Abstract
Lactose repressor protein (LacI) utilizes an allosteric mechanism to regulate transcription in E. coli, and the transition between inducer- and operator-bound states has been simulated by targeted molecular dynamics (TMD). The side chains of amino acids 149 and 193 interact and were predicted by TMD simulation to play a critical role in the early stages of the LacI conformational change. D149 contacts IPTG directly, and variations at this site provide the opportunity to dissect its role in inducer binding and signal transduction. Single mutants at D149 or S193 exhibit minimal change in operator binding, and alterations in inducer binding parallel changes in operator release, indicating normal allosteric response. The observation that the double mutant D149A/S193A exhibits wild-type properties excludes the requirement for inter-residue hydrogen bond formation in the allosteric response. The double mutant D149C/S193C purified from cell extracts shows decreased sensitivity to inducer binding, while retaining wild-type binding affinities and kinetic constants for both operator and inducer. By manipulating cysteine oxidation, we show that the more reduced state of D149C/S193C responds to inducer more similarly to wild-type protein, whereas the more oxidized state displays diminished inducer sensitivity. These features of D149C/S193C indicate that the novel disulfide bond formed in this mutant impedes the allosteric transition, consistent with the role of this region predicted by TMD simulation. Together, these results establish the requirement for flexibility of spatial relationship between D149 and S193 rather than a specific D149-S193 interaction in the LacI allosteric response to inducer.
Allosteric regulation is a major mechanism of control in many biological processes, including cell signaling as well as genetic and metabolic regulation (1, 2). Although allosteric behavior is a common feature for many regulatory proteins, a detailed atomic-level description of this mechanism remains largely elusive. Lactose repressor protein (LacI) is a prototypic genetic regulatory protein that regulates transcription of the lac operon genes (3), which encode enzymes responsible for metabolism of lactose in Escherichia coli (3, 4). High affinity binding of repressor to operator sequences within the lac operon (5) precludes transcription of the lac metabolic enzymes by RNA polymerase (4). Diminished affinity for the target operator DNA and a consequent increase in lac mRNA transcription result from a conformational change in response to binding a metabolite of lactose (1,6-allolactose) to a site within LacI distant from the DNA site (4).
LacI is a well-characterized tetrameric protein comprised of identical monomers. The structure of the monomer can be dissected into three major elements (Figure 1A and 1B): an N-terminal DNA binding domain (amino acids 1–60); a core domain (amino acids 61–330) that binds inducer and forms the dimer interface; and a C-terminal domain (amino acids 331–360) that generates the tetramer interface (6–9). The core domain of LacI is structurally homologous with the venus fly-trap (VFT) family that includes bacterial periplasmic binding proteins (PBPs) involved in transport and the extracellular domain of G-protein coupled receptors (GPCRs) as well as other receptors that transduce extracellular stimuli into intracellular signals and exhibit subdomain movements associated with function (10–12). In this larger functional context, significant efforts have been made to study allosteric behavior in LacI, which brings enormous biochemical and structural information to facilitate more detailed analysis (4, 8, 13–17).
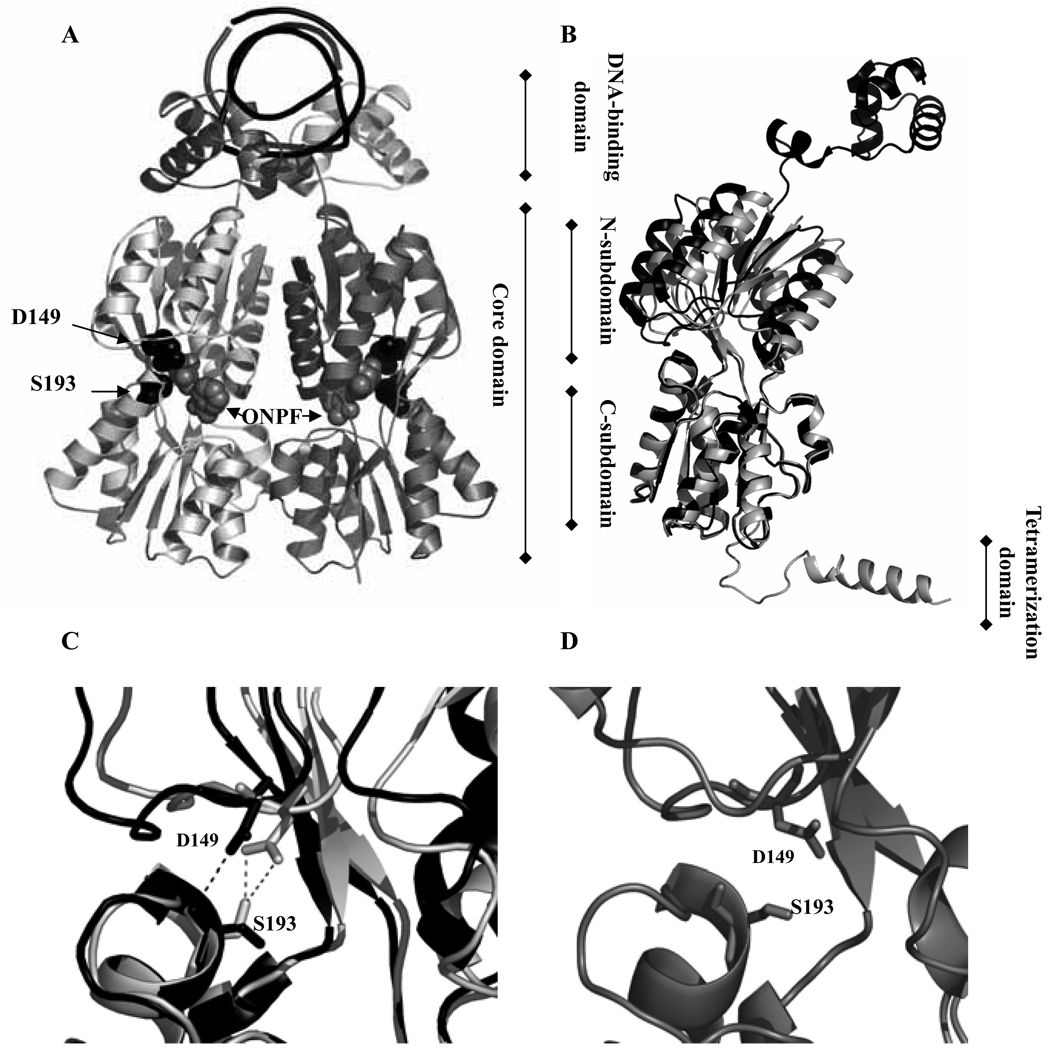
Figure 1.
(A) Structure for dimeric LacI bound to Osym DNA (top, black) and anti-inducer ONPF (gray space-filling) with positions 149 and 193 highlighted in black space-filling (dimer variant from PDB 1efa) (16). The two monomers are displayed in distinct colors (light gray, dark gray). (B) Monomer from 1efa (black) (16) and monomer from 1lbh (gray) (8) were aligned using residues in the core C-subdomain. Note the changes in the N-subdomains. (C) Detailed view of D149 and S193 in Osym•ONPF•LacI structure (1efa, black) (16) and IPTG·LacI structure (1lbh, gray) (8). Residues 149 and 193 are highlighted by sticks and hydrogen bonds are shown by dashed line. Note the movement of the backbone surrounding D149 and the altered positions of side chains of D149 and S193 in these two conformations. (D) The intermediate state simulated by TMD (18). Notice that the side chain of residue 149 is in a position different from both IPTG-bound and DNA-bound states.
The three-dimensional structures of the core domain and full-length LacI in both operator- and IPTG- bound conformations are available (7, 8, 16). The shift in LacI structure between operator- and inducer-bound conformations requires both rotation and translation of the N-subdomain within the core domain (Figure 1B) (8, 16). Amino acids D149 and S193 are located near the IPTG binding pocket at the interface of the N- and C- subdomains (Figure 1A). Shifts in the side chains at D149 and S193 as well as substantial movement of the backbone of D149 are observed between DNA- and IPTG-bound LacI structures (Figure 1C). D149 occupies a key position at the start of a flexible loop that comprises residues from 149 to 156. In the DNA-bound structure, a single bond is formed between the backbone of S193 and the side chain of D149; however, in the IPTG-bound structure, the D149 side chain forms two hydrogen bonds to the S193 side chain in each monomer (Figure 1C) (8, 16). This alteration suggests D149 and S193 might be participants in determining the distinct conformational states of the repressor.
