Summary
In addition to establishing dendritic coverage of the receptive field, neurons need to adjust their dendritic arbors to match changes of the receptive field. Here we show that dendrite arborization (da) sensory neurons establish dendritic coverage of the body wall early in Drosophila larval development and then grow in precise proportion to their substrate, the underlying body wall epithelium, as the larva more than triples in length. This phenomenon, referred to as scaling growth of dendrites, requires the function of the microRNA (miRNA) bantam (ban) in the epithelial cells rather than the da neurons themselves. We further show that ban in epithelial cells dampens Akt kinase activity in adjacent neurons to influence dendrite growth. This signaling between epithelial cells and neurons receiving sensory input from the body wall synchronizes their growth to ensure proper dendritic coverage of the receptive field.
Introduction
Animals grow extensively after establishing the body plan, and therefore face the daunting task of ensuring that growth of individual organs and tissues is coordinated with growth of the animal as a whole. This coordination of growth is particularly striking in the nervous system, where numerous types of neurons and supporting cells form a network that monitors sensory information received throughout an animal’s body and transmits signals to various tissues. At the level of a sensory neuron, dendrite arbors must grow along with their receptive field throughout development, a physical scaling process that ensures proportional enlargement of the dendrite arbor as the animal grows. How this scaling of dendrite growth is accomplished to exactly match growth of dendrites and surrounding tissue is an interesting open question of likely general relevance to the ways a neuron coordinates its dendritic field to its changing environment.
In the adult brain, the dendritic arbor and its spines are remarkably stable, as revealed by two-photon imaging of pyramidal neurons in the mature olfactory bulb and the visual, auditory, motor, and somatosensory cortices (Holtmaat et al., 2005; Majewska et al., 2006; Mizrahi and Katz, 2003; Zuo et al., 2005). Early in development, however, dendrites must grow in concert with animal growth to sustain proper connectivity and representation of the field. For example, grasshopper interneurons differentiate embryonically, but their dendrites continue to grow in proportion to overall brain size, maintaining their overall morphology and functional properties, while the brain grows nearly 3-fold in linear dimensions (Bentley and Toroian-Raymond, 1981; Bucher and Pfluger, 2000). Similarly, Manduca MN1 and MN3 motoneuron dendrites grow allometrically with the body as a whole throughout larval development (Truman and Reiss, 1988). In goldfish and zebrafish, retinal ganglion cells increase the diameter of their dendritic arbors proportional to the square root of increased diameter of the growing eye, resulting in increased resolution and accuracy in perception of visual stimulus (Lee and Stevens, 2007). Finally, mouse lumbar spinal motoneuron dendrites grow roughly in proportion to expansion of the spinal cord over a large portion of postnatal development, preserving the dendrite topology of the motoneurons (Li et al., 2005). How this proportional growth of dendrites and substrate is coordinated to ensure proper dendrite coverage is largely a mystery.
Given the important roles of miRNAs in regulating developmental timing in invertebrates (Pasquinelli and Ruvkun, 2002; Caygill and Johnston, 2008; Sokol et al., 2008) and the ability of miRNAs to regulate expression of perhaps hundreds of target genes, miRNAs are excellent candidates for temporal regulators of nervous system development. Indeed, a large proportion of vertebrate miRNAs are expressed primarily in the nervous system (Kosik and Krichevsky, 2005; Krichevsky et al., 2003) and mutations in genes required for miRNA processing, such as the RNase III Dicer, affect multiple aspects of nervous system development. For example, zebrafish dicer mutants have obvious defects in nervous system differentiation and function (Giraldez et al., 2005). Mutations in dicer or pasha, which is required for miRNA processing, cell-autonomously affect axon and dendrite targeting in Drosophila, indicating that miRNAs regulate wiring specificity (Berdnik et al., 2008). Finally, specific miRNAs have been implicated in multiple aspects of neuronal development in mammals, from regulation of neurogenesis to regulation of activity-dependent dendrite development (Gao, 2008).
Here we report our studies of scaling growth of dendrite arbors in the Drosophila peripheral nervous system (PNS). Dendrites of morphologically distinct classes of da neurons exhibit an early, rapid growth phase to enable dendrite growth to catch up with body growth as dendrites establish receptive field coverage, and a subsequent scaling phase in which dendrites grow in proportion with underlying epithelial cells and the larva as a whole to maintain receptive field coverage. From a screen of growth-defective mutants we identified the miRNA ban as a potent regulator of dendrite scaling growth in the PNS. In ban mutants, dendrites establish proper receptive field coverage, but dendrite growth fails to synchronize with growth of surrounding tissue, leading to late-stage exuberant dendrite growth. Surprisingly, ban function is not required in neurons for scaling growth of dendrites. Instead, ban function in epithelial cells is sufficient for scaling growth of class IV dendrites. In the absence of ban function, Akt expression and activity are upregulated in neurons, leading to exuberant dendrite growth. Thus, ban regulates signaling between epithelial cells and neurons essential for scaling growth of dendrites.
Results
Dendrites of da neurons grow rapidly to establish receptive field coverage then grow in proportion to larval growth to maintain this coverage
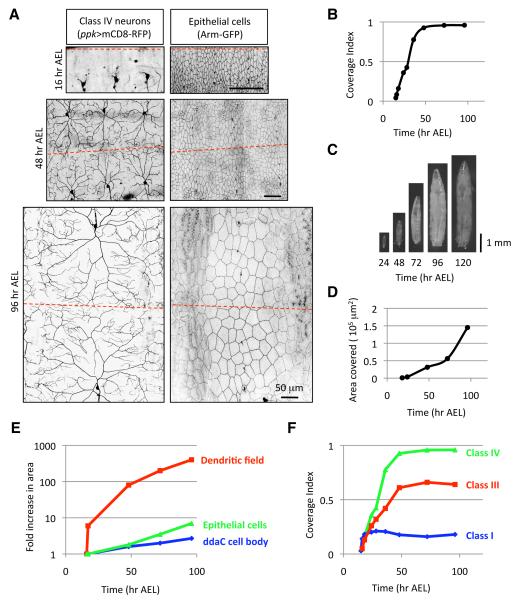
Dendrites of class IV da neurons completely and non-redundantly cover the larval body wall early in larval development, a phenomenon referred to as dendritic tiling (Grueber et al., 2002). Once field coverage is established, dendrites continue to branch and lengthen to maintain tiling as larvae grow, providing a sensitive system for analysis of how neurons first establish and later maintain coverage of the receptive field. In this study we have addressed the question of how late-stage dendrite growth is precisely coordinated with larval growth to maintain proper dendrite coverage of the body wall.
To examine this process, we used the pickpocket-EGFP (ppk-EGFP) marker (Grueber et al., 2003b) to monitor class IV dendrite growth before and after establishment of tiling. To quantitatively assess dendrite coverage we have used a metric that we refer to as the coverage index, the ratio of the territory covered by dendrites of a given da neuron, such as the class IV neuron ddaC, to the area of a hemisegment that harbors the da neurons (Figure S1 and Experimental Procedures). Dendrite outgrowth of class IV neurons begins at approximately 16 hr After Egg Laying (AEL), with class IV dendrites growing rapidly during late embryonic/early larval stages to tile the body wall between 40-48 hr AEL and subsequently maintaining this coverage until dendrites are pruned during metamorphosis (Figures 1A and 1B). Between 48hr AEL and 120 hr AEL (just prior to metamorphosis), larvae grow nearly 3-fold in length and the dorsal area of class IV receptive fields expands by more than 6-fold (Figures 1C and 1D). Therefore, class IV dendrites grow extensively and this dendrite growth must be precisely coordinated with larval growth in order to maintain proper coverage of the receptive field.
Figure 1.
Dendrites of da neurons grow rapidly to establish receptive field coverage then grow in proportion to larval growth to maintain this coverage.
(A) Live images of class IV neurons (ppk>mCD8-RFP) and epithelial cells (Arm-GFP) at 16, 48, and 96 hr AEL. The hatched red line denotes the dorsal midline in (A) and in subsequent images. Dorsal is up, anterior is left and scale bars are 50 μm.
