LSJ1 separated in the laboratory. C. elegans has a high mutation rate (~2×10−8 mutations per site per generation)(Denver et al., 2004), so npr-1 and glb-5 mutations could have occurred without directed mutagenesis.
The N2 alleles of glb-5 and npr-1 could be random mutations fixed by genetic drift, but it would seem to be an unlikely coincidence that both mutations affect O2 responses. Alternatively, one or both of these mutations might have conferred a selective advantage in the laboratory. N2 alleles of both npr-1 and glb-5 modify behaviors at 21% O2, the normal condition in the laboratory, which is higher than C. elegans’s preferred level of 5–10% O2 (Gray et al., 2004). At 21% O2, strains with the HW allele of npr-1 spend less time than N2 on good food sources, and are more easily killed by pathogenic bacteria (Gloria-Soria and Azevedo, 2008; Reddy et al., 2009; Styer et al., 2008); selection for successful behaviors at high O2 could have contributed to the fixation of npr-1 mutant alleles in N2. Whether the glb-5 polymorphism has similar effects is unknown. In addition, strains with the HW allele of npr-1 aggregate and burrow into the agar (de Bono and Bargmann, 1998), and experimentalists who isolate single animals from the agar surface would have favored solitary N2-like animals. In a distant analogy to aggregation, laboratory yeast strains have undergone mutations that favor cell separation compared to their wild ancestors (Verstrepen et al., 2003; Yvert et al., 2003).
Behavioral adaptations are likely to have occurred in many experimental species during their cultivation in the laboratory. Domesticated animals show less aggressiveness, less fearful behavior, and better mating in captivity than their wild ancestors (Grandin and Deesing, 1998); similar selections are placed on laboratory strains. If recognized, laboratory adaptations can be addressed experimentally. For example, regardless of whether N2 alleles of npr-1 and glb-5 are laboratory adaptations, the predominance of HW alleles in wild strains suggests that the HW-like response to O2 and CO2 is more ecologically relevant. Therefore, in future studies of C. elegans behavior it should be worthwhile to characterize behaviors in strains with the HW alleles of glb-5 and npr-1, particularly for behaviors that are sensitive to O2 and CO2.
Methods
Genetics and molecular biology
Strains were grown and maintained under standard conditions (Brenner, 1974). N2 is C. elegans Bristol strain N2; HW is C. elegans strain CB4856. Additional strains were isolated from two locations in California (see Supplementary material), or were acquired from the CGC or from Marie-Anne Felix, Antoine Barriere, Michael Ailion, Jody Hey, and Elie Dolgin. A complete strain list and details of strain construction for introgression lines are found in Supplementary Material. Standard molecular methods were used; genotyping primers are found in Supplementary Material.
Quantitative genetics
78 recombinant inbred advanced intercross lines from a reciprocal cross between N2 and CB4856 were analyzed. The RIAILs were inbred from a ten generation intercross employing random pair mating with equal contributions of each pair to each generation (Rockman and Kruglyak, 2008, 2009). Because of population size attrition during the intercross phase, due to failed crosses and segregating mortality and sterility, two separate inbred lines were derived from hermaphrodites in the tenth generation of the cross. The resulting RIAILs include pairs of lines that exhibit elevated relatedness; the lines are largely unique because of the random allelic fixation and breakpoint accumulation during the inbreeding phase of RIAIL construction. Nevertheless, to avoid artefactually high lod scores and low P-values from the background relatedness among these lines, we divided the dataset into two subsets, with each pair of related strains split between the subsets. Linkage scans were performed separately for the two subsets and lod scores were then summed, and p-values were estimated from 10,000 permutations performed separately for the two subsets.