Recently, a TMD simulation was utilized to simulate the conformational transition pathways in LacI (18). This prediction for the operator-bound to IPTG-bound forms shows significant movement in the flexible loop containing D149 at early stages of the conformational change. Mobility in this flexible loop allows the side chain of residue 149 to move ∼4Å, forming hydrogen bonds with the side chain at 193. These hydrogen bonds may secure the flexible loop in place (18). This event appears critical in the initial stages of propagating the message of inducer-binding through the N-subdomain of LacI to the DNA binding domain. In the intermediate state of TMD simulation (Figure 1D) (18), the side chain of residue 149 points to a position neither close to the IPTG-bound nor to the DNA-bound state. This observation further supports the hypothesis that the region around residue 149 contributes to transmitting the allosteric signal. Consistent with their predicted importance, alterations at positions 149 and 193 resulted in diminished response to IPTG binding in phenotypic screens (14). Further, because D149 is among the residues that directly contact inducer IPTG, exploring this region provides an opportunity to examine whether IPTG binding and signal transduction capacities can be uncoupled. To dissect the structural and functional influence of this specific region, multiple single substitutions at positions 149 or 193 were examined. Double substitutions, D149A/S193A and D149C/S193C, were constructed and their biochemical and biophysical characteristics examined to explore the role of specific side chain interaction and flexibility of the region in LacI allosteric response.
Materials and methods
Site-specific mutagenesis and purification
Plasmid pJC1 (9) containing the complete LacI gene was used as the expression vector in E. coli BLIM cells (19). All substitutions were derived by site-directed mutagenesis (Quickchange, Stratagene Inc.) using the corresponding oligonucleotides. Full sequencing of each LacI gene (SeqWright Inc.) verified the presence of only the expected mutation. For protein expression, cells transformed with specific plasmids were grown in 2 × YT liquid media in a shaking incubator at 37 for ∼20 hours. The cells were centrifuged and resuspended in breaking buffer (0.2 M Tris-HCl, pH 7.6, 0.2 M KCl, 0.01 M Mg acetate, 5% glucose, 0.3 mM DTT and 0.3 µM PMSF) containing lysozyme and frozen at −20. The frozen cells were slowly thawed and treated with 80 µl of 10 mg/ml DNaseI and MgCl2 (final concentration of 10 mM). Following centrifugation, the supernatant was mixed with ammonium sulfate to final 37% saturation, centrifuged, and the resuspended pellet was dialyzed overnight against 0.09 M KP buffer (0.09 M potassium phosphate, pH 7.6, 0.3 mM DTT, and 5% glucose) at 4. After removing any precipitate by centrifugation, the supernatant was applied to a phosphocellulose column equilibrated in 0.09 M KP buffer and eluted with a gradient from 0.12–0.30 M KP buffer (0.12–0.30 M potassium phosphate, pH 7.6, 0.3 mM DTT, and 5% glucose). Protein purity was assessed by using SDS-PAGE and was ≥ 90%. Stoichiometric DNA binding activity for each repressor variant was determined by filter binding assay described in the following section. Activities for purified proteins were ≥ 90%.
Operator binding
DNA binding experiments were performed by nitrocellulose filter binding assay (20, 21) with 40-bp natural operator (5' – TGTTGTGTGGAATTGTGAGCGGATAACAATTTCACACAGG – 3') (Biosource International). This assay was carried out in FB buffer (0.01 M Tris-HCl, pH 7.4, 0.15 M KCl, 0.1 mM DTT, 0.1 mM EDTA, and 5% DMSO) with 100 µg/mL of bovine serum albumin. Protein at various dilutions (1 × 10−13 to 5 × 10−9 M) was first mixed with 32P-labeled DNA. To determine DNA activity under stoichiometric conditions, the percentage of active repressor was measured with DNA concentration at least 10-fold above the Kd. To measure DNA binding affinity, DNA concentration was set at least 10-fold below the Kd. After a short incubation, the mixtures were filtered through nitrocellulose using a 96-well plate and exposed to a Fuji phosphorimaging plate overnight. The plate was read by a Fuji phosphorimager, and the radioactivity was quantified and analyzed with Igor Pro (Wavemetrics) to determine the binding affinity with the following equation:
| (1) |
where Yobs is the observed level of retained radioactivity at a given protein concentration, Ymax is the level of radioactivity measured at saturation, c is the background radioactivity detected when no repressor was present, and Kd is the equilibrium dissociation constant. The value of the Hill coefficient, n, is generally ∼1 for LacI.
IPTG binding
IPTG binding to repressor proteins was monitored by the fluorescence emission shift for intrinsic tryptophan that has been shown to accompany IPTG binding (22). The protein solution was placed in a 1-cm2 quartz cuvette at room temperature, and the fluorescence emission intensity change at wavelength > 350 nm was monitored at varying IPTG concentration (22). Protein concentration was fixed at ∼1.5 × 10−7 M monomer in fluorescence buffer containing 0.01 M Tris-HCl, 0.15 M KCl, pH 7.4, and IPTG concentration was in the range of 1 × 10−8 to 1 × 10−3 M. The experiment was performed using SLM-Aminco AB2 spectrofluorometer with an excitation wavelength of 285 nm. The emission spectra were recorded between 300 and 380 nm. The binding affinity for the protein was determined by fitting the data in Igor Pro using equation 2,
| (2) |
where Kd is the equilibrium dissociation constant; Yobs is the measured fluoresence at a given IPTG concentration; Ymax is the maximum change in fluorescence signal between zero and saturating inducer; c is a constant background coefficient; and n is the Hill coefficient, which is ∼1 in absence of operator.
Operator release
The operator release assay measures the effect of inducer sugar on DNA binding affinity (23). IPTG ranging from 10−8 to 10−3 M was added following mixture of the protein and labeled operator DNA in FB buffer with 100 µg/mL of bovine serum albumin. After incubation for ∼30 min, nitrocellulose filter binding was used to monitor the level of DNA bound in the presence of varied inducer concentration. The concentration of DNA was at least 10-fold below the Kd obtained from the operator binding assay (∼10−12 M), and the concentration of protein was set to ∼80% of maximal DNA binding for the specific repressor protein. Data were analyzed by Igor Pro using a variation of equation 2. In this case, Kd is substituted by [IPTG]mid, corresponding to the concentration of IPTG required to release 50% of the protein-DNA complex. Yobs is the measured radioactivity at a specific inducer concentration; Ymax is the maximum radioactivity differential observed, and c is the radioactivity with no protein present.
Kinetic measurements of repressor-IPTG association
The association rates for IPTG binding to LacI were measured using a stopped flow system equipped with fluorescence detection (Applied Photophysics Ltd.) (24). The association processes were monitored as the fluorescence decrease due to IPTG binding. The excitation wavelength was 285 nm, and fluorescence emission was monitored using a 350 nm cut-off filter. Time courses were measured for the association of repressor with IPTG under pseudo-first-order conditions. Final LacI concentration was 1.0 × 10−6 M, and IPTG concentrations were varied from 0.05 mM to 0.2 mM in buffer containing 0.01 M Tris-HCl, pH 7.4, 0.15 M KCl. The association rate constants were determined by fitting observed rate constant (kobs) using the equation:
| (3) |
where kassoc and kdissoc are the association and dissociation rate constants, respectively.
Kinetic measurements of repressor-operator dissociation
Operator dissociation rate analysis utilized the nitrocellulose binding assay (25). LacI (1.8 × 10−11 M) and 32P-labeled DNA (3.6 × 10−11 M) in a total volume of 10 mL were equilibrated for 15 min at room temperature in FB buffer containing 0.01 M Tris-HCl, pH 7.4, 0.15 M KCl, 0.1 mM EDTA, and 5% DMSO with 100 µg/mL of bovine serum albumin. An excess of unlabeled DNA over the initial DNA concentration (90-fold excess) was added to the equilibrated solution, immediately followed by inverting the tube 3 times gently. Duplicate aliquots (100 µl) were withdrawn and filtered on a nitrocellulose membrane at the desired times. Reactions were monitored until equilibrium was reached at ∼ 2 hr. The equilibrium value, corresponding to background retention of DNA, was subtracted from each time point. The dissociation rate constant was determined by fitting the data to a single exponential model.
Formation of oxidized- and reduced-state protein
The oxidized state of the double-cysteine mutant was obtained by utilizing a Hitrap desalting column (GE Healthcare Corp.) to remove DTT from the protein solution. After elution from the column, the solution was dialyzed in air-purged fluorescence buffer (0.01 M Tris-HCl, 0.15 M KCl, pH 7.4) for ∼1 hr to further enhance the oxidized state. The corresponding reduced state measurement was achieved by exposure of the protein to a high concentration of DTT and 6 M urea followed by rapid elution through a Hitrap desalting column using fluorescence buffer to remove excess DTT.