(B) Quantification of the coverage index of an individual ddaC class IV neuron over a developmental time course. n > 10 for each time point.
(C) Live images of Drosophila larvae over the course of larval development.
(D) Quantification of the area covered by an individual ddaC class IV neuron over a developmental time course. n > 5 for each time point.
(E) Time course showing the fold-increase in area of an individual ddaC dendritic field (red), an individual epithelial cell (green), and the ddaC cell body (blue) relative to the area of each at 16 hr AEL, shortly after dendrite outgrowth begins. n > 10 for each time point.
(F) Time course of dendrite coverage for different classes of da neurons. Class IV neurons (green) were visualized with ppk-eGFP, class III neurons (red) were visualized with ss-Gal4, UAS-mCD8-GFP (M. Kim, personal communication) and class I neurons were visualized with 2-21-Gal4, UAS-mCD8-GFP (Grueber et al., 2003a). n > 10 for each time point.
Class IV dendrites are located between muscle and epithelial cells. Cell divisions that give rise to larval cells are complete by mid-embryogenesis and larval growth is achieved by increasing cell size rather than additional proliferation (Edgar and Nijhout, 2003). Thus, all the cells that will comprise the larval body wall musculature and epithelia are in place when dendrite outgrowth begins. To simultaneously visualize growth of class IV dendrites and epithelial cells we used a protein trap line that directs GFP expression in epithelial cells and outlines their borders (Armadillo::GFP, adherens junctions, or Neuroglian::GFP, septate junctions) in combination with ppk-GAL4 driving expression of mCD8-RFP in class IV neurons (Figure 1A). Using these markers, we monitored growth of class IV dendrites and epithelial cells throughout embryonic/larval stages.
Epithelial cells grow at a nearly constant rate over the time course (Figure 1E). Likewise, the class IV neuron soma grows at a relatively constant rate. In contrast, the dendrite growth is biphasic. Initially, class IV dendrite growth outpaces growth of epithelial cells and the larva as a whole between 16 hr and 48 hr AEL, the timeframe in which class IV dendrites establish tiling (Figure 1E). Dendrite growth slows as class IV dendrite arbors achieve complete body wall coverage, and from 48 hr to 120 hr AEL class IV dendrites grow in proportion to larval growth at a rate comparable to that of epithelial cells (Figure 1E). We will refer to this late dendrite growth as scaling growth of dendrites (a phenomenon unrelated to synaptic scaling) to reflect the physical scaling of dendrite arbors as they grow precisely in proportion to surrounding cells and the larva as a whole in order to maintain proper coverage of the receptive field.
Scaling growth of dendrites is a general property of da neurons
To determine whether scaling growth is a general property of da neurons we monitored dendrite growth in class I and class III da neurons, two additional morphologically distinct classes of da neurons (Grueber et al., 2002) using the coverage index metric introduced above. Like class IV neurons, dendrites of class I and III neurons rapidly establish coverage of a characteristic region of the body wall and subsequently maintain their coverage by expanding their dendrite arbors in precise proportion to larval growth (Figures 1F and S2). Class III neurons cover their territory in the same timeframe as class IV neurons, first establishing receptive field coverage at about 48 hr AEL. In contrast, class I neurons covered their characteristic territory by 24 hr AEL. Thus, temporally distinct signals may regulate scaling of dendrite growth in class I and class III/IV neurons. Nevertheless, scaling growth of dendrites seems to be a general feature of da neuron development.
Based on the fidelity of dendrite coverage in class IV neurons we focused on these neurons for our studies of dendrite scaling. Our finding that class IV dendrites have a rapid growth phase during establishment of tiling and a scaling phase with slower dendrite growth to maintain tiling suggests that some signal(s) attenuate dendrite growth following establishment of tiling, synchronizing growth of class IV dendritic arbors with growth of surrounding tissue. We therefore set out to characterize the signaling that underlies dendrite scaling.
Scaling growth of dendrites ensures proper dendrite coverage in larva of diverse sizes and shapes
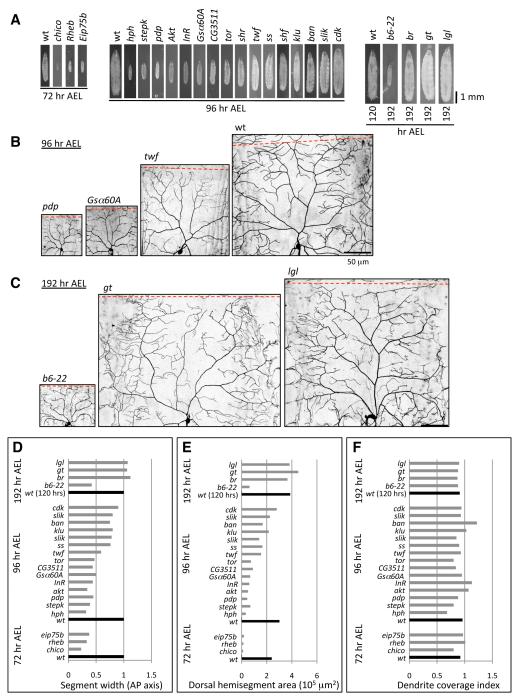
To test the capacity of dendrite scaling, we examined the effects of mutations that alter the dimensions of larvae at different developmental states on class IV dendrite growth. We chose alleles that survive until at least the 2nd larval instar, allowing us to monitor dendrite coverage by class IV neurons at a time when they should have already established tiling. Overall, we screened 35 mutant alleles that cause a range of defects in larval size, shape and growth rate, and results for a representative subset of these alleles are shown (Figure 2, Table S1). Notably, class IV dendrites properly covered the receptive field in nearly all of these mutants, accommodating a broad range of receptive field areas (ranging from 10% of wild type (wt) in chico mutants to 120% of wt in giant mutants) and shapes (Figures 2B, and 2D-2F). Dendrites also scaled properly in mutants defective in developmental rate, for example maintaining proper receptive field coverage in b6-22 mutants that develop slowly and persist as 2nd instar larvae or in broad mutants that persist as third instar larvae for days or even weeks (Figures 2C-2F). Taken together, these results demonstrate the robustness of dendrite scaling growth in class IV neurons.
Figure 2.
Screen for mutants that affect scaling growth of dendrites.
(A) Growth-defective mutants at the time point where dendrite growth was assayed. ppk>mCD8-GFP was used to visualize dendrites for 3rd chromosome and ppk-EGFP was used to visualize dendrites for 2nd chromosome mutants.
(B) Dendrites scale to accommodate diverse sizes of receptive fields, as illustrated by class IV dendrites of pdp, G-5α, twf and wt larvae at 96 hr AEL.
(C) Dendrites scale properly in mutants defective in developmental timing, such as b6-22, which persist as 2nd instar larvae (S. Younger, personal communication), or persistent 3rd instar gt or l(2)gl larvae. For (B) and (C) scale bars are 50 μm.
(D-F) Effects of growth-defective mutants on dendrite development.(D) Segment width (average length of a dorsal hemisegment along the AP axis), (E) Hemisegment area (average area of the dorsal region of a hemisegment), and (F) Coverage index of mutants at the indicated time points. Note that although the size/shape of the receptive field is changed in most cases, only ban, and to a lesser degree InR and Akt, showed a substantial increase in coverage index. Complete gene names are provided in Table S1. Measurements were taken from dendrites of at least 4 neurons for each mutant.
ban is required for scaling growth of dendrites
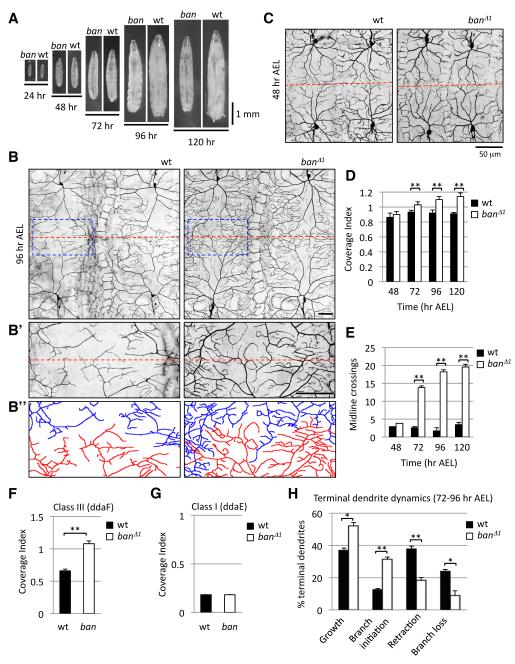
Among the few mutants that had any effect on scaling growth of dendrites, the ban mutant had the most severe dendrite overgrowth phenotype we observed (Figure 2F), with the first sign of larval growth defects at 72 hr AEL (Figure 3A). We reasoned that ban might be required for dendrite scaling but not earlier aspects of dendrite development and focused the remainder of our study on the role of ban in dendrite scaling. Notably, ban encodes a miRNA (Brennecke et al., 2003) and might represent a regulatory node for scaling of dendrite growth since miRNAs likely regulate expression of 100 or more target genes (Lim et al., 2005).