Line means plotted in Fig. 1F were used as phenotypes for nonparametric interval mapping in Rqtl (Broman et al., 2003). Lod scores were computed at each marker and at intervals of 1 cM; these genetic distances are estimated from the recombination fractions in the RIAILs as though they were observed in a backcross, using the Haldane map function. Given the presence of selection during the RIAIL construction (Seidel et al., 2008), there is no analytical method available to convert RIAIL recombination fractions into meiotic recombination rates. However, our high marker density renders the exact form of map function irrelevant. Three markers that did not vary within one of the subsets were excluded, leaving 1452 markers. QTLs were incorporated into additional rounds of interval mapping as covariates in parametric analysis under a normal model, and no additional QTLs were found. The fraction of variance explained by each QTL and their interactions were estimated using ANOVA. The structured nonparametric linkage scan for the O2 increase/CO2 decrease trait yielded two peaks on the X chromosome, the peak at npr-1 (lod score 5.2), coincident with the QTL from the O2 decrease/CO2 increase trait, and a peak 3 Mb to the right, at 7.7 Mb (lod score 5.4). However, incorporation of the npr-1 genotype as a covariate in the analysis eliminates the peak at 7.7 Mb, suggesting that this second peak is attributable to noise whose baseline is elevated by linkage to npr-1. Similarly, incorporation of glb-5 genotype as a covariate causes the X-linked QTL to localize at the npr-1 peak.
For the O2 increase/CO2 decrease response, a significant interaction effect between the two QTLs accounts for 15% of the among-line variance (F1 = 63, P < 10−10). This apparent interaction may be explained by a bounded phenotype distribution. The phenotype, turns per minute, is close to zero in an npr-1N2 background; the suppression of turning by O2 upshift/CO2 downshift could be masked by that low basal level, and easier to see in the more motile npr-1HW background.
Behavioral assays
O2 and CO2-evoked turning responses were monitored on an NGM plate seeded with OP50 bacteria (grown overnight), with 20–30 adult animals confined to a 28mm × 28mm region using Whatman filter paper dipped in 20mM CuCl2. A custom-designed Plexiglass device containing an inlet, an outlet and a 30 mm × 30 mm × .3 mm behavioral arena created laminar air flow over the animals. Gas tanks were ordered at primary mixture grade from Matheson TriGas. A standard tank at 21.2%O2/78.8%N2 was mixed 99:1 with either 100% N2 or 100% CO2 to create a tank at 21%O2/79%N2 or 21%O2/1%CO2/78%N2. A standard tank at 20.2%O2/79.8%N2 was similarly used to create tanks at 20%O2/80%N2 or 20%O2/1%CO2/79%N2. Gas mixtures were bubbled through water at a flow rate of 50 cm3/min, and a Hamilton MVP was used to switch between two gas tanks every three minutes for 60 minutes. Animals were recorded at 3 frames/second using a Zeiss microscope and Pixelink PL-A741 Monochrome Camera, and automatically tracked using Matlab software (Chalasani et al., 2007; Ramot et al., 2008).
For each assay, the first 6-minute cycle between gas mixtures was discarded, leaving 9 identical 6 minute intervals that were averaged together. Each recording was binned into 20 second windows. Data for all strains are included in Supplementary Table 1. To create data for changes in turning rate, average turning rates for 80 seconds prior to a shift in gas concentration were subtracted from the average turning rate 20–60 seconds after each shift. Averages and standard errors represent at least 3 experiments for each strain and condition, except for Fig. 1F, in which the final index for each RIAIL was averaged from two separate experiments.
Statistics were calculated using GraphPad Prism software, using either the unpaired two-tailed Student’s t-test or 1-way ANOVA with the Tukey correction for multiple comparisons between strains, as appropriate. To determine if CO2 downshifts interacted with O2 upshifts, multivariate linear regression was run in R to determine if an interaction term was significant. Full statistics are included in Supplementary Table 2.
Aggregation and bordering behaviors were measured essentially as described (de Bono and Bargmann, 1998; Gray et al., 2004). 60 young adult animals were picked onto a circular lawn of 50 μl E. coli OP50 (previously grown at room temperature for four days). After one hour, the aggregation and bordering fraction was scored. Each strain was analyzed at least three times.
