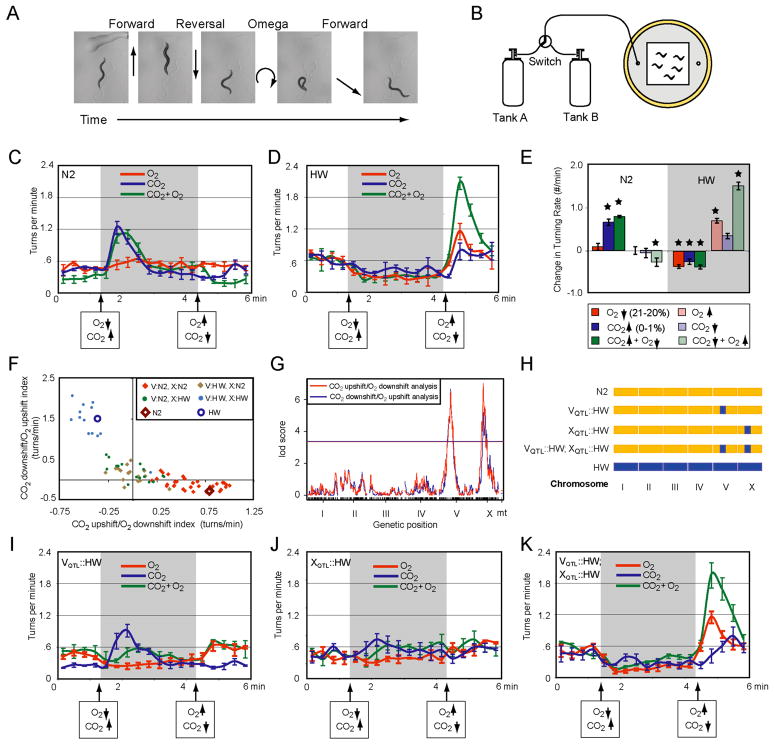
Fig. 1. Quantitative genetic analysis of O2 and CO2 responses in two C. elegans strains.
A. Forward movement interrupted by a reversal and omega turn.
B. Behavioral arena for turning assay. 20–30 animals were recorded in the presence of OP50 bacteria as O2 and CO2 concentrations were controlled by regulated gas flow.
C,D. Average turn rates of N2 and CB4856 (HW) strains across nine six-minute pulse sequences (see Methods). In all Figures, O2 changes were 21% O2->20% O2->21% O2, CO2 changes were 21% O2->21% O2/1% CO2->21% O2, and simultaneous O2 and CO2 changes were 21% O2->20% O2/1% CO2->21% O2. The balance in all mixtures was N2.
E. Average change in turning rate before and after the indicated step change (data in C and D). Stars denote significant effects of the step change (P<0.05, t-test).
F. Behavioral responses of 78 RIAILs to simultaneous O2 and CO2 step changes. Y-axis, difference in turning frequency after CO2 downshift/O2 upshift; X-axis, difference in turning frequency after CO2 upshift/O2 downshift. N2 and HW reference strains are included; each RIAIL is color-coded by genotype at the QTLs on chromosome V and X.
G. QTL analysis of data from F, showing significant QTL peaks on the left arms of V and X. Horizontal line represents the genome-wide significance threshold (P=0.05).
H. Schematic of three introgression strains with small regions of HW DNA around the QTLs on V and X introduced into an N2 background.
I–K. Turning responses of introgression strains, as in C and D.
In all figures, error bars indicate standard error of mean (SEM), and Supplementary Tables 1 and 2 have full datasets and statistics.