Figure 3.
ban is required for scaling growth of dendrites in a subset of da neurons.
(A) Growth profile of control and ban mutant larvae.
(B) Dendrite morphology of wt and ban mutant larvae at 96 hr AEL visualized with ppk-EGFP. The interface of two ddaC neurons at the dorsal midline is shown in (B’) and dendrites are depicted as camera lucida to help distinguish dendrites of adjacent neurons in (B”). Scale bars are 50 μm.
(C) Dendrite morphology of wt and ban mutant larvae at 48 hr AEL visualized with ppk-EGFP.
(D-E) Time course monitoring the dendrite coverage index (D) and midline crossing (E) of ddaC dendrites in wt or ban mutant larvae. n > 5 for all time points.
(F-G) Coverage index for (F) ddaF class III neurons and (G) ddaE class I neurons at 96 hr AEL. n = 10.
(H) Quantification of terminal dendrite dynamics measured over a 24 hr time lapse (72-96 hr AEL) in wt and ban mutant larvae. n > 1000 terminal dendrites (from 10 neurons) for each genotype.
Here and in subsequent figures, error bars represent SEM, * denotes p < 0.05 and ** denotes p < 0.001.
Dendrites of individual class IV neurons occupy a larger proportion of the body wall in ban mutant 3rd instar larvae (Figures 2F, 3B, and 3D). At 96 hr AEL, ddaC class IV neurons in ban mutants have a mean coverage index of 1.22, meaning that the receptive field of the average ddaC dendrite in ban mutant larvae is 122% of the size of the dorsal hemisegment that harbors the neuron. Thus, dendrites in ban mutants promiscuously cross boundaries that are observed by dendrites of wt neurons (Figure 3B’). For example, fewer than 2 dendrite branches cross the midline for a given wt class IV neuron, whereas more than 18 dendrite branches cross the midline in ban mutants (Figure 3E). However, although we see a coverage index of >1 for ban mutant, we do not see a significant tiling defect because branches that cross normal boundaries still avoid dendrites of neighboring class IV neurons.
The exuberant growth of dendrites in ban mutants is manifest throughout the arbor, not just at the boundaries. In addition to these defects in dendrite coverage, class IV neurons in ban mutants show significant increases in the number of dendrites, the density of dendrites, and overall dendrite length (data not shown). However, increased terminal dendrite branching is not sufficient to increase receptive field coverage. Several other mutants have been described that increase terminal dendrite branching in class IV neurons, and none of these mutants cause an overall increase in the size of the dendritic field. For example, furry mutations cause a 100% increase in the number/density of terminal dendrites (Emoto et al., 2004) without an accompanying increase in coverage index at 96hr AEL (Figure 4M). Likewise, overexpression of the small GTPase Rac drastically increases terminal dendrite branching but reduces receptive field coverage (data not shown).
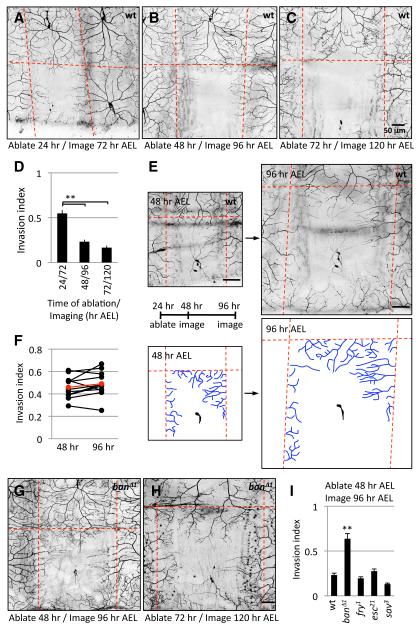
Figure 4.
ban regulates growth-inhibitory signals to ensure dendrite scaling.
(A-D) Progressive restriction of invasion capacity of class IV dendrites. Class IV neurons were ablated with a focused laser beam at the time indicated and larvae were imaged 48 hr post-ablation. Hatched red lines demarcate the dorsal midline and segment boundaries. (D) Quantification of the portion of unoccupied dendritic territory covered by invading dendrites (invasion index). n > 10 for each ablation/imaging paradigm.
(E-F) Dendrite coverage is maintained by dendrite scaling, even when dendrites establish aberrant body wall coverage. Class IV neurons were ablated at 24 hr AEL and larvae were imaged at 24 hr intervals beginning at 48 hr AEL. Images of the same territory at 48 hr and 96 hr AEL are shown. Invading dendrites from neighboring ddaC neurons are depicted in blue in the traces below each image. Hatched red lines demarcate the dorsal midline (dorsal is up) and segment boundaries. Dendrites invading the ventral portion of the field at 96 hr AEL are not traced because they are out of the field of view at 48 hr AEL. (F) The invasion index for each individual neuron monitored in this ablation paradigm is plotted at 48 and 96 hr AEL with black lines connecting the values for a given neuron. Mean values for 10 neurons are shown in red.
(G-I) Dendrites retain exuberant invasion capacity in ban mutants. Invasion activity in ban mutants (G) at 96 hr AEL in which a class IV neuron was ablated at 48 hr AEL or (H) at 120 hr AEL in a ban mutant in which a class IV neuron was ablated at 72 hr AEL. (I) Quantification of invasion activity. n > 10 for each genotype. Scale bars are 50 μm for all images.
The dendrite growth defects in ban mutants could reflect increased dendrite growth from early stages of development or defects specific to the scaling phase of dendrite growth. To distinguish between these possibilities we monitored dendrite growth over a developmental time-course, focusing on the coverage index and midline crossing events as metrics for growth of the dendrite arbor as a whole. Importantly, class IV dendrites in ban mutants are indistinguishable from wt during the early, rapid growth phase (through 48 hr AEL) as measured by coverage index, midline crossing events (Figures 3C-3E) and total dendrite branch number (data not shown). However, beginning at 72 hr AEL we noted progressively more severe defects in the coverage index and a greater number of midline crossing events in ban mutants (Figures 3D and 3E). This late-onset exuberant dendrite growth demonstrates that ban is not causing a general growth defect since ban is dispensable for establishment of dendrite coverage. Whereas a generalized defect in dendrite growth, as seen in dar mutants (Ye et al., 2007), would affect both the early (isometric) and late (scaling) phases of growth, mutations that specifically affect the scaling growth of dendrites would be dispensable for the early, rapid growth of dendritic fields. This is precisely what we see for ban mutants. Therefore, ban is specifically required for scaling of dendrite arbors, potentially by affecting growth-inhibitory signals that normally restrict dendrite growth.
To confirm that loss of ban causes these phenotypes, we conducted the following experiments. First, whereas heterozygosity for a ban null allele or deficiencies that span the ban locus show no obvious defects in dendrite scaling, placing ban mutations in trans to a deficiency that spans the locus, but not a nearby deficiency does not span the ban locus, recapitulates the dendrite defects described above (Figure S3). Second, the ban mutant dendrite defects can be fully rescued by a ban genomic rescue transgene but not a genomic transgene in which the ban locus has been deleted (Figure S3). Therefore, disrupting ban function is sufficient to cause defects in scaling growth of dendrites.