Chemical modification
Ellman’s reagent, 5,5'-dithiobis(2-nitrobenzoic acid) (DTNB), was used to determine the free sulfhydryl content in LacI (26, 27). A volume of 100 µl of the denatured protein sample (with a final monomer concentration in the range ∼1–10 µM in the presence of 6 M urea) was added to 50 µl of reaction buffer (0.1 M sodium phosphate, pH 7.6, 1 mM EDTA) and 20 µl of Ellman’s reagent. To measure the background, 100 µl of reaction buffer was utilized instead of protein solution. After incubation at room temperature to complete the reaction (∼15 min), the absorbance at 412 nm was determined against buffer background to quantify the amount of product released from the reaction between an active thiolate of protein cysteines and DTNB using an extinction coeffient ε = 14290 cm−1M−1 .
Results
Single mutations at D149 and S193
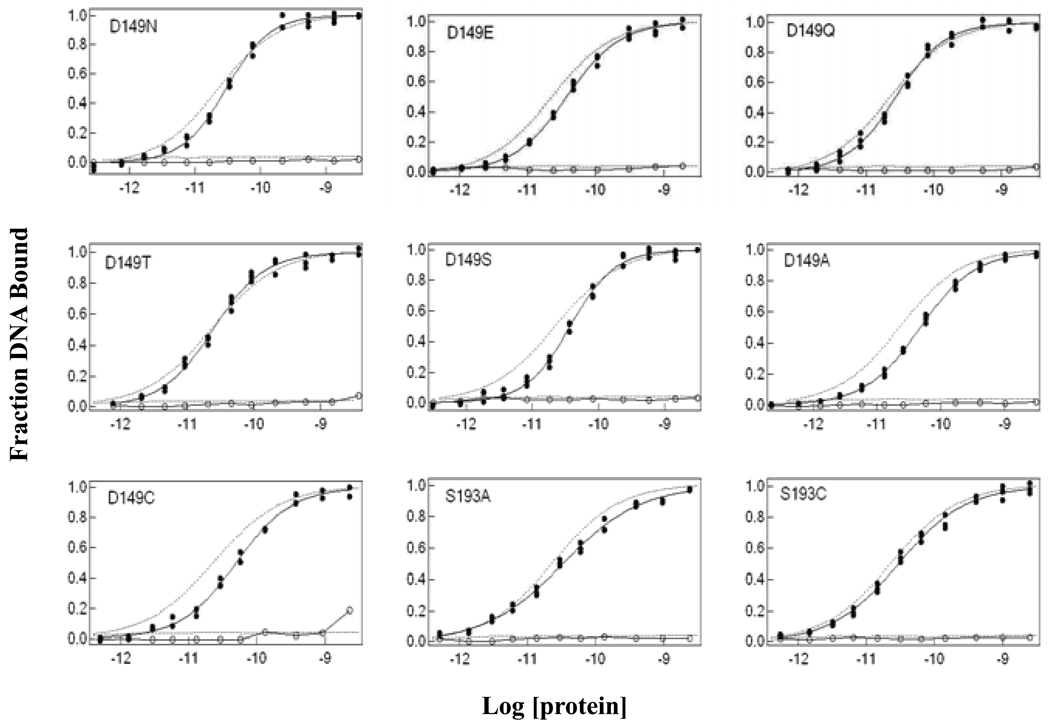
A series of single substitutions for D149 was generated to explore the functional impact of alterations in electrostatics, polarity, size, hydrogen-bonding capacity, and hydrophobic character. For operator binding affinity, D149 and S193 single mutants display values within ∼2-fold of wild-type LacI in the absence of IPTG (Figure 2; Table 1). In the presence of 1 mM IPTG, all the single mutants have sensitivity to IPTG binding similar to wild-type LacI, with the possible exception of D149C (Figure 2; Table 1). For IPTG binding, substitutions with N, S and A at residue D149 decreased binding affinity only slightly. The affinities of D149Q and D149C for IPTG were decreased by ∼4-fold, whereas those for D149T and D149E were decreased by ∼13- and ∼23-fold, respectively. Both S193A and S193C displayed comparable IPTG binding affinities to wild-type LacI (Figure 3; Table 1).
Figure 2.
Operator binding curves for wild-type LacI and single mutants. Wild-type LacI binding is represented by the dotted lines in each figure for comparison. Equilibrium dissociation constants were determined by filter binding assay as described in Materials and Methods. Protein (1 × 10−13 to 5 × 10−9) and operator DNA (∼1.5 × 10−12 M) were in FB buffer (0.01 M Tris-HCl, pH 7.4, 0.15 M KCl, 0.1 mM EDTA and 5% DMSO) with 100 µg/mL of bovine serum albumin. Fraction DNA bound in the absence of IPTG is shown by filled circles and in the presence of 1 mM IPTG by open circles. The curves shown were generated by fitting the data using Igor Pro. Results from multiple experiments are presented in Table 1.
Table 1.
Thermodynamic properties of single mutantsa
| Operator bindingbKd (M × 1011) | +IPTGcKd (M × 1011) | IPTG bindingdKd (M × 106) | Operator releasee [IPTG]mid (M × 106) | Induction Ratiof [IPTG]mid/Kd | |
|---|---|---|---|---|---|
| WTLacI | 2.2 ± 0.2 | > 10000 | 1.5 ± 0.2 | 2.2 ± 0.2 | 1.5 ± 0.2 |
| D149N | 2.5 ± 0.2 | > 10000 | 2.2 ± 0.2 | 2.5 ± 0.3 | 1.1 ± 0.2 |
| D149E | 3.4 ± 0.3 | > 10000 | 35 ± 4.0 | 110 ± 12 | 3.2 ± 0.5 |
| D149Q | 2.5 ± 0.2 | > 10000 | 7.0 ± 0.8 | 16 ± 2.3 | 2.3 ± 0.4 |
| D149T | 2.3 ± 0.2 | > 10000 | 19 ± 2.0 | 42 ± 3.6 | 2.2 ± 0.2 |
| D149S | 3.5 ± 0.3 | > 10000 | 3.2 ± 0.2 | 5.3 ± 0.5 | 1.6 ± 0.2 |
| D149A | 4.8 ± 0.3 | > 10000 | 4.2 ± 0.3 | 4.6 ± 0.5 | 1.1 ± 0.2 |
| D149C | 5.0 ± 0.4 | > 1000 | 6.0 ± 0.7 | 14 ± 1.3 | 2.3 ± 0.3 |
| S193A | 3.3 ± 0.3 | > 10000 | 1.9 ± 0.2 | 4.2 ± 0.5 | 2.2 ± 0.3 |
| S193C | 3.1 ± 0.2 | > 10000 | 2.4 ± 0.2 | 3.2 ± 0.4 | 1.3 ± 0.1 |
Each value and standard deviation were generated by fitting at least three independently generated curves. Values in bold are those significantly different from wild-type protein.
The equilibrium dissociation constants for LacI•operator DNA were measured by nitrocellulose filter binding assays in FB buffer (0.01 M Tris-HCl, pH 7.4, 0.15 M KCl, 0.1 mM DTT, 0.1 mM EDTA, and 5% DMSO) with 100 µg/mL of bovine serum albumin. DNA concentration was 1.5 × 10−12 M.
The equilibrium dissociation constants for LacI•operator DNA were measured as described in the presence of 1 mM IPTG.
The affinity of the protein for IPTG at pH 7.4 was measured by fluorescence assays in buffer containing 0.01 M Tris-HCl, pH 7.4, 0.15 M KCl. Protein concentration was fixed at 1.5 × 10−7 M.
Operator release was monitored by nitrocellulose filter binding assays in FB buffer with 100 µg/mL of bovine serum albumin. DNA concentration was at least 10-fold below Kd, and protein concentration was ∼ 80% of maximal DNA binding.
The induction ratio is given by the ratio of [IPTG]mid to the Kd for inducer binding. Repressor variants with a ratio higher than wild-type have a potential block in allosteric communication.
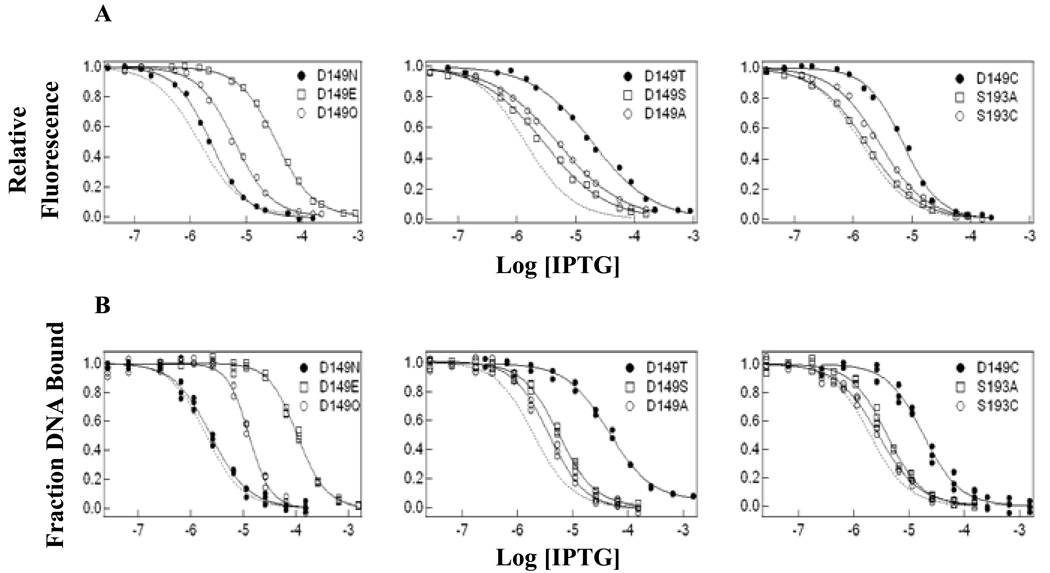
Figure 3.