Calcium imaging
Transgenic animals expressing the fluorescent calcium sensor G-CaMP1.0 in URX neurons were exposed to O2 upshifts and downshifts while trapped in a custom-fabricated PDMS device (Zimmer et al., 2009). The two-layer device allowed rapid diffusion of gas mixtures from a flow chamber into a calibrated channel containing the trapped animal. Fluorescence intensities were measured with a Nikon CoolSnap camera attached to a Zeiss Axioplan microscope while switching between different gas mixtures in the flow chamber, and analyzed with a script written in MetaMorph programming language. ΔF/F0 was calculated as the percent change in fluorescence relative to the mean basal fluorescence (F0) from 1–4 s of each recording.
Supplementary Material
Acknowledgments
We thank members of the Bargmann laboratory for critical help, advice, and comments on the manuscript, Phil Anderson, Jonathan Hodgkin, Carl Johnson, Nancy Lu, Warwick Nicholas, Patrick Phillips, and Sydney Brenner for sharing information about the historical isolation of C. elegans strains, and Marie-Anne Felix, Antoine Barriere, Michael Ailion, Jody Hey, and Elie Dolgin for wild-caught strains. This work was supported by the Howard Hughes Medical Institute (C.I.B.), the NIH (R01 HG004321 to L.K. and P50 GM071508 to the Lewis-Sigler Institute), the James S. McDonnell Foundation Centennial Fellowship (L.K.), the Damon Runyon Cancer Research Foundation (P.T.M.) and the Jane Coffin Childs Foundation (M.V.R.). C.I.B. and L.K. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Author contributions: P.T.M. designed and performed experiments, analyzed data and wrote the paper; M.V.R designed and performed experiments and analyzed data; M.Z. and H.J. performed experiments; E.Z.M. created reagents; L.K. designed experiments and analyzed data; and C.I.B. designed experiments, analyzed data and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher AJ, Busch KE, de Bono M. A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:8044–8049. doi: 10.1073/pnas.0707607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 2006;4:e274. doi: 10.1371/journal.pbio.0040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung BH, Cohen M, Rogers C, Albayram O, de Bono M. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol. 2005;15:905–917. doi: 10.1016/j.cub.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Coates JC, de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature. 2002;419:925–929. doi: 10.1038/nature01170. [DOI] [PubMed] [Google Scholar]
- de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- Egilmez NK, Ebert RH, 2nd, Shmookler Reis RJ. Strain evolution in Caenorhabditis elegans: transposable elements as markers of interstrain evolutionary history. J Mol Evol. 1995;40:372–381. doi: 10.1007/BF00164023. [DOI] [PubMed] [Google Scholar]
- Flint J. Analysis of quantitative trait loci that influence animal behavior. J Neurobiol. 2003;54:46–77. doi: 10.1002/neu.10161. [DOI] [PubMed] [Google Scholar]
- Gloria-Soria A, Azevedo RB. npr-1 regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr Biol. 2008;18:1694–1699. doi: 10.1016/j.cub.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Grandin T, Deesing MJ. Behavioral genetics and animal science. In: Grandin T, editor. Genetics and the Behavior of Domestic Animals. San Diego: CA, Academic press; 1998. pp. 1–30. [Google Scholar]
- Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- Greenway H, Armstrong W, Colmer TD. Conditions leading to high CO2 (>5 kPa) in waterlogged-flooded soils and possible effects on root growth and metabolism. Ann Bot (Lond) 2006;98:9–32. doi: 10.1093/aob/mcl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Sternberg PW. Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:8038–8043. doi: 10.1073/pnas.0707469105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, De Henau S, Dewilde S, Moens L, Couvreur M, Borgonie G, Vinogradov SN, Roy SW, Vanfleteren JR. The Caenorhabditis globin gene family reveals extensive nematode-specific radiation and diversification. BMC Evol Biol. 2008;8:279. doi: 10.1186/1471-2148-8-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, Geuens E, Dewilde S, Vierstraete A, Moens L, Vinogradov S, Vanfleteren JR. Wide diversity in structure and expression profiles among members of the Caenorhabditis elegans globin protein family. BMC Genomics. 