We next tested whether ban is required for scaling growth of dendrites in other classes of da neurons. Both class I and class III neurons establish proper dendrite coverage in ban mutants (data not shown). However, class III dendrites are defective in scaling of dendrite growth in ban mutants, showing a significant increase in dendrite coverage after 48 hr AEL (Figures 3F and S4). In contrast, larval class I dendrites show no obvious defects in dendrite coverage in ban mutants, demonstrating that ban is not required for scaling in class I neurons (Figures 3G and S4). The onset of scaling growth of dendrites differs by 24 hours in class I and class III/IV neurons (Figure 1F), thus different scaling signals may operate at the two time points with ban required for the scaling growth signal for class III/IV neurons that tile.
Next, we conducted time-lapse microscopy of single neurons to characterize the cellular basis of the ban mutant phenotype. We imaged single class IV neurons from time-matched wt or ban mutant larvae at 24 hr intervals beginning at 72 hr AEL, just after the ban phenotype is first apparent (Figure S5). We monitored dynamics of every terminal dendrite that could be unambiguously followed through the time course and measured dendrite growth, initiation of new dendrites, dendrite retraction, and branch loss. For each of these categories ban mutants differed from wt controls, exhibiting significantly more dendrite growth and branch initiation and significantly less dendrite retraction and branch loss (Figures 3H and S5). Therefore, stabilization of existing dendrites, increased dendrite growth and increased addition of new dendrites all contribute to the defect in dendrite scaling growth of the ban mutant.
Growth inhibitory signals regulate dendrite scaling
Our time-lapse studies suggest that signals normally restricting dendrite growth are largely absent in ban mutants. We set out to verify this hypothesis using laser ablation assays. Previous studies showed that following embryonic ablation of a class IV neuron, dendrites of neighboring neurons grow exuberantly to invade the unoccupied territory of the ablated neuron (Grueber et al., 2003b; Sugimura et al., 2003), with the ability of dendrites to invade unoccupied territory progressively restricted in older larvae (Sugimura et al., 2003). We therefore wanted to determine whether the timing of this restricted growth potential correlates with the onset of scaling of dendrite growth and whether ban is required for restriction of the dendrite growth potential.
Consistent with prior reports, ablating a class IV neuron at 24 hr AEL led to extensive invasion by dendrites of neighboring neurons, with 55% of the unoccupied territory covered by neighboring neurons 48 hr post-ablation (Figures 4A and 4D). This ability of dendrites to grow into unoccupied territory was severely attenuated one day later, with dendrites of neighboring neurons invading only 23% of the unoccupied territory after ablation of a class IV neuron at 48 hr AEL (Figures 4B and 4D). The extent of invasion was even further reduced when neurons were ablated at 72 hr AEL (Figures 4C and 4D). Therefore, the ability of dendrites to grow beyond their normal boundaries to invade unoccupied territory is severely restricted during larval development at a time coincident with the onset of scaling of dendrite growth.
If the restriction of dendrite growth potential in larvae is caused by scaling signals that limit dendrites to growth in proportion to body wall growth, the majority of invading activity by neighboring dendrites should be present before scaling growth ensues at 48 hr AEL. To test this prediction we ablated class IV neurons at 24 hr AEL and monitored invasion activity at 24 hr intervals over the next 72 hr (Figure 4E). By 48 hr AEL dendrites of neighboring neurons had invaded unoccupied territory, and the extent of invasion was not noticeably increased at later time-points. Instead, the entire dendrite arbor of class IV neurons, including the portion that invaded unoccupied territory, scaled with larval growth after 48 hr AEL (Figures 4E and 4F). Thus, the receptive field that is established by 48 hr AEL is maintained by scaling of dendrite growth, even in cases in which dendrites establish aberrant body wall coverage. The signals responsible for dendrite scaling growth are likely distinct from the homotypic repulsion required to establish tiling as ablation of all neighboring same-type neurons does not potentiate the ability of a class IV neuron to invade unoccupied territory (data not shown). Additionally, dendrites of class I da neurons, which do not rely on homotypic repulsion to establish their coverage, also exhibit scaling growth.
As described above, dendrite coverage is properly established in ban mutants (Figure 3C). Importantly, unlike wt controls, following ablation at 48 hr AEL, dendrites in ban mutants extensively fill unoccupied space (Figures 4G-4I), with dendrites in ban mutants invading unoccupied territory just as efficiently as dendrites in wt controls ablated at 24 hr AEL. Therefore, the receptive field boundaries of class IV neurons have not been fixed in ban mutants at 48 hr AEL. Dendrites in ban mutants invade unoccupied territory more efficiently than wt controls at later time points as well (Figure 4H). Thus, either the growth-inhibitory scaling signal is lost or dendrites are refractory to the signal in ban mutants.
To test whether machineries for dendritic tiling contribute to the progressive reduction of a dendrite’s ability to invade vacant territories, we examined mutations of furry (fry), which encodes a gene required for establishment of dendritic tiling (Emoto et al., 2004), and extra sex combs (esc) and salvador (sav), which function in a common pathway to regulate stability of terminal dendrites and, consequently, maintenance of dendrite coverage (Emoto et al., 2006; Parrish et al., 2007), for effects on dendrite invasion following neuron ablation. Unlike mutations in ban, mutations in fry, esc, or sav had no effect on the ability of dendrites to invade unoccupied territory (Figure 4I). Moreover, consistent with the scaling signal functioning in a distinct pathway, double mutant combinations of ban with fry or esc showed additive phenotypes (Figure S6). Thus, ban exerts its effects on scaling of dendrite growth independently of known pathways for establishment and maintenance of dendrite coverage.
ban functions non-autonomously to regulate scaling growth of dendrites
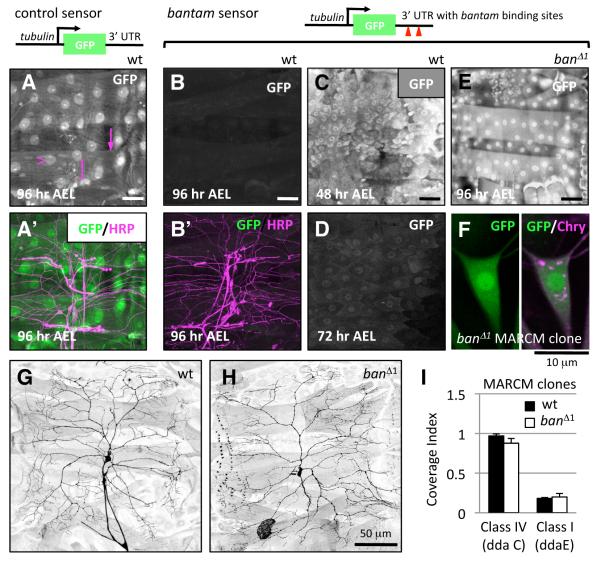
To further characterize the signaling required for scaling growth of dendrites, we wanted to determine where ban functions to regulate scaling. First, we investigated whether ban is expressed in neurons, surrounding cells, or both, by using a miRNA activity sensor as a reporter for ban expression in 3rd instar larvae (Brennecke et al., 2003). A control sensor directs ubiquitous expression of GFP, including robust GFP expression in muscle, epithelial cells, and sensory neurons (Figure 5A). The ban sensor contains two ban binding sites in the 3′UTR of the transgene, hence GFP expression is attenuated in cells that express ban. Unlike the control sensor, we detected very little, if any, GFP expression in 3rd instar muscle cells, epithelial cells, or sensory neurons using several independent transgenic fly lines with distinct insertions of the ban sensor (Figure 5B). We first observed significant attenuation of the ban sensor in larval muscle, epithelium and PNS neurons between 48 and 72 hr AEL (Figures 5C and 5D), precisely at the time when we first observed dendrite defects in ban mutants, suggesting that ban activity is more pronounced during this period than at earlier time points. Notably, the attenuation of the ban sensor was dependent on ban activity as shown by the persistent, ubiquitous expression of the sensor in ban mutant larvae (Figure 5E). Thus, ban is likely expressed in the muscle, epithelium, and PNS neurons and may be required in any of these cell types for scaling of dendrite growth.