(A) IPTG binding curves and (B) operator release curves for wild-type LacI and single mutants. Wild-type LacI behavior is shown by a dotted line in each figure for comparison. IPTG binding assays were conducted in fluorescence buffer (0.01 M Tris-HCl, pH 7.4, 0.15 M KCl) with protein concentration at 1.5 × 10−7 M monomer, and operator release experiments were performed in FB buffer with 100 µg/mL of bovine serum albumin with DNA concentration at 1.5 × 10−12 M as described in Materials and Methods. Results from multiple experiments are presented in Table 1.
To examine allosteric communication between DNA- and IPTG-binding sites, we quantified the release of DNA from the LacI•DNA complex in response to inducer. This assay determines the concentration of IPTG required to release 50% of the operator, which reflects the combined contributions of IPTG binding and the concomitant conformational change. For position 149, N, S or A substitutions yielded essentially wild-type operator release values, whereas substitution with E, Q, T and C shifted the mid-point of operator release. For D149E, which has most severe effect, [IPTG]mid for operator release was increased ∼50-fold. D149T had a ∼20-fold higher value, and values for both D149Q and D149 C increased ∼7-fold. For S193 single mutants, both S193A and S193C displayed operator release values comparable to wild-type LacI (Figure 3; Table 1). Therefore, a subset of the D149 substitutions results in diminished response to inducer binding with normal DNA affinity.
The alteration in IPTG release can derive from impaired IPTG binding or an impediment in the allosteric transition. The induction ratio, measured by the ratio of the operator release mid-point ([IPTG]mid) divided by the measured Kd for inducer binding, allows comparison between different LacI variants. Using this value, effects on allosteric communication can be distinguished from decreased IPTG binding affinity. For wild-type LacI, this ratio is ∼1.5. The ratios for all mutants are within ∼2-fold of wild-type LacI. Notice that although operator release [IPTG]mid values for D149E, D149Q, D149T and D149C are larger than that for wild-type LacI, their IPTG binding affinities are weaker than wild-type LacI, resulting in induction ratios comparable to wild-type (Table 1). These data suggest that substitutions at residue 149 do not significantly impact LacI allostery, even when inducer binding affinity is affected.
Double mutations at D149 and S193
Since single mutants at D149 or S193 did not significantly disrupt LacI allostery, two double mutants, D149A/S193A and D149C/S193C, were generated. The TMD simulation suggested that hydrogen bond formation between the side chains of D149 and S193 might be important to function (Figure 1C, 1D). Therefore, D149A/S193A was constructed to abolish the potential for hydrogen bond formation. Further, because the flexible loop that begins with D149 undergoes high mobility in the simulation, D149C/S193C was designed to constrain movement with a disulfide bond. In both operator-and IPTG- bound LacI structures, the distances between the Cα atoms of residues 149 and 193 are within a range favorable for disulfide formation as found for other systems (28).
Thermodynamic properties of double mutants
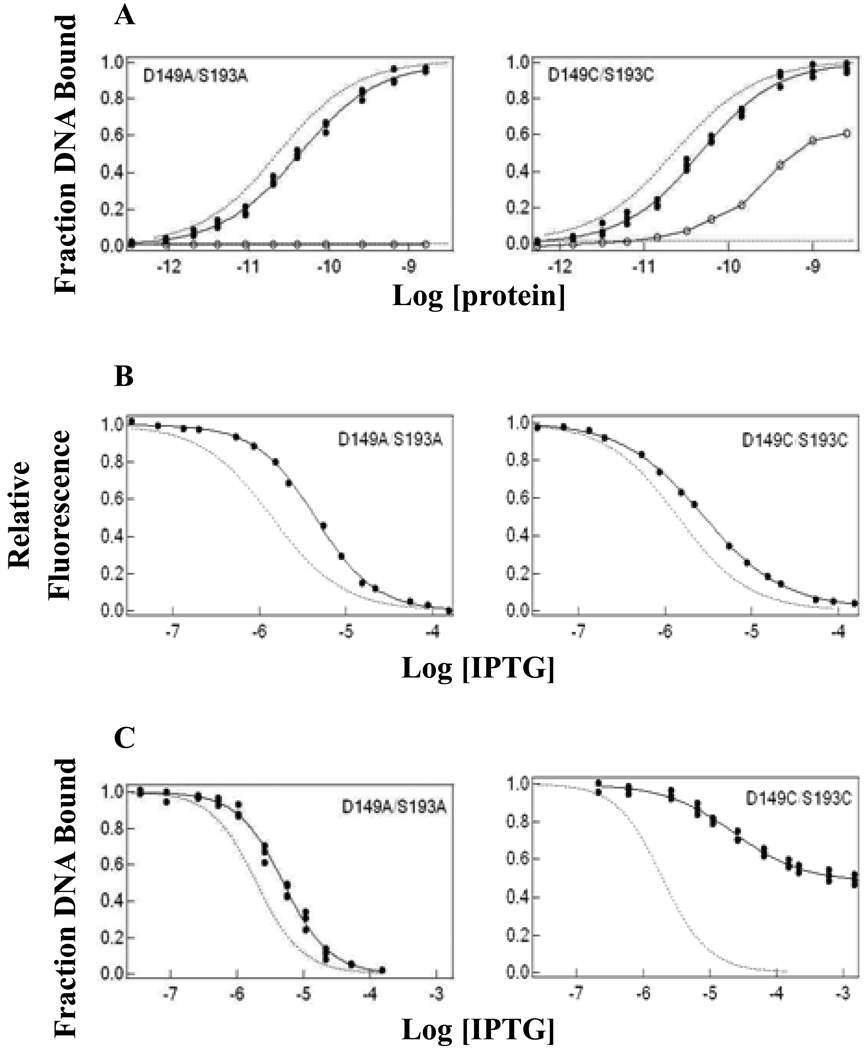
The D149A/S193A double mutant displays operator and inducer binding affinities, as well as an [IPTG]mid value for operator release, within ∼2-fold of wild-type values (Figure 4; Table 2). The inducibility for D149A/S193A is close to wild-type, demonstrating that the ability to form hydrogen bonds between residue 149 and 193 is not critical for allosteric signal transduction. Interestingly, the double mutant D149C/S193C has operator binding affinity comparable to wild-type LacI in the absence of IPTG; whereas in the presence of 1 mM IPTG, this mutant still binds to operator, reflecting decreased sensitivity to inducer binding (Figure 4; Table 2). This double-cysteine mutant exhibited IPTG binding affinity within ∼2-fold of wild-type LacI. Resistance of D149C/S193C to release of operator by inducer binding was confirmed by the operator release assay, with an over 12-fold increased [IPTG]mid value; further, the induction ratio is ∼6-fold higher than that for wild-type LacI (Table 2).
Figure 4.
(A) Operator binding, (B) IPTG binding, and (C) operator release curves for wild-type LacI and double mutant proteins. Wild-type LacI behavior is shown by a dotted line for comparison. Operator binding and release assays were carried out in FB buffer with 100 µg/mL of bovine serum albumin with DNA concentration at 1.5 × 10−12 M, whereas IPTG binding experiments were in fluorescence buffer with protein concentration at 1.5 × 10−7 M monomer as described in Materials and Methods. For operator binding curves, fraction DNA bound in the absence of IPTG is shown by filled circles and in the presence of 1 mM IPTG by open circles. Results from multiple determinations are summarized in Table 2.
Table 2.