2007;8:356. doi: 10.1186/1471-2164-8-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS. Twin studies of psychiatric illness: an update. Arch Gen Psychiatry. 2001;58:1005–1014. doi: 10.1001/archpsyc.58.11.1005. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Greenspan RJ. The nature of genetic influences on behavior: lessons from “simpler” organisms. Am J Psychiatry. 2006;163:1683–1694. doi: 10.1176/ajp.2006.163.10.1683. [DOI] [PubMed] [Google Scholar]
- Mackay TF. The genetic architecture of quantitative traits: lessons from Drosophila. Curr Opin Genet Dev. 2004;14:253–257. doi: 10.1016/j.gde.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Maydan JS, Flibotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 2007;17:337–347. doi: 10.1101/gr.5690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Flint J. Prospects for complex trait analysis in the mouse. Mammalian Genome. 2008;19:306–308. doi: 10.1007/s00335-008-9110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienhaus K, Nienhaus GU. Searching for neuroglobin’s role in the brain. IUBMB Life. 2007;59:490–497. doi: 10.1080/15216540601188538. [DOI] [PubMed] [Google Scholar]
- Osborne KA, Robichon A, Burgess E, Butland S, Shaw RA, Coulthard A, Pereira HS, Greenspan RJ, Sokolowski MB. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, Kruglyak L. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Morse TM, Lockery SR. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19:9557–9569. doi: 10.1523/JNEUROSCI.19-21-09557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramot D, Johnson BE, Berry TL, Jr, Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS ONE. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Breeding designs for recombinant inbred advanced intercross lines. Genetics. 2008;179:1069–1078. doi: 10.1534/genetics.107.083873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Recombinational landscape and population genomics of C. elegans. PLOS Genetics. 2009 doi: 10.1371/journal.pgen.1000419. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol. 2006;16:649–659. doi: 10.1016/j.cub.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Ryu WS, Samuel AD. Thermotaxis in Caenorhabditis elegans analyzed by measuring responses to defined thermal stimuli. J Neurosci. 2002;22:5727–5733. doi: 10.1523/JNEUROSCI.22-13-05727.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D, Ch’ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008;322:460–464. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvia DM, Fuhrmann JJ, Hartel PG, Zuberer DA. Principles and Applications of Soil Microbiology. Upper Saddle River: New Jersey, Prentice Hall; 1998. [Google Scholar]
- Toma DP, White KP, Hirsch J, Greenspan RJ. Identification of genes involved in Drosophila melanogaster geotaxis, a complex behavioral trait. Nat Genet. 2002;31:349–353. doi: 10.1038/ng893. [DOI] [PubMed] [Google Scholar]
- Verstrepen KJ, Derdelinckx G, Verachtert H, Delvaux FR. Yeast flocculation: what brewers should know. Appl Microbiol Biotechnol. 2003;61:197–205. doi: 10.1007/s00253-002-1200-8. [DOI] [PubMed] [Google Scholar]
- Wang L, Dankert H, Perona P, Anderson DJ. A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proceedings of the National Academy of Sciences USA. 2008;105:5657–5663. doi: 10.1073/pnas.0801327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RE, Vinogradov SN. Nonvertebrate hemoglobins: functions and molecular adaptations. Physiol Rev. 2001;81:569–628. doi: 10.1152/physrev.2001.81.2.569. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Fullerton J, Miller S, Keays DA, Brady S, Bhomra A, Jefferson A, Volpi E, Copley RR, Flint J, Mott R. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nature Genetics. 2004;36:1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Gray JM, Pokala N, Chang AJ, Karow DS, Marletta MA, Hudson ML, Morton DB, Chronis N, Bargmann CI. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron . 2009 doi: 10.1016/j.neuron.2009.02.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.