Figure 5.
ban activity is broadly distributed in late larval stages but is dispensible in neurons for dendrite scaling.
(A) A control miRNA sensor is ubiquitously expressed throughout larval development. Anti-GFP immunostaining of a control miRNA sensor 3rd instar fillet reveals expression in muscle (arrow), epithelia (open arrowhead), and PNS neurons (bracket), which are labeled by anti-HRP immunoreactivity in magenta (A’). Scale bars are 50 μm for (A-E).
(B-F) ban sensor expression in larvae. ban sensor GFP expression is not detectably expressed in 3rd instar larvae (B). In (B’) ban sensor expression (green) and HRP (magenta) are shown. (C-D) Live imaging of the ban sensor in larvae using identical microscope settings at 48 hr AEL (E) and 72 hr AEL (F) shows a marked down-regulation of GFP expression over this interval. (E) ban sensor expression remains high in ban mutant larvae, even at 96 hr AEL, demonstrating that the down-regulation of the sensor seen in (D) and (E) depends on ban function. (F) Live imaging of the ban sensor in ban△1 ddaC MARCM clones to monitor ban activity in clones. The ban sensor (green) and the CD2-Cherry used to positively label the clone (magenta) are shown.
(G-I) ban is dispensable in neurons for dendrite scaling. Control (G) or ban△1 (H) ddaC MARCM clones in dissected 3rd instar larval fillets. (I) Quantification of coverage index for control or ban△1 MARCM clones. For all categories n > 5.
To determine whether ban is required cell-autonomously for dendrite scaling we used MARCM to generate single neuron clones homozygous for a ban mutation in a heterozygous background (Lee and Luo, 1999). ban activity was effectively dampened in MARCM clones as indicated by de-repression of the ban sensor in the clones (Figure 5F). However, loss of ban function had no significant effect on dendrite coverage of class IV neurons (Figures 5G-5I). Time-lapse analysis of ban mutant class IV MARCM clones revealed no defects in dendrite coverage at any time during larval development (data not shown). Furthermore, ban is dispensable in other da neurons for dendrite scaling growth (Figure 5I, data not shown). Thus, ban function in sensory neurons is dispensable for scaling growth of dendrites.
Although scaling of dendrite growth proceeds normally, there is some reduction of overall dendrite length and the number of dendrite branches in ban mutant class IV clones (Figure S7). Therefore, ban likely acts cell-autonomously to promote dendrite growth and non-autonomously to limit dendrite growth and ensure proper scaling.
ban functions in epithelial cells to regulate scaling growth of dendrites
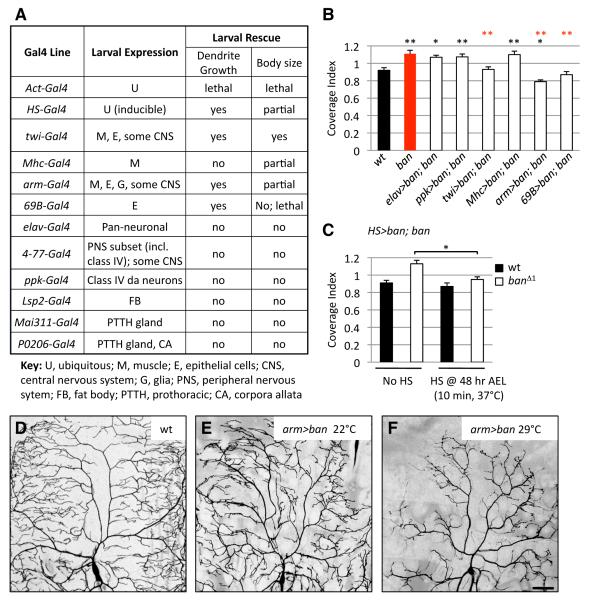
We turned to a genetic rescue assay to test the ability of transgenic expression of ban in different tissues to rescue the dendrite growth defects of ban mutants. Consistent with our MARCM results, neuronal expression of ban, using either pan-neuronal or PNS-specific Gal4 drivers, was not sufficient to rescue the scaling growth defect of ban mutants (Figures 6A, 6B and S8). Thus, ban likely functions non-autonomously in non-neuronal cells to regulate scaling of da neuron dendrite growth. Moreover, expression of ban in muscle alone could not ameliorate the dendrite defects of ban mutants. Remarkably, every time we rescued ban expression in epithelial cells, we found significant suppression of the exuberant dendrite growth of ban mutants. The three epithelial Gal4 driver lines caused reductions of dendrite growth that correlated with Gal4 expression levels in epithelial cells: arm-Gal4 caused the greatest reduction in dendrite growth and had the strongest epithelial expression whereas twi-Gal4 displayed the lowest activity and drove epithelial Gal4 expression at the lowest level. Taking advantage of the temperature-sensitive nature of Gal4 activity, we monitored rescue activity of each epithelial Gal4 line over a graded temperature series (18°C to 29°C) and found that for each driver, rescue activity was directly proportional to expression level (data not shown). Therefore, epithelial ban expression is sufficient to suppress the exuberant dendrite growth of ban mutants, and the extent of dendrite growth inhibition varies with the level of ban expression in epithelial cells.
Figure 6.
ban expression in epithelial cells is sufficient for scaling growth of dendrites.
(A) The ability of transgenic expression of ban directed by a variety of Gal4 drivers to rescue dendrite growth and larval growth defects of ban mutants was assayed at 96 hr AEL.
(B) Quantification of rescue activity of Gal4 drivers at 96 hr AEL. n > 10 neurons for each genotype. Black asterisks denote p values relative to wt and red asterisks denote p values relative to ban mutants.
(C) Quantification of activity of heat-shock-induced (HS) ban expression to rescue dendrite coverage defects of ban mutants at 96 hr AEL. n > 6 neurons for each category.
(D-F) ban levels in epithelial cells affect dendrite growth. Dendrites of class IV ddaC neurons were visualized by a 3x ppk-eGFP reporter for control (3x ppk/+; arm-gal4/+) larvae or larvae in which epithelial cells overexpressed ban (“arm>ban” = 3x ppk/+; arm-gal4/UAS-ban). Due to the temperature-sensitive nature of Gal4, raising larvae at higher temperatures (29°C vs. 22°C) results in higher levels of Gal4 activity, and therefore higher levels of ban expression in epithelial cells. Scale bar is 50 μm.
Epithelial expression of twi-Gal4 was first apparent in larval stages, suggesting that post-embryonic expression of ban in epithelia is sufficient for proper scaling of dendrite growth. Given that dendrite defects in ban mutants first appear after 48 hr AEL, we asked whether late expression of ban would suffice for dendrite scaling. To examine the temporal requirement for ban function we used a heat shock-inducible Gal4 driver to express ban during larval development. Indeed, inducing ban expression at 48 hr AEL was sufficient to rescue the dendrite defects of ban mutants (Figures 6C and S8). These findings reinforce the notion that ban is dispensable for early aspects of dendrite development.
Resupplying ban in tissues known to regulate larval growth such as the fat body (Colombani et al., 2003), prothoracic (PTTH) gland (Layalle et al., 2008; McBrayer et al., 2007), or insulin-producing cells (IPCs) (Rulifson et al., 2002) had no measurable effect on dendrite growth in ban mutants (Figure 6A, data not shown). Moreover, ablation of each of these tissues mediated by a reaper transgene caused larval growth defects without obvious dendrite growth defects (data not shown). Thus, ban function in the fat body, PTTH gland, or IPCs is not sufficient to modulate scaling of dendrite growth. Altogether, these results suggest that epithelial cells are likely the major functional sites for ban in regulation of PNS dendrite scaling.
Because ban expression in epithelial cells affects scaling growth of dendrites in a dose-dependent fashion via a mechanism that likely involves growth-inhibitory signals, we wondered whether ectopic epithelial expression of ban in a wt background could further inhibit dendrite growth and thus disrupt scaling of dendrite arbors. Indeed, overexpression of ban in epithelial cells resulted in a severe reduction in dendrite growth and induced striking defects in the pattern of dendrite growth over epithelial cells, with terminal dendrites appearing to wrap around epithelial cells (Figures 6D-6F). Consistent with ban dosage in epithelial cells regulating the strength of dendrite growth-inhibitory signals, epithelial overexpression of ban induced more robust inhibition of dendrite growth at higher temperatures (which lead to higher levels of transgene expression; Figures 6E and 6F).