Thermodynamic properties of double mutantsa
| Operator bindingbKd (M × 1011) | +IPTGcKd (M × 1011) | IPTG bindingdKd (M × 106) | Operator releasee [IPTG]mid (M × 106) | Induction Ratiof [IPTG]mid/Kd | |
|---|---|---|---|---|---|
| WTLacI | 2.2 ± 0.2 | > 10000 | 1.5 ± 0.2 | 2.2 ± 0.2 | 1.5 ± 0.2 |
| D149A/S193A | 4.0 ± 0.3 | > 10000 | 4.2 ± 0.4 | 4.6 ± 0.4 | 1.1 ± 0.1 |
| D149C/S193C | 4.2 ± 0.5 | ∼ 100 | 2.9 ± 0.2 | 27 ± 2.1 | 9.3 ± 1.0 |
Each value and standard deviation was generated by fitting at least three independently generated curves. Values in bold are those significantly different from wild-type LacI.
The equilibrium dissociation constants for LacI•operator DNA were measured by nitrocellulose filter binding assays in FB buffer (0.01 M Tris-HCl, pH 7.4, 0.15 M KCl, 0.1 mM DTT, 0.1 mM EDTA, and 5% DMSO) with 100 µg/mL of bovine serum albumin. DNA concentration was 1.5 × 10−12 M.
The equilibrium dissociation constants for LacI•operator DNA in the presence of 1 mM IPTG.
The affinity of the protein for IPTG at pH 7.4 was measured by fluorescence in buffer containing 0.01 M Tris-HCl, pH 7.4, 0.15 M KCl. Protein concentration was fixed at 1.5 × 10−7 M.
Operator release was monitored by nitrocellulose filter binding assays in FB buffer with 100 µg/mL of bovine serum albumin. DNA concentration was at least 10-fold below Kd, and protein concentration was ∼ 80% of maximal DNA binding.
The induction ratio is given by the ratio of [IPTG]mid to the Kd for inducer binding. Repressors with a ratio higher than wild-type have a potential block in allosteric communication.
To explore whether reciprocal behavior could be measured for operator impact on IPTG binding, [14C]-IPTG binding was measured in the assay buffer for operator binding. The retention of IPTG at protein concentrations just above the protein•DNA Kd and stoichiometric operator DNA was diminished by ∼2-fold for D149C/S193C compared to wild-type LacI (data not shown), consistent with expectations from DNA binding results. These distinct properties of D149C/S193C meet the criteria for an allosteric LacI mutant, and flexibility for movement in the region encompassed by D149 and/or S193 appears critical for allosteric regulation.
Kinetic measurements of repressor-IPTG association
To ensure that the differences for D149C/S193C were not kinetic in origin, wild-type and D149C/S193C association with IPTG and dissociation from operator were measured. The IPTG association rate constant for wild-type protein is 1.1 × 105 M−1s−1 at pH 7.4, consistent with data from previous work (24, 29, 30). The value for D149C/S193C is within ∼2-fold of that for wild-type LacI (Figure 5; Table 3). Dissociation rate constants were derived from the plot of kassoc versus concentration of IPTG, and the D149C/S193C value was ∼3-fold that for wild-type LacI. Thus, the disulfide bond between residues 149 and 193 does not substantially impact the rate constants for D149C/S193C binding to IPTG. The equilibrium constant for each reaction computed from the corresponding rate constants was comparable within ∼2-fold to the value for the Kd determined directly from fluorescence titration (Table 3).
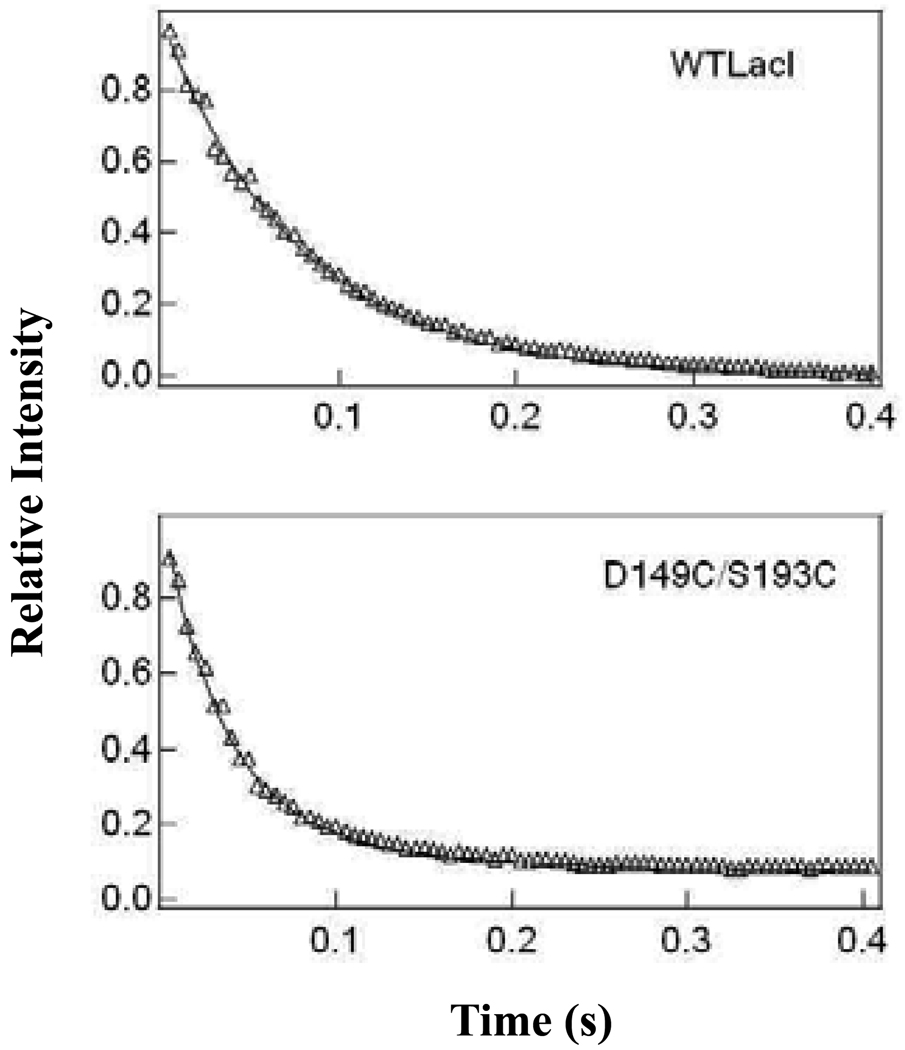
Figure 5.
Measurement of association rate constant for IPTG binding for wild-type LacI and D149C/S193C. The association rates for IPTG binding were determined by stopped flow fluorescence spectroscopy as described in Materials and Methods. The excitation wavelength was 285 nm, and a cut-off filter (350 nm) was used to monitor the decrease in emission. For each repressor, protein (1.0 × 10−6 M) was mixed with multiple IPTG concentrations (from 0.05 mM to 0.2 mM) in fluorescence buffer. Shown are data from a single experiment using 0.05 mM IPTG. Results from the complete analysis are summarized in Table 3.
Table 3.
Kinetic constants for ligand binding
| IPTG | DNA | ||||
|---|---|---|---|---|---|
| kassociationa (M−1s−1 × 10−5) | kdissociationb (s−1) | calcd kdisso/kassoc (M × 106) | Kdc (M × 106) | kdissociationd (s−1 × 103) | |
| WTLacI | 1.1 ± 0.1 | 0.3 ± 0.1 | 2.7 ± 0.3 | 1.4 ± 0.2 | 5.4 ± 0.3 |
| D149C/S193C | 2.3 ± 0.2 | 1.0 ± 0.3 | 4.4 ± 0.3 | 2.5 ± 0.2 | 5.0 ± 0.2 |
The association rate constants for IPTG binding were obtained by stopped-flow experiments at 25 °C in fluorescence buffer (0.01 M Tris-HCl, pH 7.4, 0.15 M KCl). Error values represent the standard deviation for a minimum of three measurements.
Values were calculated from the intercept of the plot of kobsd vs IPTG concentration.
Values were measured by fluorescence titration in fluorescence buffer.
The dissociation rate constants of repressors from operator DNA utilized nitrocellulose filter binding assay in FB buffer (0.01 M Tris-HCl, pH 7.4, 0.15 M KCl, 0.1 mM DTT, 0.1 mM EDTA, and 5% DMSO) with 100 µg/mL of bovine serum albumin. Protein concentration was 1.8 × 10−11 M, and labeled DNA concentration was 3.6 × 10−11 M. Unlabeled DNA was at 90-fold excess over labeled DNA.
Kinetic measurements of repressor-operator dissociation
The operator dissociation rate constants were measured by adding unlabeled operator DNA to trap released repressor (25). The dissociation rate constants for wild-type LacI and D149C/S193C measured under same conditions were essentially identical (Figure 6; Table 3). These data demonstrate that introducing a disulfide bond between residues 149 and 193 does not significantly influence kinetics for repressor-operator dissociation.