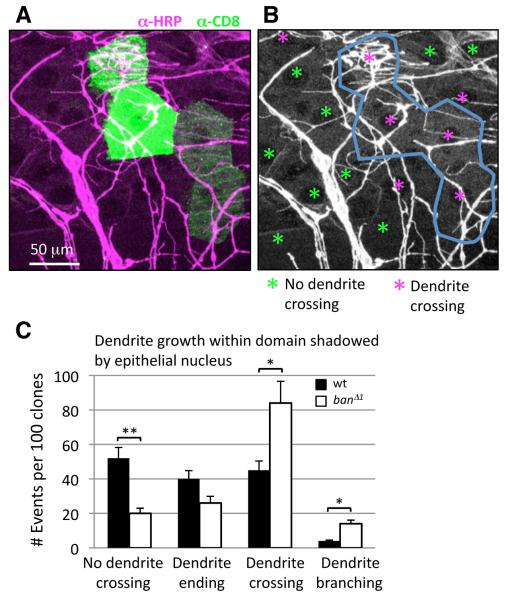
Since ban expression in epithelial cells is sufficient to ensure proper scaling, we wanted to address whether ban function in epithelial cells is necessary for scaling of dendrite growth. To this end, we used MARCM to generate ban mutant epithelial cell clones. Although it was not possible to address the contribution of epithelial ban to scaling of the entire dendrite arbor using this approach (we were only able to generate 1-4 cell epithelial clones), we monitored the pattern of dendrite growth over ban mutant or wt control epithelial clones. Class IV dendrites grow extensively over epithelial cells, with multiple dendrite branches often coursing over a single epithelial cell. We used the epithelial nucleus as a landmark and monitored dendrite growth over the epithelial cell surface shadowed by the nucleus. Although the gross morphology of epithelial cells was not obviously affected in ban mutant clones, the propensity of class IV dendrites to grow into the region shadowed by the epithelial nucleus was significantly increased for ban mutant epithelial clones when compared to wt controls or ban heterozygous epithelial cells (Figures 7A-7C). Therefore, ban is required in epithelial cells to ensure proper dendrite growth and placement over epithelial cells.
Figure 7.
ban influences dendrite distribution over the body wall epithelium.
(A) Positively labeled homozygous mutant ban△1 epithelial clones (green) were generated in a heterozygous background. PNS dendrites are labeled by HRP immunoreactivity (magenta).
(B) Distribution of dendrites over the region directly below epithelial nuclei (denoted by asterisks) was monitored for wt or ban mutant clones. Instances in which PNS dendrites cross this region are color-coded magenta and instances in which this region is devoid of dendrites are color-coded green. Note that dendrites cross this domain in a higher proportion of ban mutant epithelial cells than in neighboring heterozygous cells.
(C) Quantification of dendrite growth into the domain shadowed by the epithelial nucleus of control (Frt 2A) or ban mutant epithelial clones. Average number of events per 100 clones of each genotype is shown. n > 500 cells for each genotype.
ban regulates neuronal Akt signaling to achieve proper scaling growth of dendrites
To gain insight into the molecular mechanism underlying dendrite scaling we developed a platform for microarray-based expression profiling of dissociated, FACS-isolated PNS neurons or epithelial cells (Parrish, Kim, DeRisi, and Jan, unpublished data). We identified Akt and numerous other candidate genes that are deregulated in PNS neurons and/or epithelial cells of ban mutant larvae. Because Akt is a well-established regulator of growth (Edgar, 2006), including dendrite growth in mammalian hippocampal neurons (Jaworski et al., 2005; Kumar et al., 2005), we investigated whether ban regulates Akt as part of the scaling program.
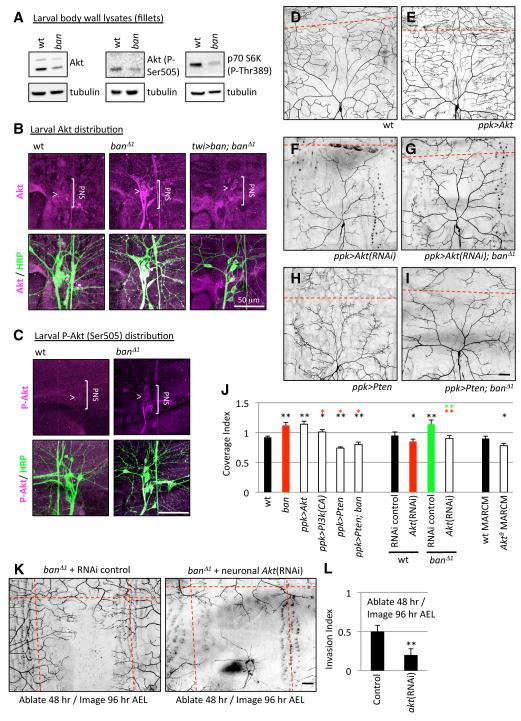
In our microarray experiments, we found that Akt expression was increased in neurons but reduced in epithelial cells of ban mutants relative to wt controls (data not shown). By monitoring Akt levels in lysates of larval fillets composed mostly of muscle and epithelial cells, we found that without ban function Akt protein levels were substantially reduced (Figure 8A). Furthermore, Akt activity was substantially reduced as shown by reductions in active, phosphorylated Akt and phosphorylated S6K, a downstream reporter of Akt activity. Therefore, Akt expression and activity are substantially reduced in ban mutant larval lysates, likely reflecting reduced Akt function in muscle/epithelia.
Figure 8.
ban regulates neuronal Akt to ensure dendrite scaling.
(A) Akt levels and activity monitored via western blotting of wt or ban larval fillet lysates.
(B and C) Expression and activity of Akt in the larval PNS. wt or ban mutant 3rd instar larvae were immunostained with antibodies to Akt / phospho-Akt (magenta) and HRP (green) to label sensory neurons and their processes. The bracket marks the position of PNS neurons and the arrowhead marks of the class IV neuron ddaC. Genotypes are indicated above the images; twi>ban denotes twist-Gal4, UAS-ban.
(D-J) Akt activity influences dendrite scaling in class IV neurons. Dendrite morphology for representative neurons of the following categories: control neuron, wt (D), class IV neuron overexpressing Akt, ppk>Akt, (E), Akt(RNAi) in a class IV neuron, ppk>Akt(RNAi) (F), Akt(RNAi) in a class IV neuron of a ban mutant larva, ppk>Akt(RNAi); ban△1 (G), overexpression of Pten in a class IV neuron, ppk>Pten (H), or overexpression of Pten in a class IV neuron of a ban mutant larva, ppk>Pten; ban (I). (J) Coverage index for class IV neurons of the indicated genotypes. Asterisks denote p values relative to color-matched controls. n > 10 for each genotype.
(K-L) Akt is required for the exuberant dendrite invasion activity in ban mutants. Class IV neurons in “ban△1 + RNAi control” (genotype: elav-Gal4/+; UAS-Dcr-2/+; ban△1, ppk-EGFP/ban△1, ppk-EGFP) and “ban△1 + neuronal Akt(RNAi)” (genotype: elav-Gal4/+; UAS-Dcr-2/+; ban△1, ppk-EGFP, UAS-Akt-RNAi/ban△1, ppk-EGFP) larvae were ablated at 48 hr AEL and imaged 48 hr post-ablation. Representative images are shown (K). Hatched red lines demarcate the dorsal midline and segment boundaries. (L) Quantification of the invasion index for the “ban△1 + RNAi control” larvae (Control) and “ban△1 + neuronal Akt(RNAi) larvae (Akt(RNAi)) from (K). n = 4 ablated/imaged neurons for each genotype.
Next we immunostained larval fillets to determine whether ban influences Akt protein levels in the PNS. In wt controls, Akt is detectible only at low levels in the soma or dendrites of the PNS (Figure 8B). By contrast, in ban mutants Akt is highly expressed in the PNS and is detectible in axons, the soma, and dendrites. Similarly, phosphorylated Akt is barely detectible in the larval PNS of wt controls but is present at high levels in the PNS of ban mutants (Figure 8C). Therefore ban regulates Akt expression and activity in the larval PNS.
To test whether this effect on Akt levels reflects a neuronal requirement for ban we monitored Akt expression levels in ban mutants in which ban expression is resupplied under the control of twist-Gal4, an experimental condition that rescues both the dendrite scaling defect and larval size defect of ban mutants (Figure 6A). We found that ban non-autonomously regulates Akt levels in da neurons since non-neuronal expression of ban (twist-Gal4) is sufficient to dampen the ectopic neuronal Akt expression normally seen in ban mutants (Figure 8B). Therefore, ban likely functions in epithelia to regulate signals that influence Akt expression and activity in neurons.
Finally, we wanted to determine whether Akt function in class IV neurons is important for scaling of dendrite growth. Based on our expression data, we predicted that increasing Akt expression/activity in class IV neurons should cause a scaling defect similar to what is seen in ban mutants. Indeed, ectopic expression of Akt, or a constitutively active form of PI3 kinase (PI3k) that leads to activation of Akt, caused a significant increase in dendrite coverage (Figures 8E and 8J), similar to ban mutants. Conversely, antagonizing Akt activity in class IV neurons by overexpressing Pten, a PIP3 phosphatase that functions as an inhibitor of Akt activity, by knocking down Akt expression via RNAi in class IV neurons, or by generating Akt null mutant class IV neuron MARCM clones caused a significant reduction in dendrite coverage (Figures 8F, 8H, and 8J). Therefore, Akt plays a critical role in regulating dendrite coverage.
If increased neuronal Akt activity underlies the dendrite defects in ban mutants, then antagonizing neuronal Akt activity should suppress the dendrite overgrowth in ban mutants. We tested this hypothesis with the following three experiments. First, we used RNAi to knock down Akt expression in class IV neurons of ban mutant larvae. On its own, Akt(RNAi) causes a reduction in dendrite growth and overall coverage of the receptive field, and this phenotype is epistatic to the dendrite overgrowth seen in ban mutants (Figures 8F, 8G, and 8J). Similarly, we overexpressed Pten in class IV neurons of ban mutant larvae and found that the Pten-mediated reduction in dendrite coverage is epistatic to the dendrite overgrowth seen in ban mutants (Figures 8H-8J). Finally, we ablated class IV neurons in ban mutants in the absence or presence of neuron-specific Akt RNAi and found that reducing neuronal Akt expression blocks the exuberant dendrite invasion activity of ban mutants (Figures 8K and 8L). Altogether, these results strongly suggest that ban functions in epithelial cells to regulate neuronal expression/activation of Akt, and deregulation of Akt leads to the dendrite growth defects of ban mutants.
Discussion
In the present study, we have shown that non-autonomous signals coordinate growth of dendrites with the growth of their substrate and the body as a whole. Dendrites of many types of neurons cover characteristic receptive fields, and growth of the dendrite arbor in synchrony with the receptive field in a process we refer to as scaling growth of dendrites allows a neuron to maintain proper dendrite coverage of the receptive field. Thus, scaling growth of dendrites is likely a general mechanism to ensure fidelity of dendrite coverage.
Dendrites of class IV neurons cover their receptive field before larval growth is complete and must maintain this coverage as the larva grows. Two properties distinguish the scaling phase of dendrite growth from the early dendrite growth when the neuron establishes receptive field coverage. During the scaling phase of growth, the dendrite arbor grows precisely in proportion to receptive field expansion (which is often achieved by animal growth). Moreover, dendrite growth is constrained by boundaries delineated when the dendrite arbor first covers the receptive field. Thus, although dendrites continue to grow, growth occurs only to maintain proportional coverage of the receptive field.
miRNAs as regulators of developmental timing in the nervous system
Dendrites of Drosophila da neurons exhibit a biphasic growth profile: dendrites establish coverage of their receptive field via an early, rapid growth phase and maintain this coverage via a late scaling growth phase in which dendrites grow in proportion with epithelial cells and the animal as a whole. The miRNA ban acts in the second phase to enable scaling of dendrite growth in da neurons, ensuring that dendrites maintain proper body wall coverage. Loss of ban disrupts epithelial-derived signaling that normally modulates dendrite growth, and, as a result, dendrites remain in the “rapid growth” phase, extending beyond their normal territories. This phenotype is reminiscent of the heterochronic phenotypes of C. elegans lin-4 and let-7 mutants in that an early developmental phase is inappropriately reiterated during a later phase.
How broadly do miRNAs regulate developmental progressions in the nervous system? Although the developmental roles of vertebrate miRNAs have been somewhat elusive because of the vast number of miRNA-encoding genes, several studies suggest that miRNAs may serve highly specialized roles in regulating developmental transitions in neuron morphogenesis. For example, completely abrogating miRNA function causes robust defects in neuron morphogenesis but not specification in zebrafish (Giraldez et al., 2005), consistent with miRNAS regulating late aspects of neuronal differentiation. Likewise, miRNAs miR9a* and miR-124 regulate the switch in subunit composition of chromatin remodeling complexes as neural progenitors differentiate into neurons in mice (Yoo et al., 2009). Additionally, a number of miRNAs function primarily at a very late step of neuron development to regulate activity-dependent dendrite growth and synaptic plasticity. For example, neuronal activity antagonizes miR-134, which normally inhibits growth of dendritic spines (Schratt et al., 2006), and promotes expression of mir-132, which promotes dendritic plasticity (Wayman et al., 2008).
ban regulates growth of proliferating and differentiated cells
Although ban is known to regulate growth in proliferating tissues in Drosophila, ban-mediated regulation of dendrite growth likely represents a distinct mode of growth control by ban for the following reasons. First, previous studies focused on autonomous regulation of tissue growth by ban (Brennecke et al., 2003; Hipfner et al., 2002; Raisin et al., 2003; Thompson and Cohen, 2006). In contrast, ban acts non-autonomously to regulate scaling of dendrite growth. Second, prior studies of ban function focused on imaginal discs where growth is achieved by increasing cell number rather than cell size. By contrast, dendrite scaling involves ban-mediated regulation of growth in differentiated, post-mitotic cells. Likewise, post-mitotic expression of ban in the larval eye disc can also regulate cell size (Raisin et al., 2003). Third, ban functions downstream of the tumor suppressor kinase Hippo to control proliferation, with Hippo activating the transcription factor Yorkie, which in turn activates ban expression (Nolo et al., 2006). Whereas hippo is required cell-autonomously for establishment and maintenance of dendrite tiling, yorkie is dispensable for dendrite growth (Emoto et al., 2006). As to the cell non-autonomous function of ban in dendrite scaling, Hippo is not required. Although these findings suggest that ban regulates growth of proliferating and differentiated tissues by different means, it is possible that in both scenarios ban is antagonizing expression of growth inhibitory factors (Edgar and Nijhout, 2003), possibly even the same factors, and removing growth inhibition has different consequences on proliferative and differentiated tissues.
Epithelial-derived signals regulate scaling growth of dendrites
We propose that ban positively regulates an epithelial-derived signal that modulates neuronal Akt expression and activity to influence dendrite growth. Several observations suggest that the signal acts over a short range, possibly even via direct adhesive interactions between dendrites and epithelia or the underlying matrix. First, ban overexpression in epithelial cells but not in muscle influences growth of dendrites. Second, removing ban function from epithelial cell clones influences the distribution of dendrites over the clone but not over adjacent wt epithelial cells. Third, in addition to inhibiting the overall rate of dendrite growth, overexpressing ban in epithelial cells induces exaggerated “wrapping” of epithelial cells by terminal dendrites. Since these signals appear to preferentially regulate dendrite growth during the scaling phase, ban may modulate dendrite/epithelial adhesion during the scaling phase of dendrite development.