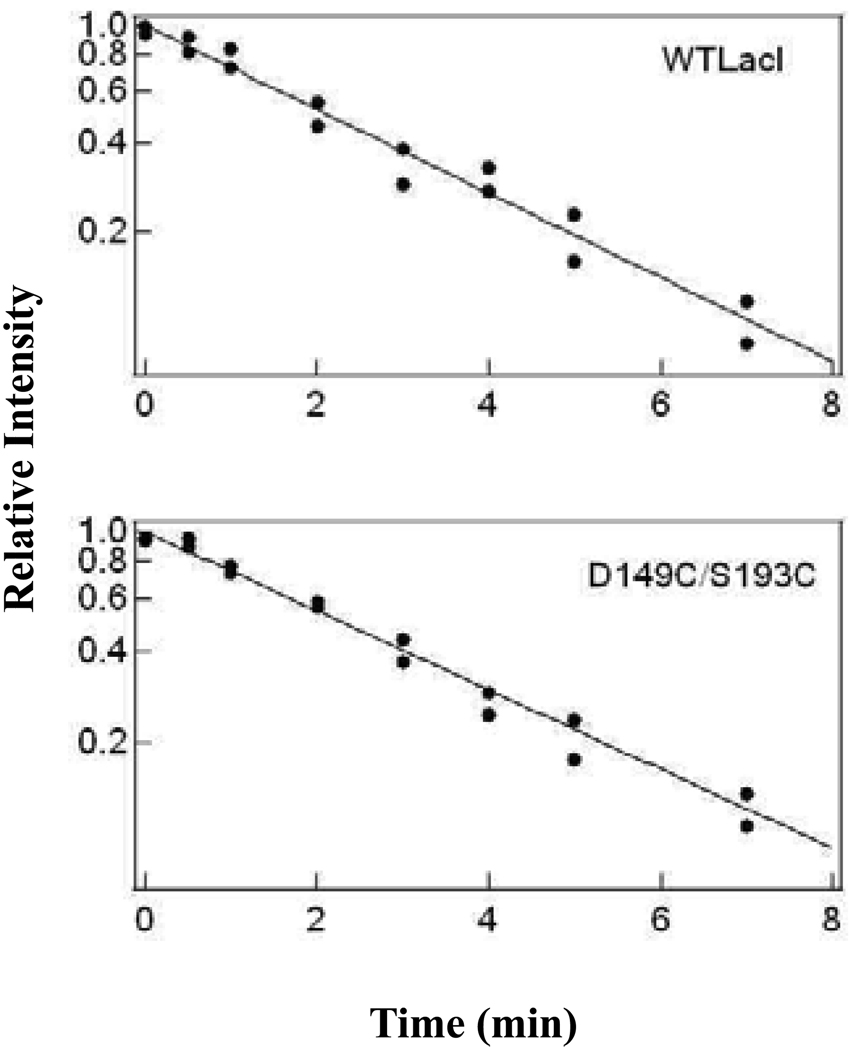
Figure 6.
Determination of dissociation rate constants for operator DNA from repressor. The Y-axis is shown in exponential format for ease of comparison. This measurement was conducted by nitrocellulose filter binding in FB buffer with 100 µg/mL of bovine serum albumin as described in Materials and Methods. Repressor (1.8 × 10−11 M) and 32P-labeled operator (3.6 ×10−11 M) were equilibrated for 15 min before a 90-fold excess of unlabeled DNA was added to the mixture at time zero. Samples were taken at time intervals for analysis. The equilibrium value measured at 2 hr was subtracted from the data at each time point to determine extent of release, and values were normalized relative to binding at time zero. The dissociation rate constants reported in Table 3 were determined by fitting the data to a single exponential model.
Determination of number of the cysteines
To assess directly the formation of the disulfide bond in D149C/S193C, the number of free cysteines per monomer in this double mutant was determined by Ellman’s reaction in 6 M urea to provide a quantitative measure of available thiols (26, 27). The results demonstrate that the ratio of thionitrobenzoate anion produced over [monomer] is 2.9 ± 0.1 for wild-type LacI, indicating that each monomer exhibits the expected 3 free cysteines (residues 107, 140 and 281) (31). For the double mutant D149C/S193C, 4.0 ± 0.1 free cysteines are detected in the native monomeric state, suggesting there is a mixture of the oxidized and reduced forms of the protein present in solution. Following treatment with DTT and rapid removal by a desalting column, 4.6 ± 0.1 free cysteines are detected, indicating more reduced D149C/S193C is obtained. Also, the D149C/S193C mutant can be pushed toward the oxidized state by removing DTT through a desalting column and dialyzing in air-bubbled buffer. Under this condition, 3.3 ± 0.1 free cysteines per monomer are detected.
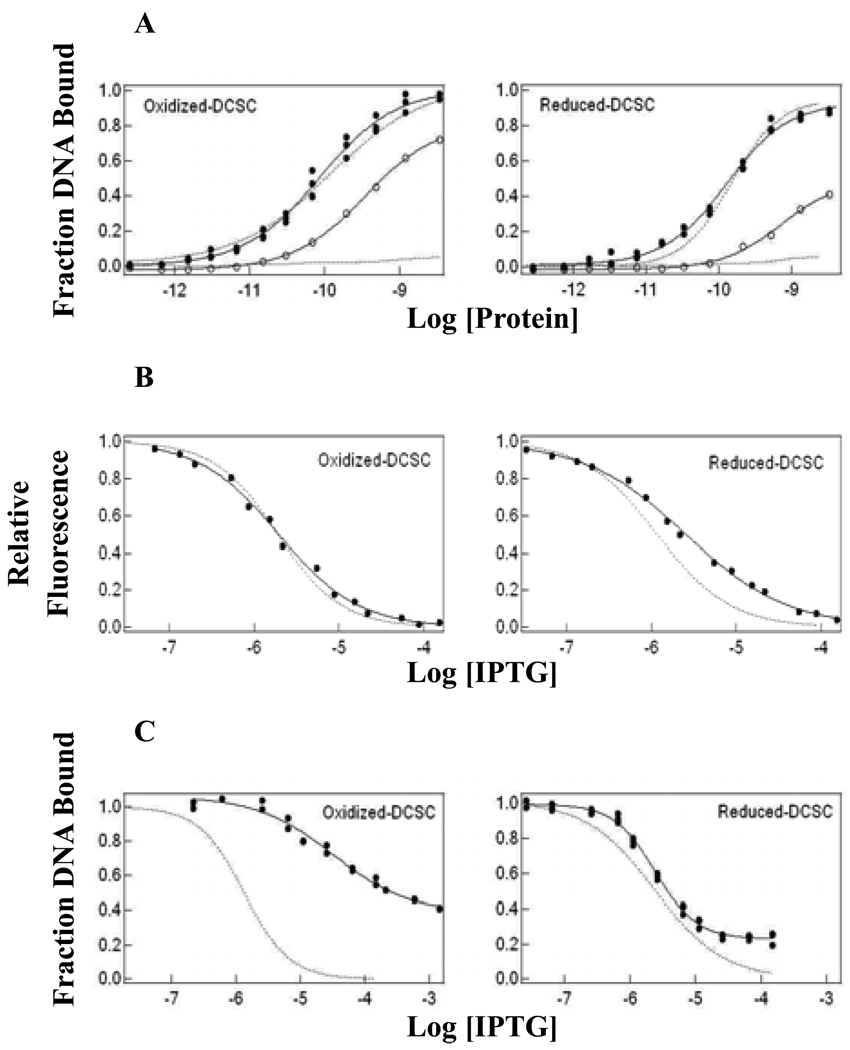
Since D149C/S193C with greater and lesser oxidation can be obtained experimentally, the impact of varied extent of disulfide bond formation was further examined. As controls, the thermodynamic properties of wild-type protein produced under the same conditions for both oxidization and reduction were also examined. The operator binding affinities for oxidized- and reduced-state D149C/S193C are decreased compared to untreated protein, consistent with results for wild-type LacI under the same conditions (Figure 7; Table 4). In the presence of a high concentration of 1 mM IPTG, D149C/S193C in both states can still bind to operator DNA with higher affinity than wild-type LacI (Figure 7; Table 4), with the oxidized protein binding ∼9-fold more tightly. IPTG binding affinities for oxidized and reduced D149C/S193C are comparable to the untreated double mutant and wild-type LacI. Importantly, the operator release data demonstrate that the oxidized and reduced forms of D149C/S193C have distinct sensitivity to IPTG, reflected in the ∼10-fold difference between their [IPTG]mid values (Table 4). Therefore, the induction ratios for oxidized and reduced D149C/S193C are > 10-fold different. The difference in response to IPTG binding between the oxidized and reduced forms of D149C/S193C confirms that the disulfide bond introduced impedes allosteric signal transduction within the LacI structure.
Figure 7.