Different signals regulate dendrite scaling in distinct types of neurons
Morphologically distinct classes of da neurons establish type-specific dendritic coverage of the body wall and maintain this coverage by means of dendrite scaling as larvae grow. However, arbors of different da neurons develop at different rates, with class I dendrites establishing their coverage one day earlier than class III and class IV dendrites. Mutations in ban disrupt scaling of dendrite growth in class III and class IV but not class I neurons, suggesting that different signals regulate dendrite scaling in distinct types of neurons. Thus, temporally distinct signals or temporally restricted sensitivity to the signals may ensure that different neurons maintain appropriate coverage of their receptive field.
Experimental Procedures
Fly Stocks
A comprehensive list of alleles used in this study is available in Table S1.
Live Imaging
Live imaging was as described (Emoto et al., 2006; Parrish et al., 2007). Embryos were collected for two hours on yeasted grape juice agar plates and were aged at 25°C in a moist chamber. At the appropriate time, a single embryo/larva was mounted in 90% glycerol under coverslips sealed with grease, imaged using a Leica SP5 microscope, and returned to grape juice agar plates between imaging sessions, if required. Details of time-lapse analysis are available in Figure S5.
Mosaic analysis
MARCM analysis was conducted as previously described (Grueber et al., 2002).
Immunohistochemistry
3rd instar larval fillets were dissected/processed as described (Grueber et al., 2002) and stained with the following antibodies: anti-HRP conjugated with Cy2 or Cy3 (1:200; Jackson labs), anti-mCD8 (1:100; Caltag), anti-Akt C67E7 (1:500; Cell Signaling), anti-phospho-D-Akt Ser505 (1:500; Cell Signaling), anti-GFP (1:2000), anti-Armadillo N27A1 (1:20; Developmenal Studies Hybridoma Bank) and secondary antibodies from Jackson labs.
Laser Ablation
A single embryo/larva was mounted, as for live imaging, and the nucleolus of a class IV neuron was targeted using a focused 337 nm pulsed nitrogen laser (Spectraphysics laser, Photonics Intstruments; 12 Hz , 10 sec) mounted on a Nikon E800 epifluorescence microscope. Following ablation, animals were recovered to grape juice agar plates and imaged live at the appropriate time.
Western Blotting
10 larvae were filleted (leaving an abdominal body wall preparation containing the epidermis, PNS neurons, and larval muscle) and homogenized in 250 ml of homogenization buffer [150 mM NaCl, 50 mM Tris pH 7.4, 2 mM EDTA, 10% glycerol, 1% Triton-X100, Complete protease inhibitors (Roche)]. Lysates were centrifuged at 25k × g for 20 min and supernatants were subjected to western blotting with anti-Phospho Drosophila Akt (Ser505; Cell Signaling; 1:1000); anti-Drosophila Akt (C67E7; Cell Signaling; 1:1000); anti-Phospho Drosophila p70 S6 Kinase (Thr398; Cell Signaling; 1:1000); anti-α-Tubulin (DM1A; AbCam; 1:2500). Blots were developed with ECL reagents (Amersham) and documented with a VersaDoc imaging system (BioRad).
Measurements
To quantify dendrite length and branchpoints, Z-series images for a given neuron were projected onto a 2D image file, dendrite arbors were traced using Neurolucida software, and features of the arbor were measured using the traces.
Dendrite field size of class I, III, and IV neurons was measured as previously described (Grueber et al., 2002). Briefly, the territory covered by dendrites of a given neuron was delineated by constructing polygons connecting nearest-neighbor terminal dendrites. The area covered by a given dendrite corresponds to the area bounded by these polygons.
For analysis of scaling of dendrite growth we used two indices defined here. The coverage index represents the portion of the larval body wall covered by dendrites of a single neuron. Measurements of the coverage index were confined to a region in dorsal hemisegments bounded by the following landmarks: muscle attachment sites (apodemes; anterior/posterior boundaries), the dorsal midline (dorsal boundary) and a line connecting a neuron of interest to the corresponding neuron in the adjacent anterior and posterior segments (ventral boundary). A dendrite that completely covers the dorsal hemisegment, but no additional territory, would have a coverage index of 1 whereas a dendrite that covered the entire dorsal hemisegment as well as territory of neighboring hemisegments would have a coverage index of > 1. A schematic explanation of the Coverage Index is available in Figure S1. The Invasion Index represents the portion of a dorsal hemisegment that is covered by dendrites of neurons from neighboring hemisegments.
Construction of pUAST-ban
A 151 bp DNA fragment containing the ban locus was PCR amplified and cloned into the pUAST vector as an EcoRI/NotI fragment to generate pUAST-ban. Standard procedures were used to generate transgenic lines.
Supplementary Material
Acknowledgements
We thank Chun Han, Peter Soba and Li Cheng for experimental advice, Joe DeRisi for use of his laboratory and equipment for microarray experiments, Susan Younger for help with genetics, Stephen Cohen, Tian Xu, Yang Hong, Chun Han, Rebecca Yang, Sabina Zimmerman and Quan Yuan for fly stocks, and members of the Jan lab for helpful comments. This work was supported by NIH grants 5K99MH084277-02 (J.Z.P) and R01NS40929 (Y.N.J). L.Y.J., and Y.N.J. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bentley D, Toroian-Raymond A. Embryonic and postembryonic morphogenesis of a grasshopper interneuron. J Comp Neurol. 1981;201:507–518. doi: 10.1002/cne.902010404. [DOI] [PubMed] [Google Scholar]
- Berdnik D, Fan AP, Potter CJ, Luo L. MicroRNA processing pathway regulates olfactory neuron morphogenesis. Curr Biol. 2008;18:1754–1759. doi: 10.1016/j.cub.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Bucher D, Pfluger H. Directional sensitivity of an identified wind-sensitive interneuron during the postembryonic development of the locust. J Insect Physiol. 2000;46:1545–1556. doi: 10.1016/s0022-1910(00)00078-0. [DOI] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Nijhout HF. Growth and Cell Cycle Control in Drosophila. In: Hall MN, Raff M, Thomas G, editors. Cell Growth: Control of Cell Size. CSHL Press; 2003. pp. 23–84. [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003a;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003b;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161:1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS, Krichevsky AM. The Elegance of the MicroRNAs: A Neuronal Perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layalle S, Arquier N, Leopold P. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev Cell. 2008;15:568–577. doi: 10.1016/j.devcel.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Lee S, Stevens CF. General design principle for scalable neural circuits in a vertebrate retina. Proc Natl Acad Sci U S A. 2007;104:12931–12935. doi: 10.1073/pnas.0705469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li Y, Brewer D, Burke RE, Ascoli GA. Developmental changes in spinal motoneuron dendrites in neonatal mice. J Comp Neurol. 2005;483:304–317. doi: 10.1002/cne.20438. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Majewska AK, Newton JR, Sur M. Remodeling of synaptic structure in sensory cortical areas in vivo. J Neurosci. 2006;26:3021–3029. doi: 10.1523/JNEUROSCI.4454-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, Thummel CS, Dauphin-Villemant C, Gilbert LI, O’Connor MB. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell. 2007;13:857–871. doi: 10.1016/j.devcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi A, Katz LC. Dendritic stability in the adult olfactory bulb. Nat Neurosci. 2003;6:1201–1207. doi: 10.1038/nn1133. [DOI] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Jan LY, Jan YN. Polycomb genes interact with the tumor suppressor genes hippo and warts in the maintenance of Drosophila sensory neuron dendrites. Genes Dev. 2007;21:956–972. doi: 10.1101/gad.1514507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- Raisin S, Pantalacci S, Breittmayer JP, Leopold P. A new genetic locus controlling growth and proliferation in Drosophila melanogaster. Genetics. 2003;164:1015–1025. doi: 10.1093/genetics/164.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Yamamoto M, Niwa R, Satoh D, Goto S, Taniguchi M, Hayashi S, Uemura T. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci. 2003;23:3752–3760. doi: 10.1523/JNEUROSCI.23-09-03752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Truman JW, Reiss SE. Hormonal regulation of the shape of identified motoneurons in the moth Manduca sexta. J Neurosci. 1988;8:765–775. doi: 10.1523/JNEUROSCI.08-03-00765.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009 doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Lin A, Chang P, Gan WB. Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron. 2005;46:181–189. doi: 10.1016/j.neuron.2005.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.