(A) DNA binding, (B) IPTG binding and (C) operator release curves for wild-type LacI and D149C/S193C exposed to oxidizing and reducing conditions. Wild-type LacI behavior for the oxidized-and reduced-states is represented by the dotted line with data shown for oxidized- and reduced-D149C/S193C. Operator binding and release assays were carried out in FB buffer with 100 µg/mL of bovine serum albumin with DNA concentration at 1.5 × 10−12 M, and IPTG binding experiments in fluorescence buffer with protein concentration at 1.5 × 10−7 M monomer as described in Materials and Methods. The curves were generated by fitting the data in Igor Pro. Results from multiple experiments are presented in Table 4.
Table 4.
Thermodynamic properties of oxidized- and reduced-state repressorsa
| Operator bindingbKd (M ×1011) | +IPTGcKd (M × 1011) | IPTG bindingdKd (M × 106) | Operator releasee [IPTG]mid (M × 106) | Induction Ratiof [IPTG]mid/ Kd | |
|---|---|---|---|---|---|
| WTLacI (Og) | 13 ± 2.0 | > 10000 | 1.9 ± 0.2 | 1.7 ± 0.2 | 0.9 ± 0.1 |
| WTLacI (Rh) | 16 ± 1.0 | > 10000 | 1.2 ± 0.1 | 2.3 ± 0.4 | 1.9 ± 0.2 |
| D149C/S193C (O) | 10 ± 1.0 | ∼ 75 | 2.0 ± 0.2 | 31 ± 2.7 | 16 ± 2.0 |
| D149C/S193C (R) | 15 ± 1.6 | ∼ 650 | 2.6 ± 0.2 | 3.0 ± 0.2 | 1.2 ± 0.1 |
Each value and standard deviation was generated by fitting at least three independently generated curves by Igor Pro. Values in bold are those significantly different from wild-type LacI.
The equilibrium dissociation constants for LacI•operator DNA were measured by nitrocellulose filter binding assays in FB buffer (0.01 M Tris-HCl, pH 7.4, 0.15 M KCl, 0.1 mM DTT, 0.1 mM EDTA, and 5% DMSO) with 100 µg/mL of bovine serum albumin. DNA concentration was 1.5 × 10−12 M.
The equilibrium dissociation constants for LacI•operator DNA in the presence of 1 mM IPTG.
The affinity of the protein for IPTG at pH 7.4 was measured by fluorescence in buffer containing 0.01 M Tris-HCl, pH 7.4, 0.15 M KCl. Protein concentration was fixed at 1.5 × 10−7 M.
Operator release was monitored by nitrocellulose filter binding assays in FB buffer with 100 µg/mL of bovine serum albumin. DNA concentration was at least 10 fold below Kd, and protein concentration was ∼ 80% of maximal DNA binding.
The induction ratio is given by the ratio of [IPTG]mid over Kd for inducer binding. Repressors with a ratio higher than wild-type have a potential block in allosteric communication.
Oxidized-state repressor
Reduced-state repressor
Discussion
Despite broad exploration of LacI as a regulatory protein, neither D149 nor S193 have been targeted for biochemical analysis of substitutions. Previous phenotypic analysis demonstrates that alterations at these sites result in proteins insensitive to IPTG binding (14). However, since D149 is among the amino acids that have direct contact with inducer IPTG, this phenotype could be caused by either diminished IPTG binding affinity or impaired allosteric transition. In this study, a series of D149 and S193 point mutations were generated to examine the effects of different amino acid side chains on function. The induction ratios for the D149 single mutants are within ∼2-fold that for wild-type protein, suggesting that coupling between inducer binding and the allosteric conformational change is not altered. In addition, both S193 single mutants, S193A and S193C, display wild-type properties. The data for single substitutions at either D149 or S193 demonstrate that substitutions at these sites either exert minimal effect on LacI binding properties or alter both inducer binding affinity and operator release, without significantly affecting the allosteric transition (i.e., there is no impact on the induction ratio).
In different ligand-bound structures, distinct interactions are observed between D149 or S193 and other surrounding amino acids. For example, the IPTG-bound LacI structure evinces at least one hydrogen bond between the R197 polypeptide backbone and the S193 backbone or side chain (8). In the operator-bound conformation, backbone hydrogen bonds are present between D149 and L128/Y126 (16). The double mutant D149A/S193A was designed to explore the importance of hydrogen bond formation by precluding the side-chain contribution at these positions. Interestingly, the double-alanine substitution does not affect binding properties of LacI, with operator binding and release properties similar to the wild-type protein. Loss of hydrogen bond capacity may be compensated by the proximal alanine-alanine hydrophobic interaction to promote allosteric signaling.
D149 is at the start of a loop (residues 149–156), for which the flexibility was postulated to be a key factor in the structural transition (18). The TMD results for LacI indicate that mobility in this loop accompanies the early steps in propagating the inducer-binding signal from the binding pocket to the DNA-binding site (18). To reduce flexibility within this region, residues that could form a disulfide bond were introduced by converting residues 149 and 193 to cysteines. This double-cysteine mutant indeed forms a disulfide bond between residues 149 and 193, and this variant binds with wild-type affinity to both operator and inducer. However, the D149C/S193C protein exhibits diminished impact of inducer binding on operator affinity compared to wild-type LacI and has an induction ratio ∼9, over 6-fold greater than wild-type LacI and other mutants, including the single substitutions D149C and S193C.
To confirm that disulfide bond formation was the basis for the change in induction ratio, D149C/S193C was exposed to conditions designed to “push” towards either the oxidized- or reduced-state. Oxidized-D149C/S193C displays an even higher induction ratio (∼16) than the protein as purified from bacterial cells (∼9), whereas the reduced-state double mutant displays an induction ratio near that for wild-type LacI. This substantial difference between oxidized- and reduced-D149C/S193C is consistent with the hypothesis that flexibility in the 149–156 region is necessary for signal transmission at the initial stage of the allosteric transition in response to inducer binding.
Limited motions at either position 149 or 193 in the double-cysteine mutant D149C/S193C may influence various interactions with other residues. The TMD simulation indicates that D149 also forms a hydrogen bond with the backbone of F161, just before the flexible loop is stabilized by interaction with S193 (18). F161 is located in the center of a hydrophobic cluster that also experiences a high degree of mobility during the simulation. If interaction with F161 is impaired, the function of the entire hydrophobic cluster during the allosteric transition may be compromised. Further, the intermediate state of TMD simulation reveals the side chain of D149 transiently flips towards residue N125, which provides the opportunity to form a hydrogen bond between side chains of D149 and N125 during the process of transmitting the IPTG binding signal to the DNA-binding domain (18). However, an interaction between resides 149 and 125 is not present in the extreme DNA- and IPTG-bound conformations (8, 16).
For S193, a hydrogen bond between side chains of residues S193 and R197 is observed in the DNA-bound structure (16). During TMD simulation, this side-chain bond breaks, and new hydrogen bonds are generated between the backbone of R197 and side chain or backbone of S193 (18). However, the interaction between these side chains is not resumed in the inducer-bound structure, indicating its importance only in the DNA-bound conformation. Interactions between D149 or S193 and other amino acids may contribute to allosteric regulation in LacI, but the double-cysteine mutant D149C/S193C limits the movement of these residues, which may consequently affect the conformational change. Which of these interactions individually or in combination is crucial for the structural shifts that are responsible for allostery is not discernible from these data. However, the importance of flexibility in this region is clearly evident.
Protein flexibility appears to play a general role in propagating a signal from a localized perturbation (e.g., ligand binding or point mutation), allowing long-range communication that is fundamental to many protein functions (e.g., 32–36). In an analysis of more than 50 pairs of protein structures, each in two different states, structural motion carries information over long distances in both sequence and space (37). Interestingly, the motions observed favor residues located in the less constrained loops and at the protein surface, and ∼2-fold greater motion is observed in allosteric proteins (37). A very recent model that describes allosteric motion for multiple proteins suggests that contact rearrangement is key to the quaternary changes that occur in LacI (38), consistent with the results reported in this study.
In LacI, substitution of other residues in this solvent-exposed flexible loop that encompasses amino acids 149–156 results in altered inducer and operator binding affinities (23). For example, despite induction ratios similar to wild-type LacI, the two mutations L148F and S151P are an “opposite” pair: L148F exhibits lowered affinity for operator and increased affinity for IPTG, whereas S151P has enhanced operator binding and diminished IPTG binding affinity (23). These results are consistent with a role for this flexible loop in regulating LacI allosteric equilibrium. Thus, residues in this flexible loop appear to make a key contribution to allosteric regulation, perhaps by impacting the range and nature of structural states available to the protein.
Overall, the mutants described herein provide specific insight into the role of D149 and S193 in LacI allosteric response. The experimental results demonstrate that impeding mobility in the D149/S193 region by disulfide bond formation interrupts allosteric signal transmission from the IPTG-binding pocket to the DNA-binding domain without significantly affecting IPTG and operator binding. Integration of biochemical examination of LacI variants with structural analysis and computational simulation provides an important avenue to complete our understanding of allosteric communication within LacI. This work provides a direct demonstration of the general utility of coupling experimental work with computational simulation for identifying regions of a protein crucial for function.
Acknowledgements
We thank members of the Matthews laboratory for helpful discussion and suggestions on experiments and Dr. Liskin Swint-Kruse for sage advice and critical reading of the manuscript.
Abbreviations
- DTNB
dithionitrobenzoic acid
- DTT
dithiothreitol
- IPTG
isopropyl-β-D-thiogalactoside
- LacI
lactose repressor protein
- TMD
Targeted Molecular Dynamics
- WT
wild-type
Footnotes
This work was supported by grants from the Robert A. Welch Foundation (C-576) and the National Institutes of Health (GM22441) to Kathleen S. Matthews.
References
- 1.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 2.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 3.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 4.Matthews KS, Nichols JC. Lactose repressor protein: Functional properties and structure. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:127–164. doi: 10.1016/s0079-6603(08)60035-5. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert W, Müller-Hill B. The lac operator is DNA. Proc. Natl. Acad. Sci. U. S. A. 1967;58:2415–2421. doi: 10.1073/pnas.58.6.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuprina VP, Rullmann JAC, Lamerichs RMJN, van Boom JH, Boelens R, Kaptein R. Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics. J. Mol. Biol. 1993;234:446–462. doi: 10.1006/jmbi.1993.1598. [DOI] [PubMed] [Google Scholar]
- 7.Friedman AM, Fischmann TO, Steitz TA. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 1995;268:1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 8.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Matthews KS. Deletion of lactose repressor carboxyl-terminal domain affects tetramer formation. J. Biol. Chem. 1992;267:13843–13850. [PubMed] [Google Scholar]
- 10.Felder CB, Graul RC, Lee AY, Merkle HP, Sadee W. The Venus flytrap of periplasmic binidng proteins: An ancient protein module present in multiple drug receptors. AAPS J. 1999;1:7–26. doi: 10.1208/ps010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Mol. Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 12.Acher FC, Bertrand HO. Amino acid recognition by Venus flytrap domains is encoded in an 8-residue motif. Biopolymers. 2005;80:357–366. doi: 10.1002/bip.20229. [DOI] [PubMed] [Google Scholar]
- 13.Miller JH. Genetic studies of the lac repressor XI. On aspects of lac repressor structure suggested by genetic experiments. J. Mol. Biol. 1979;131:249–258. doi: 10.1016/0022-2836(79)90075-5. [DOI] [PubMed] [Google Scholar]
- 14.Suckow J, Markiewicz P, Kleina LG, Miller J, Kisters-Woike B, Müller-Hill B. Genetic studies of the Lac repressor XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J. Mol. Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 15.Matthews KS, Falcon CM, Swint-Kruse L. Relieving repression. Nat. Struct. Biol. 2000;7:184–187. doi: 10.1038/73274. [DOI] [PubMed] [Google Scholar]
- 16.Bell CE, Lewis M. A closer view of the conformation of the Lac repressor bound to operator. Nat. Struct. Biol. 2000;7:209–214. doi: 10.1038/73317. [DOI] [PubMed] [Google Scholar]
- 17.Pace HC, Kercher MA, Lu P, Markiewicz P, Miller JH, Chang G, Lewis M. Lac repressor genetic map in real space. Trends Biochem. Sci. 1997;22:334–339. doi: 10.1016/s0968-0004(97)01104-3. [DOI] [PubMed] [Google Scholar]
- 18.Flynn TC, Swint-Kruse L, Kong Y, Booth C, Matthews KS, Ma J. Allosteric transition pathways in the lactose repressor protein core domains: Asymmetric motions in a homodimer. Protein Sci. 2003;12:2523–2541. doi: 10.1110/ps.03188303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wycuff DR, Matthews KS. Generation of an AraC-araBAD promoter-regulated T7 expression system. Anal. Biochem. 2000;277:67–73. doi: 10.1006/abio.1999.4385. [DOI] [PubMed] [Google Scholar]
- 20.Falcon CM, Matthews KS. Glycine insertion in the hinge region of lactose repressor protein alters DNA binding. J. Biol. Chem. 1999;274:30849–30857. doi: 10.1074/jbc.274.43.30849. [DOI] [PubMed] [Google Scholar]
- 21.O'Gorman RB, Rosenberg JM, Kallai OB, Dickerson RE, Itakura K, Riggs AD, Matthews KS. Equilibrium binding of inducer to lac repressor•operator DNA complex. J. Biol. Chem. 1980;255:10107–10114. [PubMed] [Google Scholar]
- 22.Laiken SL, Gross CA, von Hippel PH. Equilibrium and kinetic studies of Escherichia coli lac repressor-inducer interactions. J. Mol. Biol. 1972;66:143–155. doi: 10.1016/s0022-2836(72)80012-3. [DOI] [PubMed] [Google Scholar]
- 23.Swint-Kruse L, Zhan H, Fairbanks BM, Maheshwari A, Matthews KS. Perturbation from a distance: Mutations that alter LacI function through long-range effects. Biochemistry. 2003;42:14004–14016. doi: 10.1021/bi035116x. [DOI] [PubMed] [Google Scholar]
- 24.Friedman BE, Olson JS, Matthews KS. Interaction of lac repressor with inducer. Kinetic and equilibrium measurements. J. Mol. Biol. 1977;111:27–39. doi: 10.1016/s0022-2836(77)80129-0. [DOI] [PubMed] [Google Scholar]
- 25.Whitson PA, Matthews KS. Dissociation of the lactose repressor-operator DNA complex: Effects of size and sequence context of operator-containing DNA. Biochemistry. 1986;25:3845–3852. doi: 10.1021/bi00361a016. [DOI] [PubMed] [Google Scholar]
- 26.Ellman GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 27.Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman’s reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 28.Tiebel B, Aung-Hilbrich LM, Schnappinger D, Hillen W. Conformational changes necessary for gene regulation by Tet repressor assayed by reversible disulfide bond formation. EMBO J. 1998;17:5112–5119. doi: 10.1093/emboj/17.17.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakerian AE, Olson JS, Matthews KS. Thermodynamic analysis of inducer binding to the lactose repressor protein: Contributions of galactosyl hydroxyl groups and β substituents. Biochemistry. 1987;26:7250–7255. doi: 10.1021/bi00397a009. [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Moraitis M, Reedstrom RJ, Matthews KS. Kinetic and thermodynamic studies of purine repressor binding to corepressor and operator DNA. J. Biol. Chem. 1998;273:8958–8964. doi: 10.1074/jbc.273.15.8958. [DOI] [PubMed] [Google Scholar]
- 31.Yang DS, Matthews KS. Lactose repressor protein reaction with 2-chloromercuri-4-nitrophenol. J. Mol. Biol. 1976;103:433–437. doi: 10.1016/0022-2836(76)90322-3. [DOI] [PubMed] [Google Scholar]
- 32.Clarkson MW, Lee AL. Long-range dynamic effects of point mutations propagate through side chains in the serine protease inhibitor eglin c. Biochemistry. 2004;43:12448–12458. doi: 10.1021/bi0494424. [DOI] [PubMed] [Google Scholar]
- 33.Yu EW, Koshland DE., Jr Propagating conformational changes over long (and short) distances in proteins. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9517–9520. doi: 10.1073/pnas.161239298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw D, Wang SM, Villaseñor AG, Tsing S, Walter D, Browner MF, Barnett J, Kuglstatter A. The crystal structure of JNK2 reveals conformational flexibility in the MAP kinase insert and indicates its involvement in the regulation of catalytic activity. J. Mol. Biol. 2008;383:885–893. doi: 10.1016/j.jmb.2008.08.086. [DOI] [PubMed] [Google Scholar]
- 35.Dodson G, Verma CS. Protein flexibility: Its role in structure and mechanism revealed by molecular simulations. Cell. Mol. Life Sci. 2006;63:207–219. doi: 10.1007/s00018-005-5236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson NC, Daggett V. Dynameomics: Large-scale assessment of native protein flexibility. Protein Sci. 2008;17:2038–2050. doi: 10.1110/ps.037473.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daily MD, Gray JJ. Local motions in a benchmark of allosteric proteins. Proteins. 2007;67:385–399. doi: 10.1002/prot.21300. [DOI] [PubMed] [Google Scholar]
- 38.Daily MD, Gray JJ. Allosteric communication occurs via networks of tertiary and quaternary motions in proteins. PLoS Comput. Biol. 2009;5:e1000293. doi: 10.1371/journal.pcbi.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]